Reduction in ploidy in meiosis is assumed to be due to a block to the licensing step (Mcm helicase association with replication origins). When the licensing block is subverted, replication is still only partial due to inefficient elongation replication forks. This might constitute an additional level of replication regulation.
Abstract
Meiosis involves two successive rounds of chromosome segregation without an intervening S phase. Exit from meiosis I is distinct from mitotic exit, in that replication origins are not licensed by Mcm2-7 chromatin binding, but spindle disassembly occurs during a transient interphase-like state before meiosis II. The absence of licensing is assumed to explain the block to DNA replication, but this has not been formally tested. Here we attempt to subvert this block by expressing the licensing control factors Cdc18 and Cdt1 during the interval between meiotic nuclear divisions. Surprisingly, this leads only to a partial round of DNA replication, even when these factors are overexpressed and effect clear Mcm2-7 chromatin binding. Combining Cdc18 and Cdt1 expression with modulation of cyclin-dependent kinase activity, activation of Dbf4-dependent kinase, or deletion of the Spd1 inhibitor of ribonucleotide reductase has little additional effect on the extent of DNA replication. Single-molecule analysis indicates this partial round of replication results from inefficient progression of replication forks, and thus both initiation and elongation replication steps may be inhibited in late meiosis. In addition, DNA replication or damage during the meiosis I–II interval fails to arrest meiotic progress, suggesting absence of checkpoint regulation of meiosis II entry.
INTRODUCTION
Meiosis is a specialized form of cell division that generates genetically diverse haploid gametes from diploid cells. This is achieved by a single round of premeiotic S phase (meiS), followed by pairing and recombination between homologous chromosomes. Subsequently, reductional chromosome segregation occurs (meiosis I [MI]) and is followed by a final equational division (meiosis II [MII]), without an intervening S phase. This block to replication contrasts with the S and M alternation seen in the mitotic cycle. It is likely that robust mechanisms exist to prevent DNA synthesis during this MI–MII interval, since initiation from just a few origins could be a source of genetic instability in gametes. However, it is not clear whether the mechanism used simply maintains replication regulatory mechanisms used in G2 or whether additional meiotic-specific control applies.
The mechanism and regulation of meiS are broadly similar to those for mitotic S phase (mitS), and the same origins are used in both types of cell cycle (Heichinger et al., 2006; Blitzblau et al., 2012), although the duration of meiS is longer and there may be subtle differences in terms of cyclin requirements (reviewed in Forsburg, 2002; Strich, 2004). MitS is regulated at two main steps, one occurring before S phase and the second at initiation itself (for reviews see Diffley, 2004; Sclafani and Holzen, 2007; Masai et al., 2010). The first control regulates the loading of Mcm2-7 helicase onto DNA as an inactive complex, at locations where the origin recognition complex is bound (licensing/prereplicative complex formation). The second control activates helicase activity of bound Mcm2-7 and DNA synthesis and requires phosphorylation of several targets by cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK; Tanaka et al., 2007; Zegerman and Diffley, 2007; Sheu and Stillman, 2010). Although not a primary control, up-regulation of dNTP supply via ribonucleotide reductase (RNR) activation is also important to allow high fidelity of DNA synthesis (Zhao et al., 1998; Liu et al., 2005; Salguero et al., 2012).
Rereplication within S and G2 phases is prevented by blocking Mcm2-7 chromatin binding, and this is effected by overlapping mechanisms that inactivate relevant factors, such as Cdc6/Cdc18, Cdt1, and the origin recognition complex (ORC; Blow and Dutta, 2005; Arias and Walter, 2007), although the relative importance of specific control mechanisms differs among organisms (reviewed in Kearsey and Cotterill, 2003). CDK is important as a negative regulator of licensing, both via phosphorylation of the proteins involved and because direct binding to ORC may sterically inhibit Mcm2-7 chromatin binding (Tanaka and Diffley, 2002; Wuarin et al., 2002; Mimura et al., 2004; Wilmes et al., 2004). Extra rounds of replication can be achieved in G2 of the mitotic cycle by promoting Mcm2-7 chromatin binding, for example, by overexpression of Cdc18 and Cdt1 (Gopalakrishnan et al., 2001; Yanow et al., 2001) or transient inhibition of CDK (Hayles et al., 1994). Similar control mechanisms function after meiS, where inhibition of Cdk1 by stabilization of the Sic1 CDK inhibitor allows rereplication before MI (Sawarynski et al., 2009).
Preventing DNA replication between the meiotic nuclear divisions appears to be primarily due to blocking of Mcm2-7 chromatin binding. For example, in fission yeast, Mcm2-7 remains nuclear throughout meiosis, but chromatin binding does not occur between MI and MII (Lindner et al., 2002). Maintenance of CDK levels between the meiotic nuclear divisions appears to be important for preventing licensing and hence DNA replication, but this must be balanced with some reduction in CDK activity to allow disassembly of the spindle at MI. In Saccharomyces cerevisiae, this balance may be achieved via transient and partial activation of Cdc14 phosphatase by the FEAR network on exit from MI (Buonomo et al., 2003; Marston et al., 2003) rather than full activation of Cdc14 seen on mitotic exit. Prevention of licensing also involves a CDK-related, meiotic-specific kinase Ime2, which is expressed during MI–MII (Holt et al., 2007). Ime2 expression collaborates with Cdk1 to block Mcm2-7 accumulation in the nucleus during late meiosis, thus presumably helping to block licensing. Of interest, sites phosphorylated by Ime2 are resistant to Cdc14 dephosphorylation, which helps to explain how Cdc14 can allow MI exit but not promote nuclear accumulation of Mcm2-7 necessary for licensing. CDK also plays a key role in Xenopus meiosis. Maintenance of CDK activity after MI, due to incomplete degradation of cyclin B, prevents DNA replication (Furuno et al., 1994; Iwabuchi et al., 2000), presumably by blocking licensing, and depression of CDK activity by expression of the Wee1 inhibitor changes the M–M transition into an M–S–M transition (Iwabuchi et al., 2000; Nakajo et al., 2000).
There is also evidence for redundancy in mechanisms blocking inappropriate DNA replication in late meiosis, as seen for rereplication in the mitotic cycle. For instance, in S. cerevisiae, transient inhibition of Ime2 and/or Cdk1 does not lead to DNA replication between the meiotic nuclear divisions (Holt et al., 2007). In Caenorhabditis elegans, Mcm2-7 proteins become associated with chromatin from anaphase of MII, but RNA interference knockdown of geminin, which blocks licensing via inhibition of Cdt1, allows earlier licensing, from anaphase of MI (Sonneville et al., 2012). However, geminin-depleted embryos are viable, suggesting that failure to prevent licensing during MI–MII does not lead to replication of DNA.
The absence of licensing is assumed to explain the block to DNA replication in late meiosis, but this has not been formally tested. Here we address this assumption by subverting the regulation of Mcm2-7 chromatin binding. We find that the reduction of ploidy can be only partially subverted by ectopic expression of the licensing factors Cdc18 and Cdt1, even when combined with modulation of CDK activity, activation of DDK, or activation of RNR. The extent of replication appears to be constrained at least in part by limited fork movement from origins, which may represent a meiotic-specific control on DNA synthesis.
