The Rab11-family interacting proteins (Rab11-FIPs) facilitate Rab11-dependent vesicle recycling, yet it is unknown how these effectors cooperate with each other during recycling. It is found that Rab11-FIPs exhibit selective cooperation along dynamic tubular compartments to fill distinct spatiotemporal roles during recycling.
Abstract
The Rab11-family interacting proteins (Rab11-FIPs) facilitate Rab11-dependent vesicle recycling. We hypothesized that Rab11-FIPs define discrete subdomains and carry out temporally distinct roles within the recycling system. We used live-cell deconvolution microscopy of HeLa cells expressing chimeric fluorescent Rab11-FIPs to examine Rab11-FIP localization, transferrin passage through Rab11-FIP–containing compartments, and overlap among Rab11-FIPs within the recycling system. FIP1A, FIP2, and FIP5 occupy widely distributed mobile tubules and vesicles, whereas FIP1B, FIP1C, and FIP3 localize to perinuclear tubules. Internalized transferrin entered Rab11-FIP–containing compartments within 5 min, reaching maximum colocalization with FIP1B and FIP2 early in the time course, whereas localization with FIP1A, FIP1C, FIP3, and FIP5 was delayed until 10 min or later. Whereas direct interactions with FIP1A were only observed for FIP1B and FIP1C, FIP1A also associated with membranes containing FIP3. Live-cell dual-expression studies of Rab11-FIPs revealed the tubular dynamics of Rab11-FIP–containing compartments and demonstrated a series of selective associations among Rab11-FIPs in real time. These findings suggest that Rab11-FIP1 proteins participate in spatially and temporally distinct steps of the recycling process along a complex and dynamic tubular network in which Rab11-FIPs occupy discrete domains.
INTRODUCTION
Routine cell function depends on efficient trafficking between intracellular organelles and the cell surface (Hutagalung and Novick, 2011). Vesicle trafficking is generally regulated by small monomeric GTPases that operate by hydrolyzing GTP and alternating between “active” and “inactive” states, thus allowing associations with various membrane compartments (Stenmark, 2009). The discovery of the yeast secretory Sec proteins demonstrated that the membrane fusion and fission events that target membranes and proteins to various areas of a cell are guided by specific GTPases that organize this process (Novick et al., 1980). Subsequent efforts to characterize the mammalian homologues of the Sec proteins led to the discovery of Ras-like proteins from rat brain, known as Rabs (Touchot et al., 1987). Since then, >60 members of the Rab family have been identified (Segev, 2001), and despite their common mode of operation, the abundance of proteins in this subfamily allows for specificity when targeting membranes and proteins to specific areas and compartments of the cell (Pfeffer and Novick, 2010).
Early studies in neurons (Heuser and Reese, 1973), macrophages (Steinman et al., 1976), parietal cells (Forte et al., 1977), and fibroblasts (Anderson et al., 1977; Basu et al., 1981) demonstrated a need in a variety of cells to recycle membranes and proteins to the cell surface. Recycling of membranes and proteins can occur between internal compartments, such as endosomes and the Golgi apparatus (Lombardi et al., 1993), as well as between endosomes and the cell surface (Dautry-Varsat et al., 1983). In addition, recycling to and from the cell surface occurs through separate pathways primarily dependent on either Rab4 or Rab11a (Schmid et al., 1988; Sheff et al., 1999). The Rab11 family includes Rab11a, Rab11b, and Rab25 (Kikuchi et al., 1988; Chavrier et al., 1990; Goldenring et al., 1993; Bhartur et al., 2000) and operates through a centralized perinuclear compartment (Calhoun and Goldenring, 1996; Goldenring et al., 1996; Ullrich et al., 1996).
The Rab11-family interacting proteins (Rab11-FIPs) are effectors of Rab11 GTPases, which modulate Rab11a-dependent vesicle recycling (Horgan and McCaffrey, 2009). There are five members of this family, which share a conserved carboxyl-terminal Rab11-binding domain (Prekeris et al., 2000; Hales et al., 2001; Lindsay et al., 2002; Wallace et al., 2002b; Junutula et al., 2004; Jagoe et al., 2006). The Rab11-FIPs are expressed widely throughout tissues, including, but not limited to, lung, kidney, pancreas, and gastrointestinal tissues (Jin and Goldenring, 2006). The gene for Rab11-FIP1 (FIP1) encodes at least five functional transcripts (Jin and Goldenring, 2006), of which Rab11-FIP1C (also known as Rab-coupling protein [RCP]) has received the most attention. Functionally, studies implicated Rab11-FIP1C in endocytic sorting, trafficking of epidermal growth factor receptor (EGFR) and integrin subunits (Peden et al., 2004; Caswell et al., 2008), and transport between the recycling endosome and the trans-Golgi network (Jing et al., 2010). Rab11-FIP2 is involved in trafficking of a variety of receptors, channels, and transporters, including EGFR, transferrin, CXCR2, aquaporin-2, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors, norepinephrine transporter, and Niemann–Pick C1–like 1, a transmembrane protein involved in cholesterol absorption (Cullis et al., 2002; Lindsay and McCaffrey, 2002; Fan et al., 2004; Nedvetsky et al., 2007; Wang et al., 2008; Chu et al., 2009; Matthies et al., 2010). More recent investigations demonstrated that manipulation of Rab11-FIP2 perturbs caveolin localization (Lapierre et al., 2012). In addition, Rab11-FIP2 forms a ternary complex with both Rab11a and myosin Vb (Lapierre et al., 2001; Hales et al., 2002). Investigations of FIP3 demonstrated its role in the maintenance of the recycling endosome during cytokinesis and an association with dynein (Horgan et al., 2004, 2007, 2010a, b; Fielding et al., 2005; Wilson et al., 2005; Simon and Prekeris, 2008). In addition to the trafficking roles for FIP5 (Prekeris et al., 2000; Welsh et al., 2007; Oehlke et al., 2011), studies also found that FIP5 associates with the molecular motor kinesin II and Rab11a-containing vesicles (Schonteich et al., 2008). Previous investigations (Meyers and Prekeris, 2002; Wallace et al., 2002a, b) evaluated the potential for interactions between Rab11-FIPs, yet we do not know how the Rab11-FIPs cooperate with each other to organize the recycling process.
The characterization of regulatory proteins in vesicle trafficking has helped to elucidate the roles and organization of various proteins within these processes. Comparative studies of Rab5 and Rab11a demonstrated that these Rabs occupy different compartments and that the passage of transferrin through these compartments is temporally different (Trischler et al., 1999). These data were confirmed by electron microscopy studies and fluorescence imaging indicating that Rab4, Rab5, and Rab11a occupy distinct compartments with some amount of overlap and that there is a temporal difference in transferrin movement through these compartments (Sonnichsen et al., 2000). The Rab5 effector Rabenosyn-5 promotes increased association between Rab4 and Rab5, increased transferrin recycling, and reduced Rab4 and Rab11 association (de Renzis et al., 2002), suggesting that effector proteins play a significant role in modulating vesicle trafficking. Examination of the relationship between Rab11 and Rab4 by total internal reflection fluorescence microscopy indicated that cargo leaves Rab4/Rab11a-positive compartments through a Rab11a-specific compartment, further detailing the sequence of Rabs within the recycling network (Ward et al., 2005). Specialized compartments labeled by Rab11-FIPs have been proposed (Hales et al., 2001; Meyers and Prekeris, 2002; Jin and Goldenring, 2006), and recent studies implicated a Rab11-FIP2 and myosin Vb complex in time-dependent exit of Langerin from the central recycling compartment and trafficking at the plasma membrane (Gidon et al., 2012). It is clear that a greater understanding of how Rab-GTPases and their associated effector proteins are organized is required for a better understanding of how these proteins regulate vesicle trafficking.
We examined Rab11-FIP1 protein isoforms FIP1A, FIP1B, and FIP1C to determine how these proteins coordinate vesicle recycling in context with each other, as well as with other Rab11-FIPs, including FIP2, FIP3, and FIP5. Developments in live-cell imaging in the past decade have furthered the ability to visualize protein distribution and movement before fixation, adding another level of depth to our current understanding of trafficking processes (Lippincott-Schwartz et al., 2000; Lippincott-Schwartz, 2004). Therefore we used live-cell fluorescence imaging to assess compartment morphology, distribution, movement, and cargo loading into Rab11-FIP–containing compartments to gain insight into how the Rab11-FIPs come together to facilitate the recycling process. This study indicates that the Rab11-FIP1 proteins FIP1A, FIP1B, and FIP1C occupy unique compartments in live HeLa cells and also display distinct overlap with Rab11a. In addition, the loading of the compartments occupied by each of these Rab11-FIPs with transferrin is temporally distinct, pointing to time-dependent roles in the recycling process. Finally, we observed the overlap of FIP1 proteins along dynamic tubular compartments stretching from the perinuclear region to the periphery of the cell. In contrast, overlap between FIP1 proteins and other FIPs was more selective and not necessarily dependent on protein:protein interactions. The combined data suggest that Rab11-FIP1 proteins exhibit selective cooperation and participate in distinct spatiotemporal steps for transferrin recycling.
RESULTS
The Rab11-FIPs exhibit differences in distribution and movement
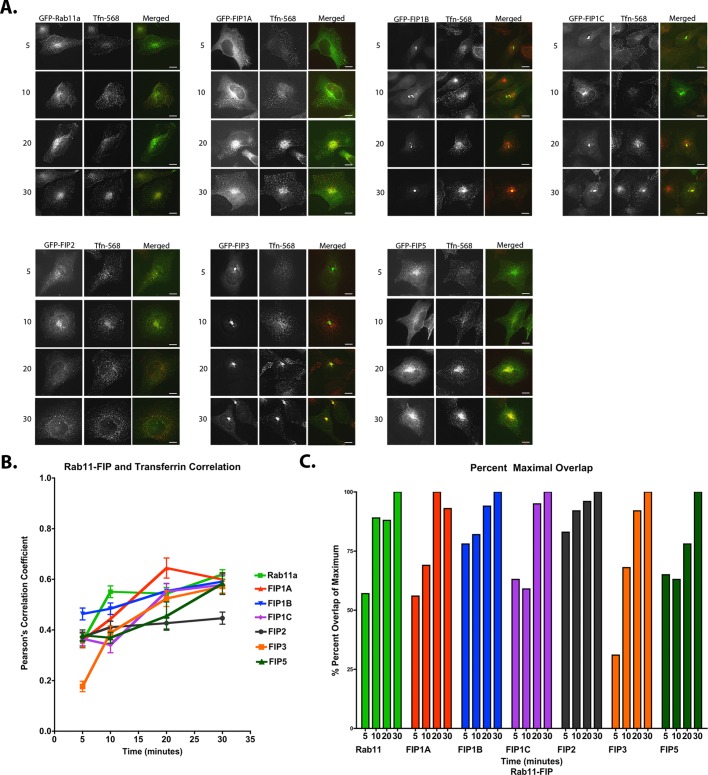
Previous studies in the field of vesicle recycling showed distinctive roles for the individual Rab11-family interacting proteins (Horgan and McCaffrey, 2009), although little is known about the interrelationship of these effector proteins with each other. We hypothesized that the Rab11-FIPs contribute to the organization of the entire recycling pathway by defining subdomains along this pathway. We reasoned that if we could discern differences in the localization and movement of the compartments labeled with these proteins in live cells, this would support different spatial roles of these proteins within the recycling system. Figure 1A shows a schematic representation of the Rab11-FIPs that we examined, including the conserved Rab11 binding domain (RBD). We expressed enhanced green fluorescent protein (EGFP)–Rab11-FIPs in HeLa cells and used live-cell deconvolution microscopy to visualize distinct differences in the localization, compartment morphology, and movement of compartments labeled with the EGFP–Rab11-FIPs. First, we observed that the compartments labeled with FIP1A, FIP2, and FIP5 maintained a wide distribution out to the periphery of the cell (Figure 1B), whereas compartments labeled with FIP1B, FIP1C, and FIP3 largely remained in the perinuclear region (Figure 1B). Structural differences, including the consensus region encoded by exon 4 in the FIP1 gene shared by FIP1A and FIP1B or the amino-terminal C2 domain shared by FIP1B and FIP1C, likely contribute to the differences observed in localization and movement (Figure 1A; Jin and Goldenring, 2006). Previous studies indicate that the C2 domain might also contribute to localization and distribution of FIP1C/RCP, FIP2, and FIP5 (Lindsay and McCaffrey, 2004). Of importance, the distributions we observed were comparable to studies using antibodies against endogenous proteins (Prekeris et al., 2000; Hales et al., 2001; Jin and Goldenring, 2006). Second, we noted that the extent to which the Rab11-FIPs occupied tubular endosomal compartments was variable. Figure 1B shows that FIP1A, FIP2, and FIP5 occupied shorter tubular domains, whereas FIP1C and FIP3 labeled longer tubules branching from the perinuclear region. FIP1B-expressing cells showed tubular structures primarily in the perinuclear region and were typically compressed into large, static compartments. In addition, we also noted that the movement of FIP1A was comparable to that of Rab11a-containing vesicles (Supplemental Video S1), whereas FIP1B and FIP1C compartments were primarily stationary, with the exception of the branching tubules (Supplemental Video S1). Taken together, the data point to two general phenotypes among the Rab11-FIPs and suggest that, whereas some might be involved in more-peripheral steps of the recycling pathway, others are likely involved in steps closely located to the central recycling endosome system adjacent to the centrioles.
