Abstract
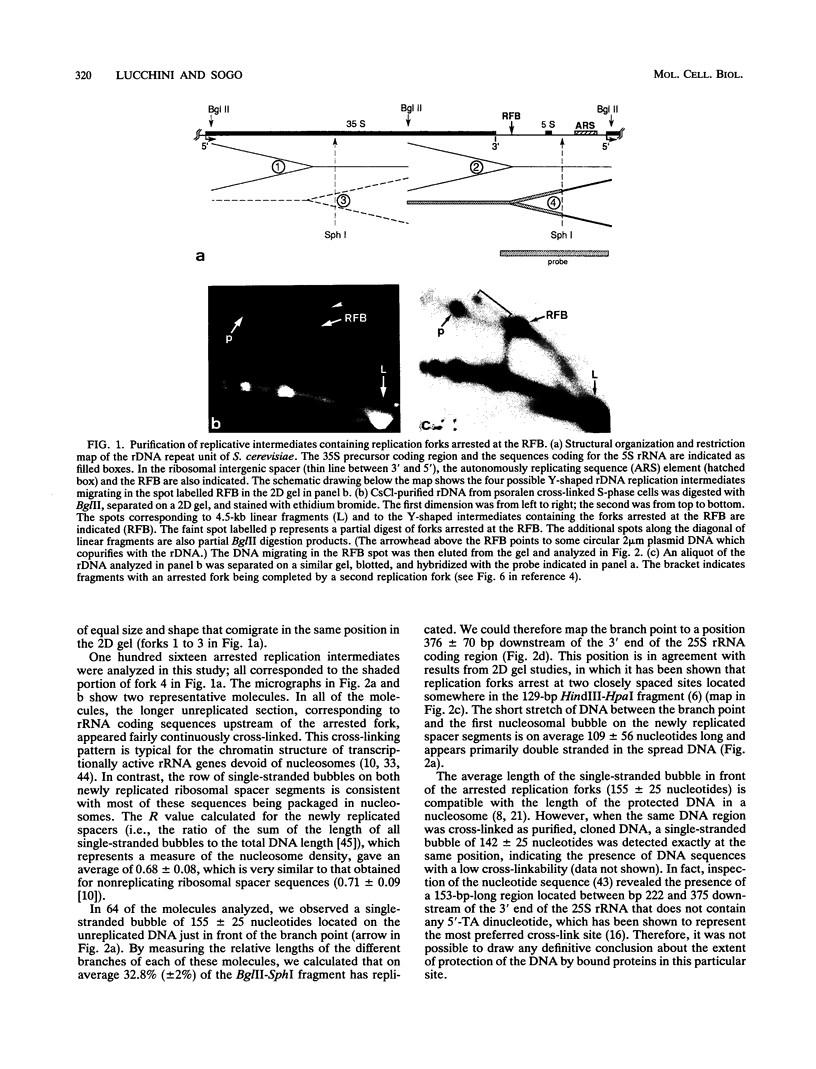
Replication intermediates containing forks arrested at the replication fork barrier near the 3' end of the yeast rRNA genes were analyzed at the chromatin level by using in vivo psoralen cross-linking as a probe for chromatin structure. These specific intermediates were purified from preparative two-dimensional agarose gels, and the extent of cross-linking in the different portions of the branched molecules was examined by electron microscopy and by using a psoralen gel retardation assay. The unreplicated section corresponding to the rRNA coding region upstream of the arrested forks appeared mostly heavily cross-linked, characteristic of transcriptionally active rRNA genes devoid of nucleosomes, whereas the replicated daughter strands representing newly synthesized spacer sequences showed a nucleosomal organization typical for bulk chromatin. The failure to detect replication forks arrested at the 3' end of inactive rRNA gene copies and the fact that most DNA encoding rRNA (rDNA) is replicated in the same direction as transcription suggest that replication forks seldom originate from origins of replication located immediately downstream of inactive genes.
