During vertebrate development, the PCP pathway controls multiple cellular processes. Loss of the gene for the PCP effector Fuzzy affects formation of primary cilia via mostly unknown mechanisms. We report that Fuzzy localizes to the primary cilia and orchestrates delivery of Rab8 and Dishevelled to the primary cilium; loss of Fuzzy affects cilia-dependent signaling.
Abstract
The planar cell polarity (PCP) pathway controls multiple cellular processes during vertebrate development. Recently the PCP pathway was implicated in ciliogenesis and in ciliary function. The primary cilium is an apically projecting solitary organelle that is generated via polarized intracellular trafficking. Because it acts as a signaling nexus, defects in ciliogenesis or cilial function cause multiple congenital anomalies in vertebrates. Loss of the PCP effector Fuzzy affects PCP signaling and formation of primary cilia; however, the mechanisms underlying these processes are largely unknown. Here we report that Fuzzy localizes to the basal body and ciliary axoneme and is essential for ciliogenesis by delivering Rab8 to the basal body and primary cilium. Fuzzy appears to control subcellular localization of the core PCP protein Dishevelled, recruiting it to Rab8-positive vesicles and to the basal body and cilium. We show that loss of Fuzzy results in inhibition of PCP signaling and hyperactivation of the canonical WNT pathway. We propose a mechanism by which Fuzzy participates in ciliogenesis and affects both canonical WNT and PCP signaling.
INTRODUCTION
The planar cell polarity (PCP) signaling pathway establishes cell polarity within an epithelial plane and governs tissue morphogenesis during development (reviewed in Goodrich and Strutt, 2011). Originally discovered in fruit flies, where it regulates the uniform orientation of extracellular projections (reviewed in Maung and Jenny, 2011), the PCP pathway has been evolutionarily conserved in vertebrates and adapted to control directional cell movements during neural tube formation or the asymmetric orientation of cell projections (e.g., stereocilia in cochlear hair cells; Goodrich and Strutt, 2011). Genetic screens of Drosophila mutants with evidence of disturbed polarity patterns led to the identification of a set of “core” PCP molecules, which include Frizzled, Van Gogh, Dishevelled (Dvl), and Flamingo (Gubb and Garcia-Bellido, 1982; Vinson and Adler, 1987; Maung and Jenny, 2011). Homozygous mutations of most core PCP genes in mice are lethal during embryogenesis, causing neural tube defects (NTDs), heart anomalies, and disorganization of cochlear neurosensory cells (Kibar et al., 2001; Goodrich and Strutt, 2011).
A second group of PCP “effector” molecules was also identified in Drosophila (Fuzzy, Inturned, Fritz, and Multiple Wing Hair; Maung and Jenny, 2011). Knockdown of either Fuzzy or Inturned gene in Xenopus laevis causes an open neural tube defect (Park et al., 2006). Of interest, knockdown of these PCP genes also results in sparse cilia on multiciliated cells of the skin and on neuroepithelial cells of the developing neural tube (Park et al., 2006), suggesting for the first time a link between the PCP genes and cilial assembly. Recently a murine gene-trap knockout of Fuzzy was shown to cause a complex phenotype, encompassing defects attributable both to disruption of PCP signaling (heart defect similar to the anomalies found in homozygous Dvl1/2, Dvl2/3, or Vangl2 mutants; Hamblet et al., 2002; Henderson et al., 2001, 2006; Etheridge et al., 2008) and disruption of developmental events regulated by the cilium (e.g., polydactyly and cranial NTD; Gray et al., 2009; Heydeck et al., 2009). As in frogs, mutant cells of the Fuzzy-knockout mouse display fewer primary cilia (Gray et al., 2009; Heydeck et al., 2009).
Primary cilia are solitary, microtubule-based organelles, which project from the apical domain of most vertebrate cells and are believed to participate in signal transduction (Gerdes et al., 2009). Ciliogenesis depends on protein delivery via a complex multistep process that includes sorting and packaging of protein cargo into vesicles, targeted traffic of vesicles to the ciliary basal body, and docking and fusion of carrier vesicles to the basal body, which anchors the ciliary axoneme (Gerdes et al., 2009; Jin et al., 2010; Nachury et al., 2010). Several lines of evidence suggest that PCP molecules may be crucial for normal ciliogenesis or for cilial function. In murine or frog cells, PCP proteins Vangl2 and Dvl are present in the basal body (Ross et al., 2005; Park et al., 2008). Zebrafish or mice with Vangl2 knockdown or double-homozygous Vangl1/Vangl2 mutations display misoriented cilia in neural tube and embryonic node cells (Borovina et al., 2010; Song et al., 2010); knockdown of Dvl in Xenopus results in sparse, misaligned cilia and abnormal apical docking of basal bodies in multiciliated epidermal cells (Park et al., 2008).
During development, cilial signals are believed to exert powerful influence on a number of signaling pathways, including canonical Wnt and noncanonical Wnt (PCP) signaling pathways (Wallingford and Mitchell, 2011). The canonical Wnt pathway relies on recruitment of cytoplasmic Dvl to block the GSK3β/axin degradation complex, allowing cytoplasmic accumulation of β-catenin and its subsequent translocation to the nucleus. There, β-catenin cooperates with transcription factors of the LEF/TCF family to stimulate expression of various gene targets (van Amerongen and Nusse, 2009). Canonical Wnt signaling is hyperresponsive in murine fibroblasts derived from mutant mice lacking the ciliary motor proteins Kif3a and Ift88, suggesting that cilia somehow restrict canonical Wnt signaling (extensively reviewed in Wallingford and Mitchell, 2011). Conversely, there is failure of noncanonical Wnt signaling: both Kif3a and IFT88 disruption lead to loss of planar polarization of stereociliary bundles in cochlear hair cells (Jones et al., 2008) or to cystic kidney disease associated with disturbed mitotic spindle orientation during renal tubular elongation (Fischer et al., 2004, 2006); the PCP pathway is known to control orientation of the mitotic spindle (Fischer et al., 2006; Fischer and Pontoglio, 2009).
Although many studies demonstrated a relationship between cilia and the regulation of Wnt signaling, the mechanisms involved are unknown (Wallingford and Mitchell, 2011). In this study, we confirm that Fuzzy is involved in assembly of the primary cilium and in regulating cilial length; we show for the first time that Fuzzy localizes to the basal body and ciliary axoneme. We demonstrate that Fuzzy is needed to organize post-Golgi vesicular traffic to the primary cilium, recruit Rab8 to carrier vesicles bound for the cilium, and deliver Rab8 to the basal body and ciliary axoneme; absence of Fuzzy blocks the ciliogenic function of Rab8. We show that Fuzzy recruits Dvl2 into Rab8(+) vesicles targeted to the ciliary axoneme, indirectly inhibiting Dvl-dependent activation of canonical Wnt pathway; loss of Fuzzy disturbs Dvl1 (Dvl2 paralogue) localization at the basal body. We also demonstrate that Fuzzy serves as an arbiter of canonical Wnt and PCP pathway activities: Fuzzy−/− mutant MEFs exhibit elevated canonical Wnt signaling and concomitant defective PCP-dependent behavior.
RESULTS
Fuzzy regulates ciliogenesis and localizes to the post-Golgi vesicles and primary cilium of mammalian cells
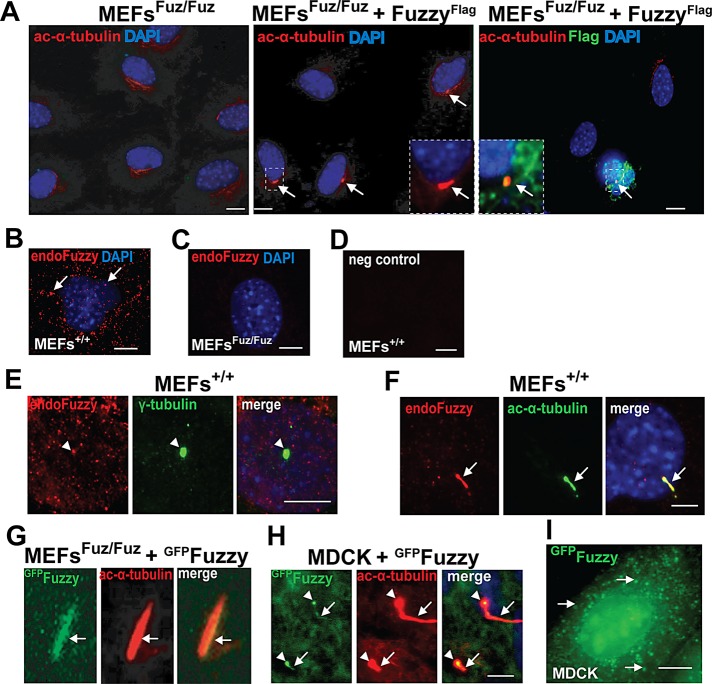
We previously described immortalized mouse embryonic fibroblast (MEF) lines established from wild-type (MEFs+/+) and homozygous Fuzzy mutant (MEFsFuz/Fuz) embryos (Seo et al., 2011). When grown for 72 h in 0.5% serum, 50–70% of the wild-type MEFs+/+ displayed primary cilia; under the same conditions, mutant MEFsFuz/Fuz remained unciliated (Figure 1A, left; Seo et al., 2011). Transient transfections of full-length human Fuzzy expression constructs fused at its N-terminus to FLAG (FuzzyFlag) or green fluorescent protein (GFP) tag (FuzzyGFP) into MEFsFuz/Fuz rescued ciliogenesis (visualized by anti–acetylated-α-tubulin antibody) in transfected cells (Figure 1A, middle); only Fuzzy-cDNA transfected cells (identified by staining with anti-FLAG antibody) were able to generate cilia (Figure 1A, middle and right). The rescue efficiencies of FLAG- and GFP-tagged Fuzzy constructs were equivalent.