FIGURE 1:
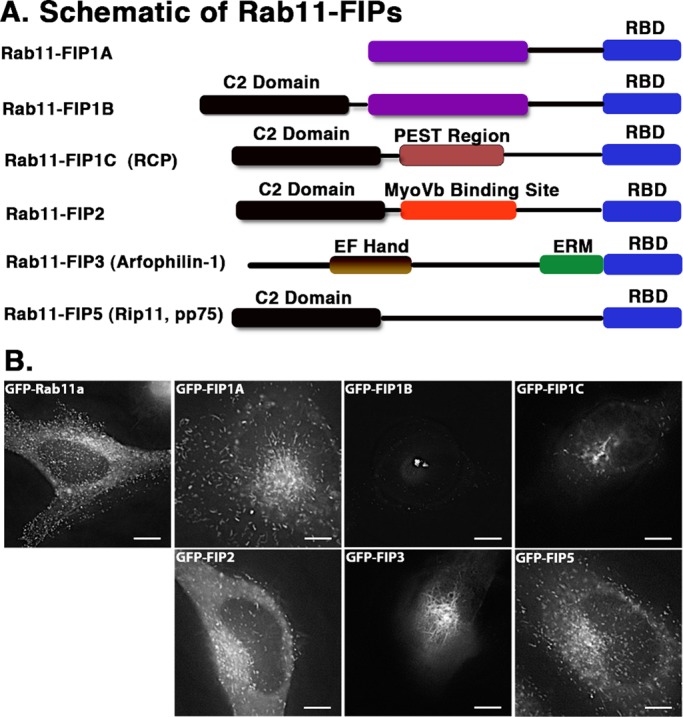
EGFP–Rab11-FIPs in live HeLa cells occupy distinct compartments. (A) Schematic representations of the Rab11-FIPs examined in the present study, the names used for each of the proteins in this and other studies, and a general outline of previously characterized regions within these proteins. Time-lapse images of each Rab11-FIP condition were collected, and representative single frames are presented for each condition in B. Corresponding time-lapse videos for the Rab11-FIP1 proteins and Rab11a are presented in Supplemental Video S1. Two general phenotypic differences were observed in FIP localization and movement. FIP1A, FIP2, and FIP5 maintain a wide distribution, similar to that observed for Rab11a. FIP1B, FIP1C, and FIP3 are primarily more centralized in localization. In addition, FIP1C and FIP3 displayed evidence of branching tubules emanating from the central perinuclear region. Data represent at least three independent experiments. Bars, 10 μm.
Rab11-FIPs influence the distribution and movement of Rab11a-containing vesicles
As a second line of evidence for differential roles of the Rab11-FIPs in vesicle recycling, we examined how these effector proteins influenced and overlapped with Rab11a. We conducted coexpression studies using live HeLa cells transfected with EGFP–Rab11-FIPs and mCherry–Rab11a. First, we confirmed that each of the Rab11-FIPs studied overlapped with Rab11a. Rab11a distribution and movement in the presence of FIP1A, FIP2, and FIP5 was comparable to Rab11a distribution and movement in single-expression studies (Figure 2 and Supplemental Video S2 of FIP1A with Rab11a). However, in the presence of FIP1B, FIP1C, and FIP3, Rab11a accumulated in large tubulovesicular compartments in and around the perinuclear region (Figure 2 and Supplemental Video S3 Rab11a with either FIP1B or FIP1C) Similar effects were observed in the case of FIP4 (Wallace et al., 2002b). In addition, compared with the perinuclear distribution observed with single expression of EGFP-FIP1B and EGFP-FIP1C (Figure 1), coexpression of mCherry-Rab11a induced a wider distribution of tubular elements throughout the cytoplasm for FIP1B and FIP1C in at least 60% of the cells imaged. These findings suggest that the availability of Rab11a might influence the dynamics of tubule formation for these two FIP1 proteins. These data indicate that Rab11-FIP1 isoforms influence Rab11a differently, suggesting different roles in targeting of key components of the vesicle-recycling system.
FIGURE 2:
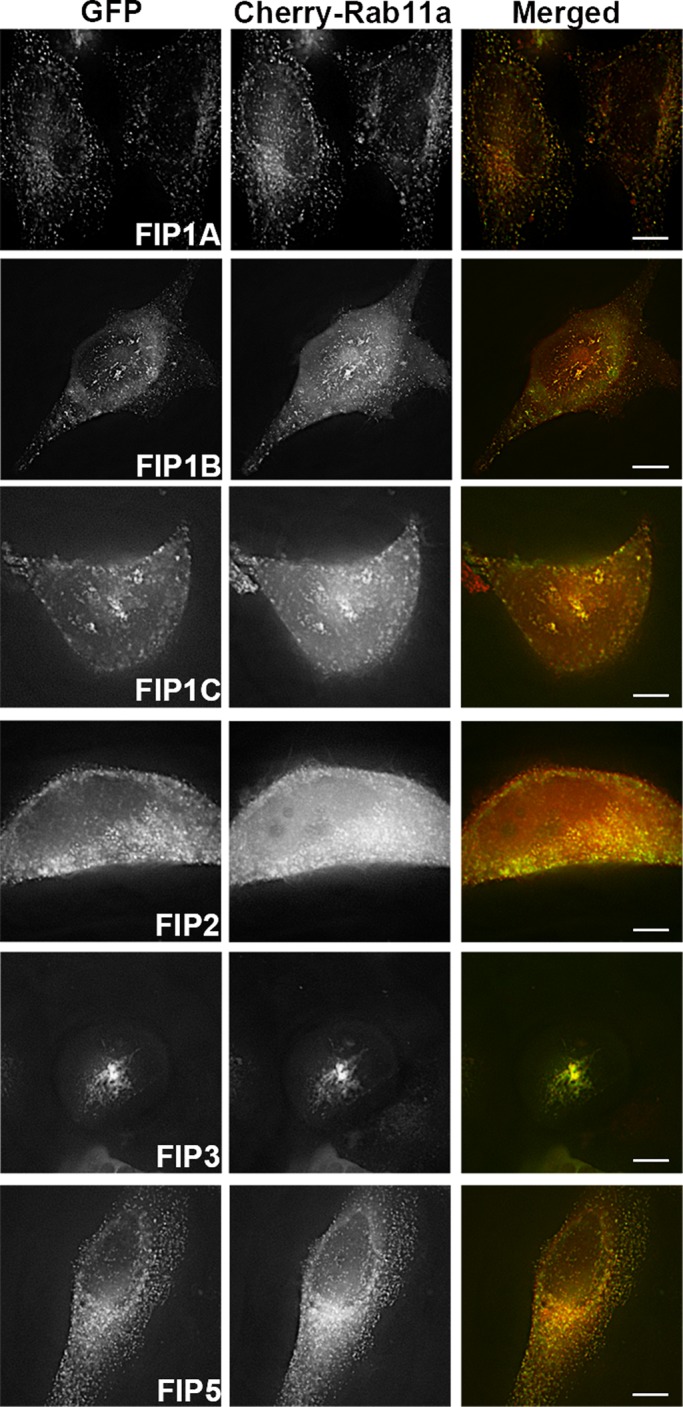
Rab11a in live HeLa cells is targeted differently in the presence of Rab11-FIPs. Single frames were collected from time-lapse imaging of Rab11-FIPs with Rab11a. FIP1A (Supplemental Video S2), FIP2, and FIP5 overlapped with Rab11a throughout the cell, whereas FIP1B and FIP1C (Supplemental Video S3), as well as FIP3, accumulate Rab11a in more centralized compartments. Data represent at least three independent experiments. Bars, 10 μm.
Rab11-FIPs participate in temporally distinct steps of the recycling process
To determine whether the Rab11-FIP1 proteins label temporally distinct compartments within the recycling pathway, we used fixed and live-cell approaches to evaluate the passage of internalized transferrin through EGFP–Rab11-FIP–labeled compartments. First, we treated HeLa cells expressing chimeric EGFP–Rab11-FIPs with fluorescently labeled transferrin–Alexa 568 after 1 h of serum starvation and examined the overlap of transferrin with EGFP–Rab11-FIPs after fixation at 5, 10, 20, and 30 min after loading. Representative images of HeLa cells transfected with EGFP–Rab11-FIPs and fixed at the indicated time points are shown in Figure 3A. Pearson's r for each Rab11-FIP protein with transferrin was determined for each condition, and the results shown represent the mean ± SEM for at least 25 cells per time point and 100 cells per condition (Figure 3B and Table 1). The mean r at each time point in each condition was taken as a percentage of the maximum mean coefficient found over the 30-min time course (Figure 3C and Table 2). We first noted that the correlation coefficients and percentages of maximal overlap for FIP1B (0.46 ± 0.02 of a 0.59 ± 0.03 max coefficient, or 78%) and FIP2 (0.37 ± 0.02 of a 0.45 ± 0.02 max coefficient, or 83%) at 5 min reached ∼80% of the total overlap observed over the 30-min time course. As a result, the overlap increased ∼20% over the remaining 25 min of the time course, suggesting that the capacity of FIP1B- and FIP2-containing compartments to accommodate transferrin was met largely in the first 5 min after transferrin uptake. We also noted that FIP1A (0.36 ± 0.03 of a 0.64 ± 0.04 max coefficient, or 56%), FIP1C (0.37 ± 0.03 of a 0.58 ± 0.04 max coefficient, or 63%), and FIP5 (0.38 ± 0.02 of a 0.58 ± 0.04 max coefficient, or 65%) demonstrated ∼60% of their respective maximal overlap at 5 min after uptake, whereas FIP3 (0.18 ± 0.02 of a 0.57 ± 0.03 max coefficient, or 31%) exhibited only ∼30% of maximal overlap at 5 min, indicating that these compartments are slower in reaching their capacity to accommodate transferrin in comparison with FIP1B and FIP2. In addition, the overlap between transferrin and FIP1A progressed earlier (0.36 ± 0.03 to 0.64 ± 0.04, or 56% between 5 and 20 min) than the overlap observed between transferrin and FIP1C (0.34 ± 0.03 to 0.58 ± 0.04, or 59% between 10 and 30 min). Of interest, FIP3 (0.18 ± 0.02 to 0.57 ± 0.03, or 31% over 30 min) and FIP5 (0.37 ± 0.02 to 0.58 ± 0.04, or 63% between 10 and 30 min) showed distinct patterns of overlap with transferrin over time. The data demonstrate observable differences in transferrin overlap in compartments containing Rab11-FIP1 isoforms and suggest that the relative involvement of each Rab11-FIP1 protein varies with time. Furthermore, the compartments containing the other Rab11-FIPs also have time-dependent loading, suggesting that Rab11-FIP proteins participate in dynamic steps in the recycling of transferrin. These data are comparable to the trend observed between Rab11a and transferrin in previous studies by others (Sonnichsen et al., 2000).
