Abstract
The Werner (WRN) gene codes for a DNA helicase that contributes to genomic stability and has been identified as the gene responsible for progeria. Recent studies have shown reduced WRN expression due to aberrant DNA hypermethylation in cancer cells. Furthermore, WRN expression is thought to affect sensitivity to DNA topoisomerase I inhibitors in cancer therapy. In this study, we examined the relationship between aberrant DNA hypermethylation of WRN and the sensitivity of cervical cancer cells to anticancer drugs. DNA was extracted from samples from 22 patients with primary cervical cancer and 6 human cervical cancer-derived cell lines. Aberrant DNA hypermethylation was analyzed by methylation-specific PCR. WRN expression in cultured cells before and after addition of 5-aza-2-deoxycytidine, a demethylating agent, was examined using RT-PCR. The sensitivity of cells to anticancer drugs was determined using a collagen gel droplet embedded culture drug sensitivity test (CD-DST). siRNA against WRN was transfected into a cervical cancer-derived cell line with high WRN expression. Changes in drug sensitivity after silencing WRN were determined by CD-DST. Aberrant DNA hypermethylation and decreased expression of WRN were detected in 7/21 cases of primary cervical cancer and in two cervical cancer-derived cell lines. These two cell lines showed high sensitivity to CPT-11, a topoisomerase I inhibitor, but became resistant to CPT-11 after treatment with 5-aza-2-deoxycytidine. Transfection of siRNA against WRN increased the sensitivity of the cells to CPT-11. Aberrant DNA hypermethylation of WRN also increased the sensitivity of cervical cancer cells to CPT-11. Therefore, epigenetic inactivation of this gene may be a biomarker for selection of drugs for the treatment of cervical cancer. This is the first report to show a relationship between the methylation of the WRN gene and sensitivity to CPT-11 in gynecological cancers.
Keywords: WRN, cervical cancer, DNA hypermethylation, CPT-11, chemosensitivity
Introduction
The incidence of cervical cancer has decreased in advanced countries due to more frequent health checkups and the development of vaccines. In the United States in 2010, there were 12,200 cases of cervical cancer and 4,210 deaths related to this disease, reflecting a decreasing tendency (1). However, cases of cervical cancer worldwide have increased from 378,000 in 1980 to 454,000 in 2010 (2). Death due to cervical cancer has decreased, but there were still about 200,000 deaths worldwide in 2010, including 46,000 females aged 15–49 years in developing countries (2).
Cervical cancer occurs due to infection with the human papillomavirus (HPV) (3). More than 100 genotypes of HPV have been detected and about 40 have been found to infect the genital tract. HPV is classified into high-risk types causing cervical cancer, including HPV16 and HPV18, and low-risk types causing conditions other than cancer, such as a polyp. HPV16, the most frequent genotype, is detected in approximately half of patients with cervical cancer (4). After infecting cells, HPV produces oncoproteins E6 and E7, which inhibit controlling the cell cycle and apoptosis, and therefore have important roles in oncogenesis. E6 is associated with p53 and induces p53 degradation by E3 ubiquitin ligase via the function of E6-associated protein (E6AP). E7 inactivates the retinoblastoma tumor suppressor gene product, pRb and its family members (5).
Epigenetic DNA methylation of tumor suppressor genes in the promoter region is generally important in carcinogenesis (6–8). In cervical cancer, carcinogenesis is related to aberrant methylation of CpG islands of p16, FHIT, retinoic acid receptor β, E-cadherin, death-associated protein kinase, HIC-1, APC, and Ras association domain family 1A genes (9–12). Methylation of CpG islands in the WRN promoter region is related to carcinogenesis in various cancers (13). The chromosomal WRN locus on the short arm of chromosome 8, is composed of 35 exons, and has a length >250 kb (14). WRN encodes the WRN protein, which is a member of the RecQ helicase family and is also an exonuclease. Loss of WRN causes abnormalities in DNA repair, replication, and telomere maintenance.
Werner syndrome (WS) is an autosomal recessive genetic disease that is caused by mutation of the WRN gene. WS symptoms include aging at an early stage and various secondary symptoms associated with aging, including bilateral cataract, skin change, short stature and graying hair; in addition, diabetes mellitus, osteoporosis, atherosclerosis and cancer also develop frequently (15). Malignant complications include sarcomas of mesenchymal origins, including soft-tissue sarcoma and osteosarcoma, suggesting that the mechanism of carcinogenesis in WS differs from that in carcinogenesis in other cancers (16). The mean age at death of WS patients is 46–54 years and one of major causes is the high prevalence of malignancy (17,18).