FIGURE 1:
Fuzzy regulates generation of cilia and ciliary length. (A) Left, mouse embryonic fibroblasts (MEFsFuz/Fuz) derived from Fuzzy−/− embryos are unciliated; middle, expression of FuzzyFlag in MEFsFuz/Fuz restores cilia visualized with anti–acetylated-α-tubulin antibody (red, arrows, and inset); right, primary cilium (red, arrow, inset) can be visualized only in the MEFsFuz/Fuz cell transfected with FuzzyFlag cDNA (identified with anti-FLAG antibody, green and inset); bar, 10 μm. (B) In unciliated MEFs+/+, endogenous Fuzzy (red, visualized with specific rabbit anti-Fuzzy antibody) is localized to the cytoplasmic vesicles; bar, 2 μm. (C) Endogenous Fuzzy protein is undetectable in MEFsFuz/Fuz; bar 2 μm. (D) Negative control: MEFs+/+ stained with only secondary antibody; bar, 2 μm. (E) In ciliated MEFs+/+, endogenous Fuzzy (red, arrowhead) is enriched at the basal body (green) detected with anti–γ-tubulin antibody; only Z-stacks at the basal body level were merged; bar, 2 μm. (F) In ciliated MEFs+/+, endogenous Fuzzy protein (red) is localized to the primary cilium (arrows, visualized with anti–acetylated-α-tubulin antibody, green staining), only Z-stacks that contain ciliary images were merged; bar, 2 μm. (G) FuzzyGFP is localized to ciliary axoneme in MEFsFuz/Fuz transfected with FuzzyGFP cDNA. (H) MDCK cells stably overexpressing FuzzyGFP produce long cilia, and FuzzyGFP is detected at the basal body (arrowhead) and ciliary axoneme (arrows); both basal body and ciliary axoneme are visualized with anti–acetylated- α-tubulin antibody in cells permeabilized with 0.4% Triton; bar, 2 μm. (I) In nonconfluent unciliated MDCK cells, FuzzyGFP is localized to the cytosplasmic vesicles (arrows); bar, 10 μm.
To better understand the mechanism of Fuzzy involvement in ciliogenesis, we examined subcellular localization of endogenous Fuzzy protein. We detected endogenous Fuzzy protein (visualized with specific anti-Fuzzy antibody, red staining) in the intracellular vesicles in unciliated MEFs+/+ (Figure 1B, arrows), enriched at the basal body (Figure 1E, arrowheads), and at the primary cilium of the ciliated wild-type MEFs+/+ (Figure 1F). In mutant MEFsFuz/Fuz (Figure 1C) or in MEFs+/+ when specific Fuzzy antibody was omitted (Figure 1D), endogenous Fuzzy protein could not be detected.
The expression pattern of exogenous GFP-tagged Fuzzy protein, FuzzyGFP, fully recapitulated the expression pattern of endogenous Fuzzy protein. In MEFsFuz/Fuz transiently transfected with FuzzyGFP cDNA, we detected FuzzyGFP fluorescence signal in the ciliary axoneme (Figure 1G, arrow). In addition, in confluent ciliated canine renal collecting duct Madin–Darby canine kidney strain II (MDCKII) cells (Gaush et al., 1966) stably expressing FuzzyGFP, the tagged protein could be visualized at the basal body (arrowheads) and ciliary axoneme (arrows; Figure 1H). We reported previously that MDCKII cells stably overexpressing FuzzyGFP protein display a significant increase in the percentage of ciliated cells and a notable increase in ciliary length (Seo et al., 2011). In proliferating nonciliated MDCK cells stably expressing FuzzyGFP, we detected the GFP signal in vesicles localized to the perinuclear zone (Figure 1I), similar to the localization of endogenous Fuzzy seen in uncliliated MEFs+/+ (Figure 1B). By using specific markers, we established that FuzzyGFP-containing vesicles did not colocalize with the endoplasmic reticulum (ER) marker calreticulin, the ER–to–cis-Golgi apparatus marker Sec23, or the cis-Golgi apparatus marker GM130 (Supplemental Figure S1). In contrast, FuzzyGFP(+) vesicles were colocalized with the trans-Golgi apparatus/late endosomal marker syntaxin 11 (Supplemental Figure S1). Cotransfection of plasmids expressing Cherry-tagged Fuzzy (FuzzyCherry) and trans-Golgi network (TGN) marker TGN38 tagged with GFP (TGN38GFP) into MDCK cells resulted in colocalization of both proteins in the perinuclear zone (Supplemental Figure S4). Treatment of the transfected cells with brefeldin A (which specifically causes redistribution of the trans-Golgi proteins to the ER and induces morphological changes in the TGN; Reaves and Banting, 1992) disturbed colocalization of FuzzyCherry and TGN38GFP and resulted in a more diffuse localization of the two proteins (Supplemental Figure S4). Taken together, our results suggest that Fuzzy participates in vesicular trafficking between the trans-Golgi network and the plasma/ciliary membrane.
Fuzzy is required for recruiting Rab8 into post-TGN vesicles and for Rab8 delivery to the basal body
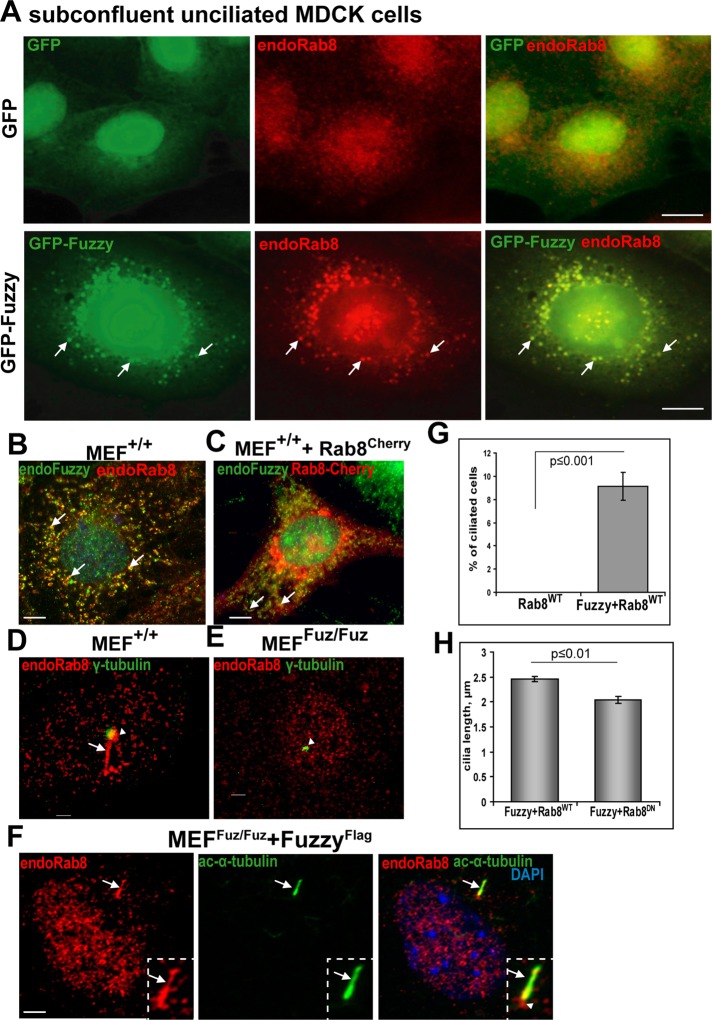
Because we detected Fuzzy in the TGN vesicles and basal body/cilium and found that Fuzzy overexpression drastically enhances ciliary length in MDCK cells (Seo et al., 2011; Supplemental Figure S3), we examined the relationship between Fuzzy and Rab8. Rab8 is a small Rab-family GTPase that is crucial for vesicular transport and ciliogenesis (Nachury et al., 2007); Rab8 is found in post-TGN vesicles, at the basal body, and at the ciliary axoneme. It is believed to catalyze docking and fusion of vesicular protein cargo with the ciliary membrane to promote ciliary growth (Nachury et al., 2007; Sato et al., 2007; Knodler et al., 2010). We found that in subconfluent unciliated MDCK cells, FuzzyGFP colocalized almost completely with endogenous Rab8 in intracellular vesicles (Figure 2A, bottom). Of importance, whereas Rab8 protein appeared as small, evenly dispersed vesicles in unciliated GFP-expressing control MDCK cells (Figure 2A, top), large Rab8(+) vesicles were evident only in FuzzyGFP-transfected MDCK cells (Figure 2A, bottom, arrows), suggesting that Fuzzy may recruit or sequester Rab8 into trafficking vesicles. We confirmed that in MEFs+/+, endogenous Fuzzy protein (red) colocalizes with endogenous Rab8 (green; Figure 2B, arrows) or with Rab8Cherry protein encoded by the transfected Rab8-Cherry cDNA (Figure 2C, arrows).
