The MIA pathway governs the localization and oxidative folding of intermembrane space proteins. This study reports that the MIA pathway is involved in the transport of mitochondrial inner membrane protein Tim22, thereby broadening the known functions of MIA to the biogenesis of inner membrane proteins.
Abstract
The mitochondrial intermembrane space assembly (MIA) pathway is generally considered to be dedicated to the redox-dependent import and biogenesis of proteins localized to the intermembrane space of mitochondria. The oxidoreductase Mia40 is a central component of the pathway responsible for the transfer of disulfide bonds to intermembrane space precursor proteins, causing their oxidative folding. Here we present the first evidence that the function of Mia40 is not restricted to the transport and oxidative folding of intermembrane space proteins. We identify Tim22, a multispanning membrane protein and core component of the TIM22 translocase of inner membrane, as a protein with cysteine residues undergoing oxidation during Tim22 biogenesis. We show that Mia40 is involved in the biogenesis and complex assembly of Tim22. Tim22 forms a disulfide-bonded intermediate with Mia40 upon import into mitochondria. Of interest, Mia40 binds the Tim22 precursor also via noncovalent interactions. We propose that Mia40 not only is responsible for disulfide bond formation, but also assists the Tim22 protein in its integration into the inner membrane of mitochondria.
INTRODUCTION
Mitochondrial proteins, with the exception of a few encoded by mitochondrial DNA and expressed within mitochondria, are synthesized on cytosolic ribosomes. Elaborate pathways have been developed in eukaryotic evolution to transport and sort mitochondrial proteins into the correct submitochondrial compartments (Neupert and Herrmann, 2007; Chacinska et al., 2009; Mokranjac and Neupert, 2009; Becker et al., 2012; Dimmer and Rapaport, 2012; Dudek et al., 2013). Virtually all mitochondrial proteins use the translocase of the outer membrane (TOM) complex for the transport across the outer mitochondrial membrane. To be targeted to the mitochondrial matrix or mitochondrial inner membrane (IM), proteins carrying a positively charged and cleavable mitochondrial signal sequence (presequence) are passed from the TOM complex to the presequence translocase of the inner mitochondrial membrane (TIM23 translocase). Another important class of IM proteins that mainly belong to the membrane-multispanning carrier family uses a different route. They are transported from the TOM complex through the intermembrane space (IMS) with the help of chaperone complexes that comprise small Tim proteins. These precursor proteins use the carrier translocase TIM22 for subsequent integration in the inner mitochondrial membrane (Rehling et al., 2003b; Neupert and Herrmann, 2007; Chacinska et al., 2009; Becker et al., 2012; Dudek et al., 2013). The core subunits of the TIM23 (Tim23 and Tim17) and TIM22 (Tim22) translocases also use the small Tim chaperone complexes in the IMS and TIM22 pathway to reach the IM (Curran et al., 2002; Truscott et al., 2002; Rehling et al., 2003a; Davis et al., 2007). The small Tim complexes of the IMS are also involved in accepting β-barrel protein precursors that emerge on the trans side of the TOM complex and their transfer to the β-barrel sorting and assembly machinery for outer membrane integration (Hoppins and Nargang, 2004; Wiedemann et al., 2004).
The IMS proteins are rich in cysteine residues, which are frequently arranged in the twin CX9C motif that is typical for the CHCH class of proteins or CX3C motif that is typical for small Tim proteins (Koehler, 2004; Gabriel et al., 2007; Longen et al., 2009). These cysteine-rich proteins use the mitochondrial intermembrane space assembly (MIA) pathway for their transport to the IMS (Endo et al., 2010; Sideris and Tokatlidis, 2010; Herrmann and Riemer, 2012; Stojanovski et al., 2012). The MIA machinery consists of two essential proteins, Mia40 and Erv1. Mia40 serves as a receptor and oxidoreductase on the trans side of the TOM complex. It specifically recognizes the IMS precursor proteins with the MISS/ITS signal and engages with them via a disulfide bond-mediated interaction (Chacinska et al., 2004; Mesecke et al., 2005; Milenkovic et al., 2009; Sideris et al., 2009; von der Malsburg et al., 2011). This interaction is a first step that initiates the transfer of disulfide bonds into IMS precursor proteins and their subsequent oxidative folding (Grumbt et al., 2007; Müller et al., 2008; Banci et al., 2009, 2010; Kawano et al., 2009; Tienson et al., 2009; Bien et al., 2010). The oxidative folding of IMS proteins catalyzed by Mia40 requires the sulfhydryl oxidase Erv1 to complete the disulfide transfer reaction (Stojanovski et al., 2008; Böttinger et al., 2012; Bourens et al., 2012). After the transfer of disulfide bonds to IMS proteins, Mia40 is released in a reduced state, and Erv1 oxidizes Mia40, thereby recharging it for the next round of precursor recognition and oxidative folding (Grumbt et al., 2007; Tienson et al., 2009; Bien et al., 2010; Banci et al., 2011). The IMS precursors leave the MIA pathway as folded and oxidized proteins that are capable of assembling into mature functional complexes (Curran et al., 2002; Vial et al., 2002; Webb et al., 2006; Gebert et al., 2008; Müller et al., 2008; Baker et al., 2009). The role of MIA in the biogenesis of soluble IMS proteins, classic MIA substrates, has been well established at the functional, mechanistic, and structural levels (Endo et al., 2010; Sideris and Tokatlidis, 2010; Herrmann and Riemer, 2012; Stojanovski et al., 2012). However, a few examples of the noncanonical substrates of MIA have been reported. Erv1 and Ccs1 (a copper chaperone involved in the IMS localization of Sod1), which do not belong to CX3C or CX9C proteins, are also the transport and oxidative folding substrates of Mia40 (Terziyska et al., 2007; Gross et al., 2011; Klöppel et al., 2011; Kallergi et al., 2012). Mia40 was shown to be involved in the import of Tim12, one of the CX3C small Tim proteins, which is not a soluble protein but is peripherally bound to the TIM22 complex in the IM (Gebert et al., 2008). In addition, the peripheral IM protein Atp23 contains a large number of cysteine residues, which are found in the oxidized disulfide state as a result of the MIA activity (Weckbecker et al., 2012). It is not known whether MIA is also involved in the biogenesis of proteins that are radically different from the aforementioned proteins.
In the present study we find that Tim22, an IM-embedded protein and core component of the carrier TIM22 translocase, is present in the cell in an oxidized state. Mia40 is found to interact with Tim22 and facilitate its transport to the IM of mitochondria. Thus we uncover a novel role of Mia40 in the biogenesis of Tim22, an important multispanning membrane protein.
RESULTS
Tim22 is oxidized upon import into mitochondria
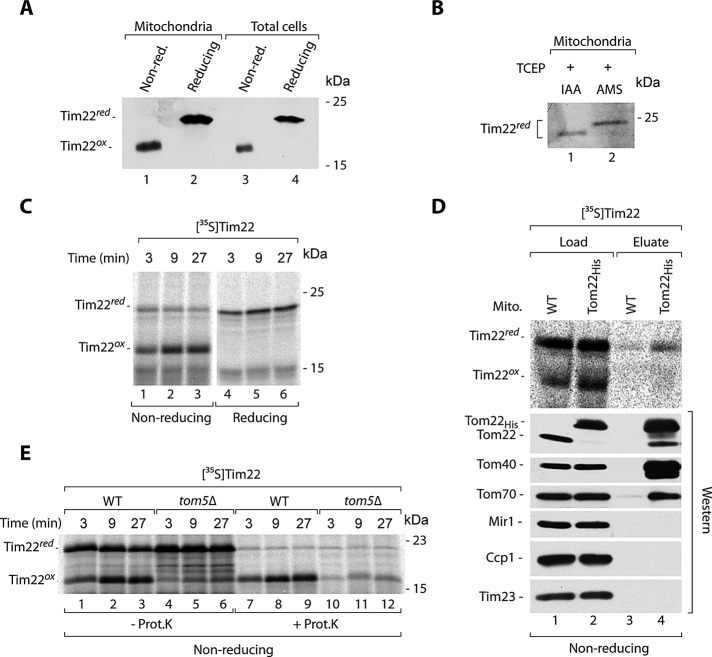
Tim22 is a main channel-forming subunit of the TIM22 translocase that serves to integrate multispanning membrane proteins, such as metabolite carriers, in the IM of mitochondria. The protein itself uses the TIM22 translocase for membrane integration to subsequently assemble into the mature translocase complex with other TIM22 subunits (the membrane proteins Tim54 and Tim18 and peripherally attached Tim9-Tim10-Tim12 complex; Kurz et al., 1999; Wagner et al., 2008). In the course of our analyses of the redox state of mitochondrial proteins, we noticed an unexpected behavior of Tim22. In isolated mitochondria, Tim22 differentially migrated on the gels under nonreducing conditions compared with standard conditions in the presence of reductant dithiothreitol (DTT; Figure 1A, lanes 1 and 2). Such a change in migration can be explained by the presence of Tim22 cysteine residues in the oxidized state. Tim22 contains two cysteine residues. The procedure of fractionation and purification of mitochondria takes several hours and may lead to the artificial oxidation of proteins in the extract. To exclude this, we analyzed the total cellular extracts obtained by rapid precipitation with trichloroacetic acid (TCA). This technique, based on lowering pH with TCA, is commonly used to preserve the redox state of protein thiols and inhibit the shuffling of existing disulfide bonds (Leichert and Jakob, 2004). Tim22 changed its migration under reducing conditions in the cellular extracts precipitated with TCA (Figure 1A, lanes 3 and 4) and thus behaved in the same way as Tim22 in isolated mitochondria. We then subjected mitochondria to the reduction of protein cysteine residues, including the cysteine residues of Tim22, and performed thiol-trapping experiments with iodoacetamide (IAA) and 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS; Figure 1B). The latter thiol-modifying agent is large, and one molecule binds to one thiol, inducing the 500-Da size shift in protein migration (Stojanovski et al., 2008; Sztolsztener et al., 2012). Subsequent to the reduction that generated free thiols from oxidized cysteine residues, Tim22 showed a specific shift when modified with AMS compared with IAA (Figure 1B). This provided additional evidence that Tim22 has its cysteine residues in an oxidized state.