Several studies have shown a relationship between WRN expression and malignancy and have indicated that epigenetic inactivation of WRN is of importance in carcinogenesis. In many tumors, loss of heterozygosity is detected in chromosome 8p, in which WRN is located, but WRN somatic mutation has not been found, suggesting that epigenetic control has a significant effect (19,20). Epigenetic DNA methylation in the promoter region of WRN and a methylation-induced decrease in WRN expression have been found in colorectal, lung, gastric, prostate and breast cancer. The methylation-induced decrease in WRN expression increases chromosomal instability (13).
Reduced WRN expression is also related to sensitivity to camptothecin (CPT-11), a topoisomerase I (Top-I) inhibitor (13,21). CPT-11 is an alkaloid found in plants such as Camptotheca acuminata. A single-strand break (SSB) occurs after Top I binds to DNA and generates a Top I-DNA cleavable complex. CPT-11 stabilizes this complex and inhibits reconnection of the SSB, resulting in inhibition of DNA synthesis (22). CPT-11 also inhibits replication fork progression, resulting in DNA double-strand breaks (DSBs) and apoptosis (23). Inactivation of WRN in cancer cells increases the effect of CPT-11 (13,21), and overall survival of patients with colorectal cancer treated with irinotecan, a camptothecin analogue, is dependent on the methylation status of CpG islands in the WRN promoter (13).
Cisplatin is a key drug in chemotherapy for cervical cancer (24). CPT-11 is a similarly important drug and has a high response rate of 24% (25). Evaluation of WRN expression as a marker of sensitivity to CPT-11 may be clinically useful in treatment of cervical cancer. Thus, in this study, the associations among cervical cancer, WRN expression, and cancer cell sensitivity to CPT-11 were investigated.
Materials and methods
Subjects and cytologic specimens
Samples were obtained from 21 cervical cancer smears collected using a ThinPrep system (Cytyc, Boxborough, MA, USA) and kept in preservation fluid (PreservCyt Solution, Cytyc) (26). Informed consent was obtained before collection. Pathological diagnosis was performed by cervical histology, and the cytological and histological results were consistent for all smears. Of the 21 cervical cancer smears, 10 were squamous carcinoma and 11 were adenocarcinoma. The histological type and stage were determined according to the General Rules for Clinical Cervical Cancer in Japan published by the Japan Society of Obstetrics and Gynecology.
Cultured cell lines
The human cervical squamous cell carcinoma-derived cell lines, SKG-I, SKG-II, SKG-IIIa and SKG-IIIb, and the human cervical adenocarcinoma-derived cell lines, HeLa and TCO-I, were used in the study. HeLa cells were incubated in DMEM (Sigma, St. Louis, MO, USA) with 10% fetal bovine serum (FBS) (Sanko Junyaku Co., Tokyo, Japan) and TCO-I cells were incubated in MEM medium (Sigma) with 10% FBS. All other cell lines were incubated in F12 medium (Sigma) with 10% FBS. Cells were incubated in 10-cm dishes at 37°C in a 5% CO2 atmosphere.
DNA extraction and methylation-specific PCR (MSP) assay of the WRN gene
DNA was extracted from 21 cervical smears and 6 cervical carcinoma-derived cell lines using a Get Pure DNA kit (Dojin Glocal, Kumamoto, Japan). DNA (1 μg) extracted from cervical smears was diluted with 50 μl of distilled water and incubated in 5.5 μl of 3 N NaOH at 37°C for 15 min. To this solution, 30 μl of 10 mM hydroquinone (Sigma) and 520 μl of 3 M sodium bisulfite (prepared at pH 5.5 with 10 N NaOH, Sigma) were added with mixing. Mineral oil was laid over the solution to prevent evaporation, and the solution was incubated overnight at 50°C. The lower layer of the reaction solution was mixed with 1 ml of Clean-up Resin (Promega, Madison, WI, USA) and then injected into a column. After rinsing with 2 ml of 80% isopropanol, the mixture was centrifuged at 15,000 rpm for 3 min to remove isopropanol. Hot (70°C) distilled water (50 μl) was added and the mixture was centrifuged at 15,000 rpm for 2 min to elute DNA. The DNA was then incubated with 5.5 μl of 2 N NaOH at 37°C for 20 min. Next, 66 μl of 5 N ammonium acetate and 243 μl of 95% ethanol were added and the mixture was incubated at −80°C for one hour and centrifuged at 15,000 rpm for 30 min to precipitate DNA. Supernatant exceeding 50 μl was removed, 1 ml of 60% ethanol was added, and the mixture was centrifuged at 15,000 rpm for 30 min and rinsed. The precipitated DNA was dried in air and dissolved in 20 μl of distilled water. DNA solution (2 μl) was used as the MSP template. In the PCR assay, AmpliTaq Gold and 10X PCR buffer/MgCl2 with dNTP (Applied Biosystems, Foster City, CA, USA) were used and the results were analyzed with a GeneAmp PCR System 9700 (Applied Biosystems).