FIGURE 3:
EGFP–Rab11-FIPs in HeLa cells overlap with transferrin at temporally distinct points. HeLa cells transfected with EGFP–Rab11-FIPs were incubated with transferrin–Alexa 568 for 5, 10, 20, and 30 min and imaged to examine overlap. FIP1B demonstrated the most overlap at 5 min and reached ∼80% of overlap during this time. FIP2 also exhibited 80% of overlap in 5 min as well, whereas FIP1A, FIP1C, FIP3, and FIP5 displayed less overlap with transferrin at 5 min, and yet the progression of overlap toward the maximum values within this group was staggered between10 and 20 min. FIP1A progressed to maximum overlap at 20 min, whereas FIP1C, FIP3, and FIP5 showed a delayed rise to maximum overlap at 30 min. (A) Representative images for each time point (25 cells) in each condition (100 cells). (B, C) Average Pearson's r (also see Tables 1 and 2) at each time point and percentage maximum overlap calculated from the average values. Results are presented as mean ± SEM. Bars,10 μm.
TABLE 1:
Pearson's r for EGFP–Rab11-FIPs with transferrin–Alexa 568.
| Time (min) | Rab11 | FIP1A | FIP1B | FIP1C | FIP2 | FIP3 | FIP5 |
|---|---|---|---|---|---|---|---|
| 5 | 0.35 ± 0.02 | 0.36 ± 0.03 | 0.46 ± 0.02 | 0.37 ± 0.03 | 0.37 ± 0.02 | 0.18 ± 0.02 | 0.38 ± 0.02 |
| 10 | 0.55 ± 0.02 | 0.44 ± 0.04 | 0.48 ± 0.02 | 0.34 ± 0.03 | 0.41 ± 0.02 | 0.39 ± 0.03 | 0.37 ± 0.02 |
| 20 | 0.54 ± 0.02 | 0.64 ± 0.04 | 0.55 ± 0.03 | 0.55 ± 0.03 | 0.43 ± 0.02 | 0.52 ± 0.03 | 0.45 ± 0.06 |
| 30 | 0.62 ± 0.02 | 0.60 ± 0.02 | 0.59 ± 0.03 | 0.58 ± 0.04 | 0.45 ± 0.02 | 0.57 ± 0.03 | 0.58 ± 0.04 |
Average Pearson's r for overlap between Rab11-FIPs and transferrin in cells treated with transferrin for 5, 10, 20, and 30 min were obtained using Imaris software, and the values presented here are mean ± SEM for at least 25 cells per time point in each condition (100 cells total). The corresponding images in Figure 3A are representative cells at each time point, and Figure 3B shows a graph of the same values over 30 min.
TABLE 2:
The percentage of maximum r observed at each time for each condition.
| Time (min) | Rab11 | FIP1A | FIP1B | FIP1C | FIP2 | FIP3 | FIP5 |
|---|---|---|---|---|---|---|---|
| 5 | 57 | 56 | 78 | 63 | 83 | 31 | 65 |
| 10 | 89 | 69 | 82 | 59 | 92 | 68 | 63 |
| 20 | 88 | 100 | 94 | 95 | 96 | 92 | 78 |
| 30 | 100 | 93 | 100 | 100 | 100 | 100 | 100 |
The average r for each Rab11-FIP and transferrin at each time point was divided by the maximum r found over the 30-min time course and multiplied by 100. The corresponding graph in Figure 3C shows these percentage values over 30 min.
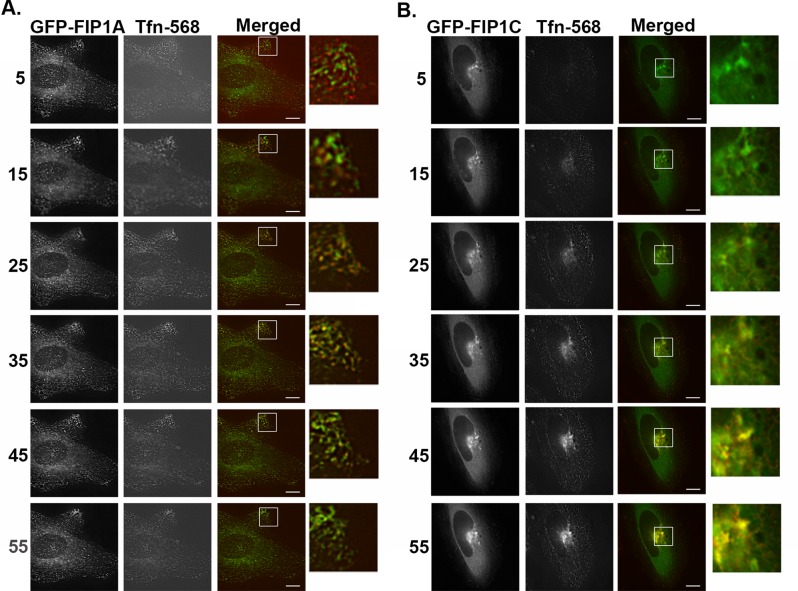
To support the data from our fixed-cell studies, we also imaged the passage of transferrin–Alexa 568 through live HeLa cells expressing EGFP–Rab11-FIP1A and EGFP–Rab11-FIP1C. In this experiment, after serum starvation, we loaded the transferrin and began imaging immediately. We then chased the labeled transferrin after 5 min with serum containing media for at least 1 h. We found a visible overlap between FIP1A and transferrin (Figure 4A and Supplemental Video S4) at 5 min that persisted until 45 min and then visibly declined. Overlap between FIP1C and transferrin was apparent at 15 min but persisted throughout the entire hour and still showed visible overlap at 55 min (Figure 4B and Supplemental Video S4). These data support the differences observed in the fixed-cell studies and confirm that Rab11-FIP1 isoforms fill different yet in some cases cooperative spatiotemporal roles in the recycling process.
FIGURE 4:
Transferrin traffics through FIP1A and FIP1C at temporally distinct time points. Live HeLa cells transfected with either EGFP-FIP1A or EGFP-FIP1C were incubated with transferrin–Alexa 568 for 5 min and then chased for 1 h. Evidence of overlap between transferrin and FIP1A (Supplemental Video S4) was observed as early as 5 min, and transferrin signal returns to baseline by 45 min (A). FIP1C (Supplemental Video S4) overlap with transferrin is evident at 15 min and persists until 55 min (B). Imaging of transferrin overlap with EGFP-FIPs was carried out on a DeltaVision deconvolution microscope. Time points (minutes) are indicated to the left of the images. Data represent at least three independent experiments. Bars,10 μm.
FIP1A associates with FIP1B and FIP1C through protein:protein interactions
We next examined whether protein:protein interactions could provide a means for Rab11-FIP1 proteins to organize vesicle recycling. Because of the similarities in FIP1B and FIP1C localization, we focused primarily on isolating FIP1A in the presence of FIP1B and FIP1C, as well as the remaining Rab11-FIPs studied here. HEK293 cells expressing EGFP–Rab11-FIPs and mCherry–Rab11-FIP1A were sheared open, and membranes were solubilized in 1% Triton X-100. The EGFP–Rab11-FIPs were isolated using an EGFP-binding protein (Rothbauer et al., 2008) bound to agarose beads. The amounts of FIP1A isolated in each condition were assayed by Western blot. We calculated the percentage of FIP1A recovered on the beads as a percentage of the total recovered on the beads and in the supernatant. Significant percentages of FIP1A were isolated with FIP1A (18.59 ± 4.45), FIP1B (13.40 ± 4.69), and FIP1C (11.55 ± 5.15) proteins in comparison to EGFP control (0.20 ± 0.10; p = 0.05) but not with FIP2 (0.22 ± 0.06), FIP3 (0.34 ± 0.26), and FIP5 (0.35 ± 0.05; Figure 5 and Table 3). The isolation of FIP1A suggested selective heteromeric associations between Rab11-FIP1 isoforms. This result prompted us to evaluate further the associations of the Rab11-FIP1 proteins in live HeLa cells by fluorescence imaging.
FIGURE 5:
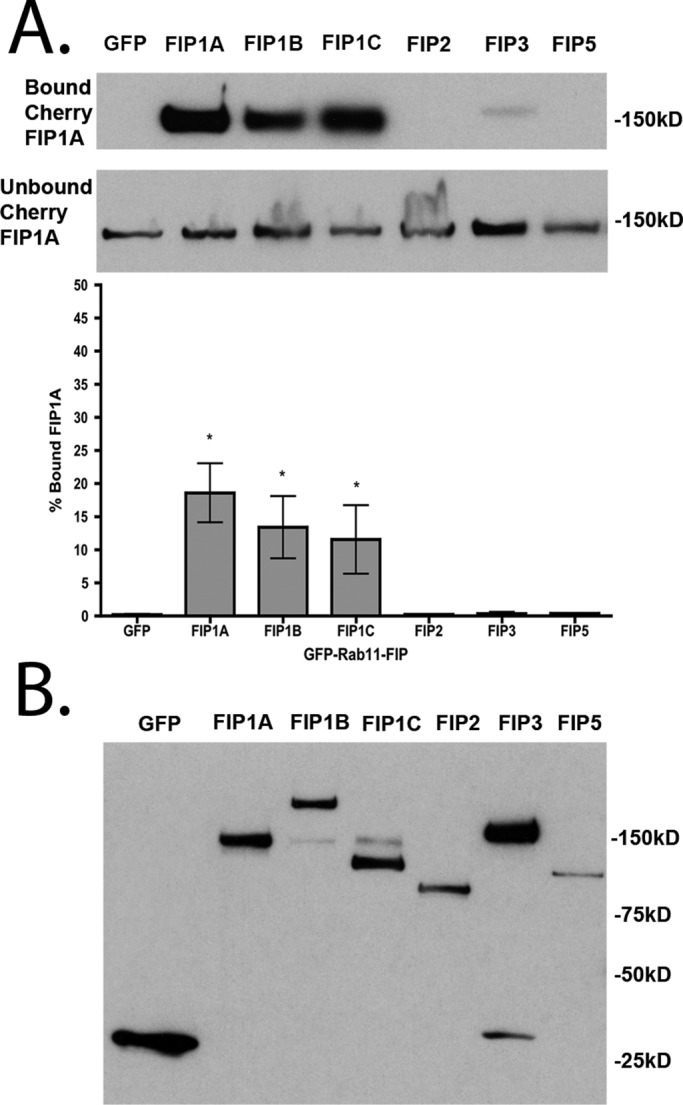
FIP1A is isolated specifically with FIP1A, FIP1B, and FIP1C in the presence of 1% Triton X-100. (A) Western blots of the bound and unbound FIP1A isolated with EGFP-FIPs from 1% Triton X-100–solubilized HEK293 cell preparations. Significant percentages of FIP1A were recovered with FIP1A (18.59 ± 4.45), FIP1B (13.40 ± 4.69), and FIP1C (11.55 ± 5.15) in comparison to the EGFP control (0.20 ± 0.10; p = 0.05). Bound material is presented as a percentage of total material recovered. (B) EGFP-FIPs isolated in each condition. Data represent at least three independent experiments. Results are presented as mean ± SEM.
TABLE 3:
Percentage values of total Cherry-FIP1A recovered with EGFP–Rab11-FIPs from HEK293 cell lysates in detergent-containing (1% Triton X100) and detergent-free conditions.
| GFP | FIP1A | FIP1B | FIP1C | FIP2 | FIP3 | FIP5 | |
|---|---|---|---|---|---|---|---|
| With Triton-X | |||||||
| Mean | 0.20 | 18.59 | 13.40 | 11.55 | 0.22 | 0.34 | 0.36 |
| SEM | 0.10 | 4.45 | 4.69 | 5.15 | 0.06 | 0.26 | 0.05 |
| Without Triton-X | |||||||
| Mean | 1.25 | 15.25 | 11.75 | 15.25 | 5.25 | 14.25 | 3.75 |
| SEM | 0.63 | 3.28 | 2.56 | 2.96 | 1.93 | 3.35 | 3.09 |
Significantly greater percentages of FIP1A were recovered with FIP1A, FIP1B, and FIP1C compared with the GFP control condition in both experiments, whereas a significantly greater percentage of FIP1A was additionally recovered with FIP3 in the detergent-free preparation. Statistical analysis was performed with a Mann–Whitney test, and significance was set at p ≤ 0.05. Figures 5 and 8 show representative Western blots and graphs of these values. Results are presented as mean ± SEM.
