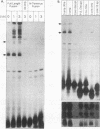
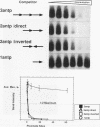
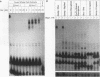
Abstract
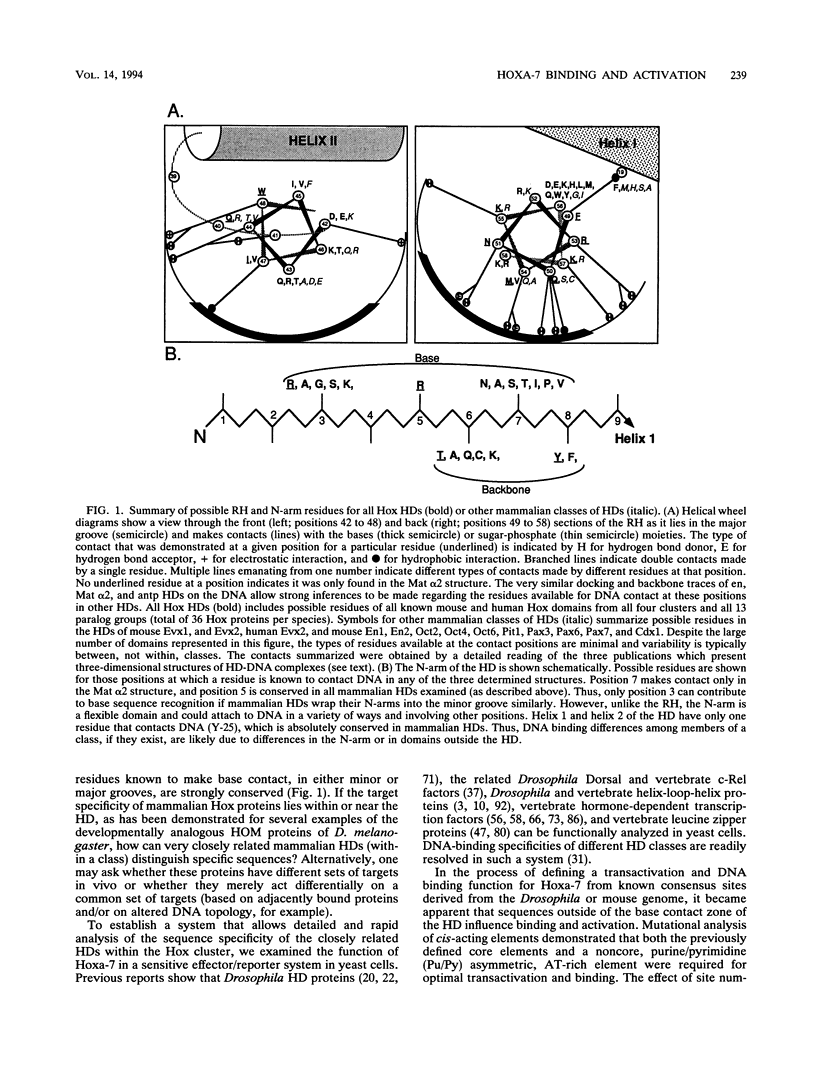
The murine developmental control gene product, Hoxa-7, was shown to function as a DNA-binding transactivator in Saccharomyces cerevisiae. The importance of the ATTA core, the preference for antp class flanking nucleotides, the importance of Asn-51 of the homeodomain (HD), and the synergism of multiple binding sites all reflect properties that have previously been described for HOM or Hox proteins in tissue culture systems. A comparison of contact positions among genes of paralog groups and classes of mammalian HDs points to a lack of diversity in positions that make base contact, suggesting that besides the combination of HD amino acid-base pair contacts, another means of recognizing differences between targets must exist if Hox genes select different targets. The HD of antennapedia is identical to the Hoxa-7 HD. The interaction of Hoxa-7 with the exact sequence used in the nuclear magnetic resonance three-dimensional structural analysis on the antennapedia HD was studied. Hoxa-7 binding and transactivation was influenced by sequences outside of the known base contact zone of this site. We conclude that Hoxa-7 protein has a second means to interact with DNA or/and that the sequences flanking the base contact zone influence HD interactions by distorting DNA within the contact zone (base or backbone). This result is discussed in terms of DNA flexure and two modes of transcription used in S. cerevisiae.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. DNA binding properties of the purified Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Amati B., Dalton S., Brooks M. W., Littlewood T. D., Evan G. I., Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992 Oct 1;359(6394):423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- Arcioni L., Simeone A., Guazzi S., Zappavigna V., Boncinelli E., Mavilio F. The upstream region of the human homeobox gene HOX3D is a target for regulation by retinoic acid and HOX homeoproteins. EMBO J. 1992 Jan;11(1):265–277. doi: 10.1002/j.1460-2075.1992.tb05049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter M., Qian Y., Otting G., Müller M., Gehring W. J., Wüthrich K. Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy. J Mol Biol. 1990 Jul 5;214(1):183–197. doi: 10.1016/0022-2836(90)90155-f. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Homeotic protein binding sites, origins of replication, and nuclear matrix anchorage sites share the ATTA and ATTTA motifs. J Cell Biochem. 1992 Oct;50(2):111–123. doi: 10.1002/jcb.240500202. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kornberg R. D. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990 Mar;10(3):887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera C. V., Alonso M. C. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991 Oct;10(10):2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. Principles of sequence-dependent flexure of DNA. J Mol Biol. 1986 Dec 20;192(4):907–918. doi: 10.1016/0022-2836(86)90036-7. [DOI] [PubMed] [Google Scholar]