FIGURE 1:
Cysteine residues of Tim22 are present in the oxidized state. (A) Isolated wild-type mitochondria and total cell extracts were separated by SDS–PAGE, followed by immunodecoration with Tim22-specific antibodies. (B) Mitochondrial proteins were reduced with 25 mM TCEP, followed by incubation with 50 mM IAA or 15 mM AMS. The samples were separated by SDS–PAGE, followed by immunodecoration with Tim22-specific antibodies. (C) Radiolabeled Tim22 precursor protein was incubated with wild-type mitochondria. Nonimported protein was removed by proteinase K. (D) Radiolabeled Tim22 precursor protein was incubated for 5 min with mitochondria isolated from WT and Tom22His yeast strains. Mitochondria were solubilized in digitonin, and protein complexes were isolated via affinity purification. Load, 4%; eluate, 100%. (E) Radiolabeled Tim22 precursor was incubated with mitochondria isolated from WT and tom5Δ yeast strains. Where indicated, nonimported protein was removed by proteinase K. WT, wild type.
We imported radiolabeled Tim22 into isolated mitochondria and observed the efficient, time-dependent generation of oxidized Tim22 in the proteinase K–protected location inside mitochondria (Figure 1C). The earliest possible location in which the proteins are resistant to externally added proteases is the IMS trans side of the TOM complex. We used a strain with histidine 10 (His10)–tagged Tom22, a core component of the TOM complex, to assess whether we can purify any of the Tim22 redox forms in association with TOM. Similar procedures have been routinely applied to investigate the association of precursor translocation intermediates with the TOM complex (Wiedemann et al., 2001; Chacinska et al., 2010). Mitochondria isolated from Tom22His and wild-type strains were incubated with radiolabeled Tim22 and subjected to solubilization with a mild detergent, digitonin, and affinity purification using nickel-nitriloacetic acid agarose (Ni-NTA; Figure 1D). By comparing the eluate to the load fraction, we observed only a slight association of reduced Tim22, but not oxidized Tim22, with the TOM complex. Tom22His, Tom40, the core component of the TOM complex, and the loosely associated receptor Tom70 were either highly enriched or present in the eluate fraction, demonstrating that the affinity purification was successful (Figure 1D). The proteins Mir1, Ccp1, and Tim23 were not eluted together with Tom22His, demonstrating the specificity of the affinity purification (Figure 1D). Taken together, results showing the resistance to externally added protease of imported Tim22 and no association between the oxidized Tim22 form and the TOM complex suggested that Tim22 oxidation occurred downstream of the TOM complex. To confirm this observation, we looked at the dependence of Tim22 oxidation on Tom5, a receptor and architectural element of the TOM complex. A deletion mutant that lacks Tom5 has been shown to express severe defects in the import of various classes of mitochondrial precursor proteins (Kurz et al., 1999; Wiedemann et al., 2001). We used mitochondria isolated from a mutant lacking Tom5 for import and oxidation experiments with Tim22. Both the import and oxidation of Tim22 were strongly inhibited in Δtom5 mitochondria compared with wild-type mitochondria (Figure 1E). Thus passage through the TOM complex was necessary for Tim22 oxidation.
Tim22 biogenesis involves interaction with Mia40 before inner membrane integration
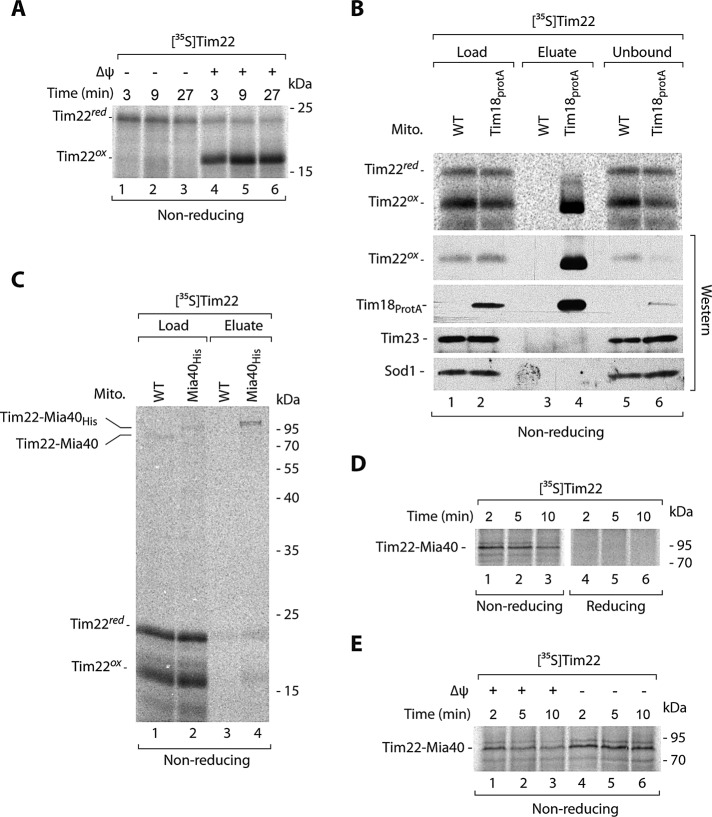
Tim22 can be oxidized in either the IMS or the IM. To differentiate between these two putative locations, we assessed the Tim22 oxidation dependence on the electrochemical potential of the IM. The IM potential is necessary for protein insertion into and translocation across the IM (Endres et al., 1999; Rehling et al., 2003a, b; Chacinska et al., 2009; Mokranjac and Neupert, 2009). Tim22 was subjected to import into energized mitochondria and mitochondria after dissipation of the electrochemical potential (Figure 2A). The oxidation of Tim22 was fully dependent on the electrochemical IM potential, suggesting that this reaction occurs at the stage of the IM. For integration into the IM, Tim22 uses the TIM22 translocase complex. We applied an affinity purification experimental schema using mitochondria with protein A–tagged Tim18 (Figure 2B), a component of the TIM22 translocase that can be used for the efficient purification of the entire TIM22 complex (Rehling et al., 2003a; Wagner et al., 2008; Gebert et al., 2011). After the import of Tim22, the extracts of Tim18ProtA and wild-type mitochondria were subjected to affinity purification. We observed an enrichment of oxidized, but not reduced, Tim22 in the eluate fraction, accompanied by a partial depletion of oxidized Tim22 in the unbound fraction (Figure 2B). This demonstrated the lack of association between reduced Tim22 and the mature translocase (Figure 2B). Tim18ProtA and mature Tim22 were efficiently eluted, whereas Tim23 and Sod1 were not eluted as expected (Figure 2B). The result on the dependence on electrochemical IM potential, supported by the presence of oxidized but not reduced Tim22 associated with TIM22, demonstrates that Tim22 oxidation depends on the IM.