RESULTS
Expression of Cdt1 and Cdc18 licensing factors results in a partial round of DNA replication during the MI–MII interval
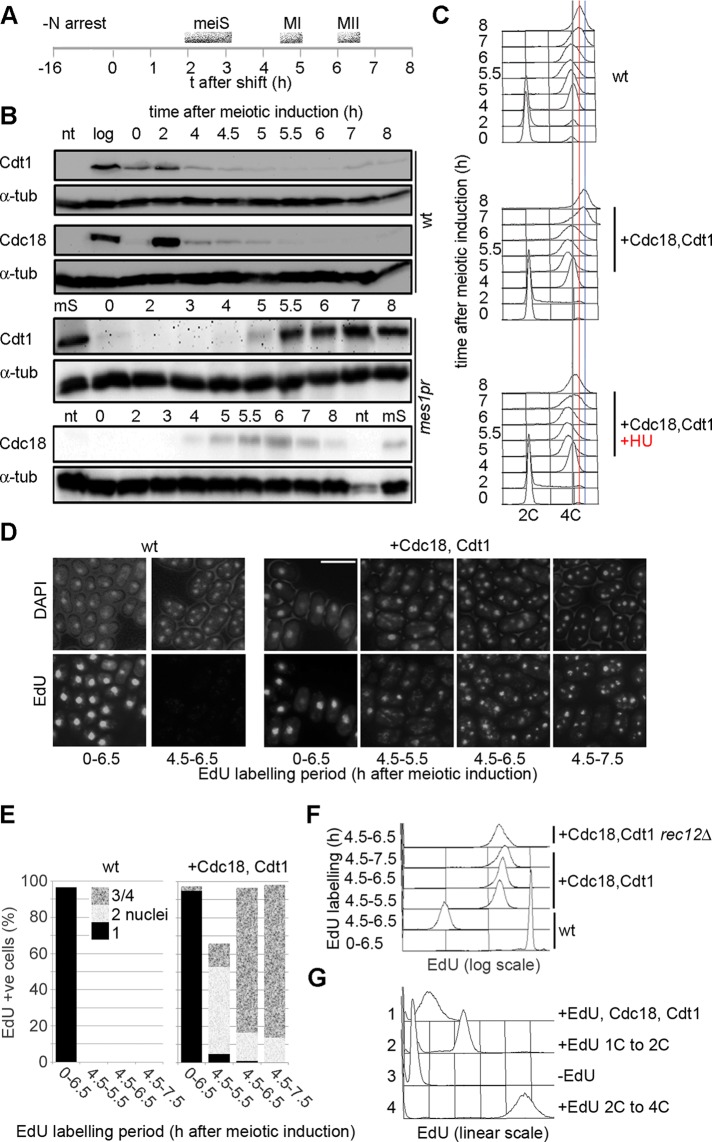
In mitS, loading of Mcm2-7 onto chromosomal origins is partly regulated by levels of Cdt1 and Cdc18. In a synchronous meiosis, induced by Pat1 inactivation, these proteins are expressed early and are subsequently degraded around the time of meiS (Figure 1, A and B, top). To determine the significance of Cdt1 and Cdc18 regulation for blocking MI–MII replication, we constructed a diploid strain in which additional integrated gene copies were expressed from the mes1 promoter, which is activated at MI (Kishida et al., 1994). Levels of Cdt1 and Cdc18 expressed in this mes1pr-cdc18, cdt1 strain increased during the MI–MII interval and were similar to physiological levels seen in meiS phase (Figure 1B, bottom; compare 5.5 h and MS). These cells also showed an increase in DNA content after MI, which was blocked by hydroxyurea (HU), suggesting that this represents a partial round of DNA replication (Figure 1C). We observed no increase in DNA content during the MI–MII interval if either Cdt1 or Cdc18 was expressed separately (unpublished data). Viability of spores generated from the mes1pr-cdc18, cdt1 strain was also significantly reduced (Supplemental Table S1).
FIGURE 1:
Expression of Cdc18 and Cdt1 in late meiosis causes DNA replication after MI. (A) Scheme of meiotic synchronization. Cells were arrested in G1 by nitrogen starvation for 16 h at 26°C; at t = 0 cells were refed and shifted to 34°C to inactivate Pat1. Timing of meiotic events is shown. (B) Levels of Cdt1 and Cdc18 in a pat1-induced meiosis. Top (wt), wild-type levels of expression (P1416, P1424); bottom (mes1pr), show expression levels of tagged Cdt1 and Cdc18 expressed from additional integrated gene copies under the control of the mes1 promoter (P2453). The ploidy of the pat1 strains does not affect the kinetics of Cdt1 and Cdc18 expression (unpublished data). Tagged proteins were used for mes1pr expression, but strains in which Cdc18 and Cdt1 function is solely provided by tagged copies are not compromised for DNA replication. α-Tubulin is shown as a loading control. log, log-phase cells; mS, cells in meiS; nt, cells with no tag. (C) DNA contents of cells executing meiosis. Top, analysis of wt (pat1-114) cells (P2454); bottom, mes1pr-cdc18, cdt1 cells (P2453), expressing Cdc18 and Cdt1 in late meiosis. Bottom, HU was added 4.5 h after meiotic induction, that is, after meiS. Gray line indicates 4C DNA content, red line indicates 4C DNA content of wt cells in late meiosis; blue line indicates the peak position at 8 h in the mes1pr-cdc18, cdt1 strain (>4C). Meiotic progression analysis is shown in Figure 2A. (D) Wild type (P3032, left) and mes1pr-cdt1, cdc18 (P3031, right) cells were induced to enter meiosis and labeled with EdU (10 μM) for the periods shown. Cells were imaged after conjugation to fluorescent azide. Bar, 10 μm. (E) Analysis of experiment shown in D, showing percentage of cells with nuclear EdU labeling and proportion of cells with one, two, or three/four nuclei. (F) Flow cytometric quantitation of EdU incorporation in cells from D. Also shown is analysis of a rec12Δ strain (P2459) expressing Cdc18 and Cdt1 in late meiosis and wt cells (P3032) incorporating EdU in meiS. (G) Quantitation of EdU incorporation in a mes1-Cdc18, Cdt1 in late meiosis (histogram 1) compared with EdU incorporation in haploid (2) or diploid (4) cells executing a complete S phase; also shown is a –EdU control (3). Comparison with these standards indicates that ∼15% of the genome has replicated in the MI–MII interval.
To confirm that replication was occurring in the MI–MII interval, we modified the strain to allow the uptake of nucleosides and labeled nascent DNA with 5-ethynyl-2′-deoxyuridine (EdU; Salic and Mitchison, 2008). In the absence of Cdt1 and Cdc18 expression in MI–MII, incorporation was only detected when EdU was added before meiS (Figure 1, D– F), and incorporation caused cells to arrest before MI. We reported previously that incorporation of EdU in mitS results in rad3-dependent G2 arrest (Hua and Kearsey, 2011), and we assume that a similar checkpoint-dependent arrest is occurring in meiosis. In the mes1pr-cdc18, cdt1 strain, EdU incorporation was detected in nearly all binucleated and tetranucleated cells when the nucleoside was added after MI (Figure 1, D– F; compare +Cdc18,Cdt1 and wt). To confirm that EdU incorporation was not occurring in meiS before MI, we added HU together with EdU after MI, since arrest of meiS with HU blocks entry into MI via operation of the DNA replication checkpoint (Murakami and Nurse, 1999). Thus, if the detected DNA replication was occurring in meiS, we would expect the addition of HU to block entry into MI, and thus cells with EdU incorporation would be uninucleate. It is significant that we still observed EdU incorporation in the nuclei of binucleated and tetranucleated cells, implying that cells had completed MI before replication responsible for incorporation was occurring (Supplemental Figure S1A). Note that we can detect EdU incorporation in this experiment because HU does not completely block the elongation step of replication (Hua and Kearsey, 2011). The extent of DNA replication was hardly affected by deletion of the rec12 gene, which is necessary for the initiation of meiotic recombination (Figure 1F); thus EdU incorporation is not dependent on homologous recombination. EdU incorporation in cells expressing Cdc18 and Cdt1 in late meiosis was quantitated by flow cytometry and compared with the signal when a complete round of replication occurs in 1C or 2C cells (Figure 1G); from this we estimate that ∼15% of the genome is replicated in the MI–MII interval. Given the extent of this synthesis and the fact that Cdt1 and Cdc18 can provoke rereplication in G2 phase of the mitotic cell cycle but are not known to activate repair synthesis, it is reasonable to conclude that a partial round of DNA replication is occurring in the MI–MII interval.