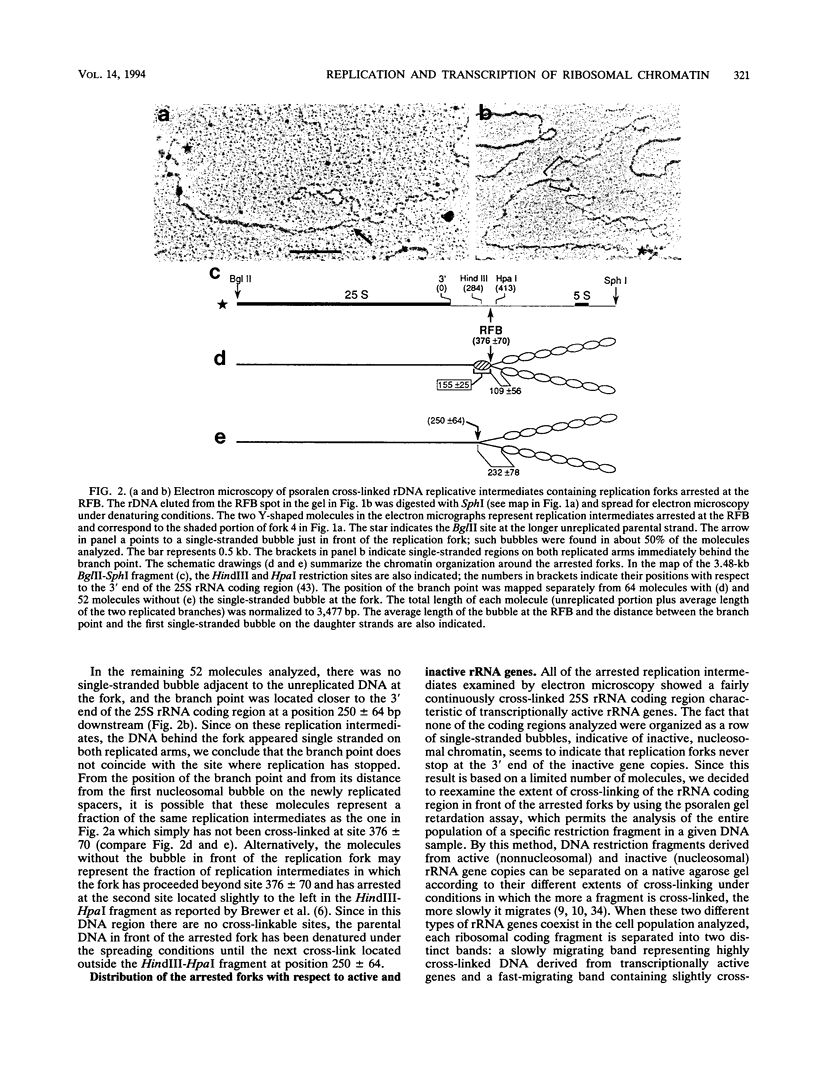
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avemann K., Knippers R., Koller T., Sogo J. M. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol Cell Biol. 1988 Aug;8(8):3026–3034. doi: 10.1128/mcb.8.8.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988 Nov 18;55(4):637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. Mapping replication origins in yeast chromosomes. Bioessays. 1991 Jul;13(7):317–322. doi: 10.1002/bies.950130702. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Lockshon D., Fangman W. L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992 Oct 16;71(2):267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Zakian V. A., Fangman W. L. Replication and meiotic transmission of yeast ribosomal RNA genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6739–6743. doi: 10.1073/pnas.77.11.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénard M., Pierron G. Mapping of a Physarum chromosomal origin of replication tightly linked to a developmentally-regulated profilin gene. Nucleic Acids Res. 1992 Jul 11;20(13):3309–3315. doi: 10.1093/nar/20.13.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Losa R., Koller T., Sogo J. M. Psoralen-crosslinking of soluble and of H1-depleted soluble rat liver chromatin. J Mol Biol. 1984 Oct 5;178(4):920–928. doi: 10.1016/0022-2836(84)90319-x. [DOI] [PubMed] [Google Scholar]
- Conconi A., Widmer R. M., Koller T., Sogo J. M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989 Jun 2;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J. M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993 May 25;21(10):2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bernardin W., Koller T., Sogo J. M. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986 Oct 5;191(3):469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Origins of DNA replication in metazoan chromosomes. J Biol Chem. 1993 Jan 5;268(1):1–4. [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Deshpande A. M., Newlon C. S. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4305–4313. doi: 10.1128/mcb.12.10.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Vaughn J. P., Hamlin J. L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991 Aug;11(8):3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito F., Brankamp R. G., Sinden R. R. DNA sequence specificity of 4,5',8-trimethylpsoralen cross-linking. Effect of neighboring bases on cross-linking the 5'-TA dinucleotide. J Biol Chem. 1988 Aug 15;263(23):11466–11472. [PubMed] [Google Scholar]
- Fangman W. L., Brewer B. J. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992 Nov 20;258(5086):1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L. Mammalian origins of replication. Bioessays. 1992 Oct;14(10):651–659. doi: 10.1002/bies.950141002. [DOI] [PubMed] [Google Scholar]
- Hanson C. V., Shen C. K., Hearst J. E. Cross-linking of DNA in situ as a probe for chromatin structure. Science. 1976 Jul 2;193(4247):62–64. doi: 10.1126/science.935855. [DOI] [PubMed] [Google Scholar]
- Heck M. M. Probing replication origins with a fork. Trends Cell Biol. 1991 Aug;1(2-3):67–70. doi: 10.1016/0962-8924(91)90092-n. [DOI] [PubMed] [Google Scholar]
- Heintz N. H. Transcription factors and the control of DNA replication. Curr Opin Cell Biol. 1992 Jun;4(3):459–467. doi: 10.1016/0955-0674(92)90012-2. [DOI] [PubMed] [Google Scholar]
