Mitochondrial fission requires recruitment of the GTPase Drp1 to mitochondria, but the molecules that mediate this recruitment have been disputed. Fis1, Mff, MiD49, and MiD51 can all recruit Drp1 to mitochondria and promote fission. MiD49 and MiD51 can promote mitochondrial fission, but their activity depends on cellular context.
Abstract
Several mitochondrial outer membrane proteins—mitochondrial fission protein 1 (Fis1), mitochondrial fission factor (Mff), mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51, respectively)—have been proposed to promote mitochondrial fission by recruiting the GTPase dynamin-related protein 1 (Drp1), but fundamental issues remain concerning their function. A recent study supported such a role for Mff but not for Fis1. In addition, it is unclear whether MiD49 and MiD51 activate or inhibit fission, because their overexpression causes extensive mitochondrial elongation. It is also unknown whether these proteins can act in the absence of one another to mediate fission. Using Fis1-null, Mff-null, and Fis1/Mff-null cells, we show that both Fis1 and Mff have roles in mitochondrial fission. Moreover, immunofluorescence analysis of Drp1 suggests that Fis1 and Mff are important for the number and size of Drp1 puncta on mitochondria. Finally, we find that either MiD49 or MiD51 can mediate Drp1 recruitment and mitochondrial fission in the absence of Fis1 and Mff. These results demonstrate that multiple receptors can recruit Drp1 to mediate mitochondrial fission.
INTRODUCTION
The opposing processes of fusion and fission (division) regulate mitochondrial morphology and function (Westermann, 2010; Chan, 2012; Youle and van der Bliek, 2012). During mitochondrial fission, the dynamin-related GTPase dynamin-related protein 1 (Drp1) is recruited from the cytosol onto the mitochondrial outer membrane, where it assembles into puncta. These puncta consist of oligomeric Drp1 complexes that wrap around and constrict the mitochondrial tubule to mediate fission (Mears et al., 2011). This proposed mechanism is analogous to how dynamin pinches off endocytic vesicles at the plasma membrane (Schmid and Frolov, 2011; Ferguson and De Camilli, 2012). Because much of Drp1 resides in the cytosol, key mechanistic issues are how Drp1 is recruited to the mitochondrial surface and what factors influence its assembly.
In mammals, four integral membrane proteins of the mitochondrial outer membrane—mitochondrial fission protein 1 (Fis1), mitochondrial fission factor (Mff), and mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51, respectively)—have been proposed to act as receptors that recruit Drp1 to the mitochondrial surface. However, there is much uncertainty concerning the precise role of these candidates and their relationship to each other. Fis1 was the first proposed receptor, based on definitive genetic and biochemical data from the budding yeast Saccharomyces cerevisiae. The yeast Drp1 orthologue, Dnm1, requires Fis1 to localize to mitochondria (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000). Fis1 physically interacts with Dnm1 via one of two molecular adaptors, Mdv1 or Caf4 (Tieu et al., 2002; Cerveny and Jensen, 2003; Griffin et al., 2005).
In mammals, however, there are no apparent orthologues of Mdv1 or Caf4, and the evidence supporting a role for Fis1 in mitochondrial fission is mixed. Supporting a role in fission, overexpression of Fis1 in mammalian cells promotes fragmentation of mitochondria (Yoon et al., 2003; Stojanovski et al., 2004; Yu et al., 2005), and inhibition of Fis1 results in elongation (Yoon et al., 2003; Lee et al., 2004; Stojanovski et al., 2004; Koch et al., 2005; Gandre-Babbe and van der Bliek, 2008). However, a recent study showed that deletion of the Fis1 gene from human colorectal carcinoma HCT116 cells does not disrupt mitochondrial morphology or Drp1 recruitment to mitochondria and suggested that mitochondrial defects from Fis1-knockdown studies may be due to off-target effects (Otera et al., 2010). Mff is the strongest candidate for a Drp1 receptor. Knockdown of Mff results in mitochondrial elongation (Gandre-Babbe and van der Bliek, 2008) and reduces the amount of Drp1 recruited to mitochondria (Otera et al., 2010).
MiD49 and MiD51 (also called mitochondrial elongation factor 1) have been proposed to be components of the mitochondrial fission machinery (Palmer et al., 2011), but there is conflicting evidence concerning their mechanism of action. One study showed that a double knockdown results in mitochondrial elongation and reduces recruitment of Drp1 to mitochondria, supporting a role in fission (Palmer et al., 2011). Paradoxically, overexpression of either MiD49 or MiD51 also causes mitochondrial elongation (Palmer et al., 2011; Zhao et al., 2011), and another study showed that knockdown of MiD51 causes mitochondrial fragmentation (Zhao et al., 2011). These latter observations led to the alternative proposal that MiD51 inhibits mitochondrial fission (Zhao et al., 2011).
To clarify the role of these candidate Drp1 receptors, we studied their function in mouse embryonic fibroblasts (MEFs). Our results support their role in mitochondrial fission and indicate that each protein is capable of recruiting Drp1 and promoting mitochondrial fission.