The primer sequences were 5′-CGGGTAGGGGTATCG TTCGC-3′ (sense) and 5′-AACGAAATCCACCGCCCGCC-3′ (antisense), 159 bp. Primer sequences for the unmethylated reaction were 5′-GTAGTTGGGTAGGGGTATTGTTTGT-3′ (sense) and 5′-AAACAACCTCCACCACCCACCCC-3′ (antisense), 165 bp. PCR was performed for 35 cycles (95, 65 and 72°C for 30 sec, respectively). DNA extracted from the cultured cell lines was prepared similarly for use in MSP analysis of the WRN gene.
RNA extraction and RT-PCR assay of WRN expression
Total RNA from 6 cervical cancer-derived cell lines was extracted using an RNeasy mini-kit (Qiagen, Valencia, CA, USA). cDNA was synthesized from 1 μg of total RNA using SuperScriptII Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). WRN expression was analyzed in an RT-PCR assay using 1 μl of first-strand cDNA as template. AmpliTaq Gold and 10X PCR buffer/MgCl2 with dNTP were used in the PCR assay, with analysis using a GeneAmp PCR System 9700 (Applied Biosystems). The primer sequences were 5′-GCATGTGTTCGGAAGAGTGTTT-3′ (sense) and 5′-TGACATGGAAGAAACGTGGAA-3′ (antisense), 258 bp. PCR was performed for 30 cycles (94, 57 and 72°C for 30 sec, respectively).
Demethylation treatment
Cervical carcinoma-derived SKGII and TCO-I cells with aberrant methylation of WRN were plated on 10-cm dishes at 106 cell/dish and incubated for 72 h. 5-Aza-2-deoxycytidine (5-aza) (Sigma), a demethylating agent, was then added at a final concentration of 1 μM in culture medium. After 48 h of incubation, 5-aza was added again and DNA and RNA were extracted 48 h after the second addition of 5-aza.
In vitro test of sensitivity to anticancer agents
The sensitivity of cervical carcinoma-derived cell lines to anti-cancer agents was determined using the collagen gel droplet-embedded culture drug sensitivity test (CD-DST) (27). Cells were pretreated with cell dispersion enzyme EZ (Nitta Gelatin Inc., Tokyo, Japan) for 2 h, followed by centrifugation to collect the cells. In a flask containing collagen gel, the cells were pre-incubated for 24 h and surviving cells that adhered to collagen gel were collected. Cellmatrix Type CD solution was added to the collected cells, and the suspension of cells and collagen gel was dropped onto a 6-well plate to prepare 3 drops of 30 μl each. The suspension was left to stand in an incubator at 37°C in a 5% CO2 atmosphere for 1 h for gelling and then overlaid with 4 ml/well of medium. Cisplatin (CDDP), doxorubicin (ADM), or CPT-11 was then added to the suspension at final concentrations of 0.2, 0.02 and 0.03 μg/ml, respectively. After 24 h, the anticancer agents were removed by rinsing and the cells were incubated without serum at 37°C in 5% CO2 for 7 days. The cells were dyed with neutral red, fixed with formalin, and dried. Images were collected by scanning using an image analyzer and the ratio of the volume of living cancer cells in the treated group (T) to that in the control group (C) (T/C ratio) was determined. In general, cells are considered to be highly sensitive to the agent when the T/C ratio is ≤50% (28).
Transfection of small interfering RNA (siRNA)
SKG-IIIb cells were plated on 60-mm dishes at 4×105 cell/dish and transfected 48 h later with siRNA using siFector (B-Bridge International Inc., Cupertino, CA, USA). In this procedure, 4.5 μl of siRNA stock solution (100 μM) and 295.5 μl of serum-free MEM were mixed in a test tube. In another tube, 13.5 μl of siFector and 286.5 μl of serum-free MEM were mixed. The solutions from the two tubes were mixed and incubated at room temperature for 30 min. Each dish containing SKG-IIIb cells was rinsed twice with 2 ml of serum-free MEM and 2.4 ml of serum-free MEM was then added. The incubated siRNA mixture was added to the dish at 0.6 ml/dish and incubated at 37°C in 5% CO2 for 6 h. After incubation, 3 ml of MEM containing 20% serum was added to the dish. A negative control siRNA was used as designed by B-Bridge International Inc. The siRNA sequence corresponding to the WRN gene was 5′-GUUCUUGUCACGUCCUCUGdTdT-3′. The expression levels of mRNA and protein were determined 48 h after siRNA addition. Anticancer agents were added 48 h after siRNA addition and the sensitivity of the cells to each agent was analyzed using the CD-DST.