FIP1A overlaps with FIP1B and FIP1C along tubular endosomal compartments
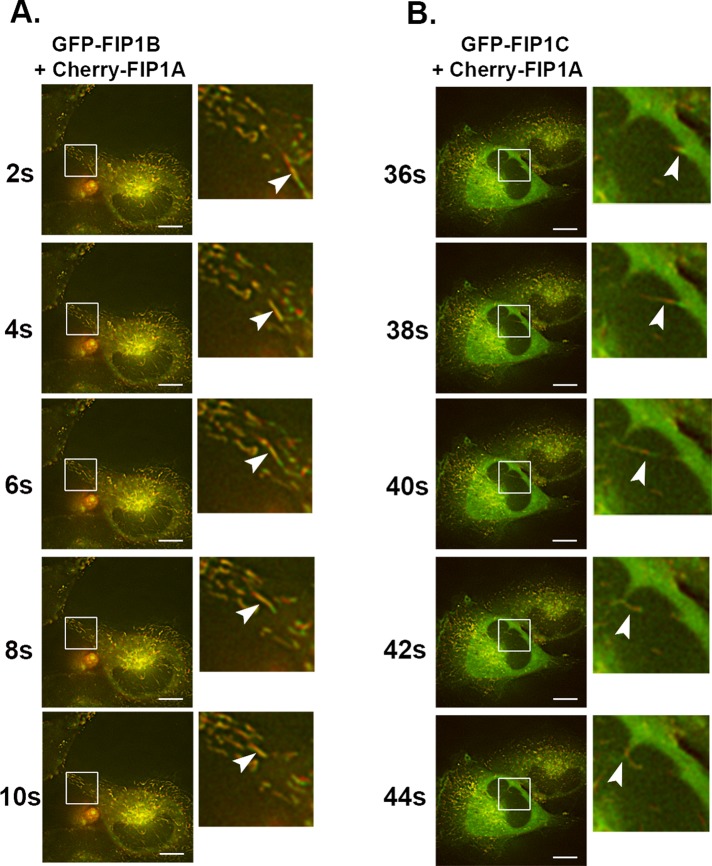
We used live-cell microscopy of HeLa cells expressing mCherry-FIP1A with either EGFP-FIP1B or EGFP-FIP1C proteins and assessed the localization, distribution, and movement of these proteins. We found that FIP1A overlapped with FIP1B and FIP1C along elaborate tubules not previously visible in single-expression studies for FIP1B and FIP1C. FIP1B and FIP1C occupied more-peripheral tubules in the presence of FIP1A (Figure 6). Furthermore, time-lapse images of these compartments highlighted the movement of FIP1A compartments in the same direction and path as FIP1B- and FIP1C-labeled compartments (Supplemental Videos S5 and S6). These data suggest that Rab11-FIP1 proteins operate along the same tubular structures, and based on the parallel movement and changes in direction of the proteins in live cells, we suspect that these proteins highlight neighboring compartments and potentially subdomains of the same compartment. Of note, we did observe a few instances in which FIP1B or FIP1C, in the presence of FIP1A, were localized to ruffles along the edge of the cell, and this pattern was reported before in the case of FIP1C in A2780 cells (Caswell et al., 2008). It is not clear whether associations between FIP1C and FIP1A could account for the phenotype in A2780 cells. We also examined overlap between coexpressed GFP-FIP1B and mCherry-FIP1C (Supplemental Figure S1 and Video S7) and found both proteins on compressed tubular compartments in the perinuclear region.
FIGURE 6:
FIP1A overlaps with FIP1B and FIP1C along tubular compartments. Time-lapse imaging of mCherry-FIP1A with EGFP-FIP1B (Supplemental Video S5) or EGFP-FIP1C (Supplemental Video S6) in live HeLa cells conducted over 2 min. (A) FIP1B on tubular compartments with FIP1A. The insets show an example tubule compartment labeled with both mCherry-FIP1A and EGFP-FIP1B moving from the perinuclear region to the periphery of the cell. (B) EGFP-FIP1C and mCherry-FIP1A along tubular compartments that move in the same direction and change directions together. Insets highlight the overlap along the compartment and the movement of an example tubule. Data represent at least three independent experiments. Bars, 10 μm.
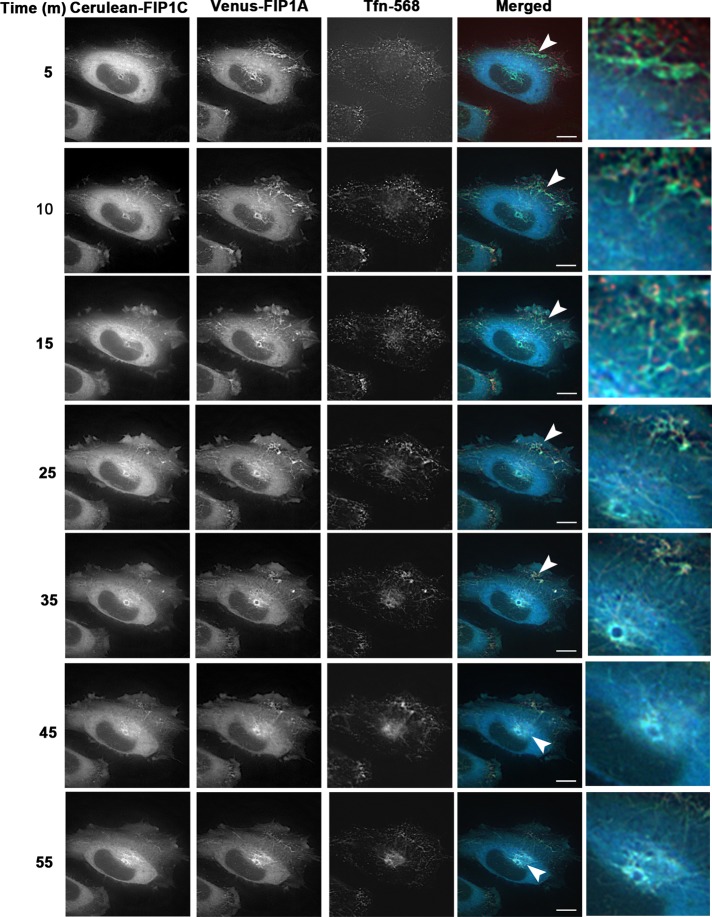
In single-transfection studies, transferrin in the presence of FIP1A was localized to the periphery of cells (Figure 4), whereas in the presence of FIP1C, transferrin was primarily localized in the perinuclear region. These findings suggested that the expression of Rab11-FIPs differentially affect the distribution of transferrin over time. Given that we observed an extensive tubulation of the recycling system out into the periphery of the cells upon coexpression of FIP1A and FIP1C, we next sought to evaluate whether this dual expression alters passage of transferrin through the recycling system. We coexpressed FIP1A and FIP1C and imaged transferrin movement over 1 h after internalization (Figure 7 and Supplemental Video S8). The live-cell images indicate that transferrin entered into the tubular system in the periphery initially. However, ultimately transferrin accumulated in tubules within the perinuclear region, similar to our observations in single-FIP1C-expression studies with transferrin. Overall the data point to a synergistic relationship between FIP1A and FIP1C in establishing a dynamic tubular recycling system, in which transferrin can enter through distal elements and then traffic progressively into more centrally located tubules.
FIGURE 7:
Cerulean-FIP1C and Venus-FIP1A coexpression induces tubules that traffic transferrin. HeLa cells expressing Cerulean-FIP1C and Venus-FIP1A and treated with transferrin-568 (Tfn-568) were imaged over 1 h every 5 min (Supplemental Video S8). Transferrin enters peripheral tubules initially and then is trafficked into perinuclear tubules. Transferrin is retained in the perinuclear region up until 55 min as observed in FIP1C single-expression studies. Insets show punctate transferrin signal along tubules containing both FIP1C and FIP1A. Data are representative of five cells imaged in two independent experiments. Bar, 10 μm.
In addition, we coexpressed EGFP-FIP1B, EGFP-FIP1C, and EGFP-FIP3 in the presence of mCherry-FIP2 (Supplemental Figure S2 and Supplemental Video S9) and found that FIP1B and FIP1C were not relocalized to tubular compartments as they were in the presence of FIP1A. Of interest, FIP1B, FIP1C, and FIP3 each accumulated FIP2 in the perinuclear region. The effects observed with FIP1A were not observed in coexpression studies with FIP2, suggesting that the effects of FIP1A are limited to protein associations with other FIP1 isoforms. The combination of the fluorescence imaging and the biochemical analysis between Rab11-FIP1 proteins suggests that the Rab11-FIP1 proteins coordinate along the same compartments at least in part through protein:protein interactions.
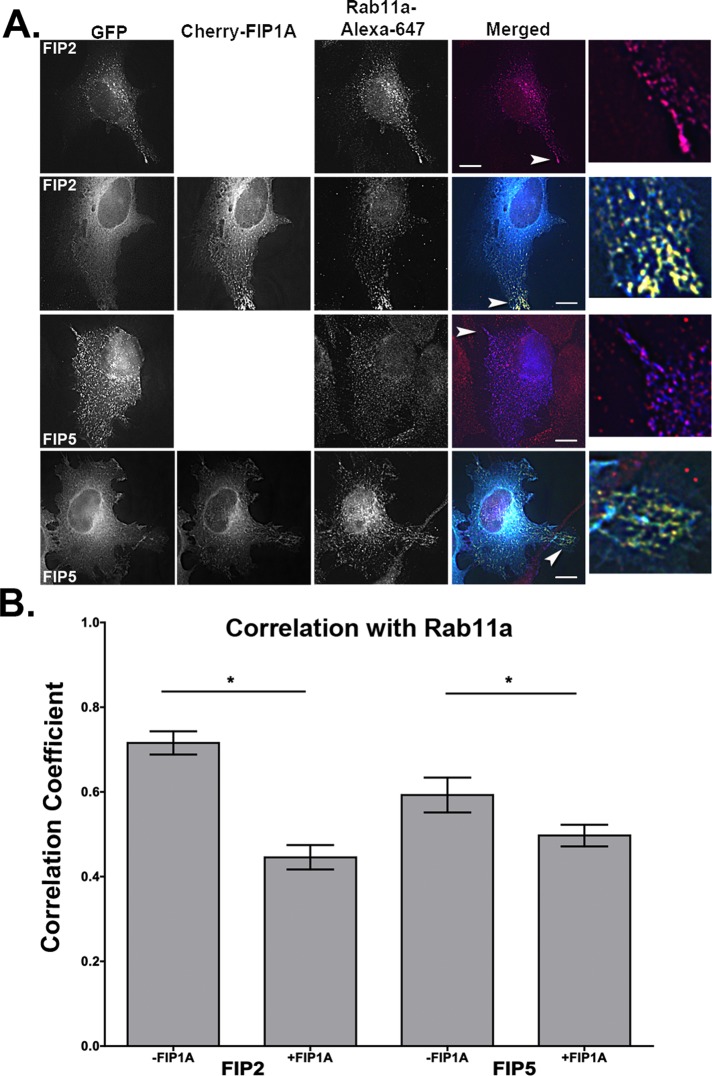
On the basis of our single-expression studies, in which FIP1A, FIP2, and FIP5 appeared to have a similar distribution and pattern of movement, we examined whether these proteins overlapped along these compartments when coexpressed in HeLa cells. Figure 8 shows that FIP1A in the periphery of HeLa cells was largely separate from FIP2 and FIP5, which had a cytosolic appearance (Figure 8 and Supplemental Video S10) in comparison to the phenotype observed in our single-expression studies. Conversely, we found that coexpressed FIP2 and FIP5 did overlap more extensively and appeared to occupy the same compartments (Figure 8 and Supplemental Video S11). We next coexpressed FIP1A with either FIP2 or FIP5 and stained for endogenous Rab11a (Figure 9). We analyzed Pearson's r to evaluate the overlap of Rab11a with FIP2 or FIP5 in the presence of FIP1A. Our data indicate that the average correlation between Rab11a with FIP2 (0.72 ± 0.03) was reduced in the presence of FIP1A (0.45 ± 0.03). Similarly, the correlation between FIP5 and Rab11a (0.59 ± 0.04) also decreased in the presence of FIP1A (0.50 ± 0.03; Figure 9). These data confirm our observations that protein:protein interactions between Rab11-FIP1 proteins and the remaining Rab11-FIPs studied here are limited, while also indicating a potential competitive relationship between FIP1A with both FIP2 and FIP5 for Rab11a.