FIGURE 2:
Tim22 interacts with Mia40. (A) Radiolabeled Tim22 precursor was imported into wild-type mitochondria in the presence or absence of the electrochemical IM potential (Δψ). Nonimported protein was removed by proteinase K. (B) Radiolabeled Tim22 precursor protein was incubated for 30 min with mitochondria isolated from WT and Tim18ProtA cells. Mitochondria were solubilized in digitonin, and protein complexes were isolated via affinity chromatography. Load, 3%; eluate, 100%; unbound 3%. (C) Mitochondria isolated from WT or Mia40His strains were incubated with radiolabeled Tim22 for 5 min. Mitochondria were solubilized in digitonin, and Mia40 conjugates were isolated by affinity purification. Load 3%; eluate 100%. (D) Radiolabeled Tim22 precursor was incubated with wild-type mitochondria and analyzed in the presence of IAA (nonreducing) or DTT (reducing). (E) Radiolabeled Tim22 precursor was incubated with wild-type mitochondria in the presence or absence of IM potential (Δψ), and Mia40 conjugates were analyzed. WT, wild type.
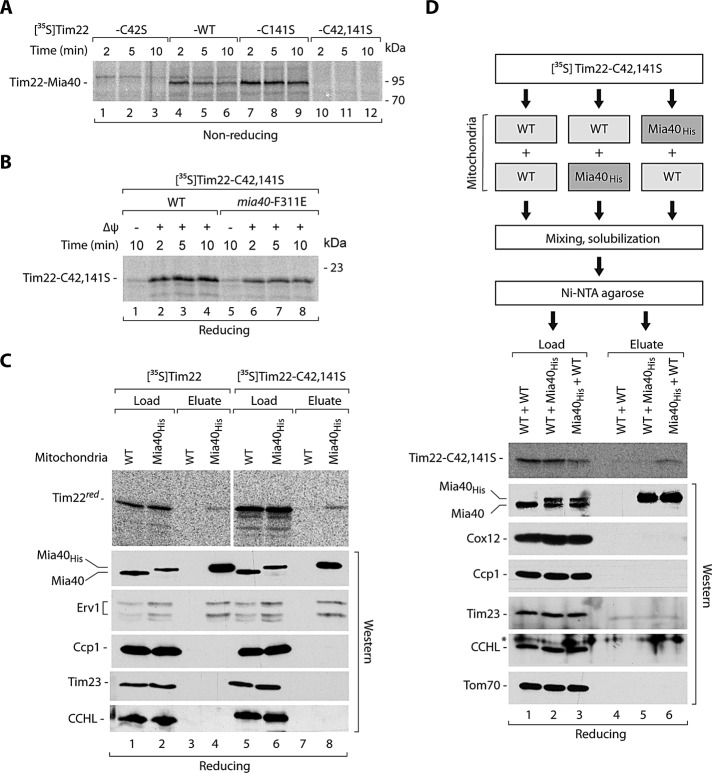
We analyzed the involvement of MIA in the Tim22 oxidation. When importing Tim22, we noticed the appearance of an additional weak band on nonreducing SDS–PAGE that could reflect the putative Tim22–Mia40 disulfide-bonded intermediate (see Figure 2C, lane 1) in analogy to the covalent intermediate complexes formed between Mia40 and its canonical substrates (Grumbt et al., 2007; Kawano et al., 2009; Milenkovic et al., 2009; Sideris et al., 2009; Bien et al., 2010; Sztolsztener et al., 2012). To provide evidence that Tim22 was indeed bound by Mia40, we imported Tim22 into Mia40His mitochondria and applied affinity purification (Figure 2C) as previously shown for Tim9 (Milenkovic et al., 2007). Tim22 formed a weak intermediate conjugate that was visible in the load fraction derived from wild-type mitochondria, and this intermediate was shifted in the extracts from mitochondria with Mia40His, consistent with an expected size shift resulting from the presence of His10 tag at Mia40 (Figure 2C, lane 2 vs. lane 1). The band representing the Tim22 conjugate was enriched in the eluate of Mia40His mitochondria, in contrast to wild-type mitochondria (Figure 2C, lane 4 vs. lane 3). The Tim22–Mia40 complex was efficiently resolved by reductant as expected (Figure 2D). Of interest, the Tim22–Mia40 complex was increased after dissipation of the IM potential (Figure 2E). This result showed that Tim22 interaction with Mia40 was independent of the electrochemical IM potential and occurred at the IMS step of Tim22 biogenesis. Because the completion of Tim22 oxidation required an electrochemical IM potential (Figure 2A), we concluded that the dissipation of an IM potential inhibited the release of Tim22 from Mia40 and thus stabilized the Tim22–Mia40 intermediate complex.
Mia40 activity is important for the biogenesis of Tim22
The temperature sensitive mutant version Mia40-3 was found to less efficiently bind classic substrates of the MIA pathway (Chacinska et al., 2004; Böttinger et al., 2012). In addition, because of premature stop codon generation, Mia40-3 is smaller than wild-type Mia40. We investigated the formation of Tim22–Mia40 intermediate upon import of Tim22 into mitochondria with Mia40-3 (Supplemental Figure S1A). Indeed, the conjugate migrated faster, confirming the presence of a smaller Mia40-3 version. However, the amount of the Tim22–Mia40 intermediate conjugate and the oxidation of imported Tim22 were not affected in the mia40-3 mitochondria compared with wild type (Supplemental Figure S1A). Tim22 assembles into the TIM22 translocase complex of 400 kDa detected using blue native electrophoresis (Rehling at al., 2003a; Wagner et al., 2008; Gebert et al., 2011). The efficiency of Tim22 assembly into the mature TIM22 complex was not affected in mia40-3 mitochondria (Supplemental Figure S1B). Thus, although Mia40 interacted with Tim22, the efficiency of Tim22 oxidation upon import into the mutant mitochondria with Mia40-3 was unchanged. We determined the steady-state levels of mitochondrial proteins in the mitochondria with Mia40-3. We noticed a decrease in the levels of the MIA substrates, including the small Tim proteins Tim9, Tim10, and Tim13 and CX9C proteins Cox17 and Pet191, whereas other mitochondrial proteins remained unchanged (Supplemental Figure S1C). Neither the mia40-3 mutation nor the observed decrease in the levels of small Tim proteins affected the import, oxidation, and assembly of Tim22.
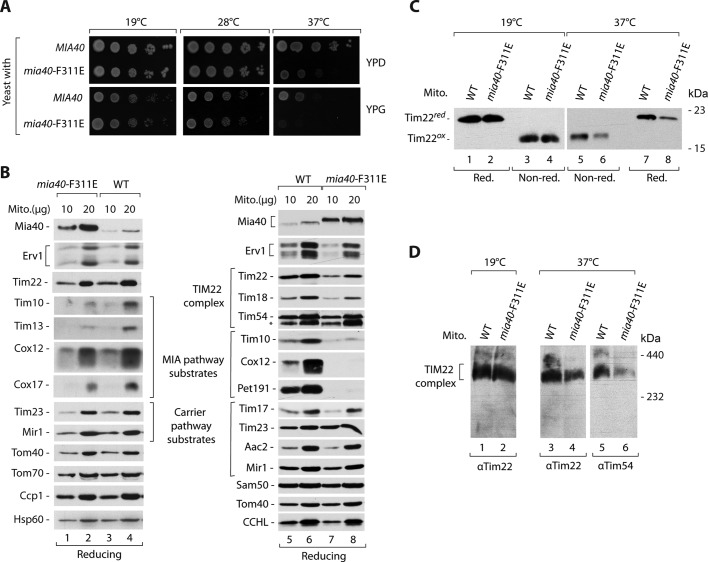
A structural analysis of Mia40 identified a phenylalanine residue at position 311 (F311) as an important part of the hydrophobic concave surface that allows Mia40 to bind its substrates (Banci et al., 2009; Kawano et al., 2009). We produced a strain with a Mia40 mutant form in which phenylalanine 311 was substituted for an aspartic acid residue (F311E) and analyzed its growth. The cells with Mia40-F311E grew normally at the lower temperature but expressed strong growth defect at the higher temperature on both carbon sources (i.e., fermentative glucose and respiratory glycerol; Figure 3A). We isolated mitochondria from the mutant cells grown under permissive conditions and analyzed the steady-state levels of mitochondrial proteins (Figure 3B). The levels of Mia40-F311E were elevated. A decrease in the levels of IMS proteins that use the MIA pathway to enter mitochondria was observed. Other mitochondrial proteins, including Tim22, were unchanged (Figure 3B, lanes 1–4). We investigated the mia40-F311E mutant mitochondria from the cells grown at the restrictive temperature (Figure 3B, lanes 5–8). We observed a decrease in the levels of Tim22, accompanied by a decrease of its partner Tim18 (Figure 3B, lanes 5–8). The other proteins that use the carrier pathway to enter the IM were not affected. The IMS proteins, including the MIA substrate Tim10, were diminished (Figure 3B, lanes 5–8). Next we analyzed the oxidation status of Tim22 in the mitochondria with Mia40-F311E from cells grown under permissive and restrictive conditions. The levels of oxidized Tim22 were unchanged under permissive conditions and were decreased under nonpermissive conditions in the mia40-F311E mutant (Figure 3C), in agreement with the observed decrease in steady-state levels. The reduced form of Tim22 was undetectable (Figure 3C). The blue native analysis also revealed a decrease in the mature TIM22 complex in the mitochondria with Mia40-F311E under restrictive conditions (Figure 3D).