Immunoblotting
siRNA-transfected SKG-IIIb cells were rinsed with PBS, trypsinized, and centrifuged at 15,000 rpm for 5 min at 4°C. Protein was extracted using a Mammalian Cell Extraction kit (BioVision Research Products, Mountain View, CA, USA). The sample (200 μg of protein) was mixed with sample buffer (Bio-Rad Laboratories, Hercules, CA, USA) containing the equivalent volume of 5% β-mercaptoethanol (Bio-Rad Laboratories) and the mixture was boiled for 5 min. After boiling, the mixture was electrophoresed on a 10% polyacrylamide gel and the proteins were transferred to nitrocellulose membranes (Bio-Rad Laboratories). The membranes were soaked in PBS containing 1% BSA and 0.1% Tween-20 and incubated at room temperature for 1 h for blocking. They were then reacted with anti-β-actin antibody (AC-74, Sigma-Aldrich, St. Louis, MO, USA; 5000-fold diluted) and anti-WRN antibody (4H12, Abcam, Cambridge, UK; 500-fold diluted) at 4°C overnight, followed by rinsing three times with PBS containing 0.1% Tween (PBS-T) for 10 min each. The anti-β-actin and anti-WRN antibodies were reacted with anti-mouse IgG antibody (PK-6102, Vector Laboratories, Burlingame, CA, USA) and anti-goat IgG antibody (BA-5000, Vector Laboratories; 250-fold diluted), respectively, at room temperature for 1 h. The membranes were rinsed with PBS-T three times and reacted with ABC complex (PK-6102, Vector Laboratories; pre-reacted at 4°C for 30 min) at room temperature for 1 h, then rinsed with PBS-T twice and PBS once, and visualized with DAB (Sigma).
Cell cycle analysis using flow cytometry
The cell cycle was evaluated 96 h after siRNA addition. Cells were trypsinized and rinsed twice with PBS. Supernatant was separated from the cell pellets by centrifugation at 1,000 rpm for 5 min, and 450 μl of PBS was added to the pellets and the mixture was pipetted well. As the mixture was vortexed, 1 ml of cool 100% ethanol was added. The mixture was then incubated at room temperature for 30 min for cell fixation. The cells were rinsed twice with PBS and 500 μl of RNase was added to the pellets after supernatant removal. The cells were then incubated at room temperature for 20 min. Subsequently, 500 μl of propidium iodide solution was added, the mixture was poured into a cell strainer, and the cell cycle was determined by flow cytometry using an Epics XL MCL (Beckman Coulter, Inc., Fullerton, CA, USA).
Results
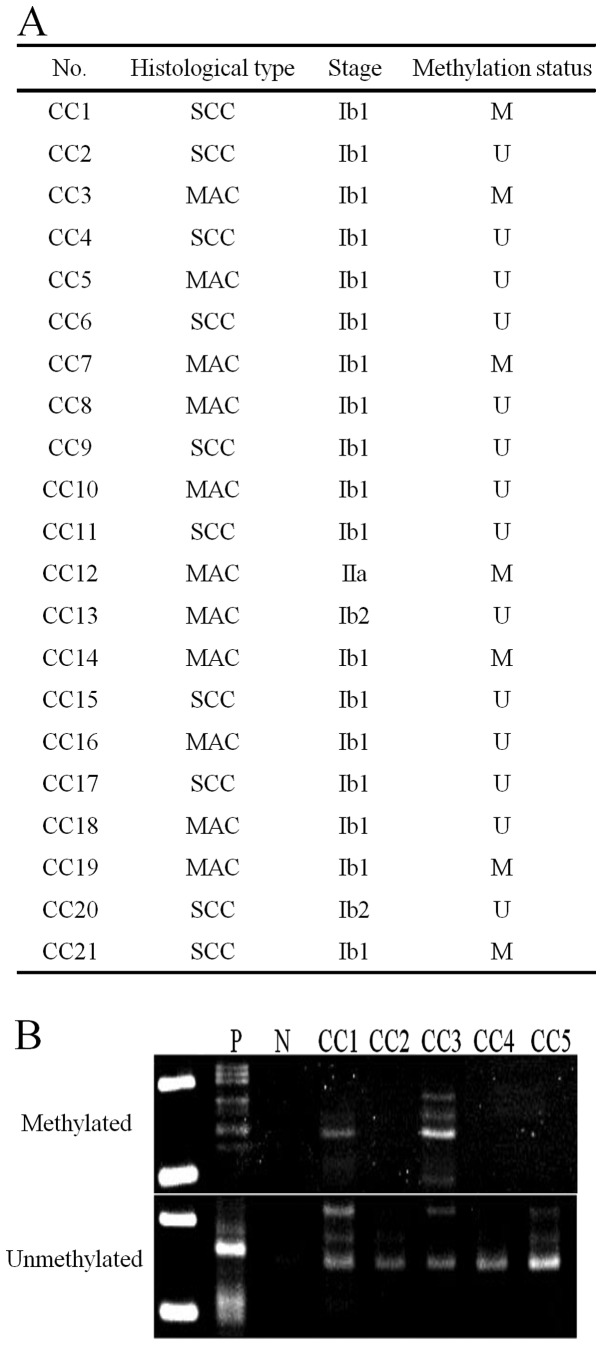
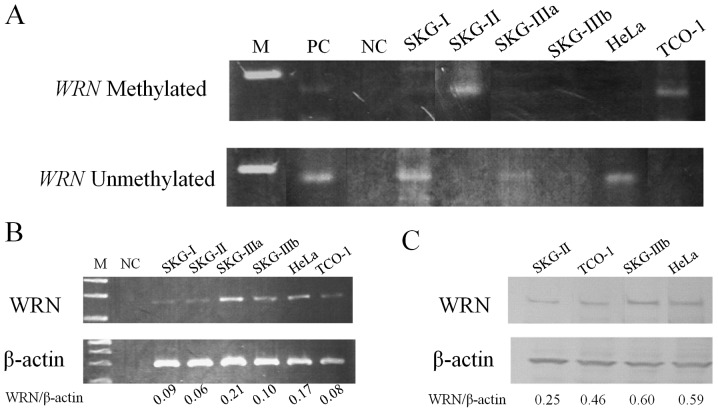