FIGURE 8:
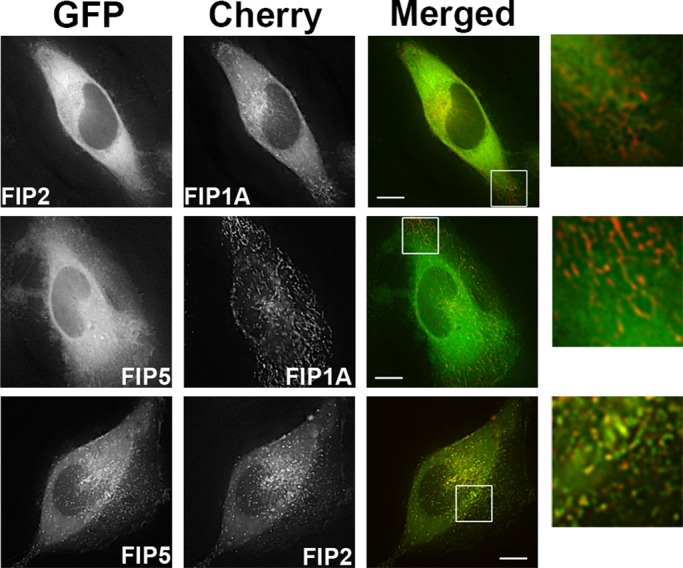
FIP1A resides on distinct compartments from FIP2 and FIP5. Time-lapse imaging of live HeLa cells transfected with EGFP-FIP2 or EGFP-FIP5 (Supplemental Video S10) and mCherry-FIP1A was conducted with deconvolution microscopy. FIP1A was found on compartments separate from FIP2 and FIP5 in peripheral regions of the cells. Time-lapse imaging of live HeLa cells transfected with EGFP-FIP5 and mCherry-FIP2 (Supplemental Video S11) was also conducted, and FIP5 and FIP2 overlapped both in the perinuclear and peripheral regions. Data are representative of at least three independent experiments. Bars, 10 μm.
FIGURE 9:
Correlation between Rab11a and FIP2 or FIP5 is decreased in the presence of FIP1A. HeLa cells transfected with GFP-FIP2 or GFP-FIP5 with or without mCherry-FIP1A were stained for Rab11a, and the correlation between Rab11a and FIP2 or FIP5 was measured for each condition using Pearson's r. The correlation between FIP2 and Rab11a (0.72 ± 0.03, 16 cells) was reduced in the presence of FIP1A (0.45 ± 0.03, 16 cells; n = 3, p < 0.0001). The correlation between FIP5 and Rab11a (0.59 ± 0.04, 14 cells) was reduced in the presence of FIP1A (0.50 ± 0.03, 16 cells; n = 3, p = 0.03). White arrowheads point to colocalization events in each condition. Bars, 10 μm.
FIP1A is additionally associated with FIP3-containing membranes
We next asked whether membrane-dependent associations would provide a platform for Rab11-FIP1A to associate with any other Rab11-FIPs. We repeated our biochemical analysis of HEK293 cells expressing EGFP-Rab11-FIPs and mCherry-FIP1A, and we isolated membranes using a detergent-free preparation, again isolating EGFP-Rab11-FIPs with GFP-binding protein (Rothbauer et al., 2008) bound to agarose beads. The amounts of FIP1A protein isolated in each condition were assessed by Western blot. We calculated the percentage of FIP1A recovered on the beads as a percentage of the total recovered on the beads and in the supernatant. The percentage of FIP1A isolated with FIP1A (15.25 ± 3.28), FIP1B (11.75 ± 2.56), FIP1C (15.25 ± 2.96), and FIP3 (14.25 ± 3.35) in membrane-intact conditions was significantly greater than that found in the EGFP control (1.25 ± 0.63; p = 0.01) condition (Figure 10 and Table 3). Again FIP2 (5.25 ± 1.93) and FIP5 (3.75 ± 3.09) did not show significant coisolation with FIP1A even in the presence of intact membranes. In conjunction with the results from detergent-solubilized preparations, these results suggest that FIP1A and FIP3 can occupy the same membrane compartments but without protein:protein interactions. In contrast, FIP1A is not present on membranes containing FIP2 or FIP5.
FIGURE 10:
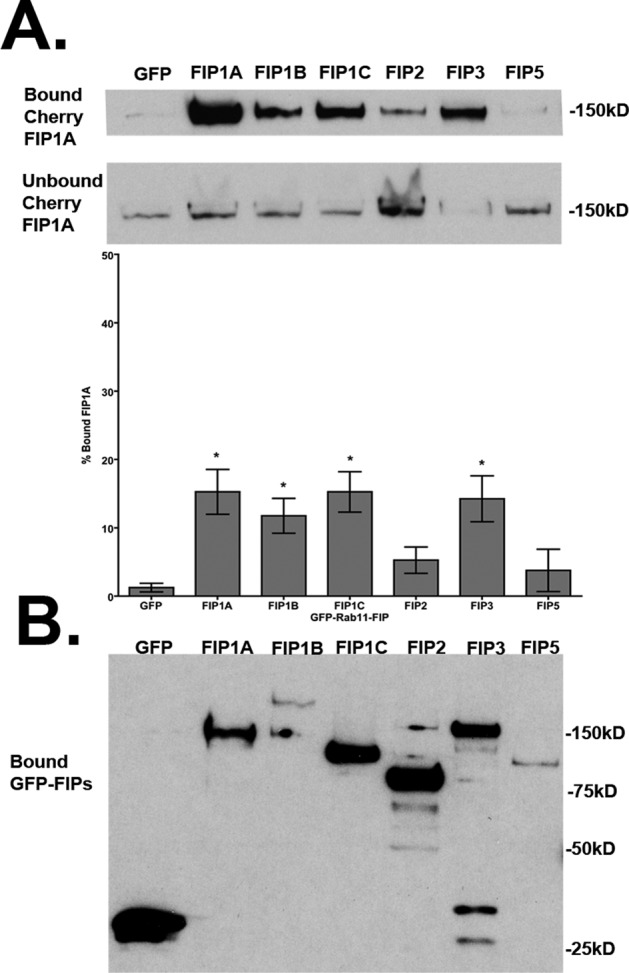
FIP1A preferentially occupies compartments similar to FIP1B, FIP1C, and FIP3. Western blots show mCherry-FIP1A recovered in bound and unbound fractions, and the amount of bound mCherry-FIP1A is shown as a percentage of the total mCherry-FIP1A recovered in each condition. The percentage of FIP1A isolated with FIP1A (15.25 ± 3.28), FIP1B (11.75 ± 2.56), FIP1C (15.25 ± 2.96), and FIP3 (14.25 ± 3.35) was significantly greater than in the EGFP control (1.25 ± 0.63; p = 0.05). (B) Isolated GFP-FIPs. Data represent at least three independent experiments. Results presented as mean ± SEM.
Rab11-FIP1 proteins, FIP1A and FIP1C, overlap with FIP3 along perinuclear tubular endosomes
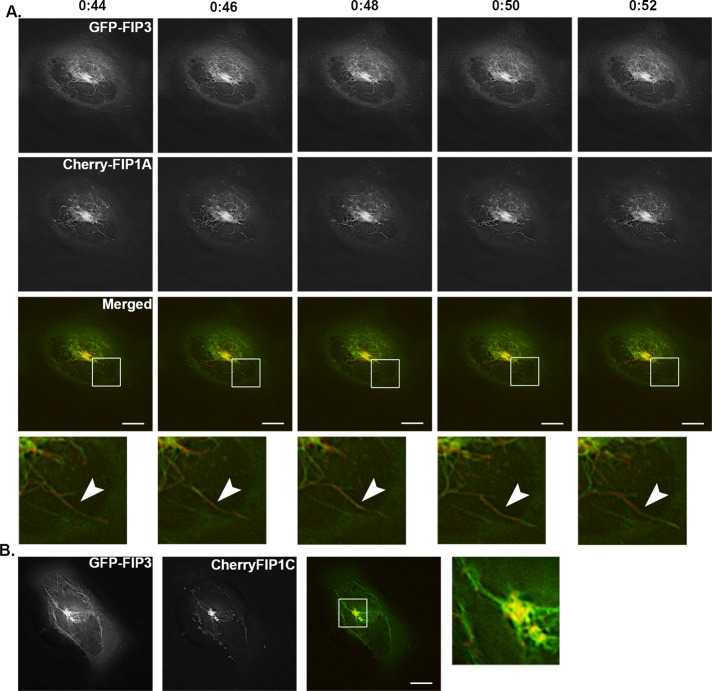
To evaluate the association of Rab11-FIP1 proteins with FIP3, we coexpressed EGFP-FIP3 with mCherry-FIP1A or mCherry-FIP1C in HeLa cells and imaged using fluorescence microscopy. Both FIP1A and FIP1C were localized to perinuclear tubular compartments overlapping with FIP3 (Supplemental Videos S12 and S13). The branching tubular morphology was similar to the morphology observed in the case of FIP3 alone (Figure 1). Figure 11A shows a time-lapse sequence of FIP1A and FIP3 expression along neighboring tubular compartments. Because we did not observe protein:protein interactions between FIP3 and FIP1A, the associations of FIP1 proteins with FIP3 more likely reflect localization in either neighboring compartments or the same compartments. The movement of FIP1A and FIP1C with FIP3 was observed in tubules restricted to the perinuclear region (Figure 11B), in contrast to the tubules extending throughout the cytoplasm observed with the coexpression of the Rab11-FIP1 proteins (Figure 6). To evaluate further the proximity of these proteins, we used structured illumination microscopy (SIM) in live HeLa cells (Shao et al., 2011) expressing EGFP-FIP3 and mCherry-FIP1A. FIP1A and FIP3 were observed on coincident tubular structures (Figure 12), and based on the resolution achieved with SIM (Gustafsson, 2000), our data now indicate the FIP3 and FIP1A proteins are within ∼200–300 nm of each other, further supporting the presence of these two proteins within the same tubular compartment. Close examination of these data suggests that FIP1A and FIP3 define patches of membrane within the same tubular structures. Previous reports suggested the presence of patches along membranes that promote formation of exclusive complexes (Meyers and Prekeris, 2002). Here we describe one such scenario in which Rab11-FIPs may define subdomains for the specific segregation of Rab11a within different regions of the same compartment. These data indicate that FIP1A and FIP3 likely occupy domains within the same dynamic membrane tubules.
FIGURE 11:
FIP1A and FIP1C overlap with FIP3 along perinuclear tubular compartments. HeLa cells expressing EGFP-FIP3 with mCherry-FIP1A (Supplemental Video S12) or mCherry-FIP1C (Supplemental Video S13) were imaged using time-lapse deconvolution microscopy. (A) Time lapse of tubules labeled with both FIP3 and FIP1A over 8 s. Time scale is above images. (B) Still image from a movie of overlap between FIP3 and FIP1C along tubules. Data represent at least three independent experiments. Bars, 10 μm.