RESULTS
Fis1 and Mff can function independently of one another to regulate mitochondrial fission in MEFs
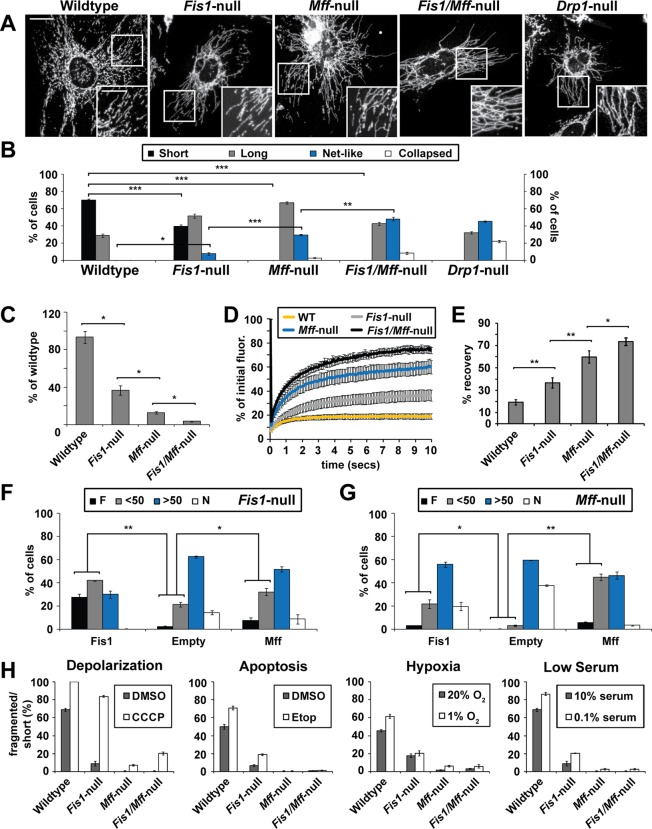
Given the disputed role of Fis1 in mitochondrial fission (Otera et al., 2010), we sought to directly compare the roles of Fis1 and Mff by generating MEF cell lines with null alleles of Fis1, Mff, or both. These cells are completely deficient for the relevant proteins (Supplemental Figure S1A). Mitochondrial morphology was assessed by immunofluorescence against the mitochondrial marker Tom20 (Figure 1A). Consistent with previous studies using Mff knockdown (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010), mitochondria in Mff-null cell lines are severely elongated and interconnected. In contrast, mitochondria in Fis1-null cell lines have more moderate elongation and interconnection (Figure 1A). We used a morphology scoring assay in which each cell was categorized as having short tubules, long tubules, net-like mitochondria, or collapsed mitochondria. For both mutants, more cells were found to have long or net-like mitochondria relative to wild-type cells (Figure 1B), but the phenotype of Mff-null cells is much more pronounced than that of Fis1-null cells. Two other independent pairs of wild-type and Fis1-null cell lines gave similar results (Supplemental Figure S1B). Of interest, mitochondrial elongation is most severe in the Fis1/Mff double mutant and approaches that of Drp1-null cells, indicating that the phenotype of Mff loss is substantially enhanced by additional removal of Fis1.
FIGURE 1:
Mitochondrial elongation in Fis1-null, Mff-null, and Fis1/Mff-null MEFs. (A) Mitochondrial morphology in wild-type and mutant cells. Mitochondria were visualized by immunofluorescence against Tom20. Scale bar, 10 μm. Inset scale bar, 5 μm. (B) Scoring of mitochondrial network morphologies for the indicated cell lines. Each cell was scored into one of four morphological categories. ***p < 0.001, **p < 0.005, *p < 0.02. (C) Morphometric analysis. Well-resolved mitochondria (Tom20) in the cell periphery were used for morphometric analysis. Total mitochondrial number and area were measured; the ratio of these two values was plotted as percentage of wild-type. Twenty ROIs from two independent experiments were analyzed. Error bars, SEM. *p < 0.0001. (D) FRAP analysis. For each of the indicated cell lines, the graph shows the fluorescence recovery over 10 s after photobleaching. Fluorescence data were collected every 200 ms postbleach. (E) Endpoint analysis of FRAP data. The fluorescence recovery ± SEM at 10 s postbleaching is shown. For D and E, 20 FRAP trials were averaged. **p < 0.001, *p < 0.01. (F, G) Fis1-null (F) and Mff-null (G) cells were transiently transfected with expression constructs for Fis1, Mff, or an empty control vector. Cells were scored into one of four morphological categories. F, fragmented; <50, <50% of mitochondria are long tubules; >50, > 50% of mitochondria are long tubules; N, net-like. **p < 0.005, *p < 0.05. Statistical testing was performed by combining the F and <50 classes, which represent cells with mostly short mitochondria. (H) Cells from the indicated cell lines were treated with 50 μM CCCP for 1 h, 100 μM etoposide for 5 h, 1% O2 for 24 h, or 0.1% serum for 3 d. Histograms show the percentage of cells with fragmented or mostly short tubular mitochondria. For B, F, G, and H, data are the averages ± SEM from three independent experiments, with 100 cells scored per experiment. All statistical testing was performed with the Student's t test.
These observations were confirmed with several independent methods. First, we used morphometric image analysis to evaluate mitochondrial length/interconnectivity. We quantified the number of discrete mitochondria and the total mitochondrial area in the cell periphery, where individual mitochondria can be readily resolved. The ratio of these two values provided a quantitative measure of mitochondrial length/interconnectivity. Significant differences were found among the wild-type cells and each of the mutants (Figure 1C). The phenotypic trend found with this approach agreed well with the manual morphological scoring.
We also used fluorescence recovery after photobleaching (FRAP) to further quantify these differences in mitochondrial morphology. A region of interest (ROI) containing multiple mitochondria with matrix-targeted Cox8-Dendra2 was photobleached, and the recovery of fluorescence over 10 s was monitored. Because of the short recovery time, the fluorescence recovery is dependent on the length and interconnectivity of the photobleached mitochondria, not on mitochondrial fusion. Each of the mutant cell lines demonstrates higher levels of recovery than wild-type cells, with Fis1/Mff-null cells having the highest level (Figure 1, D and E, and Supplemental Figure S1C). The recovery properties of Fis1-null and Mff-null cells were intermediate, with Mff-null cells showing higher levels of recovery than Fis1-null cells. To corroborate the FRAP results, we used an independent approach to measure the diffusion of Cox8-Dendra2. After photoconverting Cox8-Dendra2 with a 405-nm laser in a region of interest, we quantified the area to which the photoconverted signal immediately spread. We found that Fis1/Mff-null cells have the largest diffusion value, followed by Mff-null, then Fis1-null, and lastly wild-type cells (Supplemental Figure S1D). Taken together, these results demonstrate that both Fis1 and Mff regulate mitochondrial length and interconnectivity, with Mff playing the predominant role in MEFs.
Given that both Fis1 and Mff function to regulate mitochondrial morphology, we tested whether Fis1 and Mff could act independently of one another. When Fis1-null and Mff-null cells are transfected with Fis1 and Mff expression constructs, respectively, both show a robust rescue in mitochondrial morphology as expected (Figure 1, F and G, and Supplemental Figure S2). Of interest, when Fis1 is expressed in Mff-null cells or Mff is expressed in Fis1-null cells, there is also a substantial, albeit less complete, rescue in mitochondrial morphology. These results demonstrate that Fis1 and Mff can act independently and partially replace each other to regulate mitochondrial morphology.