FIGURE 2:
Fuzzy stimulates formation of Rab8-positive vesicles and is required for Rab8 delivery to the basal body and primary cilium. (A) Top, unciliated MDCK cells stably expressing GFP (green) and stained with anti-Rab8 antibody (red staining). Bottom, in unciliated MDCK cells, FuzzyGFP is localized to the perinuclear cargo vesicles (green, arrows) costained with endogenous Rab8 (red, arrows); bar, 10 μm. (B) In unciliated MEFs+/+, endogenous Fuzzy (green, detected with rabbit anti-Fuzzy antibody) and endogenous Rab8 (red) are colocalized in the cytoplasmic vesicles (arrows). (C) Endogenous Fuzzy (green) is colocalized with Rab8Cherry protein (red, arrows) encoded by the transfected Cherry-tagged Rab8 cDNA; bar, 5 μm. (D) In serum-starved MEFs+/+, endogenous Rab8 (red) is localized to the basal body (green, visualized with anti–γ-tubulin antibody, arrowhead) and cilium (red, arrow). (E) Rab8 localization at the basal body is barely detectable in MEFsFuz/Fuz; bar, 1 μm. (F) Transfection of MEFsFuz/Fuz with FuzzyFlag cDNA rescues ciliogenesis and restores localization of endogenous Rab8 (red) to the basal body (arrowhead). Rab8 can also be detected in the primary cilium (arrow); cilium is detected with anti–acetylated-α-tubulin antibody (green); bar, 2.5 μm. (G) Fuzzy regulates ciliogenesis in MEFs. More than 400 transfected MEFsFuz/Fuz for each transfection condition (wild-type Rab8 alone, Fuzzy plus wild-type Rab8) were scored for the presence of cilia in two independent experiments. (H) Dominant-negative Rab8Cherry affects ciliary length in MEFsFuz/Fuz, where cilia were restored by transfections with Fuzzy cDNA. Ciliary length was measured in ≥40 ciliated (positive for Rab8Cherry) cells per condition. Mean and standard errors are shown.
The function of Rab8 in ciliogenesis requires its delivery to the basal body, where it is stabilized in the active GTP-bound form by Rab8 GDP/GTP exchange factor (Rabin8; Hattula et al., 2002; Nachury et al., 2007). On the basis of our observations in MDCK cells, we hypothesized that Fuzzy may be important for Rab8 delivery to the basal body. Indeed, we identified Rab8 (red) in the basal bodies (green, visualized with anti–γ-tubulin antibody) and/or in the newly generated cilia (visible because Rab8 localizes to the ciliary axoneme) in ∼60% of MEFs+/+ (Figure 2D, arrowhead and arrow, respectively); however, in MEFsFuz/Fuz, we did not observe Rab8 enrichment in the basal bodies (Figure 2E, arrowhead, and Supplemental Figure S2). Transfection of MEFsFuz/Fuz with FuzzyFlag cDNA rescued ciliogenesis and restored Rab8 expression (red) at the basal body and the cilium (Figure 2F). Consistent with a role for Fuzzy in delivery of Rab8 to the basal body, expression of an exogenous wild-type Rab8 alone in MEFsFuz/Fuz could not rescue ciliogenesis; in contrast, cilia reappeared when mutant cells were cotransfected with FuzzyFlag cDNA and Rab8 (Figure 2G). We reasoned that if Fuzzy contributes to ciliogenesis by delivering Rab8, then loss of Rab8 function in fibroblasts expressing Fuzzy might disturb ciliogenesis. Indeed, we observed a modest but significant reduction of ciliary length when MEFsFuz/Fuz were transfected with FuzzyFlag and a dominant-negative form of Rab8 (Hattula et al., 2002; Figure 2H).
Fuzzy regulates subcellular localization of Dishevelled by interacting with Dvl2 and recruiting it to the Rab8(+) trafficking route
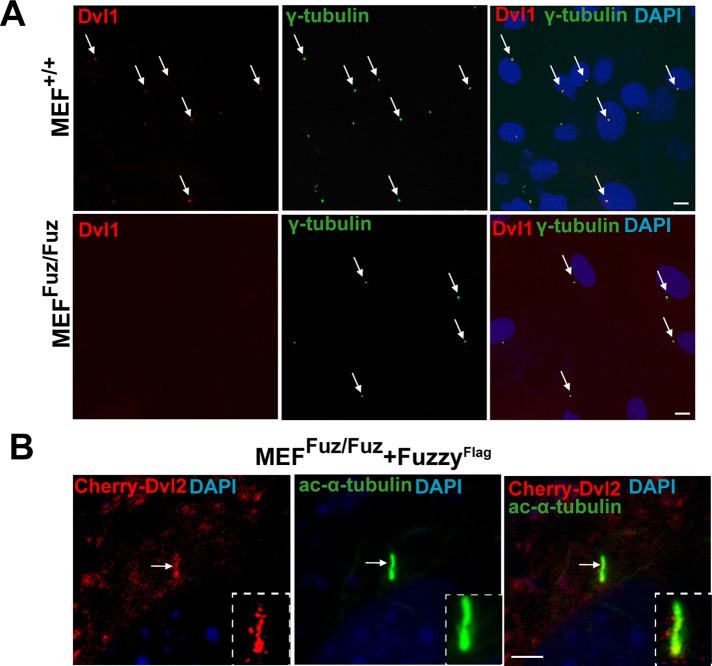
In flies, Fuzzy genetically interacts with Dvl (Lee and Adler, 2002), whereas in Xenopus, knockdown of Fuzzy (Park et al., 2006; Gray et al., 2009) or Dvl (Park et al., 2008) results in similar decrease in ciliation of skin cells, suggesting that Fuzzy and Dvl likely regulate ciliogenesis via the same signaling pathway. However, the precise relationship between the two proteins is not known. The three mammalian homologues of the Drosophila Dishevelled gene (Dvl1, 2, and 3) are known to participate in PCP signaling in mice (Wang et al., 2006; Etheridge et al., 2008). Dvl2 protein is better characterized with regard to the PCP signaling. For localization studies, we used a Dvl2 expression construct and anti-Dvl1 antibody (due to the lack of anti-Dvl2 antibody that can be used reliably for immunofluorescence microscopy). In the MEFs+/+, we identified Dvl1 at the basal bodies (Figure 3A, top), but, surprisingly, we detected almost no Dvl1 signal at the basal bodies of mutant MEFsFuz/Fuz fibroblasts (Figure 3A, bottom), suggesting that Fuzzy may regulate Dvl localization. Consistent with this hypothesis, we found that exogenous Cherry-tagged Dvl2 (Dvl2Cherry) could be detected in ciliary axoneme in ciliated MEFsFuz/Fuz when ciliogenesis was rescued by transfection with FuzzyFlag cDNA (Figure 3B).
FIGURE 3:
Fuzzy interacts with Dvl2 and regulates its subcellular localization. (A) Top, endogenous Dvl1 protein (visualized with anti-Dvl1 antibody, red staining, arrows) is detected at the basal body visualized with anti–γ-tubulin antibody (green) in MEFs+/+. Bottom, Dvl1 protein is lost at the basal body in MEFsFuz/Fuz; bar, 10 μm. (B) In MEFsFuz/Fuz cotransfected with FuzzyFlag and Dvl2Cherry, Dvl2Cherry is detected in the primary cilium (visualized with acetylated-α-tubulin, green); bar, 2.5 μm.
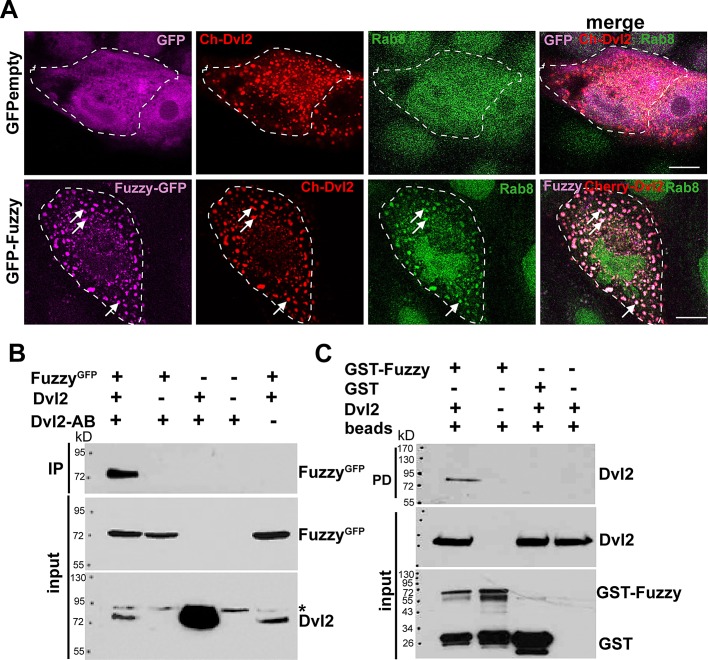
To corroborate our results in an alternative cell system, we transiently transfected Dvl2Cherry cDNA into either control MDCK stably expressing GFPempty or MDCK cells stably expressing FuzzyGFP. In the presence or absence of exogenous Fuzzy protein, Dvl2Cherry protein appeared in vesicles of various sizes throughout the cytoplasm (Figure 4A), consistent with previous reports (Schwarz-Romond et al., 2005; Smalley et al., 2005). However, in unciliated MDCK cells expressing exogenous FuzzyGFP, Dvl2Cherry completely colocalized with FuzzyGFP-containing vesicles (Figure 4A, bottom). Of importance, these Fuzzy(+) vesicles were also positive for Rab8 (Figure 4A, bottom, arrows), whereas in control MDCK/GFPempty cells, we did not detect colocalization of Dvl2Cherry and endogenous Rab8 (Figure 4B, top). Taken together, our results suggest that Fuzzy recruits Dvl2 into Rab8(+) vesicular traffic bound for the basal body and primary cilium.