FIGURE 12:
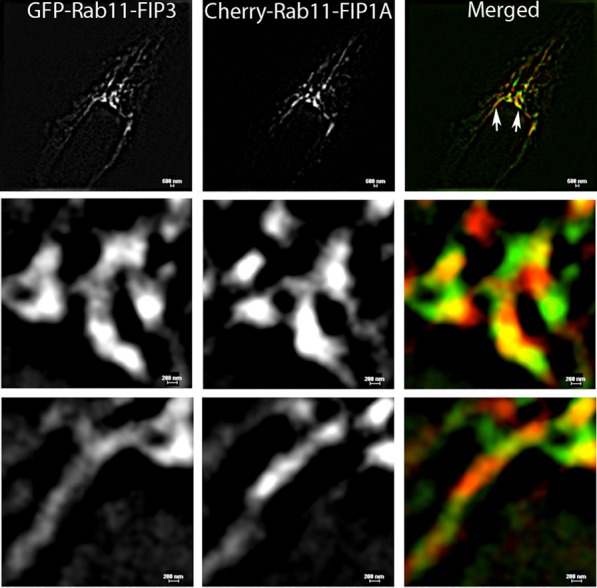
Rab11-FIP3 and Rab11-FIP1A label discrete domains along the same tubular compartments. HeLa cells transfected with EGFP-FIP3 and Cherry-FIP1A were imaged using SIM. The resolution of the SIM technology doubled the resolution of traditional deconvolution microscopy and demonstrated that the fluorescent proteins studied are within range of 200–300 nm of each other along tubular compartments. Evidence of distinct patches along the tubular compartment can be seen in the insets (bottom), where FIP3 and FIP1A overlap as well as occupy discrete domains. Bar, 500 μm (top), 200 μm (bottom).
DISCUSSION
This study is the first to demonstrate that Rab11-FIP1 proteins participate in spatially and temporally distinct steps of the recycling process coupled to differences in localization, influence over Rab11a, and selective associations between Rab11-FIP members. Single-expression studies of EGFP–Rab11-FIPs in live HeLa cells showed that the Rab11-FIP proteins studied here maintain either a wide distribution and mobile phenotype or a concentrated and relatively static phenotype. Alterations in Rab11a localization in the presence of FIP1B, FIP1C, and FIP3 indicated differential influence over a key component of the recycling process when compared with FIP1A, FIP2, and FIP5, whose overexpression did not elicit any noticeable influence on Rab11a localization or movement. Transferrin entry into compartments labeled by Rab11-FIPs underscored differences in the timing and relative involvement of these proteins during the recycling process. Finally, dual expression of Rab11-FIPs with each other highlighted points of overlap, separation, and changes in Rab11-FIP distribution, all suggesting potential points of cooperation and exchange within the dynamic recycling system.
The presence of multiple Rab11-FIPs in the same trafficking pathway provides for potential alternative routes into the same system for a variety of cargoes. In addition to the initial studies comparing Rab4, Rab5, and Rab11 (Schmid et al., 1988; Sonnichsen et al., 2000), evidence of different Rabs within the recycling system has been presented using live-cell imaging of Rab11a and Rab8a, both of which are dependent on myosin Vb (Roland et al., 2007). More recent investigations of clathrin-independent cargoes, including CD44 and CD55, showed evidence of multiple pathways into the central recycling endosome and emphasized the ability of various operators along these pathways to facilitate sorting of cargo between these routes (Eyster et al., 2009). This versatility is not exclusive to the recycling endosome, and examples exist in other trafficking processes, including endosomal-to-Golgi transport (Pfeffer, 2009), suggesting that the transport of multiple cargoes and use of a variety of Rab-GTPases or effector proteins is a common occurrence in trafficking pathways. Similar to the findings for various Rab-dependent trafficking pathways, the Rab11-FIPs are involved in the recycling of a variety of cargoes (Horgan and McCaffrey, 2009). Our data support the presence of multiple compartments in the periphery of the cell labeled with FIP1A or FIP2 and FIP5. The data might also suggest potential competitive roles for the same compartments based on the absence of FIP2 and FIP5 on the same compartments as FIP1A in the periphery of cells and a lack of protein:protein interactions. Indeed, coexpression studies of FIP2 and FIP5 with FIP1A reduced the correlation of FIP2 and FIP5 with Rab11a, further supporting competition between these Rab11-FIPs for Rab11a. The molecular basis for the competition for Rab11a by FIP proteins is not clear, but it seems likely that FIP1A might have a higher in situ affinity for Rab11a than FIP2 and FIP5. The data support the possibility of transition points between Rab11-FIPs during the recycling process. In addition, the relocalization of FIP1B and FIP1C into dispersed tubular elements in the presence of mCherry-Rab11a indicates that Rab11-FIPs may compete for limiting amounts of Rab11a within regions of the recycling system. These competing elements may in turn influence the recycling of transferrin and either hasten or delay the recycling process. Of note, coexpression of FIP1A and FIP1C induced a tubulation of recycling system membranes, but transferrin still entered initially into elements in the periphery of the cell and then trafficked subsequently into perinuclear tubules. Overall, Rab11-FIPs both cooperate in efficient vesicle recycling and compete for limited targets, such as Rab11a, that influence the trafficking of transferrin. This competition might account for the dynamic structure of the recycling system membranes.
The presence of a group of Rab11-FIPs all serving the same recycling system raises the question as to whether the Rab11-FIPs are acting in a variety of trafficking pathways. Accumulating evidence indicates that some of the Rab11-FIPs interact with multiple small GTPases. Initial studies suggested that the FIP1C protein, also known as RCP, was an interacting partner of Rab4, as well as of Rab11 (Lindsay et al., 2002). Further evidence now indicates that Rab14 might also be a potential partner for FIP1C (Kelly et al., 2010). In addition, FIP3, also known as arfophilin-1, is an effector for Arf6 and serves as a link between Rab11 and Arf-dependent trafficking systems (Shin et al., 2001; Hickson et al., 2003). Thus Rab11-FIPs ultimately might participate in multiple trafficking pathways and perhaps facilitate transitions among pathways. This idea has been well documented in the case of transitions between various Rab-containing compartments (Mizuno-Yamasaki et al., 2012). The ability to coordinate between trafficking pathways further expands the scope of Rab11-FIP function and again provides an additional explanation for our observations of reduced overlap among the more peripherally located Rab11-FIPs.
The complex tubular networks and changes in endosome morphology observed in the present study suggest internal mechanisms of regulation among the Rab11-FIP1 proteins that enable alterations in the compartments they occupy. Previous studies documented tubular networks containing EHD proteins (Naslavsky et al., 2006; Sharma et al., 2009), sorting nexins (van Weering et al., 2012), FBAR proteins (Wu et al., 2010), and Rabs and myosin motors (Roland et al., 2007; Jacobs et al., 2009). A recent investigation of FIP5 association with sorting nexin protein 18 along tubular compartments suggests that these proteins regulate the structure of tubular membrane compartments (Willenborg et al., 2011). The dynamic tubules that we observed in the present study might reflect the actions of additional effector proteins, especially the motors that complex with particular Rab11-FIPs. Investigations of FIP2 association with myosin Vb (Lapierre et al., 2001; Hales et al., 2002) and FIP5 with kinesin (Schonteich et al., 2008) exemplify the potential of the Rab11-FIPs to act as mediators of recycling system dynamics. Of interest, the associations of FIP2 and FIP5 with myosin Vb and kinesin II, respectively, also support the presence of these two Rab11-FIPs in the recycling of transferrin to the cell surface. In addition, previous studies link FIP3 with dynein-dependent transport, suggesting that FIP3 is involved with recycling steps directed toward the pericentriolar compartment (Horgan et al., 2010b). Of note, FIP2 has been shown to influence the localization of dynein (Ducharme et al., 2011). It is unclear what contribution Rab11-FIP1 proteins might make toward recruiting motor proteins, although FIP1C has been implicated in retrograde transport (Jing et al., 2010). Nevertheless, it is notable that we have observed that FIP1A could define mobile patches of membrane within FIP3-containing tubules. These results suggest that molecular motors might mediate both the extrusion of tubular elements and the mobility of defined domains within dynamic tubular membrane systems.
The goal of the present investigation was to determine whether Rab11-FIPs form an ordered assembly of compartments within the recycling system. Comparison of correlation coefficients for transferrin localization with Rab11-FIPs combined, with our findings that FIP1B and FIP1C reorganize Rab11a to larger static compartments, indicates that FIP1B and FIP1C are potentially involved in steps directed toward the pericentriolar compartment, whereas FIP1A might be involved in entry and/or exit from the pericentriolar recycling endosome. The known associations between FIP2 and myosin Vb and of FIP5 and kinesin II further support potential roles for peripherally localized Rab11-FIPs in both entry and exit steps of the Rab11a-dependent recycling pathway. On the basis of the selective overlap between Rab11-FIPs and the overlap of transferrin with each of the Rab11-FIP compartments, we conclude that transferrin enters multiple compartments simultaneously, and the magnitude of the correlations is an indication of the timing with which each of these compartments reaches its maximal capacity to accommodate transferrin. We thus propose a model (Figure 13) in which transferrin enters the Rab11a-dependent recycling system through a combination of FIP1A- and FIP2/FIP5-dependent pathways that proceeds rapidly into a pericentriolar FIP1B-containing compartment. After initial loading of the FIP1B-containing compartment, cargo would progress to adjacent tubular compartments containing FIP1C and FIP3 before recycling back to the cell surface. Our combined results indicate that Rab11-FIPs have the capacity to engage in discrete steps of the recycling process, adding to a growing body of data indicating that vesicle recycling is a continuously dynamic process that is facilitated by cooperative efforts among groups of regulatory proteins.
FIGURE 13:
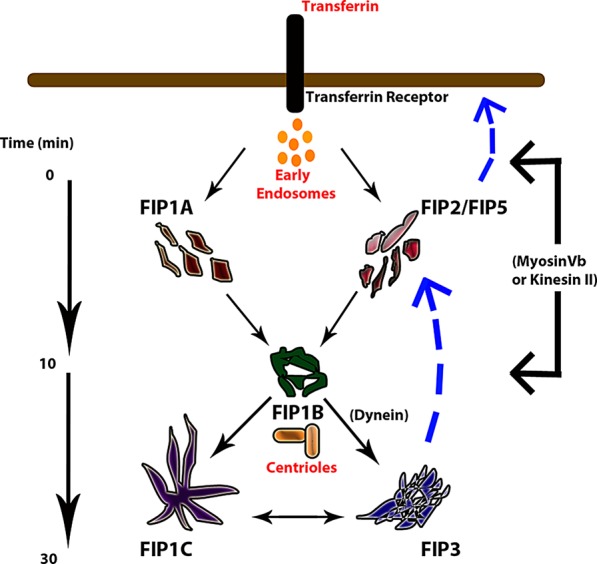
Proposed model of transferrin recycling through Rab11-FIP–containing compartments. The data suggest that transferrin enters multiple Rab11-FIP–containing compartments in the cell periphery and a central FIP1B-containing compartment in the pericentriolar region between 0 and 10 min. The overlap FIP1C and FIP3 compartments exhibited with transferrin increased between 20 and 30 min before the cargo is recycled back to the cell surface. Our investigations of transferrin overlap with Rab11-FIPs and the associations among Rab11-FIPs indicate that the Rab11-FIPs coordinate selective points of interaction and participate in time-dependent steps of the recycling pathway.
MATERIALS AND METHODS
Plasmids and expression vectors
The preparation of the following plasmids was previously described: EGFP-Rab11a (Lapierre et al., 2001), EGFP–Rab11-FIPs (FIP1A, FIP1B, FIP1C; Jin and Goldenring, 2006), and EGFP–Rab11-FIP2 and EGFP-FIP3 (Hales et al., 2001). EGFP-Rab11-FIP5 was a gift from R. Prekeris at the University of Colorado (Denver, CO). Preparation of mCherry-Rab11a was performed by cloning Rab11a from EGFP-Rab11a using EcoRI and Sal1 restriction sites. Preparation of Venus and mCherry-Rab11-FIP1A was performed by cloning FIP1A from EGFP–Rab11-FIP1A using Sal1 and BamH1 restriction sites. Preparation of the Cerulean and mCherry–Rab11-FIP1C was performed by cloning FIP1C from EGFP–Rab11-FIP1C between EcoRI and BamH1 restriction sites. Preparation of mCherry–Rab11-FIP2 was performed by cloning FIP2 from EGFP–Rab11-FIP2 using Sal1 and BamH1 restriction sites.