Aberrant methylation of the WRN gene in cervical cancer was examined using specimens collected for cytology (Fig. 1). Aberrant methylation was detected in 7 (33.3%) of 21 patients, including in 2 (20%) of 10 cases of squamous cell carcinoma and 5 (45.5%) of 11 cases of adenocarcinoma. Among 6 cervical cancer-derived cell lines, aberrant methylation was detected in 2 cell lines, SKG-II and TCO-I (Fig. 2A), and mRNA and protein levels for WRN were lower in these cells (Fig. 2B and C).
Figure 1.
Analysis of aberrant DNA hypermethylation of the WRN gene in cervical cancer cytologic specimens. (A) Cervical cancer specimens and methylation status of WRN. CC, cervical cancer; SCC, squamous cell carcinoma; MAD, mucinous adenocarcinoma; M, methylated; U, unmethylated. (B) MSP analysis of WRN in cervical cancer specimens. P, positive control; N, negative control.
Figure 2.
(A) MSP analysis of the WRN gene in cervical cancer-derived cell lines. Aberrant DNA hypermethylation of WRN was observed in SKG-II and TCO-1 cells. (B) Analysis of WRN expression in cervical cancer-derived cell lines using RT-PCR. WRN expression was decreased in SKG-II and TCO-1 cells, which had aberrant DNA hypermethylation of the WRN gene. (C) Western blot analysis of WRN in cervical cancer-derived cell lines. The WRN protein level was decreased in SKG-II and TCO-1 cells, which had aberrant DNA hypermethylation of the WRN gene.
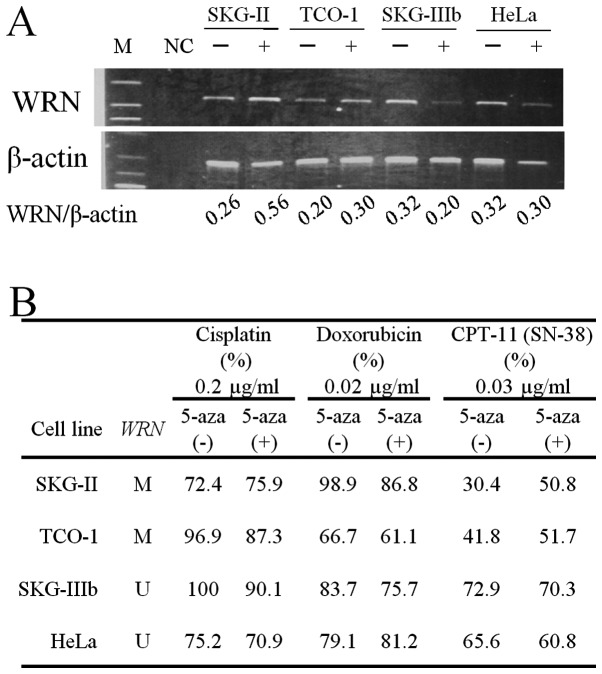
Changes in WRN mRNA levels in cervical cancer-derived cell lines were analyzed before and after treatment with 5-aza, a demethylating agent (Fig. 3A). After administration of 5-aza, WRN mRNA increased in SKG-II and TCO-I cells, in which aberrant methylation of WRN was found. Sensitivity to anticancer drugs before and after treatment with 5-aza was analyzed by CD-DST, based on the T/C ratio (Fig. 3B). Sensitivity to CDDP and ADM did not change in 4 cell lines after administration of 5-aza. For CPT-11, the T/C ratio increased to >50% in SKG-II and TCO-I cells after administration of 5-aza, showing decreased sensitivity to CPT-11.
Figure 3.
(A) Demethylation analysis of the WRN gene in cervical cancer-derived cell lines using RT-PCR. Treatment with a demethylating agent (5-aza) reactivated WRN expression in SKG-II and TCO-1 cells, which had aberrant DNA hypermethylation of the WRN gene. (B) Changes in sensitivity (T/C ratio) of cervical cancer-derived cell lines to anticancer agents after treatment with 5-aza. M, methylated; U, unmethylated.