Mitochondrial fission is induced by a number of physiological situations (Westermann, 2010; Chan, 2012). To determine whether Fis1 or Mff might selectively mediate mitochondrial fission in one of these pathways, we challenged wild-type and mutant cells with carbonyl cyanide m-chlorophenylhydrozone (CCCP; for mitochondrial membrane depolarization), staurosporine (STS) or etoposide (for apoptosis), 1% O2 (for hypoxia), or 0.1% serum (Figure 1H and Supplemental Figure S1E). After each challenge, Fis1-null cells show mitochondrial shortening, but less than in wild-type cells. In contrast, Mff-null and Fis1/Mff-null cell lines are highly resistant to mitochondrial shortening after each of these treatments. We conclude that both Fis1 and Mff are important for mitochondrial fission under basal and induced conditions, with Mff playing a larger role in both cases.
Fis1 and Mff are important for recruitment of Drp1 to mitochondria
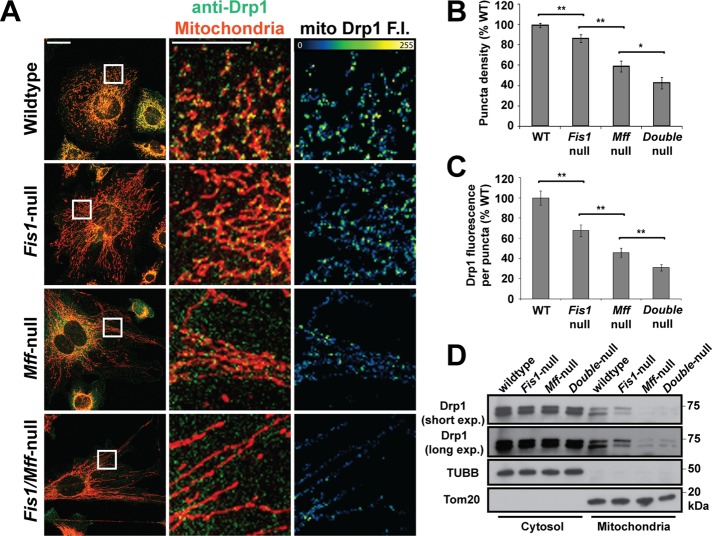
To determine whether the mutant cells are defective in recruiting Drp1, we used immunofluorescence to visualize Drp1 puncta on mitochondria in vivo. In wild-type cells, much of the Drp1 staining is diffuse in the cytosol, but a proportion can be found in punctate structures on mitochondria. This recruitment to mitochondria is reduced in cells with Mff knockdown (Otera et al., 2010). Our knockout cells have reduced numbers of mitochondrial Drp1 puncta, with Fis1/Mff-null cells having the greatest defect, followed by Mff-null cells and then Fis1-null cells (Figure 2A). We used image analysis to quantify the density (number of puncta/mitochondrial area) of Drp1 puncta on mitochondria (Figure 2B). Fis1-null cells have a small but significant decrease in the density of mitochondrial Drp1 puncta. Mff-null and Fis1/Mff-null cells have more substantial decreases. Of interest, the average total fluorescence per puncta (Figure 2, A and C), as well as the average size of puncta (Supplemental Figure S1F), also show substantial declines in the mutant cells. As expected, exogenous expression of Mff in Mff-null cells rescued mitochondrial morphology and restored mitochondrial Drp1 puncta (Supplemental Figure S2, B and D). This rescue depended on the R1 region (containing an 11-residue repeat) and the transmembrane domain, as previously shown (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010). The levels of Drp1 in purified mitochondrial fractions from Mff-null and Fis1/Mff-null cells were substantially lower than those from wild-type and Fis1-null cells (Figure 2D). The levels from Fis1-null mitochondria were only slightly less than wild-type levels. These imaging and biochemical data indicate that Fis1 and Mff facilitate Drp1 recruitment onto mitochondria, with Mff playing the predominant role.
FIGURE 2:
Drp1 recruitment and assembly on mitochondria are affected in Fis1-null and Mff-null cells. (A) Drp1 puncta in the indicated cell lines. To improve visualization of mitochondrial Drp1, the cells were briefly treated with 0.001% digitonin before fixation to reduce the level of cytosolic Drp1. Mitochondria were highlighted by immunofluorescence against Tom20. Scale bar (left), 10 μm. Center and right, magnified images of the boxed regions. Scale bar, 5 μm. Right panel, a mask corresponding to the mitochondrial channel was applied to the Drp1 channel to obtain only mitochondrial Drp1 fluorescence. The heat map reflects Drp1 fluorescence intensity (FI). (B) Density of mitochondrial Drp1 puncta. (C) Drp1 fluorescence per puncta. In B and C, the data are normalized to the wild-type control. Error bars, SEM. Twenty-five ROIs were analyzed from 10 to 12 cells for each group. *p < 0.05, **p < 0.005. Statistical testing was performed with the Student's t test. (D) Drp1 recruitment to mitochondria. Cytosol and purified mitochondrial fractions were prepared from the indicated MEF cells and analyzed by Western blotting for Drp1, Tom20 (mitochondrial marker), and β-tubulin (TUBB; cytosolic marker). The mitochondrial lanes were loaded with 30-fold more cell equivalents than with the cytosolic lanes. Both short and long exposures for the Drp1 blot are presented.
The MiDs can mediate mitochondrial fission in the absence of Fis1 and Mff
MiD49 and MiD51 play important roles in controlling mitochondrial morphology, but it is unclear whether they positively (Palmer et al., 2011) or negatively regulate mitochondrial fission (Zhao et al., 2011). One study found that simultaneous knockdown of both MiD49 and MiD51 caused mitochondrial elongation (Palmer et al., 2011), whereas another study found that knockdown of MiD51 caused mitochondrial fragmentation (Zhao et al., 2011). To examine this issue, we used short interfering RNA (siRNA) to knock down MiD49 and MiD51. Knockdown of either gene causes a similar enhancement of mitochondrial length and interconnectivity (Figure 3, A and B, and Supplemental Figure S3), suggesting that these proteins positively regulate mitochondrial fission. Simultaneous knockdown of MiD49 and MiD51 does not cause a more severe phenotype than knockdown of either gene alone. It was previously reported that knockdown of both genes is necessary to cause mitochondrial elongation (Palmer et al., 2011). Our observation that a single knockdown is sufficient may be due to a higher efficiency of knockdown. For both MiD49 and MiD51, we identified multiple siRNAs that caused a similar elongation of mitochondria (Supplemental Figure S3 B).