Single- and dual-expression live-cell imaging
HeLa cells were maintained in RPMI media (15-040-CV; Cellgro, Manassas, VA) with 10% fetal bovine serum (A15-704; PAA Laboratories, Pasching, Austria) in 35-mm glass-bottom dishes (P35G-0-14-C; MatTek Corporation, Ashland, MA) for at least 1 d. Cells were transfected using Effectene (301425; Qiagen, Valencia, CA) and 200 ng of DNA for each construct used. HeLa cells were transfected with EGFP–Rab11-FIP constructs alone or EGFP–Rab11-FIP constructs in combination with mCherry constructs containing Rab11, FIP1A, FIP1B, FIP1C, or FIP2 for at least 8 h. Cells were then imaged in a 37°C, 5% CO2 chamber using a 100× oil immersion objective (1.4 numerical aperture) on a DeltaVision deconvolution microscope (Applied Precision, Issaquah, WA) and a CoolSNAP HQ2 camera (Photometrics, Tucson, AZ). Images sizes were collected at 512 × 512 pixels, except in the case of FIP1B alone, FIP1B with Rab11a, and FIP3 with FIP1A and FIP1C, which were imaged at 1024 × 1024 pixels in order to reduce saturation from concentrated central signals and permit visualization of peripheral signals in these conditions. Images were collected a minimum of every 2 s for at least 2 min at exposures sufficient to achieve a minimum 5:1 signal-to-noise ratio such that discrete compartments were visible and expression was not ubiquitously cytosolic. Raw files were deconvolved using the Applied Precision softWoRx deconvolution package.
Transferrin trafficking
Transferrin in fixed HeLa cells.
HeLa cells transfected with EGFP–Rab11-FIPs were serum starved for at least 1 h at 37°C in serum-free RPMI (15-040-CV; Cellgro). Fluorescently labeled transferrin–Alexa 568 (T23365, 2.5 μg/ml final; Invitrogen, Carlsbad, CA) was added to cells in serum-free RPMI. For transferrin entry studies, cells were rinsed with phosphate-buffered saline (PBS) and fixed at 5, 10, 20, and 30 min with 4% paraformaldehyde for 20 min at room temperature and mounted using Prolong (P36391; Invitrogen). Exposure times were maintained at 500 ms, and sufficient signal was required to achieve a minimum 5:1 signal-to-noise ratio. Analysis of colocalization between transferrin and the individual EGFP-Rab11-FIPs was obtained using the Imaris colocalization tool using the whole field of view. Colocalization of each Rab11-FIP protein with transferrin was quantified for each condition, and the results represent the mean ± SEM of at least 25 cells per time point, for a total of 100 measurements per condition.
Transferrin passage in live cells.
For live-cell preparations, cells were mounted on the stage of a DeltaVision deconvolution microscope and identified for imaging. Cells were then treated with transferrin–Alexa 568 (2.5 μg/ml) in serum-free RPMI media (15-040-CV; Cellgro), and we immediately started imaging 1-h movies with 5-min intervals. After the 0- and 5-min time points, the medium was rinsed out with PBS, and fresh serum-containing medium was added before the start of the 10-min imaging time point. Exposure times for each EGFP–Rab11-FIP were optimized to achieve a minimum 5:1 signal-to-noise ratio and visualization of discrete compartments. Exposure times in the tetramethylrhodamine isothiocyanate (568 nm) channel were maintained at 500 ms.
Structured illumination microscopy
HeLa cells expressing EGFP–Rab11-FIP3 and mCherry–Rab11-FIP1A were prepared as described. Cells were cultured on 35-mm glass-bottom plates (P356-0-14-C; MatTek) and transfected with 200 ng of the respective DNA in RPMI (15-040-CV; Cellgro) media. Images were collected using 30-ms exposure times with a Nikon (Melville, NY) Ti-E microscope with Perfect Focus System N-SIM structured illumination microscopy, an APO 100× objective (1.49 numerical aperture), and an Andor (Belfast, United Kingdom) iXonDU-897 camera and processed using the Nikon Elements, version 3.22, software package.
Isolation and Western blotting of EGFP–Rab11-FIPs
HEK293 cells were cultured on 10-cm plates in RPMI media (15-040-CV; Cellgro) with 10% fetal bovine serum (A15-704; PAA Laboratories ) for 1 d and then cotransfected with EGFP empty vector or EGFP–Rab11-FIP chimeras (200 ng) and mCherry–Rab11-FIP1A (200 ng) for at least 16 h at 37°C and 5% CO2. Validation of transfection was determined with an EVOS microscope (Advanced Microscopy Group, Bothell, WA) at 20× magnification, and cells were scraped on ice into 1 ml of 30 mM Tris, 150 mM sodium acetate, and 20 mM magnesium acetate (buffer A). Cells were spun down at 1000 × g for 1 min at 4°C and resuspended in 500 μl of buffer A containing mammalian protease inhibitors. Cells were sheared open by passing cells 50 times through a 27-gauge needle, and debris was pelleted by centrifugation at 2000 × g for 3 min at 4°C. Lysates for detergent preparation had 1% Triton X-100 added at this time for 30 min at 4°C, and then insoluble material was centrifuged out of the mixture at 10,000 × g for 10 min at 4°C. The resulting supernatant was collected, and protein concentrations were measured using bicinchoninic acid assay (23223, 23224, 23209; Pierce, Indianapolis, IN). GFP-binding protein (Rothbauer et al., 2008) on agarose beads was equilibrated in buffer A, and a 10-μl aliquot of beads was prepared for each condition. Equal amounts of protein from each condition were added to beads and rotated at 4°C overnight. Supernatant was collected, and beads were washed at least seven times with buffer A. Beads were resuspended in 30 μl of Laemmli buffer (Laemmli, 1970). Supernatant was collected, and samples equivalent to 10% of total protein loaded on beads were collected, precipitated with an equal volume of acetone, and resuspended in 30 μl of Laemmli buffer. All samples were boiled and resolved on 10% SDS–PAGE gel, transferred to nitrocellulose membrane (10 401 196; Whatman, Piscataway, NJ) at 250 mA for 1 h at 4°C, blocked with 5% milk in 0.1% Tris-buffered saline (TBS)–Tween-20 for 1 h, and probed initially for mCherry using a rabbit anti-DsRed (632-496; Clontech, San Diego, CA) antibody at 1:2500 in 5% milk in 0.1% TBS–Tween-20 for 1 h and a goat anti-rabbit horseradish peroxidase (HRP)–conjugated secondary antibody (111-036-046; Jackson ImmunoResearch Laboratories, West Grove, PA) at a 1:5000 dilution in 5% milk in 0.1%TBS–Tween-20 for 1 h. Membranes were washed with 0.1% TBS–Tween-20 for 1 h after application of primary and secondary antibodies. Signals were detected using Pierce ECL Western Blotting Substrate (product 32106) and exposure to and development of Kodak film (864-6770; Kodak, Rochester, NY). Membranes were stripped using a 1× Reblot Plus Mild Solution reagent (Millipore, Billerica, MA 2502) and probed a second time using a primary rabbit anti-EGFP (ab290; Abcam, Cambridge, MA) antibody at 1:2500 in 5% milk in 0.1% TBS–Tween-20 and a Trueblot anti-rabbit IgG HRP (18-8816-33; eBioscience, San Diego, CA) rabbit secondary at 1:5000 in 5% milk in 0.1%TBS–Tween-20. Membranes were washed and developed as described. Band intensities were analyzed using the ImageJ densitometry plug-in (National Institutes of Health, Bethesda, MD). Intensity of bands in the bound lanes was expressed as a percentage of total intensity between bound and unbound lanes (mean ± SEM), and significance was determined by the Mann–Whitney test (p ≤ 0.05).
Immunofluorescence localization of Rab11a in Rab11-FIP coexpression studies
HeLa cells on coverslips transfected with EGFP FIP2 or EGFP-FIP5 and mCherry-FIP1A as described were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.3% Triton-X in 1× PBS for 30 min, blocked with 1% normal donkey serum (Jackson ImmunoResearch Laboratories) for 30 min, incubated with anti-Rab11a (1:100, VU57) antibodies (previously described; Ducharme et al., 2006) for 2 h at room temperature, washed 1 h with 1× PBS, incubated with Alexa 647 donkey anti-rabbit (1:200; Jackson ImmunoResearch Laboratories) for 1 h room temperature, and washed for 1 h with 1× PBS before being mounted with Prolong Gold (P36931; Invitrogen). Cells were imaged similar to the foregoing methods, using a DeltaVision deconvolution microscope, and colocalization of Rab11a with FIP2 and FIP5 was measured using SoftWoRx. Analysis was conducted on the entire field of view for each image, and correlation coefficients were used to calculate the mean ± SEM for each condition. Statistical analysis was performed using a Mann–Whitney test, and differences were significant at p ≤ 0.05.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant RO1 DK48370 to J.R.G., and N.W.B. was supported by Grant T32 CA106183. Live-cell imaging studies were performed in the Core Imaging Resource of the Vanderbilt Epithelial Biology Center. Confocal fluorescence imaging was performed through the use of the Vanderbilt University Medical Center Cell Imaging Shared Resource, supported by the Vanderbilt Digestive Disease Research Center, the Vanderbilt-Ingram Comprehensive Cancer Center, and National Institute of Health Grants CA68485, DK20593, DK58404, and HD15052. We thank Rytis Prekeris of the University of Colorado for providing the EGFP–Rab11-FIP5/Rip11 construct. We also thank Irina Kaverina of Vanderbilt University for use of the Imaris analysis software and Matt Tyska of Vanderbilt University for the use of the Nikon Elements software.
Abbreviations used:
- Rab11-FIP
Rab11-family interacting protein
- RCP
Rab-coupling protein
- SIM
structured illumination microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-09-0659) on January 2, 2013.