Partial DNA replication and DNA damage does not halt meiotic progress after meiosis I
The mes1pr-cdc18, cdt1 strain showed the same kinetics of meiotic nuclear divisions as wild-type (wt) cells, in spite of partial DNA replication (Figure 2A). Addition of HU after MI to cells expressing Cdc18 and Cdt1 during this period also did not delay the timing of MII (Figure 2A), in contrast to the effect of HU inhibition on meiS and MI entry. Whereas EdU incorporation blocks entry into MI, as seen for mitosis (Hua and Kearsey, 2011), it has no effect on the timing of MII when used to label DNA replication during the MI–MII interval (Figure 2B). To investigate whether DNA damage has any effect on the kinetics of MII, we used bleomycin to introduce double-strand breaks into chromosomes, which could be detected by pulsed-field gel electrophoresis (PFGE; Figure 2D). This treatment also did not delay MII (Figure 2C), similar to the situation for MI, for which DNA damage also does not delay meiotic division (Catlett and Forsburg, 2003; Pankratz and Forsburg, 2005; reviewed in Longhese et al., 2009). Thus neither arrest of DNA replication nor DNA damage leads to checkpoint-mediated delay of meiosis II.
FIGURE 2:
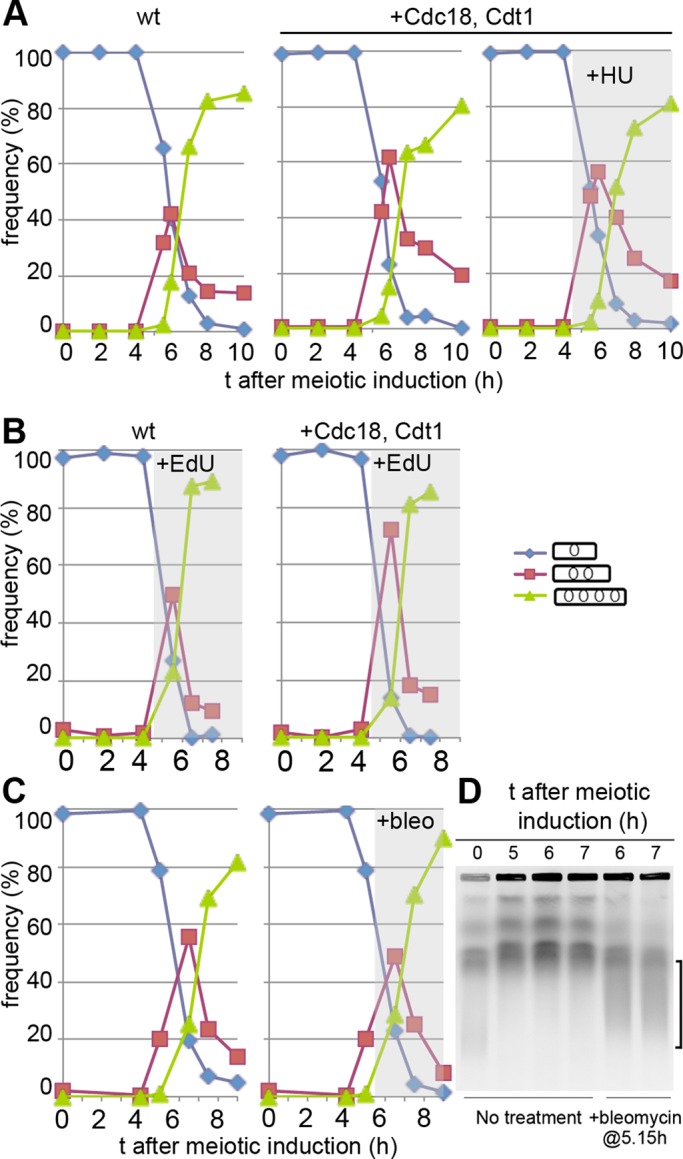
Partial or arrested DNA replication or DNA damage does not delay meiosis II. (A) Meiosis was induced by Pat1 inactivation in strains either wt (left) or expressing Cdc18 and Cdt1 in late meiosis (middle and right) and frequencies of uninucleated, binucleated, and tetranucleated cells were determined. Right, HU was added at 4.5 h after meiotic induction. The flow cytometry analyses for these experiments are shown in Figure 1C; strains used were P2454 (wt) and P2453 (+Cdc18, Cdt1). (B) As in A, but EdU was added at 4.5 h to strains modified to allow uptake of nucleosides. Strains were either wt (left) or expressed Cdc18 and Cdt1 in the MI-MII interval (right). Strains used were P3032 (wt) and P3031 (+Cdc18, Cdt1). (C) As in A, but bleomycin (30 μg/ml) was added to half the culture 5.15 h after meiotic induction. Strain used was P2456. (D) As in C, except that samples were analyzed by PFGE; the brace on the right indicates smear due to chromosome fragmentation at 6–7 h induced by bleomycin treatment.
Because MII is not delayed in the mes1pr-cdc18, cdt1 strain, it is possible that ongoing DNA replication occurring in the MI–MII interval is cut short by meiotic progression, where events such as chromatin condensation for MII or sporulation could indirectly inhibit replication. To investigate this, we deleted the spo4 gene in the mes1pr-cdc18, cdt1 strain, to prevent MII entry. Most spo4Δ cells arrest with cytoplasmic microtubules, and thus appear to have arrested before entry into MII (Nakamura et al., 2002). However, this arrest did not enhance DNA replication (Supplemental Figure S2A), implying that MII entry and sporulation-related events are not responsible for preventing a full round of chromosome replication.
Overexpression of Cdt1 and Cdc18 during MI–MII promotes Mcm2-7 chromatin binding but does not lead to a complete round of DNA replication
Physiological levels of Cdt1 and Cdc18 lead to partial DNA replication during MI–MII, implying that all components required for S phase are present but at least one replication step is inefficient. To examine licensing during MI–MII, we compared the level of Mcm2 chromatin binding during this interval with that seen around the time of meiS using a detergent extraction method that gives a global assessment of protein–chromatin interaction (Kearsey et al., 2000, 2005). Mcm2 chromatin binding was not convincingly detectable in the mes1pr-cdc18, cdt1 strain in the MI–MII interval (Figure 3A, 5.5 h), even in a mes1Δ background in which lower CDK levels in the MI–MII interval might be expected to stimulate Mcm2-7 chromatin binding (Izawa et al., 2005). Deletion of the mes1 gene also did not stimulate DNA replication during MI–MII in this strain background (unpublished data). In the mitotic cell cycle, DNA rereplication can be efficiently induced in G2 when Cdc18 and Cdt1 are overexpressed (Gopalakrishnan et al., 2001; Yanow et al., 2001). We found that when mes1-promoter driven Cdc18 and Cdt1 were expressed from plasmids to boost levels during the MI–MII interval (Figure 3B), Mcm2 chromatin binding could be clearly detected at a level similar to that seen around the time of meiS (Figure 3, C–E, compare 2.5 and 5.5 h). However, this did not result in enhanced DNA replication compared with the situation with low levels of Mcm2-7 chromatin binding (Supplemental Figure S3C), suggesting that licensing is not rate limiting for replication. We compared the levels of Cdc18 and Cdt1 achieved in late meiosis with mes1-promoter driven expression with levels in the mitotic cycle that are capable of promoting clear rereplication. These mitotic levels of the proteins were clearly lower than meiotic levels (Supplemental Figure S3, A–C), suggesting that late meiosis is a more inhibitory environment for replication compared with G2 of the mitotic cycle.