FIGURE 4:
Fuzzy recruits Dvl2 into Rab8-positive trafficking vesicles. (A) Top, in control MDCK cells stably expressing GFPempty vector (magenta color was assigned to the GFP protein), Dvl2Cherry (red) does not localize with Rab8 (green). Bottom, in the presence of FuzzyGFP (magenta color was assigned to FuzzyGFP protein), Dvl2Cherry (red staining) is recruited into vesicles together with Rab8 (green, arrows). Green color was assigned for better visibility to Rab8 protein that was visualized with mouse anti-Rab8 and anti-mouse Alexa633-conjugated secondary antibody. Bars, 10 μm. (B) Coimmunoprecipitation with anti-Dvl2 antibody reveals protein–protein interactions between Dvl2 and Fuzzy. The asterisk identifies the presence of a nonspecific protein species cross-reacting with anti-Dvl2 antibody. (C) GST-Fuzzy but not GST interacts with Dvl2 in the GST pull-down assay.
Because Fuzzy appears to colocalize with Dvl and influence its subcellular localization, we tested whether there was a biochemical link between Fuzzy and Dvl. As seen in Figure 4B, Fuzzy could be immunoprecipitated by specific anti-mouse Dvl2 antibody from cell lysates containing exogenous FuzzyGFP and Dvl2 proteins. We could not detect Fuzzy in the absence of the exogenous Dvl2 or when anti-Dvl2 antibody was omitted. (Figure 4B, lanes 2 and 5), suggesting that Fuzzy forms a specific protein complex with Dvl2. To corroborate our findings, we performed glutathione S-transferase (GST)–Fuzzy pull-down assays. We identified Dvl2 protein in the precipitates containing GST-Fuzzy protein (Figure 4C, lane 1) but not when GST alone or beads alone were mixed with the HEK293 lysates containing exogenous Dvl2 protein (Figure 4C, lanes 3 and 4, respectively).
Loss of Fuzzy results in hyperactivation of the canonical Wnt pathway
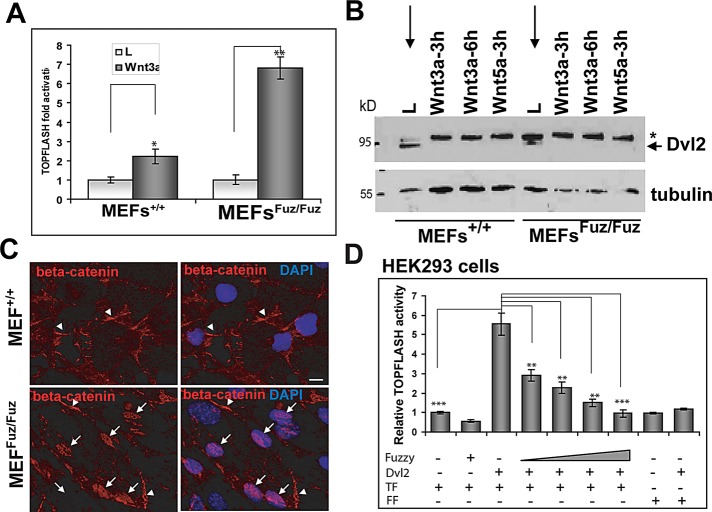
Because loss of cilia has been associated with hyperactivity of the canonical Wnt pathway in MEFs from Kif3a- and Ift88-knockout mice (Corbit et al., 2008) and we found that Fuzzy interacts with Dvl2, we tested whether Fuzzy modulates canonical Wnt signaling. MEFs+/+ and MEFsFuz/Fuz were transfected with TOPFLASH luciferase reporter and grown in the low-serum medium to induce ciliogenesis. The activation of Wnt canonical signaling was assessed in the presence of Wnt3a (Figure 5A). Whereas MEFs+/+ exhibited twofold activation by Wnt3a, MEFsFuz/Fuz displayed greater-than-sixfold stimulation of TOPFLASH activity. As in Kif3a-mutant cells (Corbit et al., 2008), hyperactivation of Wnt canonical pathway in MEFsFuz/Fuz fibroblasts was accompanied by profound hyperphosphorylation of Dvl2 (Figure 5B, star) and stabilization of β-catenin in cytoplasm (arrowheads) and nucleus (arrows) when cells were exposed to control conditioned medium from L-cells (Figure 5C). Because cytoplasmic Dvl is reported to activate TOPFLASH in a variety of cells (Smalley et al., 2005; Schwarz-Romond et al., 2007b), we tested whether Fuzzy had an effect on the Dvl-induced canonical Wnt signaling. We cotransfected HEK293 cells with TOPFLASH or FOPFLASH reporters, a Dvl2 expression construct and increasing concentrations of a Fuzzy expression vector, and/or empty pcDNA3.1 plasmid. We observed greater-than-fivefold stimulation of TOPFLASH by Dvl2; Fuzzy inhibited this stimulation in a dose-dependent manner (Figure 5D).
FIGURE 5:
Loss of Fuzzy leads to hyperactivation of canonical Wnt pathway. (A) Activation of TOPFLASH reporter transfected into MEFs+/+ and MEFsFuz/Fuz treated with control L-cell or L-Wnt3a conditioned medium; t test, *p ≤ 0.05, **p ≤ 0.001; experiments repeated three times in duplicate. (B) Dvl2 is hyperphosphorylated (asterisk) in unstimulated mutant MEFsFuz/Fuz (lane L) but not in the wild-type MEFs+/+ (lane L), in which Dvl2 becomes phosphorylated only upon treatment with Wnt3a or Wnt5a-conditioned medium. (C) β-Catenin is localized at the periphery (arrowheads) of unstimulated wild-type MEFs+/+ (top); β-catenin is found mostly in nuclei (arrows) of unstimulated mutant MEFsFuz/Fuz (bottom). Bars, 10 μm. (D) Fuzzy suppresses Dvl2-stimulated TOPFLASH activity in dose-dependent manner: 1, 2.5, 5, and 10 ng of Fuzzy were used together with 15 ng of Dvl2 and 50 ng of TOPFLASH/FOPFLASH cDNA transfected into HEK293 cells. Empty pcDNA3.1 vector was used to adjust total cDNA levels in each sample. ANOVA, followed by t test; **p ≤ 0.001, ***p ≤ 0.0001; TF-TOPFLASH, FF-FOPFLAH control reporters. Experiments were repeated three times in triplicate for each point.
Loss of Fuzzy disturbs function of the PCP pathway
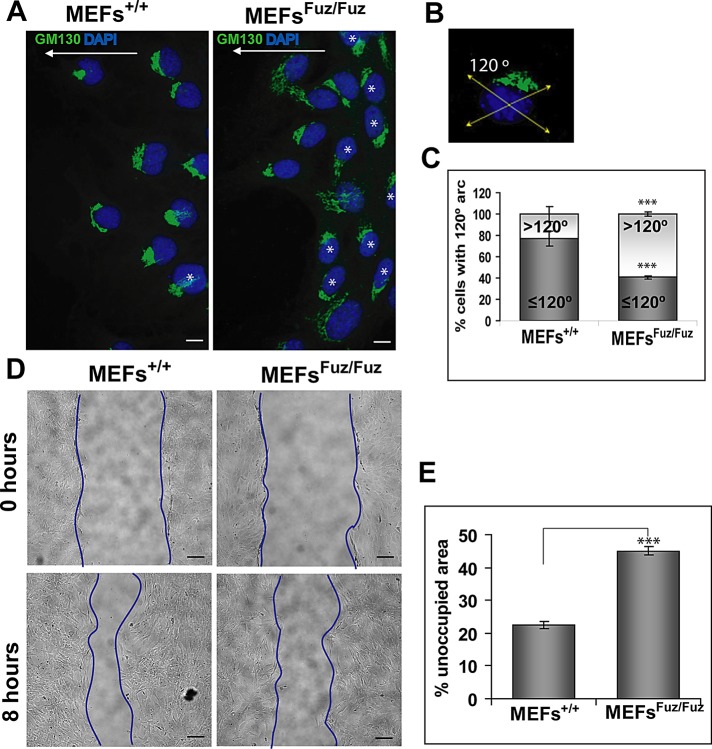
Fuzzy participates in PCP signaling during prehair initiation in the Drosophila wing cells (Gubb and Garcia-Bellido, 1982; Collier and Gubb, 1997) and in neurulating Xenopus laevis embryos (Park et al., 2006). To test whether Fuzzy acts in vertebrate PCP signaling, we analyzed whether loss of Fuzzy affects directional cell movement and cell polarization of MEFs+/+ and MEFsFuz/Fuz in a wound-healing scratch assay. Mammalian wound healing occurs as a result of coordinated cell movement into the wound and is believed to be controlled by the PCP pathway (Caddy et al., 2010). In cells at the edge of a wound, the nucleus, actin cytoskeleton, and Golgi apparatus are polarized in the axis of cell migration (Hall, 2005). Six hours after the scratch, 76.8 ± 10.0% of wild-type MEFs+/+ displayed a polarized Golgi network within a 120° arc in front of the nucleus (Figures 6, A–C). However, only 40.5 ± 2.7% of mutant MEFsFuz/Fuz exhibited polarization of the Golgi apparatus in front of the nucleus; in the remaining 59.5% of mutant cells, Golgi orientation was randomized (Figure 6A, stars). Eight hours postscratch, MEFs+/+ had moved into the scratch, leaving 22.5 ± 4.7% of unoccupied area of the initial wound surface. In contrast, MEFsFuz/Fuz had covered a significantly smaller fraction (45.2 ± 5.3% of area unoccupied by cells; p < 0.5−18) of the initial wound (Figure 6, D and E).