FIGURE 3:
Mia40 promotes the TIM22 complex assembly. (A) Wild-type (MIA40) and mia40-F311E strains were subjected to consecutive 10-fold dilutions, spotted on YPD and YPG, and grown for 3–7 d at the indicated temperatures. (B, C) Mitochondria were isolated from WT and mia40-F311E mutant strains grown on YPG liquid medium at 19°C (lanes 1–4) or shifted to 37°C (lanes 5–8). (B) Mitochondrial proteins were analyzed by SDS–PAGE, followed by Western blotting. (C) The redox state of Tim22 was analyzed in the presence of IAA (nonreducing) or DTT (reducing). (D) Mitochondria were solubilized in digitonin-containing buffer, and the TIM22 complex was analyzed by blue native electrophoresis, followed by immunodecoration with Tim22- or Tim54-specific antibodies. WT, wild type.
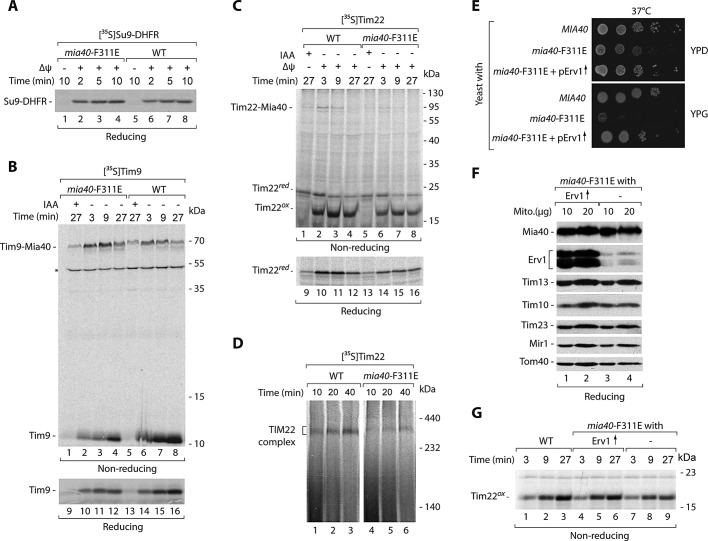
For kinetic characterization of the Tim22 biogenesis requirement for Mia40, we isolated the mia40-F311E mitochondria under permissive conditions. We determined the fitness of these mitochondria by importing Su9-DHFR, a standard IM potential–dependent precursor that follows the presequence pathway via the TIM23 translocase. Both wild-type and mia40-F311E mitochondria imported Su9-DHFR efficiently (Figure 4A), demonstrating that neither electrochemical potential nor TIM23 transport pathway was affected. We also determined whether Mia40-F311E is defective in the recognition and import of Tim9, a classic substrate of MIA (Figure 4B). Tim9 formed an intermediate with Mia40-F311E, which was more abundant in the mutant mitochondria compared with wild-type mitochondria (Figure 4B, lanes 1–8). However, the import efficiency of Tim9 decreased (Figure 4B, lanes 9–16). Thus, in the case of the canonical Tim9 substrate, the mia40-F311E mutant displayed a characteristic of another well-established mutant, mia40-4, which efficiently binds substrate proteins but is defective in the release of their oxidized forms (Chacinska et al., 2004; Stojanovski et al., 2008; Böttinger et al., 2012). These findings are consistent with the characterization of mia40-F311E mutant mitochondria reported by Kawano et al. (2009). We subjected these mitochondria to incubation with Tim22 and monitored both Tim22–Mia40 intermediate formation and oxidation under nonreducing conditions (Figure 4C, lanes 1–8) and the import of Tim22 under reducing conditions (Figure 4C, lanes 9–16). Mitochondria with Mia40-F311E were impaired in all of these functions (Figure 4C). The assembly of Tim22 into the mature TIM22 complex was less efficient in the mia40-F311E mitochondria in comparison to wild-type mitochondria (Figure 4D). Thus the interaction with Mia40 is functionally important for Tim22 biogenesis.
FIGURE 4:
Tim22 import is defective in the mia40-F311E mitochondria. (A–D) Radiolabeled precursors of Su9-DHFR, Tim9, and Tim22 were incubated with mitochondria isolated from WT and mia40-F311E mutant strains grown at 19°C. (A–C) Mitochondria were treated with proteinase K, except the Tim9 samples 1–8. (D) Mitochondria were solubilized in digitonin-containing buffer and analyzed by blue native electrophoresis. (E) Wild-type (MIA40) strain and mia40-F311E mutant transformed with the multicopy ERV1 plasmid were subjected to consecutive 10-fold dilutions, spotted on YPD and YPG, and grown for 2 d at 37°C. (F) Mitochondria were isolated from mia40-F311E– and mia40-F311E–overproducing Erv1 strain grown at 19°C and analyzed by Western blotting. (G) Radiolabeled Tim22 precursor was incubated with mitochondria isolated from the indicated strains. Mitochondria were treated with proteinase K. WT, wild type. Asterisk indicates a nonspecific band.
Involvement of Erv1 in Tim22 biogenesis
Another essential player in the MIA pathway is the sulfhydryl oxidase Erv1 (Bien et al., 2010; Banci et al., 2011; Böttinger et al., 2012). We addressed the importance of Erv1 in the biogenesis of Tim22 through analysis of the two conditional mutants of Erv1, erv1-2 and erv1-5 (Müller et al., 2008; Böttinger et al., 2012). Mitochondria isolated from mutant strains grown at the permissive temperature did not show any significant changes in the steady-state levels of mitochondrial proteins, including Mia40, Erv1, and Tim22 (Supplemental Figure S2A). Tim22 was efficiently imported into the erv1 mutant mitochondria (Supplemental Figure S2B), similar to mia40-3 and in contrast to mia40-F311E mitochondria. However, in mitochondria isolated from the erv1 mutants grown at nonpermissive conditions, a slight decrease in steady-state levels was observed for Tim22. This was accompanied by a decrease in the levels of classic MIA substrates (Tim10 and Pet191) and unchanged levels of other mitochondrial proteins, including Mia40 (Supplemental Figure S2C). The redox-state analysis confirmed the decrease of the oxidized Tim22 form in the erv1 mutants under these conditions (Supplemental Figure S2D). The effects of erv1 mutations can be indirect or can be caused by a defective cooperation of Erv1 with Mia40 in the Tim22 biogenesis. To differentiate between these two scenarios, we overproduced Erv1 in mutant cells producing Mia40-F311E. The excess of Erv1 restored the normal growth of mia40-F311E at the restrictive temperature (Figure 4E), in agreement with the previous report (Kawano et al., 2009). Mitochondria isolated from these cells contained largely increased Erv1 levels and did not show any significant changes caused by the excess of Erv1 in other mitochondrial proteins (Figure 4F). Of importance, Erv1 overproduction improved the import of Tim22 into mia40-F311E mutant mitochondria (Figure 4G). Thus Erv1 might cooperate with Mia40 in the biogenesis of Tim22.
Oxidation of cysteine residues improves the late biogenesis steps of Tim22
The oxidation of cysteine residues is blocked by the modification of thiol groups. We sought to determine how this modification affects Tim22 import efficiency. We incubated the Tim22 precursor with IAA before and during incubation with mitochondria (Figure 5A). As expected, thiol modification completely blocked the oxidation of Tim22 (Figure 5A). Furthermore, in the presence of IAA, the import of Tim22 into a protease-protected location was somewhat less efficient compared with the control reaction (Figure 5A). We also determined the efficiency of Tim22 assembly into the mature TIM22 translocase complex using native analysis. The presence of IAA during kinetic import reaction blocked Tim22 assembly into the mature complex (Figure 5B). Thus we concluded that chemical blocking of cysteine residues severely affected the late steps of Tim22 biogenesis at the level of IM integration/complex assembly.