FIGURE 3:
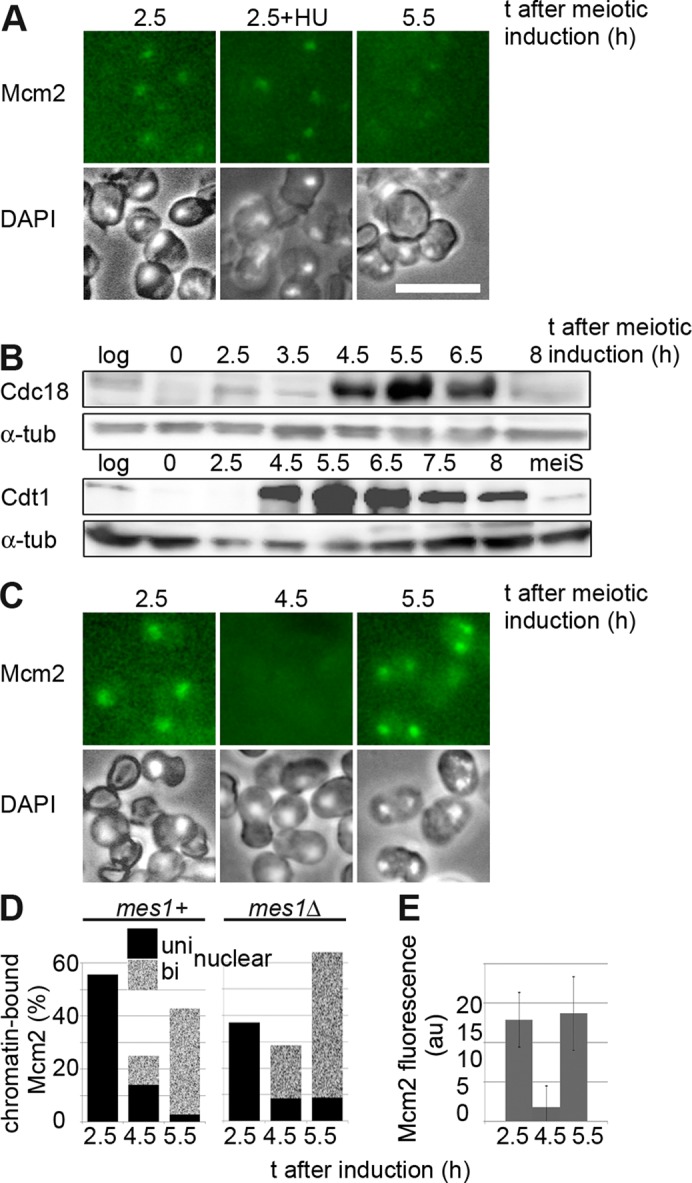
Overexpressing Cdc18 and Cdt1 in late meiosis increases loading of Mcm2 onto chromatin. (A) Cells expressing Mcm2-CFP, Cdc18, and Cdt1 in late meiosis at levels similar to those seen in meiS were detergent extracted at the time points shown and imaged to detect chromatin-bound Mcm2. Chromatin-associated Mcm2 can be detected around the time of meiS (2.5 h) both in the absence and presence of HU (to trap Mcm2 on DNA) but not at later time points. The strain used was also deleted for mes1 (P1756), but similar results were obtained for a mes1+ strain (unpublished data). Bar, 10 μm. (B) Western blot analysis of Cdc18 and Cdt1 levels in pat1-synchronized meiosis when both mes1pr-cdc18 and mes1pr-cdt1 constructs are expressed from plasmids (P2466). Levels of proteins in log-phase wt cells or meiS cells are shown for comparison. (C) Chromatin-binding assay for Mcm2 as in A, except that strain P2466 was used, overexpressing Cdc18 and Cdt1 in late meiosis. Similar results were obtained using a mes1Δ strain (unpublished data). (D) Quantitation of percentage of cells showing Mcm2-CFP chromatin binding in meiosis when Cdc18 and Cdt1 were overexpressed. At least 100 cells were counted for each time point. Strains used were P2466 (mes1+) and 2467 (mes1Δ). (E) As D, except that fluorescence intensity of chromatin-associated Mcm2-CFP in mes1+ cells (P2466) was quantitated (arbitrary units). One hundred cells were counted for each sample, and SD is shown.
We assume that when Cdt1 and Cdc18 are expressed at physiological levels during MI–MII, some Mcm2-7 chromatin binding must be occurring to be responsible for the DNA replication detected, but this is below the threshold detectable by the chromatin-binding assay. Analysis of Mcm2-7 chromatin binding at ORC has shown that this process is reiterative, and 10–40 Mcm2-7 complexes are loaded onto ORC (reviewed in Blow and Dutta, 2005). Reducing the amount of Mcm2-7 bound to one to two complexes per origin has been shown to have little effect on the efficiency of DNA replication under normal circumstances, so a very low level of Mcm2-7 chromatin binding during the MI–MII interval may be sufficient for the partial replication detected.
Effects of modulating CDK activity on replication efficiency
We investigated whether modulating CDK activity affected DNA replication during MI–MII. Inhibition followed by activation would be predicted to promote DNA replication, since inhibition should promote loading of Mcm2-7 onto chromatin, and subsequent activation should promote initiation. Of interest, just inhibiting protein synthesis in Xenopus oocytes after MI results in DNA replication via a block to cyclin B1 synthesis (Furuno et al., 1994; Narasimhachar and Coue, 2009). We carried out a similar experiment by exposing mes1pr-cdc18, cdt1 cells to cycloheximide around the time of MI, after Cdc18 and Cdt1 expression had occurred, and washing out the drug 1 h later. This treatment resulted in a marginal increase in DNA replication compared with cells not exposed to the drug (Supplemental Figure S2).
To test more specifically whether temporary inactivation of CDK could stimulate DNA replication in late meiosis, we used a Shokat allele of cdc2 (cdc2-as), which can be inhibited by an ATP analogue (Bishop et al., 2000). The allele used is slightly temperature sensitive; cells are elongated in the mitotic cell cycle and arrest in G2 rather than G1 after nitrogen starvation (unpublished data). We therefore induced meiosis from G2 as previously described (Watanabe et al., 2001); cells execute normal meiotic events when induced from G2, and cdc18 and cdt1 expression from the mes1 promoter was induced, but meiS does not occur (Supplemental Figure S4, A and B). Strains used were also deleted for the spo4 gene to arrest cells in the MI–MII interval. Inhibiting Cdc2-as activity with 1NM-PP1 after MI blocked entry into MII, although MII could occur with delayed kinetics if only a pulse of 1NM-PP1 was given (Supplemental Figure S4C); this shows that the concentration of 1NM-PP1 used is effective in inhibiting CDK. Inhibition of Cdc2-as in cells expressing Cdc18 and Cdt1 using a 1-h pulse of 1NM-PP1 resulted in a slight increase in DNA replication (Figure 4, A and B). Because Cdc2-as is temperature sensitive, we also examined the effect of reducing the temperature after the 1NM-PP1 pulse, and this resulted in increased DNA replication (Figure 4, D–F). Almost the same degree of replication can be achieved with no inhibitor, just by reducing the temperature in the MI–MII interval.
FIGURE 4:
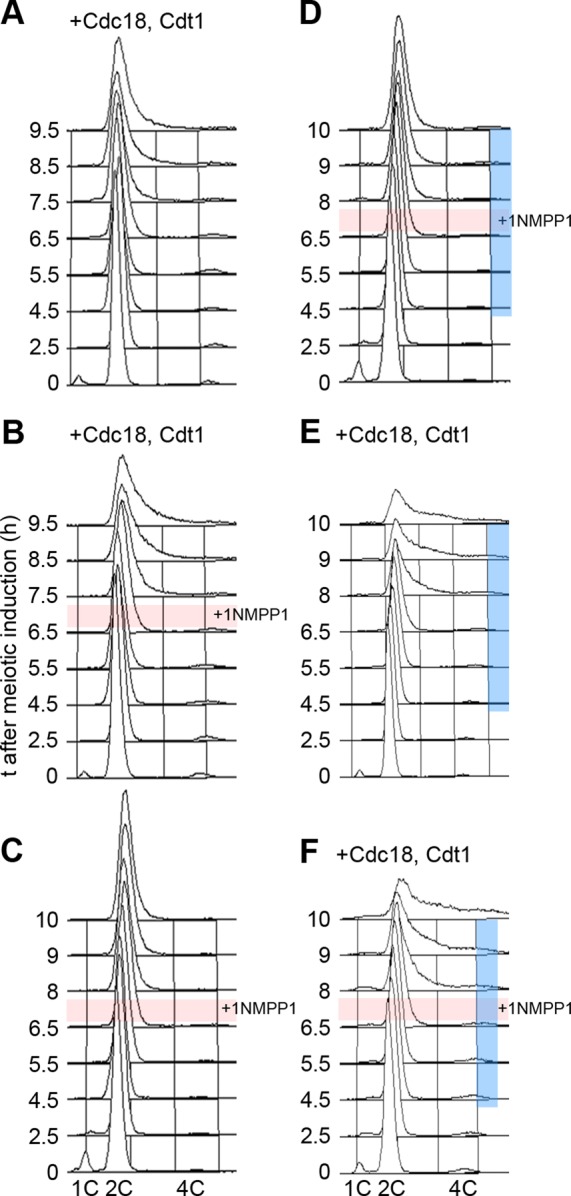
Effect of modulating CDK activity in late meiosis on DNA replication. Meiosis was induced from G2, using cdc2-as spo4Δ strains, by Pat1 inactivation. Cell DNA contents, determined by flow cytometry, are shown. Here +Cdc18, Cdt1 indicates strains expressing Cdc18 and Cdt1 at physiological levels in late meiosis; otherwise cells were wt for Cdc18 and Cdt1 expression. The pink bar indicates that a pulse of 1NM-PP1 was applied at 6.75 h after meiotic induction and washed out 1 h later. The blue bar indicates that the temperature was reduced after meiotic induction (34°C, 0–3.5 h; 32°C, 3.5–5 h; 30°C 5–6.5 h; 27°C 6.5–10 h). The timing of events in meiosis induced from G2 is given in Supplemental Figure S4. Strains used were (A, B, E, F) P3028 and (C, D) P3027.