FIGURE 6:
Loss of Fuzzy affects cell polarization and directional movements. (A) Left, most MEFs+/+ are polarized and display Golgi network (GM-130 antibody, green) within a 120º arc in front of nucleus in the direction of migration (indicated as white arrows). Right, the Golgi network position is randomized (>120º) in many MEFsFuz/Fuz (marked with white asterisk); bars, 10 μm. (B) Depiction of 120º arc. (C) Statistical analysis of MEFs+/+ and MEFsFuz/Fuz polarizations presented as percentages of cells with ≤120º and >120º arc, respectively; chi-square test, ***p ≤ 0.0001. (D) Representative images of MEFs subjected to the wound assay. Top, 0 h; bottom, 8 h; bars, 40 μm. (E) Statistical analysis of the wound assay showing percentage of unoccupied surface area 8 h after the scratch; t test. Experiments were repeated four times with two independent MEFsFuz/Fuz and 2 MEFs+/+ clones.
DISCUSSION
The role of Fuzzy in ciliogenesis was described recently; however, mechanisms of Fuzzy involvement in generating primary cilium are mostly unknown. In this study, we analyze subcellular localization of Fuzzy protein and molecular mechanisms by which Fuzzy regulates cilia formation and functions. We establish that Fuzzy localizes to TGN-originating vesicles, basal body and primary cilium, and it participates in assembly of the primary cilium by controlling localization of the key ciliary regulator, Rab8. We also show that Fuzzy arbitrates activities of both canonical Wnt/β-catenin and noncanonical Wnt/PCP pathways via its interactions with Dvl and regulation of Dvl subcellular localization.
Fuzzy is required for assembly of the cilium
Cilia are generated by Rab8-positive vesicular traffic, which carries protein cargo from the trans-Golgi network to the base of the primary cilium (Nachury et al., 2007; Das and Guo, 2011). In subconfluent, proliferating, wild-type MEFs grown in high serum-containing medium, we identified endogenous Fuzzy in Rab8(+) vesicles. In subconfluent MDCK cells, we were able to localize GFP-tagged Fuzzy to vesicles expressing markers (syntaxin 11 and Rab8) of the TGN/late endosome compartment. In confluent, ciliated cells, we identified Fuzzy in the basal body and ciliary axoneme. Taken together, these observations establish Fuzzy as a bona fide ciliary protein and suggest that Fuzzy is delivered to the primary cilium via the Rab8(+) vesicular traffic from the trans-Golgi that delivers other ciliary proteins. Of interest, in the absence of Fuzzy, ciliogenesis is compromised and Rab8 is absent in basal bodies; simple overexpression of Rab8 is insufficient to restore cilia. When exogenous Fuzzy is expressed in these cells, ciliogenesis is rescued and Rab8 is again identifiable in the primary cilium. These results suggest that Fuzzy is required for Rab8 delivery to the cilium and that defective ciliogenesis in Fuzzy-mutant cells might be attributed in part to failure of vesicular cargoes to deliver Rab8 and other proteins to their final destination.
In our experiments, MDCK cells overexpressing Fuzzy display dramatically elongated (up to 10- to 15-fold) cilia, suggesting that Fuzzy might act as a powerful regulator of ciliary elongation. Rab8 enters the ciliary axoneme and catalyzes ciliary lengthening in immortalized retinal pigmental epithelial cells (Nachury et al., 2007). Of interest, a GTP-locked, constitutively active form, Rab8Q67L, has a particularly potent effect on ciliary elongation (Nachury et al., 2007). The striking increase in the length of cilia in MDCK cells overexpressing Fuzzy suggests that Fuzzy might facilitate stabilization of active Rab8 in the ciliary axoneme. In addition to Rab8, there are several other recently identified proteins that specifically affect ciliary length (Kim et al., 2010; Wiens et al., 2010). These include small GTPases (e.g., Arl6/BBS3), actin-binding proteins (e.g., ACTR3), and GTPase activators (e.g., RGS3). The link between cilia and actin-binding proteins is particularly intriguing, given that Fuzzy disruption in flies and frogs was reported to result in profound actin mislocalization (Wong and Adler, 1993; Collier and Gubb, 1997) and/or depolymerization (Park et al., 2006).
The link between Fuzzy and Rab8 is reminiscent of the association between Rab8 and the Bardet–Biedl syndrome proteins (Nachury et al., 2007). Eight of the known 14 BBS proteins are organized into a stable multimolecular complex (the BBSome), which is believed to orchestrate polarized vesicular traffic of cargo proteins from the trans-Golgi network to the cilium (Nachury et al., 2007; Jin and Nachury, 2009). BBS1 directly interacts with Rabin8, and the BBSome facilitates Rabin8 localization to the basal body. There Rabin8 promotes activation of Rab8 so that the active GTP-bound form of the Rab8 enters the axoneme and catalyzes biogenesis of the ciliary membrane (Nachury et al., 2007; Westlake et al., 2011). Of note, the phenotype of the Fuzzy-knockout mouse (Gray et al., 2009; Heydeck and Liu, 2011; Zhang et al., 2011) overlaps with many features of the (somewhat milder) phenotypes of BBS-mutant mice (Gray et al., 2009; Heydeck et al., 2009). We therefore hypothesize that Fuzzy might participate in the same vesicle trafficking route as the BBSome. In support of this hypothesis, a known partner of Fuzzy and Inturned, Fritz, was recently shown to be the mutant protein involved in some cases of BBS and Meckel-Gruber syndrome (Kim et al., 2010). This raises the interesting possibility that Fuzzy mutations might also underlie these human diseases.
A role for Fuzzy in regulation of canonical Wnt signaling
Although the issue remains controversial (Wallingford and Mitchell, 2011), several lines of evidence support the proposal that the primary cilium restricts canonical Wnt/β-catenin signaling: 1) loss of the ciliary motor protein Kif3a stabilizes β-catenin in the cytoplasm and increases phosphorylation of Dvl2 and Dvl3 (Gerdes et al., 2007; Corbit et al., 2008); 2) loss of the ciliary protein inversin (mutated in patients with nephronophthisis type 2) leads to inappropriate activation of the canonical Wnt pathway (Simons et al., 2005); 3) canonical Wnt pathway constituents (e.g., β-catenin and Dvl) are localized to the basal body (Corbit et al., 2008); and 4) there is increased TOPFLASH reporter activity in HEK293 cells after knockdown of Bbs1, Bbs4, and Bbs6 (Gerdes et al., 2007). We found that loss of Fuzzy results in hyperactivation of TOPFLASH reporter, increases stabilization of β-catenin, and increases β-catenin translocation to the nucleus and Dvl2 phosphorylation (in mutant vs. wild-type MEFs). Zhang et al. (2011) recently observed β-catenin stabilization, canonical Wnt reporter hyperactivation, and excessive proliferation in cells of oral tissues and Meckel cartilage in Fuz−/− embryos. They explained the Fuzzy effect on canonical Wnt pathway by the existence of a negative feedback mechanism by which β-catenin directly binds to the Fuzzy promoter and regulates Fuzzy expression. However, the mechanism of how Fuzzy is able to modulate the canonical Wnt pathway remained unclear (Zhang et al., 2011). Our observations suggest an alternative novel mechanism by which Fuzzy might enhance canonical Wnt activity. Dvl is known to form nonvesicular cytoplasmic protein aggregates; this correlates with the ability of Dvl to stimulate canonical Wnt signaling (Schwarz-Romond et al., 2005, 2007a). We found that Fuzzy is required for sequestration of Dvl2 into Rab8(+) cargo vesicles targeted to the cilium, and loss of Fuzzy results in loss of Dvl1 at the basal body. Thus, we propose that Fuzzy accomplishes two related tasks: 1) it depletes the cytoplasmic Dvl pool available for the canonical Wnt signaling pathway activity; and 2) it brings Dvl proteins to the basal body/ciliary axoneme, where they participate in PCP signaling. In the absence of Fuzzy, a larger intracellular pool of Dvl proteins is available to facilitate canonical Wnt/β-catenin signaling.