- Hernández P., Martín-Parras L., Martínez-Robles M. L., Schvartzman J. B. Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J. 1993 Apr;12(4):1475–1485. doi: 10.1002/j.1460-2075.1993.tb05791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968 Mar 14;32(2):327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987 Nov 6;51(3):473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Zhu J. G., Davis L. R., Newlon C. S. Close association of a DNA replication origin and an ARS element on chromosome III of the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14A):6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka M., Nishizawa M., Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992 Jun;233(3):355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- Lang W. H., Reeder R. H. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. The two faces of higher eukaryotic DNA replication origins. Cell. 1990 Sep 7;62(5):845–847. doi: 10.1016/0092-8674(90)90258-g. [DOI] [PubMed] [Google Scholar]
- Lucchini R., Pauli U., Braun R., Koller T., Sogo J. M. Structure of the extrachromosomal ribosomal RNA chromatin of Physarum polycephalum. J Mol Biol. 1987 Aug 20;196(4):829–843. doi: 10.1016/0022-2836(87)90408-6. [DOI] [PubMed] [Google Scholar]
- Lucchini R., Sogo J. M. Different chromatin structures along the spacers flanking active and inactive Xenopus rRNA genes. Mol Cell Biol. 1992 Oct;12(10):4288–4296. doi: 10.1128/mcb.12.10.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Bustin M., Miller O. L., Jr Electron microscopic analysis of chromosome metabolism in the Drosophila melanogaster embryo. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):741–754. doi: 10.1101/sqb.1978.042.01.075. [DOI] [PubMed] [Google Scholar]
- Mestel R., Yip M., Holland J. P., Wang E., Kang J., Holland M. J. Sequences within the spacer region of yeast rRNA cistrons that stimulate 35S rRNA synthesis in vivo mediate RNA polymerase I-dependent promoter and terminator activities. Mol Cell Biol. 1989 Mar;9(3):1243–1254. doi: 10.1128/mcb.9.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow B. E., Johnson S. P., Warner J. R. The rRNA enhancer regulates rRNA transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Feb;13(2):1283–1289. doi: 10.1128/mcb.13.2.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer L. D., Miller O. L., Jr Electron microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol Cell Biol. 1986 Apr;6(4):1148–1157. doi: 10.1128/mcb.6.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. C., Choe S. Y., Reeder R. H. In vitro definition of the yeast RNA polymerase I enhancer. Mol Cell Biol. 1993 May;13(5):2644–2654. doi: 10.1128/mcb.13.5.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J. B., Adolph S., Martín-Parras L., Schildkraut C. L. Evidence that replication initiates at only some of the potential origins in each oligomeric form of bovine papillomavirus type 1 DNA. Mol Cell Biol. 1990 Jun;10(6):3078–3086. doi: 10.1128/mcb.10.6.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Ness P. J., Widmer R. M., Parish R. W., Koller T. Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J Mol Biol. 1984 Oct 5;178(4):897–919. doi: 10.1016/0022-2836(84)90318-8. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Stahl H., Koller T., Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986 May 5;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Insertion of a genetic marker into the ribosomal DNA of yeast. Plasmid. 1979 Oct;2(4):536–554. doi: 10.1016/0147-619x(79)90053-2. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Veinot-Drebot L. M., Singer R. A., Johnston G. C. rRNA transcription initiation is decreased by inhibitors of the yeast cell cycle control step "start". J Biol Chem. 1989 Nov 25;264(33):19528–19534. [PubMed] [Google Scholar]
- Yang L., Botchan M. Replication of bovine papillomavirus type 1 DNA initiates within an E2-responsive enhancer element. J Virol. 1990 Dec;64(12):5903–5911. doi: 10.1128/jvi.64.12.5903-5911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Newlon C. S., Huberman J. A. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]