FIGURE 5:
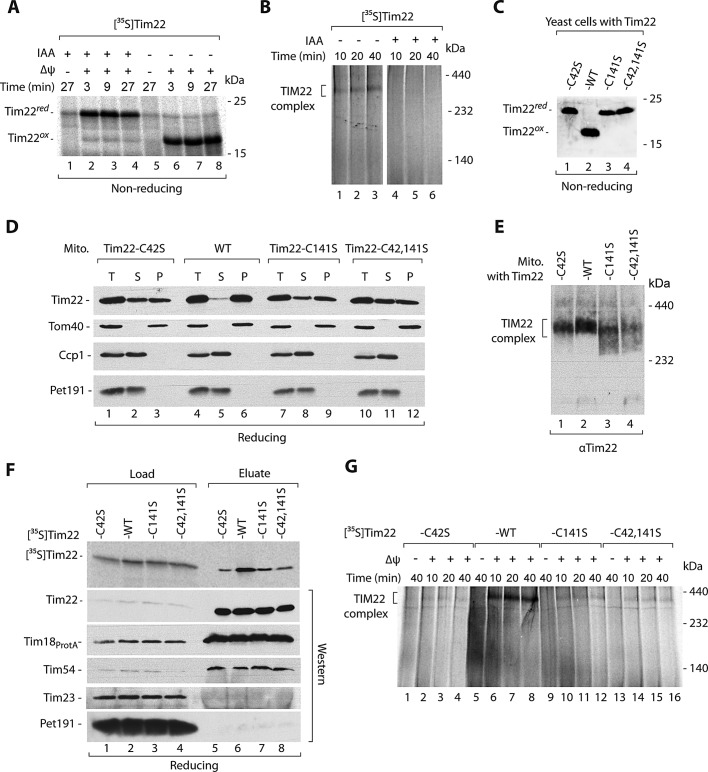
Cysteine residues of Tim22 are important for the Tim22 biogenesis. (A, B) Radiolabeled Tim22 precursor protein was preincubated for 5 min with or without 50 mM IAA. (A) Nonimported protein was removed by proteinase K. (B) Samples were solubilized in digitonin-containing buffer and analyzed by blue native electrophoresis. (C) Total protein extracts prepared from WT and tim22 cysteine mutant cells were separated on SDS–PAGE, followed by immunodecoration with Tim22-specific antibody. (D) Mitochondria isolated from WT and tim22 cysteine-residue mutants were subjected to alkaline carbonate extraction. T, total mitochondrial protein extract; S, supernatant; P, pellet of mitochondrial membranes. (E) Mitochondria isolated from WT and tim22 cysteine-residue mutants shifted to 37°C for 6 h were analyzed by blue native electrophoresis. (F) Radiolabeled wild-type Tim22 and Tim22 cysteine-mutant precursor proteins were incubated for 30 min with mitochondria isolated from Tim18ProtA yeast strains. Mitochondria were solubilized in digitonin-containing buffer, and protein complexes were isolated. Load, 3%; eluate, 100%. (G) Radiolabeled wild-type Tim22 and Tim22 cysteine-mutant precursor proteins were incubated with wild-type mitochondria. Mitochondria were solubilized in digitonin-containing buffer and analyzed by blue native electrophoresis. WT, wild type.
To assess directly the importance of Tim22 cysteine residues for Tim22 biogenesis, we constructed strains in which wild-type Tim22 was replaced by mutant forms with single cysteine residues exchanged for serine (Tim22-C42S and Tim22-C141S) and also by a double-cysteine mutant form that lacked both cysteine residues (Tim22-C42,141S). We analyzed the growth of these strains and noticed that they did not express any growth defects (Supplemental Figure S3A). We checked the redox state of mutated versions of Tim22. As expected, all of them were found in forms that migrated similarly to reduced Tim22 (Figure 5C). This confirmed that the formation of oxidized Tim22 indeed required the presence of both Tim22 cysteine residues. Furthermore, we did not detect any changes in the steady-state levels of various mitochondrial proteins, including Tim22, in the mitochondria isolated from the tim22 mutant strains (Supplemental Figure S3B). We decided to address the integrity of mature Tim22 by using carbonate extraction assay (Figure 5D). Of interest, Tim22-C42S, Tim22-C141S, and Tim22-C24,142S were found to be partially extracted from the mitochondrial membranes, in contrast to wild-type Tim22, which remained fully bound to the mitochondrial membranes (Figure 5D). The integral outer membrane Tom40 protein was also found in the mitochondrial membranes, whereas two IMS proteins, Ccp1 and Pet191, were efficiently extracted from the mitochondria, demonstrating that the carbonate extraction was successful (Figure 5D). The native analysis of mitochondria with the Tim22 cysteine-residue mutant forms isolated from the cells grown at the higher temperature revealed a loss of integrity of the mature TIM22 translocase complex (Figure 5E).
We also performed a set of kinetic assays based on the import of Tim22 and its mutant forms. Glutathione was shown to improve the oxidative folding of IMS-targeted proteins governed by the MIA pathway (Bien et al., 2010); however, we did not observe a clear stimulatory effect in the case of Tim22 import (Supplemental Figure S3C). We detected only a small decrease in the import of Tim22 cysteine-residue mutant forms compared with wild-type Tim22 protein (Supplemental Figure S3D). Next we analyzed the integration of imported Tim22 mutant versions with the TIM22 translocase using two different assays—affinity purification via Tim18ProtA and native electrophoresis. The imported cysteine-residue mutant forms of Tim22 were not efficiently coeluted together with Tim18ProtA compared with wild-type Tim22 (Figure 5F). The efficiency of Tim18ProtA purification was confirmed by the presence of equal levels of the TIM22 components Tim22 and Tim54 in the eluate fractions (Figure 5D). It is striking that the cysteine-residue mutant forms of Tim22 were defective in assembly to the mature TIM22 translocase (Figure 5G), in line with the results obtained upon thiol blocking (Figure 5B). These findings demonstrate the important role of Tim22 cysteine residues and their redox state in the late events of Tim22 biogenesis, that is, assembly into the mature complex.
Noncovalent interactions contribute to binding of Tim22 to Mia40
We also examined the importance of Tim22 cysteine residues for Mia40 binding. First, we imported radiolabeled Tim22, Tim22-C42S, Tim22-C141S, and Tim22-C42,141S into the wild-type mitochondria and performed analysis under nonreducing conditions (Figure 6A). As expected, Tim22 lacking cysteine residues could not form the covalent intermediate complex with Mia40. Of interest, the formation of Tim22–Mia40 intermediate was inhibited in the case of Tim22-C42S and increased in the case of Tim22-C141S compared with wild-type Tim22 (Figure 6A). Thus Mia40 likely engages with Cys42 of Tim22.
FIGURE 6:
Mia40 binds the incoming Tim22 precursor in a covalent and noncovalent manner. (A) Radiolabeled wild-type Tim22 and Tim22 cysteine-mutant precursor proteins were incubated with wild-type mitochondria. (B) Radiolabeled Tim22-C42,141S precursor was incubated with the mitochondria isolated from WT and mia40-F311E mutant cells. Mitochondria were treated with proteinase K. (C) Mitochondria isolated from WT or Mia40His strains were incubated with radiolabeled Tim22 or Tim22-C42,141S for 5 min. Mitochondria were solubilized in digitonin buffer, and Mia40 complexes were isolated by affinity purification. Load, 3%; eluate, 100%. (D) Radiolabeled Tim22-C42,141S was incubated with WT or Mia40His mitochondria. Mitochondria were solubilized in digitonin with wild-type or Mia40His mitochondria as indicated and subjected to affinity purification. Load, 0,5%; eluate, 100%. (A–D) The samples were analyzed by digital autoradiography or Western blotting. WT, wild type. Asterisk indicates a nonspecific band.
Our examination of the effects observed for the mia40-F311E mutant and for the tim22 cysteine-residue mutant forms revealed that they were not identical. Thus we imported Tim22-C42,141S into mitochondria with Mia40-F311E (Figure 6B). Unexpectedly, mia40-F311E mitochondria showed the same defect for Tim22 lacking the cysteine residues as for wild-type Tim22 (Figure 6B). This raised the possibility that the interaction of Tim22 with Mia40 did not absolutely require disulfide bond formation. Thus we sought to determine whether disulfide bond formation is important for binding to Mia40. We imported both Tim22 and Tim22-C42,141S to Mia40His and performed affinity purification (Figure 6C). A fraction of Tim22 was eluted together with Mia40His, as shown in the previous experiment (see Figure 2C). Tim22-C42,141S was also eluted together with Mia40His, albeit with lower efficiency (Figure 6C, compare the eluate and load fractions). As expected, Mia40His and its partner Erv1 were found in the eluate fractions, but other control mitochondrial proteins did not coelute together with Mia40His, demonstrating the specificity of Mia40His affinity purification (Figure 6C). To exclude the possibility of an interaction at the postlysis stage between Tim22 and Mia40, we performed a subsequent experiment. We imported radioactively labeled Tim22-C42,141S into wild-type and Mia40His mitochondria (Figure 6D). When the Mia40His mitochondria with preimported Tim22-C42,141S were mixed and solubilized with wild-type mitochondria and subjected to affinity purification, Tim22-C42,141S was found to be present in the elution fraction (Figure 6D, lane 6). In parallel, Tim22-C42,141S was imported into wild-type mitochondria, which were subsequently mixed and solubilized with Mia40His mitochondria. In this case, Tim22-C42,141S was not found in the eluate as anticipated for the interaction that occurs in intact mitochondria (Figure 6D, lane 5). The efficiency and specificity of the purification were confirmed by the immunodecoration for Mia40 and other control mitochondrial proteins (Figure 6D). Thus the interaction between Tim22-C42,141S and Mia40 was specific and occurred under native conditions in the intact mitochondria. In summary, Tim22 did not absolutely require its cysteine residues to be recognized and bound by Mia40.