A role for Fuzzy in regulation of the noncanonical (PCP) Wnt pathway
Our studies show that loss of Fuzzy in mammalian cells leads to defective cell polarization and reduced directional mobility, the hallmarks of defective PCP signaling. Our data are consistent with early reports that loss of Fuzzy leads to typical PCP defects such as randomization of hair orientation on Drosophila wing cells (Collier and Gubb, 1997; Lee and Adler, 2002) and open neural tube in Xenopus laevis (Park et al., 2006). We propose that defective PCP signaling is a consequence of both abnormal ciliogenesis and mislocalization of Dvl1 and Dvl2. Dvl proteins are critical for transducing the noncanonical Wnt (PCP) signal (Wallingford and Harland, 2001; Wallingford and Habas, 2005). Although no robust genetic interactions were detected between Fuzzy and Vangl2 genes during neural tube formation (Heydeck and Liu, 2011), Fuzzy−/− mice exhibit the same type of heart outflow defect seen in Dvl2 and Looptail (Vangl2) mouse mutants and were reported to display a decreased expression of PCP genes (Zhang et al., 2011). Of importance, we recently identified Fuzzy mutations in a subset of patients with neural tube defects (Seo et al., 2011), defects often attributable to the defective PCP signaling. Although Fuzzy was originally envisioned as a “downstream effector” for core PCP proteins in the context of unciliated fruitfly cells, our data are consistent with the view that in mammalian ciliated cells, Fuzzy might control the polarized subcellular redistribution of some core PCP proteins such as Dvl1 and Dvl2.
Model of Fuzzy action in the cell
On the basis of our experimental observations, we propose a model in which Fuzzy recruits Rab8 into the post–trans-Golgi vesicles that deliver key protein cargoes to the primary cilium (Figure 7).
FIGURE 7:
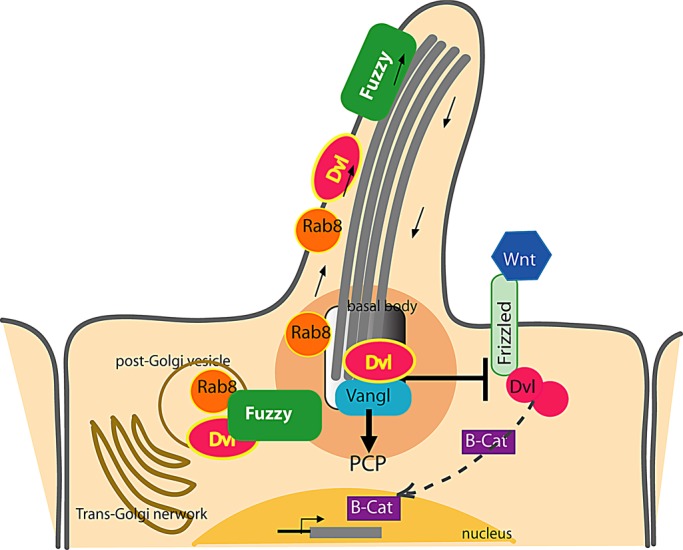
Fuzzy delivers Dvl to the cilium via Rab8(+) vesicular traffic and arbitrates relative activity of canonical and noncanonical (PCP) Wnt pathways. In ciliated cells, Fuzzy is required to recruit Rab8 and Dvl2 to the polarized trafficking route, which brings both molecules to the primary cilium. Sequestration of Dvl2 into vesicles depletes intracellular Dvl2 available for canonical Wnt signaling. At the cilium of wild-type cells, Dvl2 coordinates basal body polarization and facilitates PCP signaling. In the absence of Fuzzy, the Rab8-trafficking route is disrupted, ciliogenesis is compromised, and Dvl2 recruitment to the basal bodies is disturbed. Although noncanonical Wnt signaling is defective, the canonical Wnt/β-catenin signaling is hyperactive due to the greater availability of intracellular Dvl2.
MATERIALS AND METHODS
Animal care
This study was conducted according to the guidelines of the Canadian Council on Animal Care, and the protocol was approved by the McGill University Animal Care committee (Protocol 5423).
Fuzzy mutant mouse
We generated Fuzzy transgenic mice from the Gene Trap ES cell AN0439 (www.genetrap.org) at the McGill Transgenic Core Facility. The phenotype of the Fuzzy-knockout mice was similar to that of mice described in Heydeck et al. (2009). Homozygous Fuzzygt/gt embryos were obtained by brother:sister mating of heterozygous Fuzzygt/+ mice. Genotyping was carried out with the following primers: wild-type allele, mFuzzy-Forward, 5′-CACCTCTG AGCTGAGGCTGG-3′, and mFuzzy-Reverse, 5′-CTCAATTCTTTCTTCAGTCTTTC-3′; trapped allele, bGEO-1F, 5′-TTATCGATGAGCGTGGTGGTTATGC-3′, and bGEO-2R, 5′-GCGCGTACATCGGGCAAATAATATC-3′.
cDNA constructs
The human Fuzzy cDNA with ATG or lacking ATG initiation site and containing FLAG tag at its C-terminus was synthesized by GenScript (Piscataway, NJ) using GenBank NM_025129 as a reference. The sequential KpnI and EcoRI sites were added at the N-terminus, and the HindIII site was incorporated at the C-terminus to facilitate further cloning. KpnI-HindIII Fuzzy cDNA was cloned in-frame into pEGFP or pCherry (both Clontech, Mountain View, CA) to generate FuzzyGFP and FuzzyCherry, where EGFP or Cherry tag, respectively, is fused to the N-terminus of full-length Fuzzy protein. The EcoRI-HindIII Fuzzy cassette (containing ATG) was cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA) to generate pcFuzzy or into pGEX4T vector (Promega, Madison, WI) to generate GST-Fuzzy. Human wild-type Rab8 (Gene Bank NM_0053670) and a dominant-negative mutant Rab8T22N were synthesized by GenScript. The KpnI and BamHI flanking sequences were added to facilitate cloning into pCherry (Clontech). Integrity of all constructs was confirmed by sequencing, and expression was confirmed by immunoblotting. Mouse Dvl2 mammalian expression construct was previously described (Torban et al., 2004). Cherry-mouse Dvl2 was kindly provided by P. Gros (McGill University, Montreal, Canada). GFP-tagged TGN38 (TGN38GFP) was kindly provided by S. Gruenheid (McGill University).
Mouse embryonic fibroblasts
MEFs were established from E12.5 litters of Fuzzy−/− and Fuzzy+/+ genotypes. Briefly, embryos were microdissected, washed with DMEM medium supplemented with 1% penicillin/streptomycin/Fungizone (all from Wisent, St. Bruno, Canada), minced, and incubated in 0.25% trypsin (Invitrogen) overnight at 4ºC, followed by 30 min of incubation at 37ºC. Tissues were triturated by pipetting with 10- and 5-ml pipettes in fresh DMEM medium supplemented with antibiotics and 10% fetal bovine serum (FBS; Wisent) and transferred into T175 tissue culture flasks. Cells grown to 80–90% confluency (passage 1) were split into T75 flasks. Primary MEFs from passage 2 were immortalized with E6/E7 antigen (kindly provided by Paul Goodyer, McGill University) as described in Hawley-Nelson et al. (1989). Frozen stocks were prepared from immortalized MEFs at passage 9 and used for transfections. Primary and immortalized MEFs were prepared from at least two embryos for each genotype.
Transfections
Immortalized mutant MEFsFuz/Fuz or wild-type MEFs+/+ were grown on cover slips in 12-well plates for 24 h before transfection. A 750-ng amount of pEGFP, pcDNA3.1, pEGFP-Fuzzy, or pcFuzzy-FLAG was transiently transfected into MEFs by using Fugene HD as recommended by the manufacturer (Roche, Indianapolis, IN). We routinely achieved ∼10–20% transfection efficiency. In some experiments, MEFsFuz/Fuz were transiently transfected either with 750 ng of wild-type Rab8-Cherry alone or with 750 ng of pcFuzzy-FLAG in combination with either 750 ng of wild-type or dominant-negative Cherry-Rab8.
MDCKII cells were split into six-well plates 24 h before transfection and grown in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin. A 0.5-μg amount of either pEGFP or pEGFP-Fuzzy was transfected into MDCK cells with Lipofectamine 2000 as recommended by the manufacturer (Invitrogen). Transfected cells were grown for 2 wk in medium supplemented with 800 μg/ml neomycin G418 (Invitrogen). Individual neomycin-resistant clones were picked out and expanded, and pEGFP or pEFGP-Fuzzy expression was confirmed by immunoblotting.
For colocalization studies, MDCK cells were grown in 12-well plates and cotransfected (as described) with 0.6 μg of FuzzyCherry and 0.3 μg of TGN38GFP, followed by treatment with 0.5 μg/ml brefeldin A (Sigma-Aldrich, St. Louis, MO) for 24 h as described in Hoffmeister et al. (2011); cells were fixed and processed for microscopy as described later.