We interpret these results to indicate that when Cdc18 and Cdt1 are expressed at physiological levels in late meiosis, some increased DNA replication during MI–MII can be achieved by transiently reducing CDK activity, most likely by increasing the efficiency of Mcm2-7 chromatin loading. The requirement for reactivation of CDK activity presumably reflects the positive role of the kinase in the initiation step. Nevertheless, modulation of CDK activity combined with Cdc18 and Cdt1 expression resulted in only partial DNA replication, indicating that there is an additional constraint on DNA replication.
Regulation of Hsk1-Dfp1 and RNR activity in meiosis
Previous studies showed that levels of the Dfp1 regulator of Hsk1 (orthologous to Dbf4-Cdc7, i.e., DDK) are reduced around the time of anaphase of MI (Ogino et al., 2006), consistent with its APC-mediated ubiquitylation in mitosis (Cheng et al., 1999; Weinreich and Stillman, 1999; Ferreira et al., 2000), and this potentially constitutes a block to late meiotic replication. To investigate this possibility, we constructed a Dfp1m strain in which the N-terminally localized APC destruction box (RXXL) is deleted and found that this mutated protein is stabilized in the MI-MII interval but not subsequently (Figure 5, A and B; compare 5.5 h, Dfp1m, and wt). However, when combined with late meiotic expression of Cdc18 and Cdt1, no increase in DNA replication is seen (Figure 5D).
FIGURE 5:
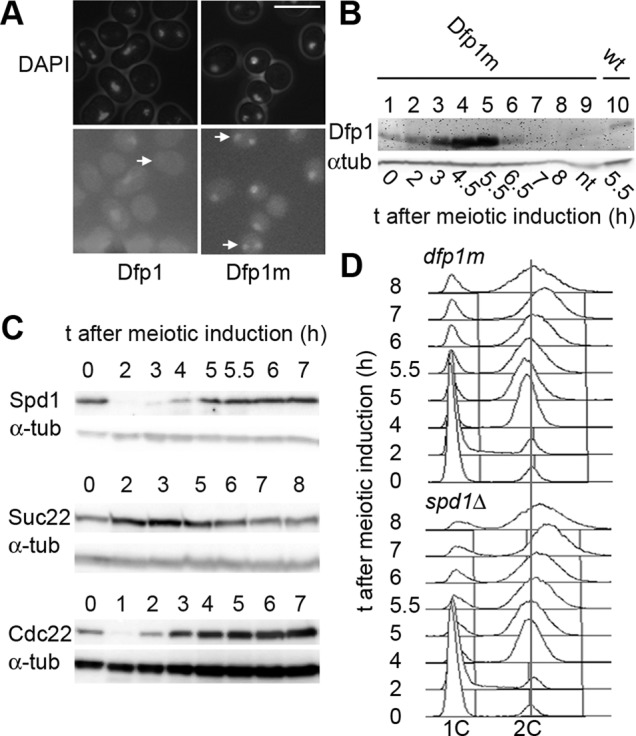
Stabilizing the Dfp1 regulator of DDK or inactivating the RNR inhibitor Spd1 does not promote further DNA replication between meiosis I and II. (A) Dfp1m-CFP, where the RXXL motif in the N-terminal region is deleted, is stabilized after MI. Dfp1m-CFP can be detected in the nuclei of binucleated cells (arrows), having completed MI (P3025), in contrast to the wt protein (P3026). Bar, 10 μm. (B) Analysis of RXXL-mutated Dfp1m levels in pat1-synchronized meiosis; levels of mutant and wt proteins are compared 5.5 h after meiotic induction (compare lanes 5 and 10). Strains used were P3025 and P3026. (C) Levels of Spd1 and RNR subunits Suc22 and Cdc22 in a pat1-synchronized meiosis; strains used were P2383, P3017, and P3018. (D) Analysis of cell DNA contents during a pat1-synchronized meiosis. Cells overexpressed Cdc18 and Cdt1 during MI–MII and also expressed either the stabilized version of Dfp1 (Dfp1m; top, P3014) or were deleted for the spd1 gene (bottom, P3021).
We also considered whether dNTP supply by RNR could be relevant to meiotic DNA replication, since meiS is very sensitive to stabilization of the Spd1 inhibitor of RNR (Holmberg et al., 2005), and although RNR subunits are present in late meiosis, Spd1 levels recover after proteolysis during meiS (Figure 5C). However, deleting the spd1 gene did not stimulate DNA replication in the mes1pr-cdc18, cdt1 strain (Figure 5D).
Analysis of replication by molecular combing shows that the elongation step of DNA replication is inefficient
To gain insight into the rate-limiting replication step in late meiosis, we labeled nascent DNA in cells expressing Cdc18 and Cdt1 after MI using 5-bromo-2′-deoxyuridine (BrdU) and analyzed incorporation by molecular combing. These experiments were carried out in a rec12Δ background to avoid detection of DNA synthesis associated with recombination. BrdU was added before most cells had entered MI, so that replication tracks should indicate the contiguous region replicated from a single origin. In a rec12Δ strain with no late Cdc18 and Cdt1 expression, ∼1% of DNA tracks were labeled, possibly reflecting mitochondrial DNA synthesis or cells still in meiS (Figure 6A). In contrast, in strains expressing Cdc18 and Cdt1 in late meiosis, the fraction of labeled DNA increased significantly (Figure 6, B and C) and is approximately consistent with quantitation of DNA synthesis using EdU. Overexpression of Cdc18 and Cdt1 resulted in a similar level of replication (11% of fibers BrdU positive) to that obtained with expression at physiological levels (13%). In both cases, most DNA fibers labeled with BrdU were short (median, 17–18 kb), which is only about twice the track length seen when the elongation step of a normal S phase is inhibited with HU (Patel et al., 2006), and only ∼7% of labeled tracks were >50 kb. This indicates that either the elongation rate is very slow or elongating replication forks terminate prematurely and provides an explanation for the inefficient replication in this interval when the licensing block is subverted. Only 9% of fibers had more than one labeled region, and the data set was too small to allow reliable estimation of the average interorigin distance.