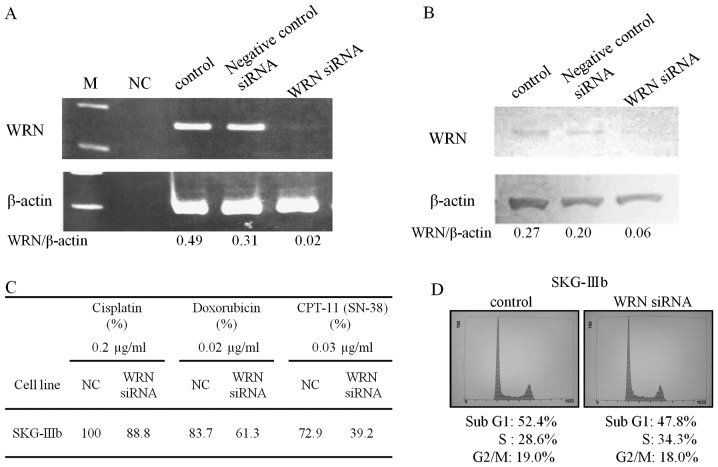
Introduction of siRNA for WRN in SKG-IIIb cells decreased the levels of WRN mRNA and protein (Fig. 4A and B). The sensitivity of the cells to CPT-11 was increased by siRNA treatment based on the marked decrease in the T/C ratio in the CD-DST (Fig. 4C). Flow cytometry indicated that the percentage of S-phase cells increased from 28.6 to 34.3% after siRNA for WRN was introduced into SKG-IIIb cells (Fig. 4D).
Figure 4.
(A) RT-PCR after WRN knockdown by siRNA in SKG-IIIb cells. (B) Western blot analysis after WRN knockdown by siRNA in SKG-IIIb cells. (C) siRNA-induced changes in sensitivity (T/C ratio) of SKG-IIIb cells to anticancer agents. After suppression of WRN expression, only sensitivity to CPT-11 was increased. (D) Cell cycle analysis of SKG-IIIb cells using flow cytometry. After suppression of WRN expression, the percentage of cells in the S-phase increased.
Discussion
This study provided the first evidence of a relationship of WRN promoter methylation with cervical cancer, with aberrant methylation of WRN detected in 33.3% of specimens of cervical cancer and in two cervical cancer-derived cell lines. Decreased WRN mRNA and protein levels were also found in both cell lines. These results suggested that aberrant methylation of WRN plays an important role in cervical cancer. The sensitivity to CPT-11 of cervical cancer cells with aberrant methylation of WRN was decreased by treatment with a demethylating agent. This treatment also increased the level of WRN mRNA, consistent with the general effect of demethylating agents on expression of many genes.
Selective downregulation of WRN expression using siRNA increased the sensitivity of cervical cancer cells to CPT-11, but not to other anticancer agents. Several previous studies have shown that WRN inactivation increases the anticancer effect of CPT-11 (13,21). CPT-11 acts on the covalent complex of topoisomerase I (Top-I) and DNA, and inhibits DNA replication and causes strand breaks (29–31). CPT-11 acts in S-phase and causes activation of S-phase checkpoint function via the ATR (ataxia and Rad-related protein)-CHK1 (checkpoint kinase-1) pathway (32–34). WRN is involved in the ATR-CHK1 pathway by recognizing the Top-I-DNA complex and detecting replication-derived DNA structures or unresolved positive supercoils (35,36). Thus, if ATR, CHK1 and WRN have reduced activity, cells are hypersensitive to CPT-11, and administration of CPT-11 to inactivated WS cells increases S-phase DSBs and unresolved recombination structures. A similar effect does not occur with etoposide, a topoisomerase II inhibitor (37). The S-phase checkpoint is activated upon DNA damage and is regulated by ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein (ATR) kinases. WRN is related to both kinases (38,39). Cells with inactivated WRN proceed to S-phase earlier than wild-type cells; however, the slow progress in late S-phase for cells with inactivated WRN causes the final S-phase lengths to be equal (40). In this study, siRNA for WRN produced a small, but insignificant, increase in the number of S-phase cells.
In chemotherapy for cervical cancer, the response rate to cisplatin is 20–30% (24) and that to monotherapy with CPT-11 has been found to be 24% (25). A combination of cisplatin and CPT-11 has a good response rate of 59% and is effective therapy (41). Taxanes also play an important role in chemotherapy for cervical cancer, with response rates of 17% for taxane monotherapy (42) and 46% for cisplatin and paclitaxel (TP) combination therapy for recurrent or advanced cervical cancer, making this regimen the current standard of care (43). We previously showed a relationship between aberrant methylation of CHFR and sensitivity to taxanes, and suggested that aberrant methylation of CHFR could serve as a molecular marker for the sensitivity of cervical cancer to anticancer drugs (44). Thus, examination of aberrant methylation of CHFR and WRN in specimens collected for cytology may be useful for prediction of treatment outcome before administration of anticancer drugs.