For protein interaction studies, human embryonic kidney HEK293 cells were plated in 100-mm Petri dishes and transiently transfected using Lipofectamine 2000 with various expression constructs: 2 μg of pEGFP-Fuzzy or pcDNA-Fuzzy, 2 μg of Dvl2; pcDNA3.1empty was used to adjust total DNA to 4 μg per dish. Cells were lysed at 48 h posttransfection and used for interaction studies.
Ciliogenesis
MEFsFuz/Fuz and MEFs+/+ were plated at 105 cells per well in 12-well plates containing coverslips and cultured in DMEM medium as described. At 24 h later, the FBS content was reduced to 0.5% to induce ciliogenesis. Cells were analyzed 72 h later for the presence of a primary cilium as described later. MDCK clones for each stable pEGFP or pEGFP-Fuzzy were plated into 12-well plates on glass cover slips coated with 0.01% collagen I (Sigma-Aldrich). Cells were grown for 10 d in DMEM medium containing 10% FBS, fixed in 4% paraformaldehyde (PFA), and analyzed for the presence of a primary cilium as described later. Z-stacks were acquired on Axio Observor-100 (Zeiss, Jena, Germany) or LSM780 (Zeiss) using AxioVision Ref 4.8 software (Zeiss). Images were processed using Photoshop (Adobe, San Jose, CA).
Luciferase assay
Human embryonic kidney HEK293 cells were plated in 24-well plates and transfected in triplicate with 50 ng of TOPFLASH or FOPFLASH (provided by Paul Goodyer), 15 ng of Dvl2, and 1–10 ng of pcFuzzy. pcDNA3.1 plasmid was added to all samples to adjust the total amount of transfected DNA to 75 ng per sample. Cells were lysed 48 h posttransfection, and luciferase activity was assessed with the Dual Luciferase Reporter Assay Kit (Promega) as recommended by the manufacturer; TOPFLASH activity was normalized for the activity of Renilla luciferase. Experiments were repeated three times.
Immortalized MEFsFuz/Fuz and MEFs+/+ were plated in triplicate in 24-well plates at 0.5 × 104 per well. At 24 h later cells were transfected with 900 ng of TOPFLASH or FOPFLASH and 100 ng of Renilla luciferase using Fugene HD as recommended by the manufacturer (Roche). At 6 h posttransfection, fresh medium containing 0.5% FBS was added. At 36 h later, the medium was supplemented with 200 ng/ml of recombinant Wnt3a (R&D Systems, Minneapolis, MN) (where indicated); 18 h later, cells were lysed, and luciferase activity was analyzed as described.
Immunodetection
MDCK cells stably expressing pEGFP, pEFGP-Fuzzy, or HEK293 cells transfected with pEGFP-Fuzzy and/or Dvl2 were lysed in PLC buffer containing 50 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.5), 150 mm NaCl, 10% glycerol, 0.5% Triton X-100, 1.5 mm MgCl2, 1 mm ethylene glycol tetraacetic acid, 2 mM Na2 vanadate, 10 mM Na2 pyrophosphate, 25 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (BioShop, Burlington, Canada). A 50-μg amount of the total cell lysate was resolved on 10% PAGE–SDS gel and immunoblotted with monoclonal anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA), anti-FLAG (Sigma-Aldrich), or anti-Dvl2 (Santa Cruz Biotechnology) antibodies. Secondary anti-mouse horseradish peroxidase (HRP)–conjugated antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used. West Pico ECL kit (Thermo Scientific, Waltham, MA) was used to detect proteins of interest.
To detect endogenous expression of β-catenin and Dvl2 in MEFsFuz/Fuz and MEFs+/+, we plated cells at 105 per cm2 into six-well plates and induced ciliogenesis as described. In some experiments, MEFs were exposed to conditioned medium from control L-cells (#CRL-2648; American Type Culture Collection, Manassas, VA) or l-cells expressing Wnt3a (#CRL-2647; American Type Culture Collection) or Wnt5a (#CRL-2814; American Type Culture Collection) for 3 and 6 h. MEFs were lysed in PLC buffer; 40 μg of each lysate were resolved on an 8% SDS–PAGE gel. The proteins were detected with rabbit polyclonal anti-Dvl2 (Cell Signaling Technology, Beverly, MA) or anti β-catenin (Santa Cruz Biotechnology) antibodies, followed by HRP-conjugated anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch Laboratories) and the West Pico ECL kit. Loading was monitored by expression of α-tubulin visualized by anti–α-tubulin antibody (Sigma-Aldrich). Protein band intensity was quantified using Alpha Innotech AlphaImager system and AlphaEase FC32-bit software (ThermoFisher Scientific, Waltham, MA). The experiments were repeated four times.
Immunofluorescence microscopy
To visualize cilia, we grew MEFs or MDCK clones as described above. Cells were fixed in 4% PFA for 15 min and permeabilized in 0.5% Triton X-100 in phosphate-buffered saline (PBS). Cells were blocked in 10% normal goat serum (Jackson ImmunoResearch Laboratories) and 0.1% Triton in PBS. Cilia were detected with mouse- or rabbit acetylated-α-tubulin (Sigma-Aldrich and Abcam [Cambridge, MA], respectively). For immunofluorescence detection of other proteins, the following primary antibodies were used: mouse anti-Dvl1 (Santa Cruz Biotechnology), rabbit anti–β-catenin (Santa Cruz Biotechnology), rabbit anti-Rab8 (Sigma-Aldrich), rabbit or mouse anti–γ-tubulin (Abcam and Sigma-Aldrich, respectively), mouse and rabbit anti-FLAG (Sigma-Aldrich and GenScript, respectively), rabbit anti-GM130 (BD Transduction Laboratories, Lexington, KY), mouse anti-calreticulin (BD Transduction Laboratories), mouse anti-syntaxin11 (BD Transduction Laboratories), rabbit anti-Fuzzy (Sigma-Aldrich), and rabbit anti–ZO-1 (Zymed, San Francisco, CA). All primary antibodies were visualized with secondary donkey Alexa Flour 546, Alexa Fluor 395, or Alexa Fluor 647 anti-mouse or Alexa Fluor 488 and Alexa Fluor 546 anti-rabbit secondary antibodies (all from Molecular Probes, Eugene, OR). Nuclei were stained with 4′,6-diamidino-2-phenylindole (Invitrogen), and the slides were mounted in Prolong Gold Antifade (Molecular Probes). For all images, Z-stack projections were acquired by using an Axio Observer-100 microscope with AxioVision, release 4.8, software, or LSM-1 or LSM780 confocal microscope (both from Zeiss); x-z image projections were analyzed in MetaMorph (Molecular Devices, Sunnyvale, CA) and processed in Photoshop. All figures were assembled in Illustrator (Adobe).
Protein interaction assays
Coimmunoprecipitations and GST pull-down assays were performed as described previously (Torban et al., 2004). Briefly, pEGFP-Fuzzy or pcFuzzy-FLAG and mouse Dvl2 (Torban et al., 2004) cDNAs were transiently expressed in HEK293 cells and lysed in PLC buffer. One-twentieth of each sample was reserved as an input fraction. The lysates (1 mg of protein per sample) were incubated overnight with anti-Dvl2 (Santa Cruz Biotechnology) monoclonal antibody and agarose A beads (Santa Cruz Biotechnology). The protein–antibody complexes were washed in the high-stringency PLC buffer (4×) containing 500 mM NaCl, resolved on 8% PAGE–SDS, and immunoblotted with antibodies as indicated in Figure 3. The full-length human Fuzzy cloned into pGEX-5X-2 was purified from BL21 Escherichia coli on Sepharose 4B beads (Amersham Biosciences, Piscataway, NJ); GST pull downs were carried out as described (Torban et al., 2004). Briefly, GST-Fuzzy or GST proteins on beads were mixed with mouse Dvl2 protein produced by transient transfection into HEK293 cells. Mixtures were incubated at 4ºC for 4 h and then washed (5×) with PLC buffer and eluted with 2× Laemmli sample buffer (Invitrogen). Proteins were resolved on 8% SDS–PAGE buffer and analyzed by immunoblotting as described with anti-Dvl2 antibody. All experiments were repeated three times. The film images were sized using Photoshop.
Wound assay
Primary or immortalized MEFsFuz/Fuz and MEFs+/+ were grown to confluence in the six-well plate coated with collagen I as described above. A wound was scratched on the culture surface with a 10-μl pipette tip, and cells were photographed at 0 and 8 h after the start of the experiment. Images were acquired using an Axio Observor-100 microscope. Twelve pictures per time point per well were taken to measure the surface area within a wound unoccupied by the cells after 8 h, using the Outline Spline (μm2) Interactive Measurement program (Zeiss). The experiment was repeated four times in duplicate for each genotype. To ascertain cell polarity, MEFs were grown on coverslips and fixed 6 h after the scratch; orientation of the Golgi apparatus was revealed with anti-GM130 antibody as described in Caddy et al. (2010) by using the Axio Observor-100 microscope Interactive Measurement package (Zeiss).
Statistical analysis
The two-tailed, two-sample, unequal-variance t test was conducted in all experiments in which two samples were compared. One-variance analysis of variance (ANOVA) test followed by t test was conducted for Figure 5B. A chi-square test was used to compare the ratio of nucleated/elongated cilia in MDCK or in MEFs+/+ versus MEFsFuz/Fuz displaying polarized orientation of the trans-Golgi network. Mean and standard errors are shown in all graphs.