FIGURE 6:
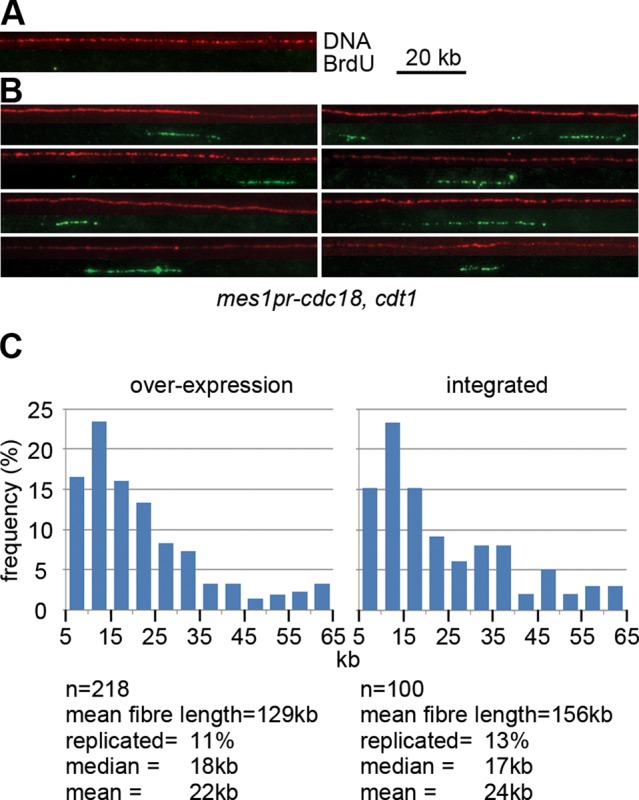
Analysis of DNA replication in the meiosis I–II interval by molecular combing. (A, B) DNA combing analysis of DNA fibers from cells in late meiosis. Representative images of the combed DNA fibers from (A) control strain (P3023) and (B) a strain expressing Cdc18 and Cdt1 during late meiosis (P2458, P2463). BrdU was added to cells 4.5 h after meiotic induction, and cells were harvested and processed 1.5 h later. Each panel shows a DNA fiber (shown in red, detected with anti-DNA antibody) and the corresponding image for BrdU detection (shown in green, detected with anti-BrdU). (C) Distribution of BrdU track lengths in cells expressing Cdc18 and Cdt1 in late meiosis from plasmids (left, P2463) or integrated genes (right, P2458). The relationship 10 μm = 20 kb was used for calculating replication track lengths.
DISCUSSION
We investigated whether the inhibition of Mcm2-7 chromatin binding between the meiotic nuclear divisions is solely responsible for replication inhibition during this interval. As in the mitotic cycle, Mcm2-7 chromatin binding in meiosis is regulated by proteolysis of Cdc18 and Cdt1 during meiS. Ectopic expression of these factors during the MI–MII interval leads to a partial round of DNA replication (∼15% of the genome is replicated, estimated from EdU labeling). When Cdc18 and Cdt1 are expressed at levels similar to those seen in meiS, the level of Mcm2-7 on chromatin is lower than in meiS, suggesting that there are additional regulatory mechanisms inhibiting licensing. When Cdc18 and Cdt1 were overexpressed, this resulted in a higher level of Mcm2-7 chromatin binding, comparable to levels seen in meiS, but the overall extent of DNA replication is not higher than that seen with physiological levels of expression. This is in contrast to the situation seen in the mitotic cell cycle, in which overexpression of these factors leads to dramatic rereplication (Gopalakrishnan et al., 2001; Yanow et al., 2001). We also showed that levels of Cdc18 and Cdt1 comparable to those achieved during the MI–MII interval were efficient at driving rereplication in G2 of the mitotic cycle. This suggests that there are additional constraints after Mcm2-7 chromatin binding affecting initiation or elongation that restrict the extent of DNA replication.
CDK activity is important for regulating DNA replication as a negative regulator of Mcm2-7 chromatin binding, acting on both ORC (Vas et al., 2001; Wuarin et al., 2002) and Cdc18 (Lopez-Girona et al., 1998) in fission yeast. It also phosphorylates initiation factors and activates replication (Tanaka et al., 2007; Zegerman and Diffley, 2007); thus transient inhibition of CDK is usually required to drive rereplication in the mitotic cycle. We did not observe any effect of inactivation of Mes1 on Mcm2-7 chromatin binding, and since this allows Cdc13 (cyclin B) degradation in the MI–MII interval by relieving inhibition of specific APC coactivators (Izawa et al., 2005; Kimata et al., 2011), this implies that Cdc13–Cdc2 activity is not solely responsible for inhibition of Mcm2-7 chromatin binding. However, other, possibly meiotic-specific, cyclins, such as Rem1 (Malapeira et al., 2005), could be differently regulated and help maintain some Cdc2 activity in late meiosis. We directly modulated Cdc2 activity using a Shokat allele, and transient inhibition increased DNA replication in the MI–MII interval, compared with the situation in which just Cdc18 and Cdt1 were expressed, and with some cells showing 4C DNA contents. However, most cells still had DNA contents substantially less than 4C, again suggesting that regulation of CDK, Cdc18, and Cdt1 is not the only factor blocking replication during this interval.
We found no evidence that regulation of the Hsk1-Dfp1 kinase or of the dNTP supply via RNR regulation contributes to preventing replication after MI. It was recently shown that the Hsk1-Dfp1 requirement for DNA replication is suppressed by high temperatures (Matsumoto et al., 2011), and it is possible that the temperature shift used to achieve Pat1 inactivation and meiosis in our experiments lessened any regulatory role for this protein kinase. In addition, there may be a reduced requirement for DDK for DNA replication in meiosis (Wan et al., 2006). Inactivating the Spd1 inhibitor of RNR, which is essential for allowing meiS progression (Holmberg et al., 2005), also did not enhance late meiotic replication.
In the mitotic cycle, rereplication activates checkpoint mechanisms that delay mitotic entry, and this G2 arrest appears to promote further rereplication (Melixetian et al., 2004; Zhu et al., 2004; Zhu and Dutta, 2006; Lin and Dutta, 2007; Klotz-Noack et al., 2012). In contrast, we show here that partial replication in the MI–MII interval does not inhibit MII entry. This replication may be more akin to a normal S phase rather than rereplication and may not activate checkpoint mechanisms (Torres-Rosell et al., 2007), whereas rereplication in G2 may generate abnormal structures that effect checkpoint activation (Davidson et al., 2006). However, inhibiting MI–MII replication with HU also does not block MII entry, in contrast to inhibition of meiS, which blocks MI entry. In addition, EdU incorporation blocks entry into MI, probably by activating a checkpoint response (Hua and Kearsey, 2011), but not MII. These results indicate that progress into MII is less stringently regulated by aberrations in chromosome structure than MI.
The partial round of replication seen on Cdc18 and Cdt1 expression could be due to extensive elongation from very infrequent initiation sites or more frequent initiation and limited elongation. Using molecular combing, we showed that short replication tracks (median, 17–18 kb) result from late expression of Cdc18 and Cdt1, suggesting slow elongation or premature fork termination. This helps to explain why even overexpression of Cdc18 and Cdt1 or modulation of CDK levels, which affects initiation, does not lead to extensive replication. This inefficiency could arise indirectly; for example, the environment for elongation may be suboptimal during MI–MII, as appears to be the case to a lesser extent for rereplicating forks in the mitotic cell cycle (Melixetian et al., 2004), and forks may consequently move more slowly or stall. During the MI–MII interval, it is possible that chromatin does not fully decondense, which may be a constraint on fork movement. In addition, the lack of checkpoint-mediated arrest of meiotic progression may cause replication to be inhibited by chromosome condensation on MII entry, although we did not observe enhanced replication when cells were arrested in the MI–MII interval.