The standard treatment for cervical cancer is concurrent chemoradiotherapy from stages IB bulky to IIB (45). Neoadjuvant chemotherapy (NAC) is a promising approach for reduction of tumor size to increase the number of patients indicated for surgery and reduce distant metastasis through an effect on micrometastasis (46). It has been suggested that NAC could be used in stages IB2 to IIIB (47), but the efficacy of NAC remains inconclusive (48). A disadvantage of NAC is that the tumor may grow prior to the main therapy in cases that are non-responsive to NAC. However, this concern may be avoided by choice of the most effective chemotherapy based on evaluation of methylation of CHFR and WRN in specimens collected before initiation of NAC. This approach may represent a new therapeutic strategy for cervical cancer.
In conclusion, our data suggest that aberrant methylation of WRN plays an important role in carcinogenesis and sensitivity to CPT-11 of cervical cancer. This is the first report to show a relationship between the methylation of the WRN gene and sensitivity to CPT-11 in gynecologic cancer.
Acknowledgements
The authors gratefully acknowledge grant support from the Japan Society for the Promotion of Science (JSPS) through a Grant-in-Aid for Scientific Research (KAKENHI), a Grant-in-Aid for Scientific Research (B) (22390313), a Grant-in-Aid for Scientific Research (C) (22591866), and a Grant-in-Aid for Young Scientists (B) (21791573); the Ichiro Kanehara Foundation; Kobayashi Foundation for Cancer Research; and the Keio University Medical Science Fund through a Research Grant for Life Sciences and Medicine.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 3.zur Hausen H. Papillomaviruses in anogenital cancer as a model to understand the role of viruses in human cancers. Cancer Res. 1989;49:4677–4681. [PubMed] [Google Scholar]
- 4.Munoz N, Bosch FX, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 5.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 7.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16:50–59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 9.Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clin Cancer Res. 2001;7:584–589. [PubMed] [Google Scholar]
- 10.Wong Y, Chung TK, Cheung T, et al. Methylation of p16INK4A in primary gynecologic malignancy. Cancer Lett. 1999;136:231–235. doi: 10.1016/s0304-3835(98)00327-9. [DOI] [PubMed] [Google Scholar]
- 11.Dong SM, Kim H-S, Rha S-H, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7:1982–1986. [PubMed] [Google Scholar]
- 12.Kuzmin I, Liu L, Dammann R, et al. Inactivation of RAS association domain family 1A gene in cervical carcinomas and the role of human papillomavirus infection. Cancer Res. 2003;63:1888–1893. [PubMed] [Google Scholar]
- 13.Agrelo R, Cheng W-H, Setien F, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci USA. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu CE, Oshima J, Wijsman EM, et al. Mutations in the consensus helicase domains of the Werner syndrome gene. Werner’s Syndrome Collaborative Group. Am J Hum Genet. 1997;60:330–341. [PMC free article] [PubMed] [Google Scholar]
- 15.Muftuoglu M, Oshima J, Kobbe C, Cheng W-H, Leistritz DF, Bohr VA. The clinical characteristics of Werner syndrome: molecular and biochemical diagnosis. Hum Genet. 2008;124:369–377. doi: 10.1007/s00439-008-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 17.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Huang S, Lee L, Hanson NB, et al. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaheen AC, Malcolm CC, Neil RJC, et al. Two novel regions of interstitial deletion on chromosome 8p in colorectal cancer. Oncogene. 1999;18:657–665. doi: 10.1038/sj.onc.1202340. [DOI] [PubMed] [Google Scholar]
- 20.Armes JE, Hammet F, de Silva M, et al. Candidate tumor-suppressor genes on chromosome arm 8p in early-onset and high-grade breast cancers. Oncogene. 2004;23:5697–5702. doi: 10.1038/sj.onc.1207740. [DOI] [PubMed] [Google Scholar]
- 21.Futami K, Takagi M, Shimamoto A, Sugimoto M, Furuichi Y. Increased chemotherapeutic activity of camptothecin in cancer cells by siRNA-induced silencing of WRN helicase. Biol Pharm Bull. 2007;30:1958–1961. doi: 10.1248/bpb.30.1958. [DOI] [PubMed] [Google Scholar]
- 22.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 23.Hsiang YH, Lihou MG, Liu LF. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 24.Thigpen T. The role of chemotherapy in the management of carcinoma of the cervix. Cancer J. 2003;9:425–432. doi: 10.1097/00130404-200309000-00013. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi S, Dobashi K, Fujimoto S, et al. A late phase II study of CPT-11 on uterine cervical cancer and ovarian cancer. Research Groups of CPT-11 in Gynecologic Cancers. Gan To Kagaku Ryoho. 1991;18:1681–1689. (In Japanese) [PubMed] [Google Scholar]
