Abstract
microRNAs (miRNAs) are 20–24 nucleotide RNAs that regulate a variety of developmental and metabolic processes. The accumulation of miRNAs in vivo can be controlled at multiple levels. In addition to miRNA biogenesis, mechanisms that lead to RNA degradation, such as 3′ uridylation and 3′ truncation, also affect the steady-state levels of miRNAs. On the other hand, 2’-O-methylation in plant miRNAs protects their 3′ ends from truncation and uridylation. The recent identification of HESO1 as the key enzyme responsible for miRNA uridylation in Arabidopsis was a first step toward a full understanding of the mechanisms underlying miRNA turnover. Analyses of the heso1 mutant predicted the existence of another uridylation activity and a previously unknown nuclease that act on miRNAs. The future identification of these enzymes will enrich our understanding of miRNA turnover.
Keywords: argonaute, HEN1, HESO1, methylation, microRNA, miRNA stability, miRNA turnover, uridylation
Introduction
miRNAs are small RNAs of 20–24 nucleotides (nt) that play crucial roles in numerous developmental and metabolic processes in plants and animals by regulating gene expression primarily at post-transcriptional levels.1,2 They bind to Argonaute proteins to form functional effector complexes to engage in target recognition through sequence complementarity and repression of target gene expression by mRNA degradation and translational repression.2,3 Plant miRNAs regulate genes encoding various types of proteins, including transcription factors or other regulatory proteins that function in plant development or signal transduction.4 The critical roles of miRNAs in patterning, signaling and metabolism necessitate the proper control of their steady-state levels, which are presumably influenced by the opposing activities of miRNA biogenesis and degradation. Here, we discuss the mechanisms that impact miRNA stability in plants, with an emphasis on the opposing effects of methylation and uridylaton.
miRNA Biogenesis
miRNAs are the final products of non-coding RNA genes. Following transcription, the primary transcript (pri-miRNA) is processed into the hairpin-structured precursor (pre-miRNA), which is further processed into the mature miRNA, by RNase III-type nucleases.5 In animals, the maturation of the miRNA from the pri-miRNA involves the nuclear enzyme Drosha6 and the cytoplasmic enzyme Dicer.7,8 In Arabidopsis, the formation of the pre-miRNA from the pri-miRNA and the processing of the pre-miRNA to the mature miRNA are both performed by the Dicer-like protein, DCL1.9-11 The processing of the pre-miRNA by Dicer or DCL1 yields the miRNA/miRNA* duplex, which has a 2-nt overhang at the 3′ end of each strand, as well as a 5′ phosphate (P) and a 3′ OH on each strand.7,10 miRNA accumulation in Arabidopsis requires a protein named HEN1.11 HEN1 is a methyltransferase that acts on the miRNA/miRNA* duplex to deposit a methyl group onto the 2’ OH of the 3′ terminal ribose on each strand.12 In vitro methylation assays12 and structures of HEN1 in complex with a miRNA/miRNA* duplex13 indicate that the two termini of the miRNA/miRNA* duplex are separately and independently methylated. Therefore, HEN1 does not distinguish the miRNA strand from miRNA* or contribute to strand selection. After methylation, the miRNA/miRNA* duplex is loaded into the RNA-induced silencing complex (RISC) that contains the ARGONAUTE1 (AGO1) protein as the core component, where the miRNA, after shedding of the miRNA* strand, guides the cleavage or translation repression of its target mRNAs through base-pairing with specific targets.14,15,17
2’-O-Methylation Increases miRNA Stability
The abundance of a certain miRNA in cells is governed by its rates of biogenesis and degradation. Studies on one of the biogenesis steps in plants, miRNA 2’-O-methylation, provided the first glimpse into processes that lead to miRNA degradation. Methylation on the ribose of the last nucleotide by HEN1 is a universal step in the biogenesis of miRNAs in plants.12,18 In Arabidopsis, almost all miRNAs are fully 2’-O-methylated at their 3′ ends in vivo. In order to understand the function of miRNA methylation, northern blots were conducted to examine various miRNAs in wild type and hen1 mutants. It was found that miRNAs are reduced in abundance and heterogeneous in size when they are unmethylated.19 Primer extension experiments showed that the size heterogeneity resides in the 3′ ends of miRNAs.19 Cloning and sequencing (by the traditional Sanger method) of miR173 and miR167 in wild type and hen1 mutants revealed that miRNAs undergo 3′ truncation and 3′ uridylation, the addition of a short U-rich tail.19 This led to the hypothesis that the 2’-O-methyl moiety in plant miRNAs promotes their stability by protecting them from a 3′-to-5′ exonucleolytic activity and a uridylation activity.19 In our recent study, high-throughput sequencing was employed to examine a large number of miRNAs in wild type and hen1 mutants. Results confirmed conclusions from our previous study19 and showed that all annotated miRNAs present in the libraries undergo 3′ truncation and 3′ uridylation in the absence of methylation.20 In addition to miRNAs, siRNAs in plants and Drosophila and piRNAs in animals also undergo 2’-O-methylation by HEN1 or HEN1 orthologs.19,22-29 In both plants and animals, the lack of small RNA methylation is often associated with small RNA instability as inferred from reduced accumulation, 3′ truncation, and 3′ uridylation.19-22,29,30