Alternatively, inefficiency in the elongation step of replication could represent a meiotic mechanism to promote genome stability, for example, via modification of chromatin or inactivation of an elongation factor. As far as we have established, initiation/elongation factors are present in late meiosis. We analyzed levels of initiation/elongation factors (ORC, Mcm, Cut5, Cdc45, Sld2, Sld3, Cdc23), and these are present at levels higher than or equal to those seen in meiS (unpublished data); in addition, SILAC analysis indicates that replicative polymerases and polymerase-associated factors are also present (J. Gregan, unpublished results). Although control at this step might seem to be biologically irrelevant, given regulation at the earlier licensing step, it is an established feature of mitS phase regulation that multiple overlapping controls are used to improve the fidelity of control that would be achieved with just a single regulatory mechanism (for review see Diffley, 2011). In the mitotic cell cycle, these regulatory devices operate in parallel on licensing, but in meiosis, inefficient elongation could act as a fail-safe device operating sequentially with earlier licensing regulation to limit any replication occurring from rogue initiation events. This might seem unlikely, since partially rereplicated DNA can lead to gene amplification (Green et al., 2010), but it would be preferable to having unrestrained forks, and rereplicated DNA could be subjected to subsequent repair by nascent strand displacement or exonuclease activity. A precedent for this kind of mechanism exists in Drosophila polytene chromosomes, in which the SUUR protein represses fork movement, possibly via an effect on chromatin structure involving the repressive H3K27me3 modification (Sher et al., 2012). Maintaining genome stability in gamete formation is important, particularly in multicellular organisms, which may have hundreds of thousands of replication origins, and high-fidelity licensing control may fail to block completely illegal initiation events. It will be interesting to establish whether there are extra dimensions to meiotic replication control compared with the situation in somatic cells and whether breakdown of such mechanism(s) is a source of mutations in gametes.
MATERIALS AND METHODS
Yeast strain constructions and growth conditions
Strains were constructed using standard methods (Moreno et al., 1991) and are listed in Supplemental Table S2. Details of strain constructions, together with the oligonucleotides used (Supplemental Table S3), are given in the Supplemental Information. Homozygous diploids were constructed by centrifuging haploid cells at 15,000 × g for 15 min and plating on phloxin B–containing YES plates to allow selection of diploid colonies from colony color (Carazo-Salas and Nurse, 2006). Ploidy was checked by flow cytometry. HU was used at 12 mM, cycloheximide at 200 μg/ml, thiamine at 15 μM, and 1NM-PP1 at 1 μM.
Meiosis induction
To induce synchronized meiosis, pat1-114 cells were first arrested in G1 phase by growth in EMM lacking NH4Cl (EMM-N) at 26°C for 16 h and then refed in EMM medium at 34ºC. For induction of meiosis from G2, cdc2as cells were grown to log phase at 26°C and then transferred to EMM-N medium for 16 h at the same temperature. Meiosis was induced after transferring to EMM medium by shifting to 34°C. DNA replication was monitored by flow cytometry or EdU incorporation as previously described (Hua and Kearsey, 2011).
Chromatin-binding assays
Mcm2-CFP chromatin-binding assays were carried out by digesting cells with zymolyase and extracting with a Triton X-100–containing buffer (Kearsey et al., 2005). Cells were imaged by fluorescence microscopy to assess Mcm2-CFP retention.
Flow cytometry
Cells were fixed in 70% ethanol and analyzed after RNase digestion and SYTOX Green staining as previously described (Kearsey et al., 2005).
Pulsed-field gel electrophoresis
PFGE was carried out using 0.8% agarose/TAE gels (40 mM Tris-acetate, 2 mM EDTA, pH 8.3) in a CHEF III apparatus (Bio-Rad, Hercules, CA) with settings 2 V/cm for 48 h; switch time, 30 min; and angle, 106º.
EdU incorporation and detection
Procedures to allow incorporation and detection of EdU (Life Technologies, Carlsbad, CA) by fluorescence microscopy and flow cytometry were as described previously (Hua and Kearsey, 2011). EdU was used at a concentration of 10 μM.
Protein analysis
Protein extracts were made by TCA extraction and analyzed by Western blotting as described previously (Ralph et al., 2006). Tandem affinity purification–tagged proteins were detected with peroxidase–antiperoxidase–soluble complex (P1291; Sigma-Aldrich, St. Louis, MO). CFP/GFP/YFP-tagged proteins were detected using anti-GFP antibody (11814460001; Roche, Indianapolis, IN). Cdt1-myc was detected using anti-myc antibody (M5546; Sigma-Aldrich), and α-tubulin was used as loading control and detected with antibody T5168 (Sigma-Aldrich). Anti-Cdt1 (gift from H. Nishitani [University of Hyogo, Japan]) and anti-Cdc18 antibodies (gift from O. Harris and J. Hayles [London Research Institute, Cancer Research UK]) were used in some experiments.
DNA combing
DNA combing was carried out as previously described (Michalet et al., 1997; Patel et al., 2006). BrdU was used at a concentration of 3 μM. BrdU was detected using rat anti-BrdU (BU1/75; Sera Lab, Haywards Heath, United Kingdom), and DNA was detected using mouse anti-DNA (MAB3034; Chemicon, Temecula, CA). Secondary antibodies used were goat anti-rat Alexa 488 (A11006; Molecular Probes, Eugene, OR) and goat anti-mouse Alexa 546 (A11030; Molecular Probes). Images were collected using μManager software source (Edelstein et al., 2010) controlling a Zeiss Axioplan 2 microscope (100× objective, numerical aperture 1.4; Carl Zeiss, Jena, Germany) coupled to a Hamamatsu ORCA ER camera (Hamamatsu, Hamamatsu, Japan). Only BrdU-positive fibers >2.5 μm were taken into account as real incorporation patches and were disregarded unless DNA staining was observed for at least 2.5 μm at each end of BrdU region.
Fluorescence microscopy
Cells were viewed live or fixed in methanol/acetone and mounted in 1.2% agarose containing 50 ng/ml 4′,6-diamidino-2-phenylindole (DAPI). Images were collected as for DNA combing slides. For quantitative measurements of Mcm2-CFP fluorescence, images were collected in the same session, and samples were only illuminated with the excitation lamp during data collection to minimize bleaching. ImageJ (National Institutes of Health, Bethesda, MD) was used for quantitative measurements of fluorescence intensity. Nuclei were identified from the DAPI channel, allowing positioning of a mask of defined pixel diameter to measure average pixel intensity in the CFP channel, and the average background intensity was subtracted. Several hundred nuclei were counted for each sample.
Supplementary Material
Acknowledgments
This work was supported by Austrian Science Fund Grants F3403 and P23609 (J.G.) and by Cancer Research UK (S.E.K). M.N. was supported by a Cancer Research UK Studentship. We thank Tony Carr, Oliver Harris, Damien Hermand, and Hideo Nishitani for strains and antibodies. We are grateful to Estrella Guarino, Israel Salguero, Marianne Shepherd, and Larissa Wakefield for comments.
Abbreviations used:
- BrdU
5-bromo-2′-deoxyuridine
- CDK
cyclin-dependent kinase
- DDK
Dbf4-dependent kinase
- EdU
5-ethynyl-2′-deoxyuridine
- MI
meiosis I
- MII
meiosis II
- meiS
premeiotic S phase
- mitS
mitotic S phase
- ORC
origin recognition complex
- PFGE
pulsed-field gel electrophoresis
- RNR
ribonucleotide reductase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0825) on January 9, 2013.
Present addresses: *State Key Laboratory of Proteomics, Beijing Proteome Research Center, Beijing Institute of Radiation Medicine, Beijing 102206, China
†Cell Biology Program, Sloan-Kettering Institute for Cancer Research, Memorial Sloan-Kettering Cancer Center, New York, NY 10065.