DISCUSSION
The role of the MIA pathway in facilitating the oxidative biogenesis of soluble intermembrane space proteins is well established. As a result of MIA activity, precursor proteins adopt an oxidized state that is frequently a prerequisite for assembly into functional complexes. Thus classes of mitochondrial proteins, such as the catalysts of the MIA pathway Mia40 and Erv1 and substrate proteins of the MIA pathway, are present in the intermembrane space in an oxidized state (Endo et al., 2010; Sideris and Tokatlidis, 2010; Herrmann and Riemer, 2012; Stojanovski et al., 2012), despite the fact that the IMS is a reducing cellular environment (Kojer et al., 2012). Our finding that Tim22, a multispanning IM component and the main subunit of the TIM22 translocase, was found in an oxidized state extends the role of disulfide bond formation in the biogenesis of mitochondrial proteins. Tim22 possesses only two cysteine residues, and it is tempting to propose that they form a disulfide bond. This proposal is supported by the following observations. First, the observed modification was reversible upon treatment with a reductant (Figure 1). Second, the single-cysteine mutants of Tim22 migrated in exactly the same way as chemically reduced wild-type Tim22 (Figure 5).
We present two main lines of evidence to support a direct role of Mia40 in Tim22 oxidation and biogenesis. First, Mia40 formed an intermediate with Tim22, similar to the intermediate complexes formed by Mia40 and other client proteins (Mesecke et al., 2005; Grumbt et al., 2007; Milenkovic et al., 2009; Sideris et al., 2009; Bien et al., 2010). Second, a specific mutation, mia40-F311E, which affects the hydrophobic surface of Mia40 (Kawano et al., 2009), caused the Tim22 biogenesis defect. This defect was not mediated by decreased levels of small Tim proteins (also the substrates of MIA) that act as chaperones in assisting membrane proteins, such as Tim22 (Koehler, 2004; Endo et al., 2010; Sideris and Tokatlidis, 2010; Herrmann and Riemer, 2012; Stojanovski et al., 2012). Our rejection of the influence of small Tim proteins is based on the comparison of the two mia40 mutant strains. The mitochondria from the well-established mia40-3 mutant did not show any defect in Tim22 binding and oxidation, despite the significant reduction of the levels of small Tim proteins. In contrast, under permissive conditions, the mitochondria with Mia40-F311E exhibited a similar defect in the accumulation of small Tim proteins but a considerable slowdown in the kinetics of Tim22 oxidation, biogenesis, and mature complex assembly. Our findings on the role of Mia40 in Tim22 biogenesis also explained the decrease in the Tim22 amount observed in the mitochondria of another hydrophobic surface mutant, mia40-F334E (Kawano et al., 2009).
Our data demonstrated that hydrophobic interactions that are disrupted in the Mia40-F311E mutant are important for Tim22 binding by Mia40. This behavior of Mia40-F311E toward Tim22 is different from that to Tim9, a classic Mia40 substrate, which interacted with Mia40-F311E efficiently but was delayed in the release from Mia40 and oxidative biogenesis. This suggests a difference in the mechanism by which the oxidative biogenesis of classic soluble MIA substrates and Tim22 is catalyzed. This notion is further supported by our observation that Mia40 was also able to bind Tim22 via hydrophobic interactions without any involvement of covalent disulfide bonds, albeit with the reduced efficiency. Of interest, the atypical MIA substrate Atp23 was recently reported to bind Mia40 via hydrophobic amino acid residue stretches without the necessity for disulfide bond formation (Weckbecker et al., 2012).
An interesting issue is the involvement of the sulfhydryl oxidase Erv1, another essential component of the MIA pathway. On one hand, Erv1 in excess was shown to rescue several mia40 mutants, including mia40-F311E (Stojanovski et al., 2008; Kawano et al., 2009). We show that in the case of mia40-F311E, Erv1 not only restored the growth, but also partially improved the oxidative biogenesis of Tim22. In addition, a small decrease in the levels of oxidized Tim22 was observed in the conditional erv1 mutants grown under nonpermissive conditions. However, under these conditions Mia40 becomes inactive in the client binding (Böttinger et al., 2012). Furthermore, mitochondria isolated from erv1 mutants were not defective in Tim22 import. The mode of cooperation between Erv1 and Mia40 is complex. On one hand, Erv1 is involved in recharging Mia40 after the release of a client protein (Grumbt et al., 2007; Tienson et al., 2009; Bien et al., 2010; Banci et al., 2011). On the other hand, Erv1 directly helps Mia40 in the efficient oxidation of classic substrates by the formation of ternary complex with Mia40 and protein substrates (Stojanovski et al., 2008; Böttinger et al., 2012; Bourens et al., 2012). In addition, Erv1 is involved in the biogenesis of cytosolic iron–sulfur cluster proteins (Lange et al., 2001). Moreover, the human homologue of Erv1, ALR, was recently shown to be involved in the oxidative folding and IMS trapping of human MIA40 by a yet-unknown mechanism (Sztolsztener et al., 2012). Thus the understanding of Erv1 function and mechanism, including its putative role in the Tim22 biogenesis, requires further research.
The present results favor the following model for Tim22 biogenesis. After passage through the TOM channel, reduced Tim22 is recognized by Mia40 in the IMS. This interaction is strengthened via the intermolecular disulfide bond between Tim22 and Mia40 and eventually leads to the transfer of the disulfide bond into Tim22 and release. Tim22 release from Mia40 is coupled with the integration of Tim22 into the IM and complex assembly. Thus the Tim22 interaction with Mia40 and disulfide bond formation contribute to the efficient assembly of Tim22 into the mature functional form embedded in the IM. In summary, the identification of Tim22 as a substrate of MIA expands the function of the MIA pathway and disulfide bond transfer beyond the assembly of mitochondrial IMS proteins.
MATERIALS AND METHODS
Yeast strains and plasmid construction
Strains generated and used in this study are derivatives of Saccharomyces cerevisiae YPH499 (MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801; Sikorski and Hieter, 1989) or BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; European Saccharomyces cerevisiae Archive for Functional Analysis [EUROSCAF]; Institute for Molecular Biosciences, Johann Wolfgang Goethe-University Frankfurt, Frankfurt, Germany). The temperature sensitive mia40-3 (YPH-BG-fomp2-8; 178), erv1-2 (YBG-0702b; 235), erv1-5 (YPH-BGErv1ts-C159S; 318), and strains harboring Mia40His (yAC53-64; 277), Tom22His (31) and Tim18protA (574) were described previously (Meisinger et al., 2001; Rehling et al., 2003a; Chacinska et al., 2004; Milenkovic et al., 2007; Müller et al., 2008; Böttinger et al., 2012). The tom5Δ deletion strain (569) in the BY4741 genetic background was purchased from EUROSCARF. Mutated versions of TIM22 were generated by site-directed mutagenesis using pRS414-TIM22 (pLIW3a; 60p) plasmid as a template, resulting in generation of pRS414-TIM22-C42S (pLIW31; 61p), pRS414-TIM22-C141S (pLIW32; 62p), and pRS414-TIM22-C42,141S (pLIW321; 63p). The plasmids were transformed into the YPH499 derivative with the chromosomal tim22::ADE2 deletion and with wild-type TIM22 allele on a centromeric URA3-containing plasmid (pTIM22WT-URA3) (gift of Bernard Guiard [Centre de Génétique Moléculaire, CNRS, Gif-sur-Yvette, France]; 644). The transformants were subjected to a plasmid-shuffling procedure in the presence of 5-fluoroorotic acid for URA3 plasmid elimination as described earlier (Chacinska et al., 2008). As a result, yeast strains with a wild-type version of Tim22 (640) or Tim22-Cys42Ser (641); Tim22-Cys141Ser (642); Tim22-Cys42Ser,Cys141Ser (643) substitutions were obtained. The mia40-F311E mutant was generated using site-directed mutagenesis. pFL39-MIA40-TRP1 (pGB8220) served as a template. The obtained construct pFL39-MIA40-F311E (pMS101; 12p) was transformed into the yeast strain (182) with chromosomal mia40::ADE2 deletion in the presence of wild-type allele of MIA40 on URA3-containing plasmid (pGB8210) for plasmid shuffling. To overproduce Erv1, mia40-F311E mutant strain was transformed with the multicopy plasmid p9014-8 harboring the ERV1 gene under the control of its endogenous promoter (Stojanovski et al., 2008).