FIGURE 3:
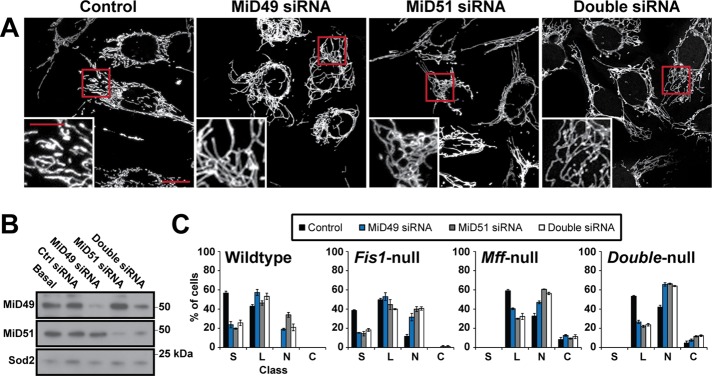
Knockdown of MiD49 or MiD51 causes mitochondrial elongation and enhances the Fis1-null, Mff-null, and Fis1/Mff-null phenotypes. (A) Mitochondrial morphology in wild-type MEFs treated with control siRNA, siRNA against MiD49, siRNA against MiD51, or both. Scale bar, 10 μm. Insets are magnified images of the boxed regions. Scale bar, 5 μm. Mitochondria were highlighted by expression of Cox8-DsRed. (B) Western blot of cell lysates containing single and double knockdown of the MiDs. SOD2 is a loading control. (C) Scoring of mitochondrial network morphologies for knockdown experiments in wild-type, Fis1-null, Mff-null, and Fis1/Mff-null cells. Data were obtained from three independent experiments, with 100 cells scored per experiment. Error bars, SEM. C, collapsed; L, long; N, net-like; S, short.
The mitochondrial phenotype of Fis1/Mff-null cells is not as severe as that of Drp1-null cells (Figure 1, A and B), suggesting that residual mitochondrial fission exists in these cells. We found that knockdown of either MiD49 or MiD51 enhances the mitochondrial connectivity of Fis1/Mff-null cells, as well as in Fis1-null and Mff-null cells (Figure 3C and Supplemental Figure S3B). Simultaneous knockdown of MiD49 and MiD51 again does not cause a more severe phenotype than knockdown of either gene alone.
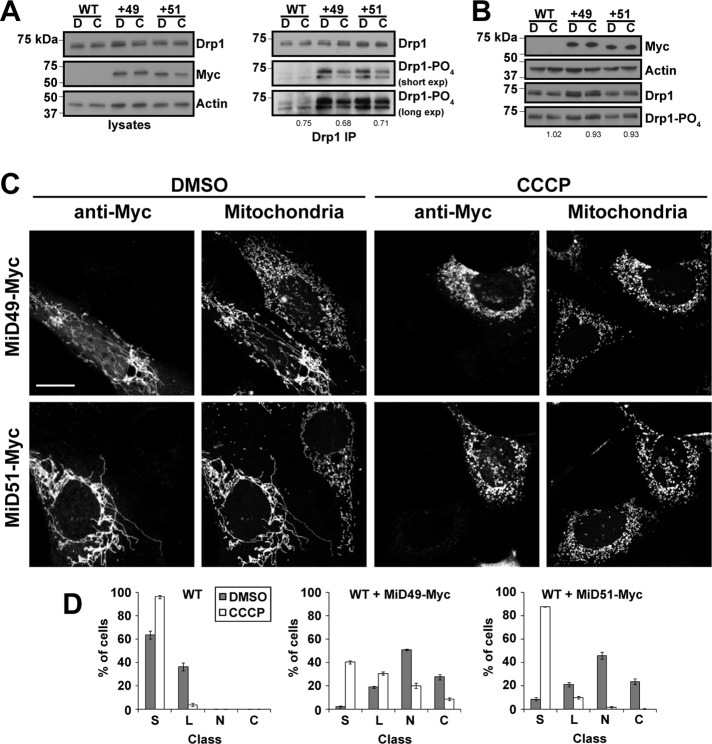
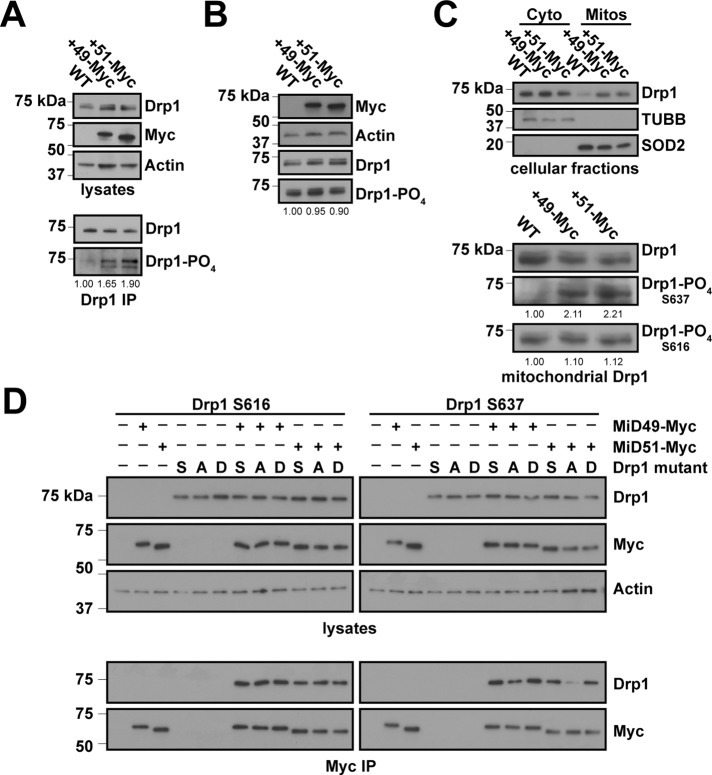
If MiD49 and MiD51 are involved in mitochondrial fission, it is puzzling that overexpression of either protein leads to extreme mitochondrial elongation (Palmer et al., 2011; Zhao et al., 2011). We also found similar elongation of mitochondria when MiD49 or MiD51 is overexpressed (see Figure 5C later in the paper). This effect is associated with enhanced Drp1 levels on mitochondria (Figure 4C and Supplemental Figure S4C). We wondered whether these proteins might be recruiting an inactive form of Drp1. Phosphorylation of Drp1 at S637 has been shown to negatively regulate Drp1 function (Chang and Blackstone, 2007; Cribbs and Strack, 2007; Cereghetti et al., 2008). Drp1 is also phosphorylated at S616, and this modification has been shown to positively regulate Drp1 function during mitosis (Taguchi et al., 2007; Kashatus et al., 2011). Using phosphospecific Drp1 antibodies that detect phosphorylation on S616 or S637, we found that levels of Drp1 S637-PO4, but not S616-PO4, are significantly enhanced in cell lines constitutively overexpressing either MiD49 or MiD51 (Figure 4, A and B, and Supplemental Figure S4A). We also found enhanced levels of Drp1 S637-PO4, but not S616-PO4, in the mitochondrial fractions of these cells (Figure 4C, bottom). These results suggest that the MiDs may preferentially bind to the S637-PO4 form of Drp1. To test this idea, we cotransfected 293T cells with Myc-tagged forms of MiD49 or MiD51 in combination with wild-type Drp1, an S → A (phospho-null) mutant or an S → D (phosphomimetic) mutant of Drp1 at S616 or S637. Cells were treated with a reversible cross-linker to capture the MiD–Drp1 interaction, and cell lysates were subjected to anti-myc immunoprecipitation. Wild-type Drp1 and Drp1S637D were coimmunoprecipitated more efficiently than Drp1S637A (Figure 4D). This preference is specific for residue S637 and is not found for S616.