- Chen W., Tabor S., Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987 Sep 25;50(7):1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- Chouinard S., Kaufman T. C. Control of expression of the homeotic labial (lab) locus of Drosophila melanogaster: evidence for both positive and negative autogenous regulation. Development. 1991 Dec;113(4):1267–1280. doi: 10.1242/dev.113.4.1267. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Desplan C., Theis J., O'Farrell P. H. The sequence specificity of homeodomain-DNA interaction. Cell. 1988 Sep 23;54(7):1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessain S., Gross C. T., Kuziora M. A., McGinnis W. Antp-type homeodomains have distinct DNA binding specificities that correlate with their different regulatory functions in embryos. EMBO J. 1992 Mar;11(3):991–1002. doi: 10.1002/j.1460-2075.1992.tb05138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., Young K. E., von Kessler D. P., Beachy P. A. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991 May;10(5):1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., von Kessler D. P., Beachy P. A. Differential DNA sequence recognition is a determinant of specificity in homeotic gene action. EMBO J. 1992 Nov;11(11):4059–4072. doi: 10.1002/j.1460-2075.1992.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick V. D., Ingles C. J. The Drosophila fushi tarazu polypeptide is a DNA-binding transcriptional activator in yeast cells. Nature. 1989 Feb 16;337(6208):666–668. doi: 10.1038/337666a0. [DOI] [PubMed] [Google Scholar]
- Florence B., Handrow R., Laughon A. DNA-binding specificity of the fushi tarazu homeodomain. Mol Cell Biol. 1991 Jul;11(7):3613–3623. doi: 10.1128/mcb.11.7.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Gibson G., Schier A., LeMotte P., Gehring W. J. The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell. 1990 Sep 21;62(6):1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Thomas G. H., Elgin S. C. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989 Sep 29;245(4925):1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- Gross M. K., Merrill G. F. Regulation of thymidine kinase protein levels during myogenic withdrawal from the cell cycle is independent of mRNA regulation. Nucleic Acids Res. 1988 Dec 23;16(24):11625–11643. doi: 10.1093/nar/16.24.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueneberg D. A., Natesan S., Alexandre C., Gilman M. Z. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992 Aug 21;257(5073):1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- Han K., Levine M. S., Manley J. L. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989 Feb 24;56(4):573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. A genetic model for interaction of the homeodomain recognition helix with DNA. Science. 1991 Jan 25;251(4992):426–430. doi: 10.1126/science.1671176. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989 Jun 30;57(7):1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Scott M. P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990 Nov 30;63(5):883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Austin R. H. Importance of DNA stiffness in protein-DNA binding specificity. Nature. 1987 Sep 17;329(6136):263–266. doi: 10.1038/329263a0. [DOI] [PubMed] [Google Scholar]
- Kalb V. F., Jr, Bernlohr R. W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977 Oct;82(2):362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- Kamens J., Brent R. A yeast transcription assay defines distinct rel and dorsal DNA recognition sequences. New Biol. 1991 Oct;3(10):1005–1013. [PubMed] [Google Scholar]