REFERENCES
- Anderson RG, Brown MS, Goldstein JL. Role of the coated endocytic vesicle in the uptake of receptor-bound low density lipoprotein in human fibroblasts. Cell. 1977;10:351–364. doi: 10.1016/0092-8674(77)90022-8. [DOI] [PubMed] [Google Scholar]
- Basu SK, Goldstein JL, Anderson RG, Brown MS. Monensin interrupts the recycling of low density lipoprotein receptors in human fibroblasts. Cell. 1981;24:493–502. doi: 10.1016/0092-8674(81)90340-8. [DOI] [PubMed] [Google Scholar]
- Bhartur SG, Calhoun BC, Woodrum J, Kurkjian J, Iyer S, Lai F, Goldenring JR. Genomic structure of murine Rab11 family members. Biochem Biophys Res Commun. 2000;269:611–617. doi: 10.1006/bbrc.2000.2334. [DOI] [PubMed] [Google Scholar]
- Calhoun BC, Goldenring JR. Rab proteins in gastric parietal cells: evidence for the membrane recycling hypothesis. Yale J Biol Med. 1996;69:1–8. [PMC free article] [PubMed] [Google Scholar]
- Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–155. doi: 10.1083/jcb.200804140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P, Vingron M, Sander C, Simons K, Zerial M. Molecular cloning of YPT1/SEC4-related cDNAs from an epithelial cell line. Mol Cell Biol. 1990;10:6578–6585. doi: 10.1128/mcb.10.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu BB, Ge L, Xie C, Zhao Y, Miao HH, Wang J, Li BL, Song BL. Requirement of myosinVb.Rab11a.Rab11-FIP2 complex in cholesterol-regulated translocation of NPC1L1 to the cell surface. J Biol Chem. 2009;284:22481–22490. doi: 10.1074/jbc.M109.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renzis S, Sonnichsen B, Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat Cell Biol. 2002;4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- Ducharme NA, Hales CM, Lapierre LA, Ham AJ, Oztan A, Apodaca G, Goldenring JR. MARK2/EMK1/Par-1Balpha phosphorylation of Rab11-family interacting protein 2 is necessary for the timely establishment of polarity in Madin-Darby canine kidney cells. Mol Biol Cell. 2006;17:3625–3637. doi: 10.1091/mbc.E05-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme NA, Ham AJ, Lapierre LA, Goldenring JR. Rab11-FIP2 influences multiple components of the endosomal system in polarized MDCK cells. Cellular Logist. 2011;1:57–68. doi: 10.4161/cl.1.2.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyster CA, Higginson JD, Huebner R, Porat-Shliom N, Weigert R, Wu WW, Shen RF, Donaldson JG. Discovery of new cargo proteins that enter cells through clathrin-independent endocytosis. Traffic. 2009;10:590–599. doi: 10.1111/j.1600-0854.2009.00894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan GH, Lapierre LA, Goldenring JR, Sai J, Richmond A. Rab11-family interacting protein 2 and myosin Vb are required for CXCR2 recycling and receptor-mediated chemotaxis. Mol Biol Cell. 2004;15:2456–2469. doi: 10.1091/mbc.E03-09-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 2005;24:3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte TM, Machen TE, Forte JG. Ultrastructural changes in oxyntic cells associated with secretory function: a membrane-recycling hypothesis. Gastroenterology. 1977;73:941–955. [PubMed] [Google Scholar]
- Gidon A, et al. A Rab11A/myosin Vb/Rab11-FIP2 complex frames two late recycling steps of langerin from the ERC to the plasma membrane. Traffic. 2012;13:815–833. doi: 10.1111/j.1600-0854.2012.01354.x. [DOI] [PubMed] [Google Scholar]
- Goldenring JR, Shen KR, Vaughan HD, Modlin IM. Identification of a small GTP-binding protein, Rab25, expressed in the gastrointestinal mucosa, kidney, and lung. J Biol Chem. 1993;268:18419–18422. [PubMed] [Google Scholar]
- Goldenring JR, Smith J, Vaughan HD, Cameron P, Hawkins W, Navarre J. Rab11 is an apically located small GTP-binding protein in epithelial tissues. Am J Physiol. 1996;270:G515–G525. doi: 10.1152/ajpgi.1996.270.3.G515. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Hales CM, Vaerman JP, Goldenring JR. Rab11 family interacting protein 2 associates with myosin Vb and regulates plasma membrane recycling. J Biol Chem. 2002;277:50415–50421. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- Heuser JE, Reese TS. Evidence for recycling of synaptic vesicle membrane during transmitter release at the frog neuromuscular junction. J Cell Biol. 1973;57:315–344. doi: 10.1083/jcb.57.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GR, Matheson J, Riggs B, Maier VH, Fielding AB, Prekeris R, Sullivan W, Barr FA, Gould GW. Arfophilins are dual Arf/Rab 11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Mol Biol Cell. 2003;14:2908–2920. doi: 10.1091/mbc.E03-03-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan CP, Hanscom SR, Jolly RS, Futter CE, McCaffrey MW. Rab11-FIP3 binds dynein light intermediate chain 2 and its overexpression fragments the Golgi complex. Biochem Biophys Res Commun. 2010a;394:387–392. doi: 10.1016/j.bbrc.2010.03.028. [DOI] [PubMed] [Google Scholar]
- Horgan CP, Hanscom SR, Jolly RS, Futter CE, McCaffrey MW. Rab11-FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J Cell Sci. 2010b;123:181–191. doi: 10.1242/jcs.052670. [DOI] [PubMed] [Google Scholar]
- Horgan CP, McCaffrey MW. The dynamic Rab11-FIPs. Biochem Soc Trans. 2009;37:1032–1036. doi: 10.1042/BST0371032. [DOI] [PubMed] [Google Scholar]
- Horgan CP, Oleksy A, Zhdanov AV, Lall PY, White IJ, Khan AR, Futter CE, McCaffrey JG, McCaffrey MW. Rab11-FIP3 is critical for the structural integrity of the endosomal recycling compartment. Traffic. 2007;8:414–430. doi: 10.1111/j.1600-0854.2007.00543.x. [DOI] [PubMed] [Google Scholar]
- Horgan CP, Walsh M, Zurawski TH, McCaffrey MW. Rab11-FIP3 localises to a Rab11-positive pericentrosomal compartment during interphase and to the cleavage furrow during cytokinesis. Biochem Biophys Res Commun. 2004;319:83–94. doi: 10.1016/j.bbrc.2004.04.157. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs DT, Weigert R, Grode KD, Donaldson JG, Cheney RE. Myosin Vc is a molecular motor that functions in secretory granule trafficking. Mol Biol Cell. 2009;20:4471–4488. doi: 10.1091/mbc.E08-08-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagoe WN, Lindsay AJ, Read RJ, McCoy AJ, McCaffrey MW, Khan AR. Crystal structure of rab11 in complex with rab11 family interacting protein 2. Structure. 2006;14:1273–1283. doi: 10.1016/j.str.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Jin M, Goldenring JR. The Rab11-FIP1/RCP gene codes for multiple protein transcripts related to the plasma membrane recycling system. Biochim Biophys Acta. 2006;1759:281–295. doi: 10.1016/j.bbaexp.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Jing J, Junutula JR, Wu C, Burden J, Matern H, Peden AA, Prekeris R. FIP1/RCP binding to golgin-97 regulates retrograde transport from recycling endosomes to the trans-Golgi network. Mol Biol Cell. 2010;21:3041–3053. doi: 10.1091/mbc.E10-04-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem. 2004;279:33430–33437. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- Kelly EE, Horgan CP, Adams C, Patzer TM, Ni Shuilleabhain DM, Norman JC, McCaffrey MW. Class I Rab11-family interacting proteins are binding targets for the Rab14 GTPase. Biol Cell. 2010;102:51–62. doi: 10.1042/BC20090068. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Yamashita T, Kawata M, Yamamoto K, Ikeda K, Tanimoto T, Takai Y. Purification and characterization of a novel GTP-binding protein with a molecular weight of 24,000 from bovine brain membranes. J Biol Chem. 1988;263:2897–2904. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapierre LA, Ducharme NA, Drake KR, Goldenring JR, Kenworthy AK. Coordinated regulation of caveolin-1 and Rab11a in apical recycling compartments of polarized epithelial cells. Exp Cell Res. 2012;318:103–113. doi: 10.1016/j.yexcr.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, Provance DW, Jr, Mercer JA, Bahler M, Goldenring JR. Myosin vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem. 2002;277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- Lindsay AJ, McCaffrey MW. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J Biol Chem. 2002;277:27193–27199. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- Lindsay AJ, McCaffrey MW. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J Cell Sci. 2004;117:4365–4375. doi: 10.1242/jcs.01280. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J. Dynamics of secretory membrane trafficking. Ann NY Acad Sci. 2004;1038:115–124. doi: 10.1196/annals.1315.019. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies HJ, Moore JL, Saunders C, Matthies DS, Lapierre LA, Goldenring JR, Blakely RD, Galli A. Rab11 supports amphetamine-stimulated norepinephrine transporter trafficking. J Neurosci. 2010;30:7863–7877. doi: 10.1523/JNEUROSCI.4574-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers JM, Prekeris R. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J Biol Chem. 2002;277:49003–49010. doi: 10.1074/jbc.M205728200. [DOI] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedvetsky PI, et al. A role of myosin Vb and Rab11-FIP2 in the aquaporin-2 shuttle. Traffic. 2007;8:110–123. doi: 10.1111/j.1600-0854.2006.00508.x. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Oehlke O, Martin HW, Osterberg N, Roussa E. Rab11b and its effector Rip11 regulate the acidosis-induced traffic of V-ATPase in salivary ducts. J Cell Physiol. 2011;226:638–651. doi: 10.1002/jcp.22388. [DOI] [PubMed] [Google Scholar]
- Peden AA, Schonteich E, Chun J, Junutula JR, Scheller RH, Prekeris R. The RCP-Rab11 complex regulates endocytic protein sorting. Mol Biol Cell. 2004;15:3530–3541. doi: 10.1091/mbc.E03-12-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Multiple routes of protein transport from endosomes to the trans Golgi network. FEBS Lett. 2009;583:3811–3816. doi: 10.1016/j.febslet.2009.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR, Novick PJ. Membrane traffic. Curr Opin Cell Biol. 2010;22:419–421. doi: 10.1016/j.ceb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Klumperman J, Scheller RH. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol Cell. 2000;6:1437–1448. doi: 10.1016/s1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–2837. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Schmid SL, Fuchs R, Male P, Mellman I. Two distinct subpopulations of endosomes involved in membrane recycling and transport to lysosomes. Cell. 1988;52:73–83. doi: 10.1016/0092-8674(88)90532-6. [DOI] [PubMed] [Google Scholar]
- Schonteich E, Wilson GM, Burden J, Hopkins CR, Anderson K, Goldenring JR, Prekeris R. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J Cell Sci. 2008;121:3824–3833. doi: 10.1242/jcs.032441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N. Ypt/rab gtpases: regulators of protein trafficking. Sci STKE. 2001;2001:re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- Shao L, Kner P, Rego EH, Gustafsson MG. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff DR, Daro EA, Hull M, Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin OH, Couvillon AD, Exton JH. Arfophilin is a common target of both class II and class III ADP-ribosylation factors. Biochemistry. 2001;40:10846–10852. doi: 10.1021/bi0107391. [DOI] [PubMed] [Google Scholar]
- Simon GC, Prekeris R. The role of FIP3-dependent endosome transport during cytokinesis. Commun Integr Biol. 2008;1:132–133. doi: 10.4161/cib.1.2.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Brodie SE, Cohn ZA. Membrane flow during pinocytosis. A stereologic analysis. J Cell Biol. 1976;68:665–687. doi: 10.1083/jcb.68.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Touchot N, Chardin P, Tavitian A. Four additional members of the ras gene superfamily isolated by an oligonucleotide strategy: molecular cloning of YPT-related cDNAs from a rat brain library. Proc Natl Acad Sci USA. 1987;84:8210–8214. doi: 10.1073/pnas.84.23.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trischler M, Stoorvogel W, Ullrich O. Biochemical analysis of distinct Rab5- and Rab11-positive endosomes along the transferrin pathway. J Cell Sci. 1999;112:4773–4783. doi: 10.1242/jcs.112.24.4773. [DOI] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering JR, Verkade P, Cullen PJ. SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic. 2012;13:94–107. doi: 10.1111/j.1600-0854.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. The novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem Biophys Res Commun. 2002a;292:909–915. doi: 10.1006/bbrc.2002.6736. [DOI] [PubMed] [Google Scholar]
- Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem Biophys Res Commun. 2002b;299:770–779. doi: 10.1016/s0006-291x(02)02720-1. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ES, Martinez C, Vaccaro C, Zhou J, Tang Q, Ober RJ. From sorting endosomes to exocytosis: association of Rab4 and Rab11 GTPases with the Fc receptor, FcRn, during recycling. Mol Biol Cell. 2005;16:2028–2038. doi: 10.1091/mbc.E04-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh GI, Leney SE, Lloyd-Lewis B, Wherlock M, Lindsay AJ, McCaffrey MW, Tavare JM. Rip11 is a Rab11- and AS160-RabGAP-binding protein required for insulin-stimulated glucose uptake in adipocytes. J Cell Sci. 2007;120:4197–4208. doi: 10.1242/jcs.007310. [DOI] [PubMed] [Google Scholar]
- Willenborg C, Jing J, Wu C, Matern H, Schaack J, Burden J, Prekeris R. Interaction between FIP5 and SNX18 regulates epithelial lumen formation. J Cell Biol. 2011;195:71–86. doi: 10.1083/jcb.201011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, Frey AM, Peden AA, Gould GW, Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Huang B, Graham M, Raimondi A, Heuser JE, Zhuang X, De Camilli P. Coupling between clathrin-dependent endocytic budding and F-BAR-dependent tubulation in a cell-free system. Nat Cell Biol. 2010;12:902–908. doi: 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.