FIGURE 5:
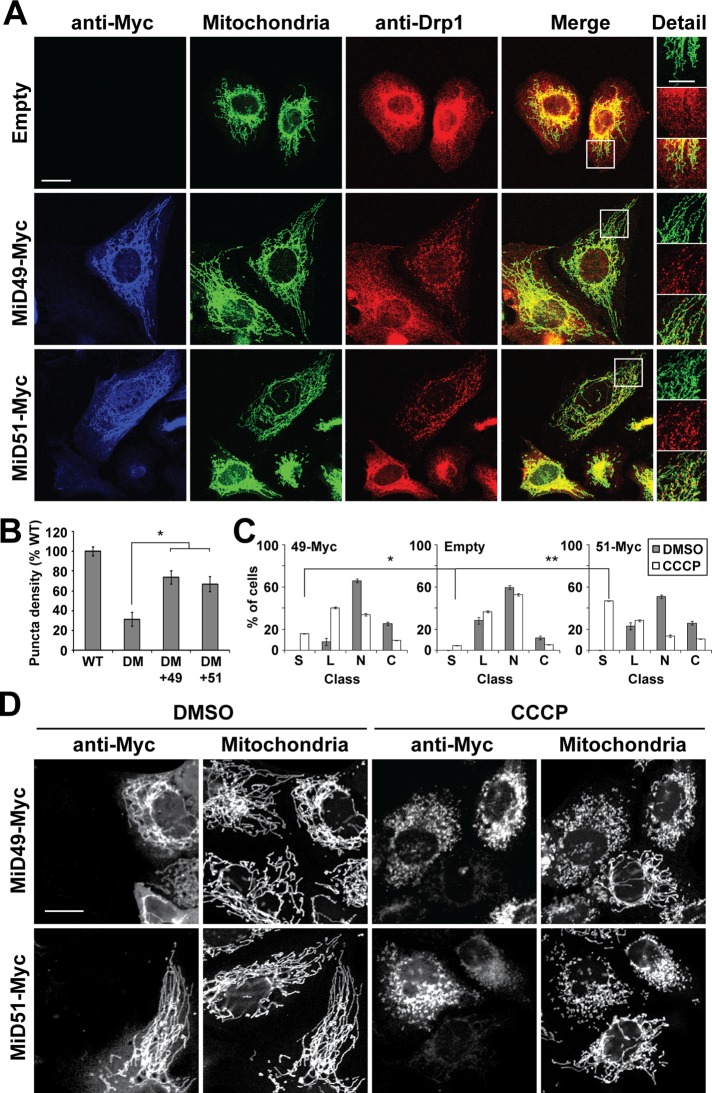
Mitochondrial elongation and increased Drp1 S637-PO4 caused by MiD overexpression can be reversed by CCCP. (A) Reduction of Drp1 S637-PO4 levels in CCCP treated cells. Left, lysates from control HeLa cells or HeLa cells expressing MiD49-Myc or MiD51-Myc were analyzed by Western blotting for Drp1. Cells were treated with dimethyl sulfoxide (DMSO; indicated by D) or CCCP (indicated by C). Right, Drp1 S637-PO4 was analyzed as in Figure 4A. Both short and long exposures for the Drp1 S637-PO4 blot are presented. (B) Drp1 S616-PO4 levels do not change in CCCP-treated cells. Lysates from control HeLa cells or HeLa cells expressing MiD49-Myc or MiD51-Myc were analyzed as in Figure 4B. Cells were treated with DMSO (indicated by D) or CCCP (indicated by C). Densitometry was performed on the S616-PO4 and Drp1 S637-PO4 blots and was normalized to the total Drp1 in each lane. Values are presented as ratios of the CCCP/DMSO values for each group. (C) CCCP reverses mitochondrial elongation in MiD-overexpressing MEFs. Cells were transfected with MiD49-Myc or MiD51-Myc and subsequently treated with vehicle (DMSO) or CCCP. MiD-expressing cells were identified by Myc immunofluorescence, and mitochondria were visualized with Tom20 immunofluorescence. In the DMSO-treated samples, note that MiD-Myc expression causes mitochondrial elongation. In the CCCP-treated samples, note that MiD-Myc–expressing cells have fragmented mitochondria. Scale bar, 10 μm. (D) Scoring of mitochondrial network morphologies for MEFs transfected with MiD49-Myc, MiD51-Myc, or empty vector and treated with DMSO or CCCP. The data are from three independent experiments, each with 100 cells scored. Error bars, SEM. C, collapsed; L, long; N, net-like; S, short.