Growth of yeast and isolation of mitochondria
The cells were grown on liquid or solid (2.5% [wt/vol] agar) yeast extract/peptone/glycerol (YPG) or yeast extract/peptone/dextrose (YPD) medium containing 1% (wt/vol) yeast extract and 2% (wt/vol) bactopeptone either with 3% (wt/vol) glycerol or 2% (wt/vol) glucose as a carbon source, respectively. The selective medium without l-tryptophan (0.67% yeast nitrogen base, 0.074% complete supplement mixture minus l-tryptophan, 2% glucose) was used for the selection of transformants. To obtain mitochondria by differential centrifugation (Meisinger et al., 2006), yeast strains were grown at 19°C for 24 h or were shifted to 37°C for last 6 h. Isolated mitochondria were resuspended in SM buffer (250 mM sucrose, 10 mM 3-(N-morpholino)propanesulfonic acid [MOPS]–KOH, pH 7.2). After denaturation at 65°C for 15 min, samples were analyzed by SDS–PAGE, followed by Western blotting. To check the accessibility of free thiol groups of Tim22 protein, mitochondria were solubilized in digitonin-containing buffer (1% [wt/vol] digitonin, 10% [wt/vol] glycerol, 20 mM Tris-HCl, pH 7.4, 100 mM NaCl) in the presence of 25 mM Tris(2-carboxyethyl)phosphine (TCEP) for 20 min at room temperature. After precipitation, proteins were resuspended in Laemmli buffer containing either 50 mM IAA or 15 mM AMS and incubated at 30°C for 15 min. To analyze membrane integration of Tim22, mitochondria were incubated with 0.1 M Na2CO3 (pH 10.8) for 30 min on ice. Total (T) samples were separated into pellet (P) and supernatant (S) by ultracentrifugation for 1 h, 100,000 × g. After denaturation at 65°C for 15 min, samples were analyzed by reducing SDS–PAGE, followed by Western blotting.
In vitro import of precursor proteins
Radiolabeled precursor proteins were synthesized in rabbit reticulocyte lysate (Promega, Madison, WI) in the presence of [35S]methionine (GE Healthcare, Piscataway, NJ). Import of radiolabeled precursor proteins into the isolated mitochondria was performed at 25 or 30°C in import buffer (3% bovine serum albumin, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM methionine, 10 mM MOPS-KOH, 10 mM KH2PO4, pH 7.2) containing 2 mM ATP and 2 mM NADH, 0.1 mg/ml creatine kinase, and 5 mM creatine phosphate. Import reactions were stopped by placing samples on ice in the presence of 50 mM IAA and/or VOA (1 μM valinomycin, 20 μM oligomycin, 8 μM antimycin). Samples were treated with 50 μg/ml proteinase K for 15 min on ice. The activity of proteinase K was inhibited by 2 mM phenylmethylsulfonyl fluoride (PMSF). Samples were centrifuged (20,000 × g, 15 min, 4°C), washed with SM buffer (250 mM sucrose, 10 mM MOPS-KOH, pH 7.2, 2 mM PMSF). After denaturation at 65°C for 15 min, samples were separated by SDS–PAGE. For blue native analysis of protein complexes, mitochondrial pellets were resuspended in digitonin solubilization buffer (1% [wt/vol] digitonin, 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 10% [wt/vol] glycerol, 0.1 mM EDTA, 1 mM PMSF; Meisinger et al., 2001; Rehling et al., 2003a; Chacinska et al., 2004; Müller et al., 2008; Gebert et al., 2011). Soluble fraction was separated on a 6–16.5% gradient gel at 4°C. Radiolabeled proteins were detected by digital autoradiography (PhosphorImager Storm 820; Amersham Biosciences, Piscataway, NJ) and analyzed by ImageQuant 5.0 software (Molecular Dynamics, Sunnyvale, CA).
Affinity chromatography
Radiolabeled Tim22 precursors were imported into isolated mitochondria containing either a carboxy-terminal His10-tagged Mia40 (Mia40His), a carboxy-terminal His10-tagged Tom22 (Tom22His), or a protein A–tagged Tim18 (Tim18ProtA). Import reactions were performed at 25°C for 5–30 min. The isolated mitochondria were solubilized in digitonin-containing buffer (1% [wt/vol] digitonin, 10% [wt/vol] glycerol, 20 mM Tris-HCl, pH 7.4, 50–100 mM NaCl, 20 mM imidazole, 50 mM IAA, and 1 mM PMSF) for 20 min on ice. After clarifying spin (20,000 × g, 15 min, 4°C), supernatants were incubated with Ni-NTA (Qiagen, Valencia, CA) or immunoglobulin G (IgG)–Sepharose (GE Healthcare) for 1 h at 4°C. Ni-NTA was washed two times with washing buffer (20 mM Tris-HCl, pH 7.4, 50–100 mM NaCl, 20 mM imidazole), followed by the elution of protein complexes with elution buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 400 mM imidazole, and 50 mM IAA or 50 mM dithiothreitol). The IgG–Sepharose column was washed three times (10% [wt/vol] glycerol, 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 0.5 mM EDTA, 1 mM PMSF), followed by elution with Laemmli buffer. If necessary, eluted proteins were precipitated with StrataClean resin (Agilent Technologies, Santa Clara, CA). Samples were subjected to SDS–PAGE, followed by autoradiography and Western blotting.
Total protein cell extraction
Total protein extracts were prepared from yeast cultures of OD600 = 3.0. Cells were resuspended in lysis buffer (100 mM NaCl, 20 mM Tris-HCl, pH 7.4, 0.1% SDS, 10% TCA, 2 mM PMSF) and agitated with ∼200 μl of glass beads (Sigma-Aldrich, St. Louis, MO) for 30 min at 4°C. Collected supernatant was centrifuged (20,000 × g,15 min, 4°C), washed with ice-cold acetone, dried, and analyzed by SDS–PAGE, followed by Western blotting.
Miscellaneous
Mitochondrial proteins were resuspended in Laemmli buffer containing either 50 mM DTT (reducing conditions) or 50 mM IAA (nonreducing conditions). Proteins were separated by SDS–PAGE on 15% gels. Primary antibodies were raised in rabbits. Specific protein bands were visualized using secondary anti-rabbit antibodies conjugated with horseradish peroxidase (Sigma-Aldrich) and chemiluminescence. Signals were detected with x-ray films (Foton-Bis, Bydgoszcz, Poland). In some figures, nonrelevant gel parts were excised digitally. Protein concentration was measured by the Bradford method and with bovine serum albumin as protein standard.
Supplementary Material
Acknowledgments
We are grateful to Bernard Guiard and Peter Rehling for discussions and materials. We thank Krzysztof Kucharczyk and BioVectis (Warsaw, Poland) for sharing their expertise. The research of A.C. is supported by the Foundation for Polish Science Welcome Program, cofinanced by the European Union through the European Regional Development Fund, a European Molecular Biology Organization Installation Grant, and grants from the Ministry of Science and Higher Education in Poland (N N301 298337). L.W. and A.T. were supported by stipends from the Welcome Program.
Abbreviations used:
- AMS
4-aceto-4′-maleimidylstilbene-2,2′-disulfonic acid
- DTT
dithiothreitol
- IAA
iodoacetamide
- IM
inner membrane
- IMS
intermembrane space
- MIA
mitochondrial intermembrane space assembly
- PMSF
phenylmethylsulfonyl fluoride
- TCEP
Tris(2-carboxyethyl)phosphine
- TIM
translocase of inner membrane
- TOM
translocase of outer membrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-09-0649) on January 2, 2013.