- 26.Susumu N, Aoki D, Noda T, et al. Diagnostic clinical application of two-color fluorescence in situ hybridization that detects chromosome 1 and 17 alterations to direct touch smear and liquid-based thin-layer cytologic preparations of endometrial cancers. Int J Gynecol Cancer. 2005;15:70–80. doi: 10.1111/j.1048-891x.2005.14424.x. [DOI] [PubMed] [Google Scholar]
- 27.Kawaguchi M, Banno K, Susumu N, et al. Successful analysis of anticancer drug sensitivity by CD-DST using pleural fluid and ascites from patients with advanced ovarian cancer: case reports. Anticancer Res. 2005;25:3547–3551. [PubMed] [Google Scholar]
- 28.Kawamura M, Gika M, Abiko T, et al. Clinical evaluation of chemosensitivity testing for patients with unresectable non-small cell lung cancer (NSCLC) using collagen gel droplet embedded culture drug sensitivity test (CD-DST) Cancer Chemother Pharmacol. 2007;59:507–513. doi: 10.1007/s00280-006-0292-8. [DOI] [PubMed] [Google Scholar]
- 29.Leppard JB, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 30.Covey JM, Jaxel C, Kohn KW, Pommier Y. Protein-linked DNA strand breaks induced in mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 1989;49:5016–5022. [PubMed] [Google Scholar]
- 31.Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan S, Capasso H, Walworth NC. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast. 1999;15:821–828. doi: 10.1002/(SICI)1097-0061(199907)15:10A<821::AID-YEA422>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Cliby WA, Lewis KA, Lilly KK, Kaufmann SH. S phase and G2 arrests induced by topoisomerase I poisons are dependent on ATR kinase function. J Biol Chem. 2002;277:1599–1606. doi: 10.1074/jbc.M106287200. [DOI] [PubMed] [Google Scholar]
- 34.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol. 2007;27:5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koster DA, Palle K, Bot ESM, Bjornsti M-A, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 36.Patro BS, Frøhlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. J Cell Sci. 2011;124:3967–3979. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christmann M, Tomicic MT, Gestrich C, Roos WP, Bohr VA, Kaina B. WRN protects against topo I but not topo II inhibitors by preventing DNA break formation. DNA Repair. 2008;7:1999–2009. doi: 10.1016/j.dnarep.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 39.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 40.Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiyama T, Yakushiji M, Noda K, et al. Phase II study of irinotecan and cisplatin as first-line chemotherapy in advanced or recurrent cervical cancer. Oncology. 2000;58:31–37. doi: 10.1159/000012076. [DOI] [PubMed] [Google Scholar]
- 42.McGuire WP, Blessing JA, Moore D, Lentz SS, Photopulos G. Paclitaxel has moderate activity in squamous cervix cancer. A Gynecologic Oncology Group study. J Clin Oncol. 1996;14:792–795. doi: 10.1200/JCO.1996.14.3.792. [DOI] [PubMed] [Google Scholar]
- 43.Rose PG, Blessing JA, Gershenson DM, McGehee R. Paclitaxel and cisplatin as first-line therapy in recurrent or advanced squamous cell carcinoma of the cervix: a gynecologic oncology group study. J Clin Oncol. 1999;17:2676–2680. doi: 10.1200/JCO.1999.17.9.2676. [DOI] [PubMed] [Google Scholar]
- 44.Banno K, Yanokura M, Kawaguchi M, et al. Epigenetic inactivation of the CHFR gene in cervical cancer contributes to sensitivity to taxanes. Int J Oncol. 2007;31:713–720. [PubMed] [Google Scholar]
- 45.NCCN Clinical Practice Guidelines in Oncology. Cervical cancer guideline (Version 1. 2012) [Google Scholar]
- 46.Japan Society of Gynecologic Oncology. Kanehara & Co; 2011. Treatment Guidelines for cervical cancer. [Google Scholar]
- 47.Benedetti-Panici P, Greggi S, Colombo A, et al. Neoadjuvant chemotherapy and radical surgery versus exclusive radiotherapy in locally advanced squamous cell cervical cancer: results from the Italian multicenter randomized study. J Clin Oncol. 2002;20:179–188. doi: 10.1200/JCO.2002.20.1.179. [DOI] [PubMed] [Google Scholar]
- 48.Chang TC, Lai CH, Hong JH, et al. Randomized trial of neoadjuvant cisplatin, vincristine, bleomycin, and radical hysterectomy versus radiation therapy for bulky stage IB and IIA cervical cancer. J Clin Oncol. 2000;18:1740–1747. doi: 10.1200/JCO.2000.18.8.1740. [DOI] [PubMed] [Google Scholar]