FIGURE 4:
Overexpression of MiD49 or MiD51 causes enhanced phosphorylation of Drp1 on S637. (A) Increased Drp1 S637-PO4 in cells overexpressing MiD49 or MiD51. Top, lysates from control HeLa cells or HeLa cells expressing MiD49-Myc or MiD51-Myc were analyzed by Western blotting for Drp1 and the Myc-tagged MiD. Bottom, Drp1 was immunoprecipitated, and the levels of Drp1 S637-PO4 were detected with a phosphospecific Drp1 antibody. Drp1 was used as a loading control for the immunoprecipitated samples. (B) Drp1 S616-PO4 levels in cells overexpressing MiD49 or MiD51. Lysates from control HeLa cells or HeLa cells expressing MiD49-Myc or MiD51-Myc were analyzed by Western blotting for Drp1, Drp1 S616-PO4, and the Myc-tagged MiD. Actin was used as a loading control. (C) Increased Drp1 S637-PO4 recruitment to mitochondria in cells overexpressing MiD49 or MiD51. Top, cytosol and crude mitochondrial fractions were prepared from the indicated HeLa cells and analyzed by Western blotting for Drp1, superoxide dismutase 2 (SOD2; mitochondrial), and β-tubulin (TUBB; cytosolic). The mitochondrial lanes were loaded with 20-fold more cell equivalents than with the cytosolic lanes. Bottom, the loading of mitochondrial fractions was normalized for the total Drp1 level. The relative levels of Drp1 S616-PO4 and Drp1 S637-PO4 on mitochondria were assessed by Western blotting with phosphospecific antibodies. In A–C, densitometry was performed on the S616-PO4 and Drp1 S637-PO4 blots and was normalized to the total Drp1 in each sample. Values are presented as proportions of wild type. (D) Binding of MiD49 and MiD51 to phospho-mutants of Drp1. 293T cells were cotransfected with Myc-tagged MiD and Drp1 mutants as indicated. Cells were treated with cross-linker and solubilized. Top, expression of Drp1 and Myc-MiD in the lysates. Bottom panel, anti-Myc immunoprecipitates were analyzed for Drp1. Note that this assay only detects transfected Drp1 (compare lanes 2, 3–7, and 10). A, phospho-null mutant; D, phosphomimetic mutant; S, wild-type Drp1.
Dephosphorylation at S637 is activated by CCCP and STS (Cribbs and Strack, 2007; Cereghetti et al., 2008). Treatment of MiD-overexpressing cells with CCCP caused a reduction in Drp1 S637-PO4 levels (Figure 5A) and further enhancement of Drp1 recruitment to mitochondria (Supplemental Figure S4C). In contrast, phosphorylation at S616 was not modulated during CCCP treatment in wild-type cells or cells constitutively overexpressing either MiD49 or MiD51 (Figure 5B). Of interest, mitochondria in cells overexpressing MiD49 or MiD51 showed rapid and robust shortening upon treatment with CCCP or STS (Figure 5, C and D, and Supplemental Figure S4B). Thus the mitochondrial elongation caused by MiD overexpression is associated with enhanced Drp1 S637-PO4 levels and can be reversed by treatments that cause dephosphorylation of Drp1 at this site.
With a method to activate the pro-fission mode of the MiDs, we investigated whether they are capable of mediating mitochondrial fission in the absence of Fis1 and Mff. Expression of either MiD protein partially rescues recruitment of Drp1 to the mitochondria of Fis1/Mff-null cells (Figure 6, A and B). In spite of this Drp1 recruitment, the mitochondria of Fis1/Mff-null cells expressing MiDs remain extremely elongated (Figure 6, A, C, and D). As noted earlier, Fis1/Mff-null cells are highly resistant to CCCP-induced mitochondrial fragmentation (Figure 1H). However, when these cells overexpress MiD49 or MiD51 and are treated with CCCP, their mitochondria undergo rapid shortening, in contrast to nontransfected cells or cells transfected with an empty vector (Figure 6, C and D). Taken together, our experiments demonstrate that, under certain cellular contexts, the MiDs can function in the absence of Fis1 and Mff to recruit Drp1 to mitochondria and mediate mitochondrial fission.
FIGURE 6:
MiD49 and MiD51 can restore CCCP-induced mitochondrial fragmentation in Fis1/Mff-null cells. (A) Restoration of Drp1 puncta in Fis1/Mff-null cells. Fis1/Mff-null cells were transfected with MiD49-Myc, MiD51-Myc, or empty vector. Cells were analyzed for MiD-Myc expression (anti-Myc), mitochondrial morphology (Cox8-Dendra2), and Drp1. Scale bar, 10 μm. Right, magnified images of the boxed regions. Scale bar, 5 μm. (B) Density of Drp1 puncta on mitochondria. Twenty ROIs were analyzed in 8–10 cells per group. Data are normalized to wild-type cells. Error bars, SEM. *p < 0.001. DM, Fis1/Mff double mutant. (C) Scoring of mitochondrial network morphologies for Fis1/Mff-null cells transfected with MiD49-Myc, MiD51-Myc, or empty vector and treated with DMSO or CCCP. Data are from three independent experiments, each with 100 cells scored. Error bars, SEM. C, collapsed; L, long; N, net-like; S, short. **p = 0.002, *p = 0.02. (D) Representative micrographs of Fis1/Mff-null cells transfected with MiD49-Myc or MiD51-Myc and treated with DMSO or CCCP. MiD-Myc–expressing cells were analyzed for mitochondrial morphology (Tom20). With CCCP treatment, note that the MiD-Myc–transfected cells have fragmented mitochondria, whereas the nontransfected cells are resistant to CCCP-induced fragmentation. Scale bar, 10 μm. Statistical testing was performed with the Student's t test.
DISCUSSION
Our work clarifies several issues concerning the recruitment of Drp1 to mitochondria. Analysis of Fis1-null MEFs indicates a clear, although minor, role of Fis1 in Drp1 recruitment and mitochondrial fission. Consistent with previous results (Otera et al., 2010), its role appears to be substantially less important than that of Mff. It is possible that the relative importance of Fis1 versus Mff depends on the particular cell type. Cell-type specificity might explain why we found a clear mitochondrial morphology defect in Fis1-null MEFs versus a previous study of Fis1-null HCT116 cells (Otera et al., 2010). It is also possible that the relative importance of these proteins depends on which signaling pathway is activated, although we have not yet identified conditions that selectively promote Fis1-mediated fission (Figure 1H).
Our results suggest that Fis1 and Mff, beyond merely recruiting Drp1, might have a role in promoting Drp1 assembly. In the absence of Fis1, Mff, or both, the remaining Drp1 puncta associated with mitochondria are notably smaller in size and lower in intensity. In this regard, these Drp1 receptors may play a role similar to yeast Mdv1, which promotes self-assembly of Dnm1 (Lackner et al., 2009). Future biochemical studies are needed to test this model.