- Keleher C. A., Passmore S., Johnson A. D. Yeast repressor alpha 2 binds to its operator cooperatively with yeast protein Mcm1. Mol Cell Biol. 1989 Nov;9(11):5228–5230. doi: 10.1128/mcb.9.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M., Drescher U., Gruss P. The murine homeo domain protein Hox 1.1. Ann N Y Acad Sci. 1987;511:88–100. doi: 10.1111/j.1749-6632.1987.tb36240.x. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Ko H. S., Fast P., McBride W., Staudt L. M. A human protein specific for the immunoglobulin octamer DNA motif contains a functional homeobox domain. Cell. 1988 Oct 7;55(1):135–144. doi: 10.1016/0092-8674(88)90016-5. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Saffman E. E., Kornfeld K., Hogness D. S. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989 Jun 16;57(6):1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- Kuziora M. A., McGinnis W. A homeodomain substitution changes the regulatory specificity of the deformed protein in Drosophila embryos. Cell. 1989 Nov 3;59(3):563–571. doi: 10.1016/0092-8674(89)90039-1. [DOI] [PubMed] [Google Scholar]
- Käs E., Izaurralde E., Laemmli U. K. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J Mol Biol. 1989 Dec 5;210(3):587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- Käs E., Poljak L., Adachi Y., Laemmli U. K. A model for chromatin opening: stimulation of topoisomerase II and restriction enzyme cleavage of chromatin by distamycin. EMBO J. 1993 Jan;12(1):115–126. doi: 10.1002/j.1460-2075.1993.tb05637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991 Dec 3;30(48):11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Lin L., McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992 Jun;6(6):1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- Losa R., Omari S., Thoma F. Poly(dA).poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res. 1990 Jun 25;18(12):3495–3502. doi: 10.1093/nar/18.12.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D. P., Sluka J. P., Shin J. A., Griffin J. H., Simon M. I., Dervan P. B. Orientation of the putative recognition helix in the DNA-binding domain of Hin recombinase complexed with the hix site. Biochemistry. 1990 Jul 17;29(28):6561–6567. doi: 10.1021/bi00480a003. [DOI] [PubMed] [Google Scholar]
- Malicki J., Schughart K., McGinnis W. Mouse Hox-2.2 specifies thoracic segmental identity in Drosophila embryos and larvae. Cell. 1990 Nov 30;63(5):961–967. doi: 10.1016/0092-8674(90)90499-5. [DOI] [PubMed] [Google Scholar]
- Mann R. S., Hogness D. S. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990 Feb 23;60(4):597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- McGinnis N., Kuziora M. A., McGinnis W. Human Hox-4.2 and Drosophila deformed encode similar regulatory specificities in Drosophila embryos and larvae. Cell. 1990 Nov 30;63(5):969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. Homeobox genes and axial patterning. Cell. 1992 Jan 24;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Metzger D., White J. H., Chambon P. The human oestrogen receptor functions in yeast. Nature. 1988 Jul 7;334(6177):31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- Müller M., Affolter M., Leupin W., Otting G., Wüthrich K., Gehring W. J. Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J. 1988 Dec 20;7(13):4299–4304. doi: 10.1002/j.1460-2075.1988.tb03328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z., Tsai M. J., McDonnell D. P., O'Malley B. W. Identification of novel steroid-response elements. Gene Expr. 1992;2(1):39–47. [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Odenwald W. F., Garbern J., Arnheiter H., Tournier-Lasserve E., Lazzarini R. A. The Hox-1.3 homeo box protein is a sequence-specific DNA-binding phosphoprotein. Genes Dev. 1989 Feb;3(2):158–172. doi: 10.1101/gad.3.2.158. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y., Horikoshi M., Roeder R. G., Desplan C. Engrailed, a homeodomain protein, can repress in vitro transcription by competition with the TATA box-binding protein transcription factor IID. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2289–2293. doi: 10.1073/pnas.87.6.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Struhl K. Analysis of Saccharomyces cerevisiae his3 transcription in vitro: biochemical support for multiple mechanisms of transcription. Mol Cell Biol. 1990 Jun;10(6):2832–2839. doi: 10.1128/mcb.10.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalsky M. L., Sharif M., Yamamoto K. R. The viral erbA oncogene protein, a constitutive repressor in animal cells, is a hormone-regulated activator in yeast. Cell. 1990 Dec 21;63(6):1277–1286. doi: 10.1016/0092-8674(90)90423-c. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöpperl H., Featherstone M. S. An autoregulatory element of the murine Hox-4.2 gene. EMBO J. 1992 Oct;11(10):3673–3680. doi: 10.1002/j.1460-2075.1992.tb05452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Otting G., Furukubo-Tokunaga K., Affolter M., Gehring W. J., Wüthrich K. NMR structure determination reveals that the homeodomain is connected through a flexible linker to the main body in the Drosophila Antennapedia protein. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10738–10742. doi: 10.1073/pnas.89.22.10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M. L., Jackson-Grusby L., Brent R. Gene activation and DNA binding by Drosophila Ubx and abd-A proteins. Cell. 1989 Jun 16;57(6):1045–1052. doi: 10.1016/0092-8674(89)90342-5. [DOI] [PubMed] [Google Scholar]
- Schena M., Yamamoto K. R. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 1988 Aug 19;241(4868):965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack J. M., Tannahill D. Mechanism of anteroposterior axis specification in vertebrates. Lessons from the amphibians. Development. 1992 Feb;114(2):285–302. doi: 10.1242/dev.114.2.285. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. The JUN oncoprotein, a vertebrate transcription factor, activates transcription in yeast. Nature. 1988 Apr 14;332(6165):649–650. doi: 10.1038/332649a0. [DOI] [PubMed] [Google Scholar]
- Ullmann A. One-step purification of hybrid proteins which have beta-galactosidase activity. Gene. 1984 Jul-Aug;29(1-2):27–31. doi: 10.1016/0378-1119(84)90162-8. [DOI] [PubMed] [Google Scholar]
- Vershon A. K., Johnson A. D. A short, disordered protein region mediates interactions between the homeodomain of the yeast alpha 2 protein and the MCM1 protein. Cell. 1993 Jan 15;72(1):105–112. doi: 10.1016/0092-8674(93)90054-t. [DOI] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. E., Fahrner T. J., Johnston M., Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991 May 31;252(5010):1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- Winslow G. M., Hayashi S., Krasnow M., Hogness D. S., Scott M. P. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell. 1989 Jun 16;57(6):1017–1030. doi: 10.1016/0092-8674(89)90340-1. [DOI] [PubMed] [Google Scholar]
- Winter E., Varshavsky A. A DNA binding protein that recognizes oligo(dA).oligo(dT) tracts. EMBO J. 1989 Jun;8(6):1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Yoon C., Privé G. G., Goodsell D. S., Dickerson R. E. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappavigna V., Renucci A., Izpisúa-Belmonte J. C., Urier G., Peschle C., Duboule D. HOX4 genes encode transcription factors with potential auto- and cross-regulatory capacities. EMBO J. 1991 Dec;10(13):4177–4187. doi: 10.1002/j.1460-2075.1991.tb04996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos A. S., Gyuris J., Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993 Jan 29;72(2):223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]