Supplementary Material
Acknowledgments
We thank Paul Goodyer, Samantha Gruenheid, and Philippe Gros (McGill University, Montreal, Canada) for providing E6/E7, TOPFLASH, FOPFLASH, Dvl2, TGN38-GFP constructs, and reagents and antibodies against calreticulin, GM130, Sec23, and syntaxin11. This work was supported by an operating grant from the Canadian Institute of Health Research to E.T. E.T. is a recipient of a Chercheur Boursier Junior I award from the Fonds de la Recherche du Québec–Santé. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used:
- Dvl
Dishevelled
- PCP
planar cell polarity
- TGN
trans-Golgi network
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-06-0437) on January 9, 2013.
The authors declare no conflict of interest.
REFERENCES
- Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat Cell Biol. 2010;12:407–412. doi: 10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- Caddy J, et al. Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev Cell. 2010;19:138–147. doi: 10.1016/j.devcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Gubb D. Drosophila tissue polarity requires the cell-autonomous activity of the fuzzy gene, which encodes a novel transmembrane protein. Development. 1997;124:4029–4037. doi: 10.1242/dev.124.20.4029. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- Das A, Guo W. Rabs and the exocyst in ciliogenesis, tubulogenesis and beyond. Trends Cell Biol. 2011;21:383–386. doi: 10.1016/j.tcb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge SL, et al. Murine dishevelled 3 functions in redundant pathways with Dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 2008;4:e1000259. doi: 10.1371/journal.pgen.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E, Gresh L, Reimann A, Pontoglio M. Cystic kidney diseases: learning from animal models. Nephrol Dial Transplant. 2004;19:2700–2702. doi: 10.1093/ndt/gfh533. [DOI] [PubMed] [Google Scholar]
- Fischer E, Legue E, Doyen A, Nato F, Nicolas JF, Torres V, Yaniv M, Pontoglio M. Defective planar cell polarity in polycystic kidney disease. Nat Genet. 2006;38:21–23. doi: 10.1038/ng1701. [DOI] [PubMed] [Google Scholar]
- Fischer E, Pontoglio M. Planar cell polarity and cilia. Semin Cell Dev Biol. 2009;20:998–1005. doi: 10.1016/j.semcdb.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Gaush CR, Hard WL, Smith TF. Characterization of an established line of canine kidney cells (MDCK) Proc Soc Exp Biol Med. 1966;122:931–935. doi: 10.3181/00379727-122-31293. [DOI] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Strutt D. Principles of planar polarity in animal development. Development. 2011;138:1877–1892. doi: 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RS, et al. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat Cell Biol. 2009;11:1225–1232. doi: 10.1038/ncb1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubb D, Garcia-Bellido A. A genetic analysis of the determination of cuticular polarity during development in Drosophila melanogaster. J Embryol Exp Morphol. 1982;68:37–57. [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Hamblet NS, Lijam N, Ruiz-Lozano P, Wang J, Yang Y, Luo Z, Mei L, Chien KR, Sussman DJ, Wynshaw-Boris A. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 2002;129:5827–5838. doi: 10.1242/dev.00164. [DOI] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Arffman A, Peranen J. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell. 2002;13:3268–3280. doi: 10.1091/mbc.E02-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DJ, Conway SJ, Greene ND, Gerrelli D, Murdoch JN, Anderson RH, Copp AJ. Cardiovascular defects associated with abnormalities in midline development in the Loop-tail mouse mutant. Circ Res. 2001;89:6–12. doi: 10.1161/hh1301.092497. [DOI] [PubMed] [Google Scholar]
- Henderson DJ, Phillips HM, Chaudhry B. Vang-like 2 and noncanonical Wnt signaling in outflow tract development. Trends Cardiovasc Med. 2006;16:38–45. doi: 10.1016/j.tcm.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Heydeck W, Liu A. PCP effector proteins inturned and fuzzy play nonredundant roles in the patterning but not convergent extension of mammalian neural tube. Dev Dyn. 2011;240:1938–1948. doi: 10.1002/dvdy.22696. [DOI] [PubMed] [Google Scholar]
- Heydeck W, Zeng H, Liu A. Planar cell polarity effector gene Fuzzy regulates cilia formation and Hedgehog signal transduction in mouse. Dev Dyn. 2009;238:3035–3042. doi: 10.1002/dvdy.22130. [DOI] [PubMed] [Google Scholar]
- Hoffmeister H, Babinger K, Gurster S, Cedzich A, Meese C, Schadendorf K, Osten L, de Vries U, Rascle A, Witzgall R. Polycystin-2 takes different routes to the somatic and ciliary plasma membrane. J Cell Biol. 2011;192:631–645. doi: 10.1083/jcb.201007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Nachury MV. The BBSome. Curr Biol. 2009;19:R472–R473. doi: 10.1016/j.cub.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Roper VC, Foucher I, Qian D, Banizs B, Petit C, Yoder BK, Chen P. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet. 2008;40:69–77. doi: 10.1038/ng.2007.54. [DOI] [PubMed] [Google Scholar]
- Kibar Z, Underhill DA, Canonne-Hergaux F, Gauthier S, Justice MJ, Gros P. Identification of a new chemically induced allele (Lp(m1Jus)) at the loop-tail locus: morphology, histology, and genetic mapping. Genomics. 2001;72:331–337. doi: 10.1006/geno.2000.6493. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Adler PN. The function of the frizzled pathway in the Drosophila wing is dependent on inturned and fuzzy. Genetics. 2002;160:1535–1547. doi: 10.1093/genetics/160.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maung SM, Jenny A. Planar cell polarity in Drosophila. Organogenesis. 2011;7:165–179. doi: 10.4161/org.7.3.18143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier. Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet. 2006;38:303–311. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves B, Banting G. Perturbation of the morphology of the trans-Golgi network following brefeldin A treatment: redistribution of a TGN-specific integral membrane protein, TGN38. J Cell Biol. 1992;116:85–94. doi: 10.1083/jcb.116.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AJ, et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- Sato T, et al. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 2007;448:366–369. doi: 10.1038/nature05929. [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007a;14:484–492. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T, Merrifield C, Nichols BJ, Bienz M. The Wnt signalling effector Dishevelled forms dynamic protein assemblies rather than stable associations with cytoplasmic vesicles. J Cell Sci. 2005;118:5269–5277. doi: 10.1242/jcs.02646. [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T, Metcalfe C, Bienz M. Dynamic recruitment of axin by Dishevelled protein assemblies. J Cell Sci. 2007b;120:2402–2412. doi: 10.1242/jcs.002956. [DOI] [PubMed] [Google Scholar]
- Seo JH, Zilber Y, Babayeva S, Liu J, Kyriakopoulos P, De Marco P, Merello E, Capra V, Gros P, Torban E. Mutations in the planar cell polarity gene, Fuzzy, are associated with neural tube defects in humans. Hum Mol Genet. 2011;20:4324–4333. doi: 10.1093/hmg/ddr359. [DOI] [PubMed] [Google Scholar]
- Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley MJ, Signoret N, Robertson D, Tilley A, Hann A, Ewan K, Ding Y, Paterson H, Dale TC. Dishevelled (Dvl-2) activates canonical Wnt signalling in the absence of cytoplasmic puncta. J Cell Sci. 2005;118:5279–5289. doi: 10.1242/jcs.02647. [DOI] [PubMed] [Google Scholar]
- Song H, Hu J, Chen W, Elliott G, Andre P, Gao B, Yang Y. Planar cell polarity breaks bilateral symmetry by controlling ciliary positioning. Nature. 2010;466:378–382. doi: 10.1038/nature09129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torban E, Wang HJ, Groulx N, Gros P. Independent mutations in mouse Vangl2 that cause neural tube defects in looptail mice impair interaction with members of the Dishevelled family. J Biol Chem. 2004;279:52703–52713. doi: 10.1074/jbc.M408675200. [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- Vinson CR, Adler PN. Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature. 1987;329:549–551. doi: 10.1038/329549a0. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Harland RM. Xenopus Dishevelled signaling regulates both neural and mesodermal convergent extension: parallel forces elongating the body axis. Development. 2001;128:2581–2592. doi: 10.1242/dev.128.13.2581. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Mitchell B. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 2011;25:201–213. doi: 10.1101/gad.2008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hamblet NS, Mark S, Dickinson ME, Brinkman BC, Segil N, Fraser SE, Chen P, Wallingford JB, Wynshaw-Boris A. Dishevelled genes mediate a conserved mammalian PCP pathway to regulate convergent extension during neurulation. Development. 2006;133:1767–1778. doi: 10.1242/dev.02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci USA. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens CJ, et al. Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J Biol Chem. 2010;285:16218–16230. doi: 10.1074/jbc.M109.070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LL, Adler PN. Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J Cell Biol. 1993;123:209–221. doi: 10.1083/jcb.123.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wlodarczyk BJ, Niederreither K, Venugopalan S, Florez S, Finnell RH, Amendt BA. Fuz regulates craniofacial development through tissue specific responses to signaling factors. PLoS One. 2011;6:e24608. doi: 10.1371/journal.pone.0024608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.