Previous studies yielded perplexing observations of MiD49 and MiD51 overexpression versus knockdown. In particular, it was unclear why MiD overexpression causes mitochondrial elongation (Palmer et al., 2011; Zhao et al., 2011). We find that when these proteins are overexpressed, recruitment of Drp1 is enhanced, but there is also increased inhibitory phosphorylation of Drp1 on S637. With CCCP treatment, this phosphorylation is reduced, and mitochondrial fission ensues. We cannot rule out that additional mechanisms might also inhibit Drp1 function upon MiD overexpression. Our results suggest the potential for regulation, in which the outcome of MiD activity depends on the physiological state of the cell. For example, we speculate that MiDs might recruit Drp1 but maintain it in an inactive state until a cellular signal triggers fission.
Finally, our results indicate that Fis1, Mff, MiD49, and MiD51 can each recruit Drp1 and promote mitochondrial fission. In particular, the MiDs are able to promote fission in the absence of both Fis1 and Mff. It remains possible that these proteins can also function together to mediate fission. Future studies will clarify whether these proteins preferentially operate in specific cell types or cellular circumstances.
MATERIALS AND METHODS
Materials
Antibody sources were as follows: Drp1 (BD Biosciences, San Diego, CA), Drp1 S616-PO4 and S637-PO4 (Cell Signaling Technology, Beverly, MA), superoxide dismutase 2 (SOD2; Abcam, Cambridge, MA), β-tubulin (TUBB; Imgenex, San Diego, CA), Tom20 (Santa Cruz Biotechnology, Santa Cruz, CA), MiD49 (also known as SMCR7; Proteintech Group, Chicago, IL), MiD51 (also known as SMCR7L; Proteintech Group), actin (Millipore, Billerica, MA), Fis1 (Alexis/Axxora, San Diego, CA), Mff (Sigma-Aldrich, St. Louis, MO; and gift from A. van der Bliek, University of California, Los Angeles, Los Angeles, CA), and Myc (mouse monoclonal 9E10 from Covance, Berkeley, CA; and rabbit polyclonal from Sigma-Aldrich). CCCP (Sigma-Aldrich) was used at 50 μM. Staurosporine (Sigma-Aldrich) was used at 1 μM. Etoposide (Sigma-Aldrich) was used at 100 μM. Z-VAD-FMK (BD Biosciences) was used at 50 μM. Cells were grown in LabTek chambered glass slides (Nunc, Rochester, NY) for fixed- and live-cell imaging.
Immunofluorescence and imaging
For immunofluorescence, cells were fixed in 4% formaldehyde for 10 min at 37°C, permeabilized with 0.1% Triton X-100 at room temperature, and incubated with antibodies in 5% fetal calf serum. For Drp1 immunofluorescence, cells were permeabilized in a low-concentration digitonin buffer (0.001% digitonin, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 150 mM NaCl, 2 mM MgCl2, 2 mM EDTA, 320 mM sucrose, pH 7.4) for 1.5 min at 37°C and then immediately fixed. Cells were then processed as described.
Scoring of mitochondrial network morphology was performed blind to genotype and treatment. All quantification was done in triplicate, and 100 cells were scored per experiment.
All fluorescence imaging was performed using a Plan-Apochromat 63×/1.4 oil objective on a Zeiss LSM 710 confocal microscope driven by Zen 2009 software (Carl Zeiss, Jena, Germany). Image cropping was performed using ImageJ software (National Institutes of Health, Bethesda, MD). Global adjustments to brightness and contrast were performed using Photoshop (Adobe, San Jose, CA). We used 488- and 561-nm lasers to excite unconverted and photoconverted Dendra2, respectively, and a 405-nm laser to photoconvert Dendra2. Live-cell imaging was done on a stage-top heated platform maintained at 37°C.
FRAP and diffusion assays
Cells constitutively expressing matrix targeted Cox8-Dendra2 were used for both assays. For FRAP, mitochondria in a 13 × 2 μm region were photobleached using a 405-nm laser at 10% laser power. Imaging was performed in a 15 × 9 μm region. Bleaching was performed to ∼10% of original fluorescence for all trials. For photoconversion of Cox8-Dendra2, a 405-nm laser at 4% laser power was used to photoconvert mitochondria in a 13 × 2 μm region. After photoconversion, a Z-stack was collected of the entire cell. The area of the signal was measured using ImageJ and normalized to the original photoconverted area of mitochondria in the 13 × 2 μm region.
Image analysis
All image analysis was performed using ImageJ software. For mitochondrial morphometric analysis, Z-stacks were collected from cells labeled with Tom20, and summed projections were generated. Analysis was limited to regions of interest in the periphery of cells, where individual mitochondria are readily resolved, and images were thresholded to select mitochondria. From the thresholded fluorescence, binary images were generated, and the total mitochondrial area and number of discrete mitochondria were measured. The morphometric ratio was generated by dividing the number of mitochondria by the total mitochondrial area.
Mitochondrial Drp1 puncta were analyzed in the periphery of cells by first creating a binary mask of the mitochondrial channel (Tom20 or Cox8-Dendra2) and using it to subtract all extramitochondrial Drp1 fluorescence. To select mitochondrial Drp1 puncta for analysis, mitochondrial Drp1 fluorescence was thresholded. The thresholding value was determined as the average threshold value needed to select mitochondrial Drp1 puncta in wild-type cells. To count mitochondrial puncta and measure their area, the thresholded image was converted to a binary image. To measure Drp1 puncta fluorescence, puncta identified by thresholding were analyzed for fluorescence intensities.
Generation of mutant MEFs
Mouse embryonic stem (ES) cell lines containing gene trap disruptions of Fis1 (line S9-7D1) and Mff (line AZ0438) were available at the Mutant Mouse Regional Resource Centers. The ES cell lines were injected into blastocysts to generate mouse lines. From matings of heterozygous animals, we established MEF lines from midgestation embryos as previously described (Chen et al., 2003). Western blot analysis of the Fis1-null, Mff-null, and Fis1/Mff-null cells confirmed complete loss of the relevant proteins (Supplemental Figure S1A).
Cell culture
Cell lines stably expressing Cox8-Dendra2, MiD49-Myc, and MiD51-Myc were generated via retroviral transduction. All cell lines were cultured in DMEM containing 10% fetal bovine serum and supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine.
For hypoxia experiments, cells were placed into a Billups-Rothenberg modular incubator chamber, and the atmosphere was exchanged with 1% O2/5% CO2. The O2 levels were monitored using a Sperian Toxipro single-gas detector (Honeywell Analytics, Lincolnshire, IL). Cells were incubated at 37°C for 24 h.