REFERENCES
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Chan CS, Hochwagen A, Bell SP. Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLoS Genet. 2012;8:e1002643. doi: 10.1371/journal.pgen.1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Rabitsch KP, Fuchs J, Gruber S, Sullivan M, Uhlmann F, Petronczki M, Toth A, Nasmyth K. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev Cell. 2003;4:727–739. doi: 10.1016/s1534-5807(03)00129-1. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol. 2006;8:1102–1107. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- Catlett MG, Forsburg SL. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol Biol Cell. 2003;14:4707–4720. doi: 10.1091/mbc.E03-05-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Collyer T, Hardy CF. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol Cell Biol. 1999;19:4270–4278. doi: 10.1128/mcb.19.6.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IF, Li A, Blow JJ. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell. 2006;24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–R786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Quality control in the initiation of eukaryotic DNA replication. Phil Trans R Soc Lond. B Biol Sci. 2011;366:3545–3553. doi: 10.1098/rstb.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using manager. Curr Protocols Mol Biol. 2010:14.20.11–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MF, Santocanale C, Drury LS, Diffley JF. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol Cell Biol. 2000;20:242–248. doi: 10.1128/mcb.20.1.242-248.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. Only connect: linking meiotic DNA replication to chromosome dynamics. Mol Cell. 2002;9:703–711. doi: 10.1016/s1097-2765(02)00508-7. [DOI] [PubMed] [Google Scholar]
- Furuno N, Nishizawa M, Okazaki K, Tanaka H, Iwashita J, Nakajo N, Ogawa Y, Sagata N. Suppression of DNA replication via Mos function during meiotic divisions in Xenopus oocytes. EMBO J. 1994;13:2399–2410. doi: 10.1002/j.1460-2075.1994.tb06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Simancek P, Houchens C, Snaith HA, Frattini MG, Sazer S, Kelly TJ. Redundant control of rereplication in fission yeast. Proc Natl Acad Sci USA. 2001;98:13114–13119. doi: 10.1073/pnas.221467598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BM, Finn KJ, Li JJ. Loss of DNA replication control is a potent inducer of gene amplification. Science. 2010;329:943–946. doi: 10.1126/science.1190966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006;25:5171–5179. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 2005;19:853–862. doi: 10.1101/gad.329905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Hutti JE, Cantley LC, Morgan DO. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol Cell. 2007;25:689–702. doi: 10.1016/j.molcel.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H, Kearsey SE. Monitoring DNA replication in fission yeast by incorporation of 5-ethynyl-2′-deoxyuridine. Nucleic Acids Res. 2011;39:e60. doi: 10.1093/nar/gkr063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi M, Ohsumi K, Yamamoto TM, Sawada W, Kishimoto T. Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M-M transition in Xenopus oocyte extracts. EMBO J. 2000;19:4513–4523. doi: 10.1093/emboj/19.17.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature. 2005;434:529–533. doi: 10.1038/nature03406. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Brimage L, Namdar M, Ralph E, Yang X. In situ assay for analyzing the chromatin binding of proteins in fission yeast. Methods Mol Biol. 2005;296:181–188. doi: 10.1385/1-59259-857-9:181. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Cotterill S. Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Mol Cell. 2003;12:1067–1075. doi: 10.1016/s1097-2765(03)00441-6. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Montgomery S, Labib K, Lindner K. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 2000;19:1681–1690. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Kitamura K, Fenner N, Yamano H. Mes1 controls the meiosis I to meiosis II transition by distinctly regulating the anaphase-promoting complex/cyclosome coactivators Fzr1/Mfr1 and Slp1 in fission yeast. Mol Biol Cell. 2011;22:1486–1494. doi: 10.1091/mbc.E10-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida M, Nagai T, Nakaseko Y, Shimoda C. Meiosis-dependent mRNA splicing of the fission yeast Schizosaccharomyces pombe mes1+ gene. Curr Genet. 1994;25:497–503. doi: 10.1007/BF00351668. [DOI] [PubMed] [Google Scholar]
- Klotz-Noack K, McIntosh D, Schurch N, Pratt N, Blow JJ. Re-replication induced by geminin depletion occurs from G2 and is enhanced by checkpoint activation. J Cell Sci. 2012;125:2436–2445. doi: 10.1242/jcs.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Dutta A. ATR pathway is the primary pathway for activating G2/M checkpoint induction after re-replication. J Biol Chem. 2007;282:30357–30362. doi: 10.1074/jbc.M705178200. [DOI] [PubMed] [Google Scholar]
- Lindner K, Gregan J, Montgomery S, Kearsey SE. Essential role of MCM proteins in premeiotic DNA replication. Mol Biol Cell. 2002;13:435–444. doi: 10.1091/mbc.01-11-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 2005;24:3940–3951. doi: 10.1038/sj.emboj.7600854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair. 2009;8:1127–1138. doi: 10.1016/j.dnarep.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol Biol Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malapeira J, Moldon A, Hidalgo E, Smith GR, Nurse P, Ayte J. A meiosis-specific cyclin regulated by splicing is required for proper progression through meiosis. Mol Cell Biol. 2005;25:6330–6337. doi: 10.1128/MCB.25.15.6330-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Lee BH, Amon A. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev Cell. 2003;4:711–726. doi: 10.1016/s1534-5807(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how. Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Hayano M, Kanoh Y, Masai H. Multiple pathways can bypass the essential role of fission yeast Hsk1 kinase in DNA replication initiation. J Cell Biol. 2011;195:387–401. doi: 10.1083/jcb.201107025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melixetian M, Ballabeni A, Masiero L, Gasparini P, Zamponi R, Bartek J, Lukas J, Helin K. Loss of geminin induces rereplication in the presence of functional p53. J Cell Biol. 2004;165:473–482. doi: 10.1083/jcb.200403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- Mimura S, Seki T, Tanaka S, Diffley JF. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–1123. doi: 10.1038/nature03024. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami H, Nurse P. Meiotic DNA replication checkpoint control in fission yeast. Genes Dev. 1999;13:2581–2593. doi: 10.1101/gad.13.19.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajo N, Yoshitome S, Iwashita J, Iida M, Uto K, Ueno S, Okamoto K, Sagata N. Absence of Wee1 ensures the meiotic cell cycle in Xenopus oocytes. Genes Dev. 2000;14:328–338. [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Nakamura-Kubo M, Shimoda C. Novel fission yeast Cdc7-Dbf4-like kinase complex required for the initiation and progression of meiotic second division. Mol Cell Biol. 2002;22:309–320. doi: 10.1128/MCB.22.1.309-320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhachar Y, Coue M. Geminin stabilizes Cdt1 during meiosis in Xenopus oocytes. J Biol Chem. 2009;284:27235–27242. doi: 10.1074/jbc.M109.008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, Arai KI, Masai H. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc Natl Acad Sci USA. 2006;103:8131–8136. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz DG, Forsburg SL. Meiotic S-phase damage activates recombination without checkpoint arrest. Mol Biol Cell. 2005;16:1651–1660. doi: 10.1091/mbc.E04-10-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell. 2006;17:308–316. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E, Boye E, Kearsey SE. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 2006;7:1134–1139. doi: 10.1038/sj.embor.7400827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salguero I, Guarino E, Shepherd ME, Deegan TD, Havens CG, MacNeill SA, Walter JC, Kearsey SE. Ribonucleotide reductase activity is coupled to DNA synthesis via proliferating cell nuclear antigen. Curr Biol. 2012;22:720–726. doi: 10.1016/j.cub.2012.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarynski KE, Najor NA, Kepsel AC, Brush GS. Sic1-induced DNA rereplication during meiosis. Proc Natl Acad Sci USA. 2009;106:232–237. doi: 10.1073/pnas.0809731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 2012;22:64–75. doi: 10.1101/gr.126003.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Querenet M, Craig A, Gartner A, Blow JJ. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J Cell Biol. 2012;196:233–246. doi: 10.1083/jcb.201110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R. Meiotic DNA replication. Curr Top Dev Biol. 2004;61:29–60. doi: 10.1016/S0070-2153(04)61002-7. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Diffley JF. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2–7 during G1 phase. Nat Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, Pasero P, Lisby M, Haber JE, Aragon L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–1415. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Zhang C, Shokat KM, Hollingsworth NM. Chemical inactivation of cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 2006;174:1767–1774. doi: 10.1534/genetics.106.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, Cross FR. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Buck V, Nurse P, Millar JB. Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell. 2002;111:419–431. doi: 10.1016/s0092-8674(02)01042-5. [DOI] [PubMed] [Google Scholar]
- Yanow SK, Lygerou Z, Nurse P. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 2001;20:4648–4656. doi: 10.1093/emboj/20.17.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Dutta A. An ATR- and BRCA1-mediated Fanconi anemia pathway is required for activating the G2/M checkpoint and DNA damage repair upon rereplication. Mol Cell Biol. 2006;26:4601–4611. doi: 10.1128/MCB.02141-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.