REFERENCES
- Baker MJ, Webb CT, Stroud DA, Palmer CS, Frazier AE, Guiard B, Chacinska A, Gulbis JM, Ryan MT. Structural and functional requirements for activity of the Tim9-Tim10 complex in mitochondrial protein import. Mol Biol Cell. 2009;20:769–779. doi: 10.1091/mbc.E08-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Calderone V, Cefaro C, Ciofi-Baffoni S, Gallo A, Kallergi E, Lionaki E, Pozidis C, Tokatlidis K. Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc Natl Acad Sci USA. 2011;108:4811–4816. doi: 10.1073/pnas.1014542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, et al. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc Natl Acad Sci USA. 2010;107:20190–20195. doi: 10.1073/pnas.1010095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- Becker T, Böttinger L, Pfanner N. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci. 2012;37:85–91. doi: 10.1016/j.tibs.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Bien M, Longen S, Wagener N, Chwalla I, Herrmann JM, Riemer J. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol Cell. 2010;37:516–528. doi: 10.1016/j.molcel.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Böttinger L, Gornicka A, Czerwik T, Bragoszewski P, Loniewska-Lwowska A, Schulze-Specking A, Truscott KN, Guiard B, Milenkovic D, Chacinska A. In vivo evidence for cooperation of Mia40 and Erv1 in the oxidation of mitochondrial proteins. Mol Biol Cell. 2012;23:3957–3969. doi: 10.1091/mbc.E12-05-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M, Dabir DV, Tienson HL, Sorokina I, Koehler CM, Barrientos A. Role of twin Cys-Xaa9-Cys motif cysteines in mitochondrial import of the cytochrome c oxidase biogenesis factor Cmc1. J Biol Chem. 2012;287:31258–31269. doi: 10.1074/jbc.M112.383562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Guiard B, Müller JM, Schulze-Specking A, Gabriel K, Kutik S, Pfanner N. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J Biol Chem. 2008;283:29723–29729. doi: 10.1074/jbc.M805356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuán Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, et al. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol Cell Biol. 2010;30:307–318. doi: 10.1128/MCB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Oppliger W, Koehler CM. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 2002;21:942–953. doi: 10.1093/emboj/21.5.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Alder NN, Jensen RE, Johnson AE. The Tim9p/10p and Tim8p/13p complexes bind to specific sites on Tim23p during mitochondrial protein import. Mol Biol Cell. 2007;18:475–486. doi: 10.1091/mbc.E06-06-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Rapaport D. Unresolved mysteries in the biogenesis of mitochondrial membrane proteins. Biochim Biophys Acta. 2012;1818:1085–1090. doi: 10.1016/j.bbamem.2011.08.027. [DOI] [PubMed] [Google Scholar]
- Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta. 2013;1833:274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S. Structural basis for the disulfide relay system in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13:1359–1373. doi: 10.1089/ars.2010.3099. [DOI] [PubMed] [Google Scholar]
- Endres M, Neupert W, Brunner M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 1999;18:3214–3221. doi: 10.1093/emboj/18.12.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K, Milenkovic D, Chacinska A, Müller J, Guiard B, Pfanner N, Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J Mol Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- Gebert N, Chacinska A, Wagner K, Guiard B, Koehler CM, Rehling P, Pfanner N, Wiedemann N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 2008;9:548–554. doi: 10.1038/embor.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, et al. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell. 2011;44:811–818. doi: 10.1016/j.molcel.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Gross DP, Burgard CA, Reddehase S, Leitch JM, Culotta VC, Hell K. Mitochondrial Ccs1 contains a structural disulfide bond crucial for the import of this unconventional substrate by the disulfide relay system. Mol Biol Cell. 2011;22:3758–3767. doi: 10.1091/mbc.E11-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt B, Stroobant V, Terziyska N, Israel L, Hell K. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J Biol Chem. 2007;282:37461–37470. doi: 10.1074/jbc.M707439200. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J Biol Chem. 2012;287:4426–4433. doi: 10.1074/jbc.R111.270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins SC, Nargang FE. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem. 2004;279:12396–12405. doi: 10.1074/jbc.M313037200. [DOI] [PubMed] [Google Scholar]
- Kallergi E, et al. Targeting and maturation of Erv1/ALR in the mitochondrial intermembrane space. ACS Chem Biol. 2012;7:707–714. doi: 10.1021/cb200485b. [DOI] [PubMed] [Google Scholar]
- Kawano S, Yamano K, Naoé M, Momose T, Terao K, Nishikawa S, Watanabe N, Endo T. Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc Natl Acad Sci USA. 2009;106:14403–14407. doi: 10.1073/pnas.0901793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel C, Suzuki Y, Kojer K, Petrungaro C, Longen S, Fiedler S, Keller S, Riemer J. Mia40-dependent oxidation of cysteines in domain I of Ccs1 controls its distribution between mitochondria and the cytosol. Mol Biol Cell. 2011;22:3749–3757. doi: 10.1091/mbc.E11-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM. The small Tim proteins and the twin Cx3C motif. Trends Biochem Sci. 2004;29:1–4. doi: 10.1016/j.tibs.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Kojer K, Bien M, Gangel H, Morgan B, Dick TP, Riemer J. Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 2012;31:3169–3182. doi: 10.1038/emboj.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz M, Martin H, Rassow J, Pfanner N, Ryan MT. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol Biol Cell. 1999;10:2461–2474. doi: 10.1091/mbc.10.7.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Lisowsky T, Gerber J, Mühlenhoff U, Kispal G, Lill R. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2001;2:715–720. doi: 10.1093/embo-reports/kve161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J. Systematic analysis of the twin cx(9)c protein family. J Mol Biol. 2009;393:356–368. doi: 10.1016/j.jmb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Müller H, Meyer HE, Wagner R, Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol Cell Biol. 2001;21:2337–2348. doi: 10.1128/MCB.21.7.2337-2348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Gabriel K, Guiard B, Schulze-Specking A, Pfanner N, Chacinska A. Biogenesis of the essential Tim9-Tim10 chaperone complex of mitochondria: site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J Biol Chem. 2007;282:22472–22480. doi: 10.1074/jbc.M703294200. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Ramming T, Müller JM, Wenz LS, Gebert N, Schulze-Specking A, Stojanovski D, Rospert S, Chacinska A. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell. 2009;20:2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W. Thirty years of protein translocation into mitochondria: unexpectedly complex and still puzzling. Biochim Biophys Acta. 2009;1793:33–41. doi: 10.1016/j.bbamcr.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Müller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. Precursor oxidation by Mia40 and Erv1 promotes vectorial transport of proteins into the mitochondrial intermembrane space. Mol Biol Cell. 2008;19:226–236. doi: 10.1091/mbc.E07-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 2003a;299:1747–1751. doi: 10.1126/science.1080945. [DOI] [PubMed] [Google Scholar]
- Rehling P, Pfanner N, Meisinger C. Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J Mol Biol. 2003b;326:639–657. doi: 10.1016/s0022-2836(02)01440-7. [DOI] [PubMed] [Google Scholar]
- Sideris DP, Petrakis N, Katrakili N, Mikropoulou D, Gallo A, Ciofi-Baffoni S, Banci L, Bertini I, Tokatlidis K. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J Cell Biol. 2009;187:1007–1022. doi: 10.1083/jcb.200905134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris DP, Tokatlidis K. Oxidative protein folding in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13:1189–1204. doi: 10.1089/ars.2010.3157. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Bragoszewski P, Chacinska A. The MIA pathway: a tight bond between protein transport and oxidative folding in mitochondria. Biochim Biophys Acta. 2012;1823:1142–1150. doi: 10.1016/j.bbamcr.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Milenkovic D, Müller JM, Gabriel K, Schulze-Specking A, Baker MJ, Ryan MT, Guiard B, Pfanner N, Chacinska A. Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J Cell Biol. 2008;183:195–202. doi: 10.1083/jcb.200804095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztolsztener ME, Brewinska A, Guiard B, Chacinska A. Disulfide bond formation: sulfhydryl oxidase ALR controls mitochondrial biogenesis of human MIA40. Traffic. 2012 doi: 10.1111/tra.12030. doi:10.1111/tra.12030. [DOI] [PubMed] [Google Scholar]
- Terziyska N, Grumbt B, Bien M, Neupert W, Herrmann JM, Hell K. The sulfhydryl oxidase Erv1 is a substrate of the Mia40-dependent protein translocation pathway. FEBS Lett. 2007;581:1098–1102. doi: 10.1016/j.febslet.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Tienson HL, Dabir DV, Neal SE, Loo R, Hasson SA, Boontheung P, Kim SK, Loo JA, Koehler CM. Reconstitution of the mia40-erv1 oxidative folding pathway for the small tim proteins. Mol Biol Cell. 2009;20:3481–3490. doi: 10.1091/mbc.E08-10-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Müller H, Meisinger C, Pfanner N, Guiard B. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol. 2002;22:7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial S, Lu H, Allen S, Savory P, Thornton D, Sheehan J, Tokatlidis K. Assembly of Tim9 and Tim10 into a functional chaperone. J Biol Chem. 2002;277:36100–36108. doi: 10.1074/jbc.M202310200. [DOI] [PubMed] [Google Scholar]
- von der Malsburg K, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Wagner K, Gebert N, Guiard B, Brandner K, Truscott KN, Wiedemann N, Pfanner N, Rehling P. The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol Cell Biol. 2008;28:4251–4260. doi: 10.1128/MCB.02216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Weckbecker D, Longen S, Riemer J, Herrmann JM. Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J. 2012;31:4348–4358. doi: 10.1038/emboj.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Ryan MT. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.