Cloning and RNA interference
Fis1, Mff isoform 5, MiD49, MiD51, and Drp1 variant 2 transcripts were amplified from a MEF cDNA library using PCR. MiD49 and MiD51 were cloned into the XhoI and BamHI sites of a pcDNA3.1(–) plasmid containing a C-terminal 4xMyc tag. Fis1, Fis1ΔTMD (transmembrane domain deleted), Mff isoform 5, MffΔR1 (amino acid repeat 1 [residues 20–31] deleted), MffΔTMD, and Drp1 variant 2 were cloned into the BamHI and XhoI sites of pcDNA3.1(+). The entire open reading frames were confirmed by DNA sequencing. For generation of retroviral vectors, MiD49/51-Myc, Mff, MffΔR1, and MffΔTMD sequences were cloned into the pQCXIP-Puromycin vector (Invitrogen, Carlsbad, CA). Cells transduced with these viral constructs were selected with 1 μg/ml puromycin (Invitrogen) for 4–5 d.
Oligonucleotides for siRNAs were purchased from Integrated DNA Technologies (Coralville, IA). The MiD49 siRNA oligonucleotides were based on the following sequences:
5′-ACACCTAAGTTCAGCACTATAGCAC-3′ (MiD49siRNA #1)
5′-GCCATGCCTTGAAGATGTGAATAAA-3′ (MiD49siRNA #2)
The MiD51 siRNA oligonucleotides were based on the following sequences:
5′-AGATTCCAAATGTCCTAAATCACAG-3′ (MiD51siRNA #1)
5′-GGAATAAGACAGTATTTAGGTTTCC-3′ (MiD51siRNA #2)
Results presented in Figure 3 were obtained with MiD49 siRNA #1 and MiD51 siRNA #1. Results presented in Supplemental Figure S3 were obtained with MiD49 siRNA #2 and MiD51 siRNA #2. Control siRNA was targeted against a noncoding sequence in the mouse genome:
5′-CGTTAATCGCGTATAATACGCGTAT-3′
siRNA nucleotides and plasmids were transfected using Lipofectamine 2000 (Invitrogen). Cells transfected with plasmids were assessed 24 h posttransfection. Fis1 and Mff plasmids were cotransfected with either Cox8-TagRFP or Cox8-DsRed at a ratio of 5:1. MiD49/51-Myc–positive cells were visualized with an anti-Myc antibody. Transfection of siRNA oligonucleotides was performed when cells were plated and at 24 and 48 h postplating. Mitochondrial morphology and protein levels were assessed at 72 h.
Drp1 immunoprecipitation and cell fractionation
To assess levels of Drp1 S637-PO4, we immunoprecipitated Drp1 (anti-DLP1; BD Biosciences) from 5 million cells lysed in IP buffer (1% Triton X-100, 5% glycerol, 150 mM NaCl, 25 mM Tris-HCl, 1 mM EDTA, pH 7.4) supplemented with a protease inhibitor cocktail (HALT; Thermo-Pierce, Rockford, IL). The lysates were cleared with a 21,000 × g spin at 4°C for 10 min. Immune complexes were captured with protein A/G agarose (Thermo-Pierce), and the beads were washed with IP buffer.
For cell fractionation, cells were collected by trypsinization and washed once with phosphate-buffered saline (PBS). Cells were resuspended in 1.5 ml of mitochondria isolation buffer (10 mM HEPES, 220 mM mannitol, 70 mM sucrose, 80 mM KCl, 0.5 mM EDTA, 2 mM Mg acetate, pH 7.4) supplemented with a protease inhibitor cocktail and lysed using a nitrogen bomb (Parr, Moline, IL) at 250 psi for 10 min, followed by mechanical homogenization with a glass Dounce homogenizer. Lysates were centrifuged at 700 × g three times for 10 min to obtain a postnuclear supernatant. The postnuclear supernatant was centrifuged at 10,000 × g for 10 min to obtain a crude mitochondrial pellet. The resultant supernatant was centrifuged at 21,000 × g for 30 min to obtain the cytosol fraction. To obtain purified mitochondria, the crude mitochondrial pellet was placed in a discontinuous Percoll gradient consisting of 80, 52, and 26% Percoll diluted in mitochondria isolation buffer. Centrifugation was performed at 42,500 × g for 45 min. Purified mitochondria were recovered from the 52 and 26% Percoll interface.
Myc coimmunoprecipitation assay
MiD-Drp1 interactions were assessed by transfecting 293Ts with Myc-tagged MiD constructs and Drp1 mutant constructs. For mouse Drp1 variant 2, residues S579 and S600 correspond to human Drp1 residues S616 and S637, respectively. Human nomenclature is presented in Figure 4D for consistency. Cells were harvested by trypsinization 24 h posttransfection, and cross-linked with 250 μM dithiobis(succinimidylpropionate) (Thermo-Pierce) in PBS for 30 min at room temperature. Cross-linker was quenched by adding Tris-HCl, pH 7.5, to a final concentration of 150 mM and incubating for an additional 10 min. Cells were pelleted, the quenched cross-linker solution was removed, and pellets were solubilized in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, and 1% NP-40) supplemented with a protease inhibitor cocktail. Myc immunoprecipitation was performed with rabbit anti-Myc agarose beads (Sigma-Aldrich). Cross-link bonds were reversed by solubilizing samples in Laemmli buffer (50 mM Tris-HCl, pH 6.8, 1% SDS, 5% glycerol, and 0.005% bromophenol blue) containing 10% β-mercaptoethanol and boiling for 5 min.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM062967). O.C.L. was supported by an R. L. Kirschstein National Research Service Award (5F31GM089327). The Drp1-null cells were a generous gift from Katsuyoshi Mihara (Kyushu University, Fukuoka, Japan).
Abbreviations used:
- CCCP
carbonyl cyanide m-chlorophenylhydrozone
- DMSO
dimethyl sulfoxide
- Drp1
dynamin-related protein 1
- Fis1
mitochondrial fission protein 1
- FRAP
fluorescence recovery after photobleaching
- MEF
mouse embryonic fibroblast
- Mff
mitochondrial fission factor
- MiD49
51, mitochondrial dynamics proteins of 49 and 51 kDa, respectively
- ROI
region of interest
- siRNA
short interfering RNA
- STS
staurosporine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-10-0721) on January 2, 2013.
*Present address: College of Life Sciences, Wuhan University, Wuhan, Hubei 430072, China.
REFERENCES
- Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny KL, Jensen RE. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol Biol Cell. 2003;14:4126–4139. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes P, Shepard KA, Yaffe MP. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annu Rev Cell Dev Biol. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev. Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.