SDNs Degrade Mature miRNAs
It is now known that multiple enzymes, such as 3′ and 5′ exonucleases, can engage in miRNA degradation. For example, in C. elegans, XRN-2 (a 5′-to-3′ exonuclease) is involved in the degradation of mature miRNAs.31 SDNs, a family of 3′-to-5′ exonucleases encoded by the SMALL RNA DEGRADING NUCLEASE (SDN) genes, degrade mature miRNAs in Arabidopsis.32 SDN1 has specificity for short, single-stranded RNAs and does not digest small RNA duplexes, pre-miRNAs, or longer RNAs in vitro.32 Knock-down of multiple SDN genes in Arabidopsis results in elevated miRNA levels and pleiotropic developmental abnormalities.32 This suggests that maintaining proper miRNA levels through miRNA turnover is crucial to plant development. Are SDNs the enzymes responsible for the production of the 3′ truncated species in hen1 mutants? This has not yet been determined. If they were responsible for the production of these 3′ truncated species, knocking down SDNs in a hen1 mutant background would reduce the levels of 3′ truncated species, which can be determined through small RNA high-throughput sequencing. It is known, however, that the SDNs impact the steady-state levels of miRNAs. Knocking down multiple members of the SDN gene family using an artificial miRNA results in elevated miRNA accumulation.32 Since miRNA species are nearly all methylated in wild type,12,19 the effect of SDN knockdown on miRNA accumulation in the wild-type background suggests that SDNs can degrade methylated miRNAs. Biochemical assays show that SDN1 can degrade 2’-O-methylated miRNAs in vitro, although at a lower efficiency than unmethylated miRNAs.32
Uridylation by HESO1 Destabilizes Unmethylated miRNAs
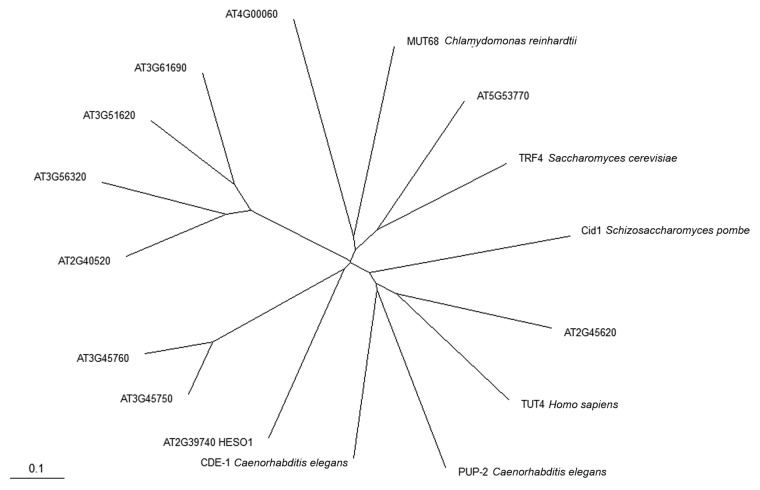
miRNA 3′ uridylation is a widespread phenomenon in hen1 mutants in plants and animals.19-22,29,30 Our recent study identified the HEN1 SUPPRESSOR1 (HESO1) gene to be responsible for small RNA uridylation in hen1 mutants in Arabidopsis.20,21 High-throughput sequencing of miRNAs in hen1 heso1 and hen1 showed that the heso1 mutation greatly reduces uridylation. In vitro, HESO1 exhibits nucleotidyl transferase activity that adds tens of non-templated Us to an RNA oligonucleotide, and its activity is completely inhibited by 2’-O-methylation on the RNA’s 3′ terminal ribose. HESO1 prefers U relative to the other three nucleotides. These biochemical properties are consistent with the genetic data showing that miRNAs gain a U-rich tail in the absence of methylation. The heso1–1 mutation is likely a null allele, yet uridylation, mostly monouridylation, is still present in this mutant, suggesting that another nucleotidyl transferase can uridylate unmethylated miRNAs. This activity is likely also inhibited by 2'-O-methylation since monouridylation does not occur as frequently in wild type as it does in hen1 heso1. BLAST searches using known nucleotidyl transferases from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Chlamydomonas reinhardtii, Caenorhabditis elegans and human revealed at least ten potential nucleotidyl transferase genes, including HESO1, in Arabidopsis (Fig. 1). It is possible that one of the other nine genes is responsible for monouridylation of miRNAs in the absence of HESO1 activity.
Figure 1. A Phylogenetic tree of the nucleotidyl transferase (NT) domain from polyA polymerases and terminal uridylyl transferases. Ten Arabidopsis proteins and homologous proteins from other eukaryotes were used in the analysis. The Arabidopsis proteins all have the “AT” prefix, and HESO1 is At2g39740. Evolutionary distance is indicated by the scale bar.
Uridylation leads to miRNA degradation, as miRNA levels are increased in hen1 heso1 relative to hen1. We note that miRNAs remain unmethylated in hen1 heso1, but the increased levels of miRNAs result in more effective repression of miRNA target genes in hen1 heso1 relative to hen1.20,21 This indicates that unmethylated miRNAs are functional in target regulation. Therefore, the methylation of miRNAs probably only serves to stabilize miRNAs instead of impacting their functionality. How does uridylation trigger the degradation of miRNAs? We presume that the U-rich tail serves as a preferred substrate for a nuclease. Intriguingly, the nuclease is most likely distinct from that causing the 3′ truncation of miRNAs in hen1 mutants because the abundance of 3′ truncated miRNA species was similar in hen1 heso1 and hen1, although the former genotype has greatly reduced uridylation.20,21 Although the nature of this enzyme is currently unknown, we predict that, if it were an exonuclease, it would differ from the one generating the 3′ truncated species in that it is highly processive such that it completely degrades uridylated miRNAs without leaving many truncated intermediates.
So far, it is not clear whether HESO1 is also involved in the degradation of 2’-O-methylated miRNAs. Loss-of-function in HESO1 did not affect miRNA accumulation in the wild-type HEN1 background, which suggests that HESO1 does not affect miRNA stability in wild type, in which miRNAs are fully methylated. This is consistent with the fact that HESO1 activity is completely inhibited by 2’-O-methylation on its substrate. However, it is possible that HESO1 acts to degrade miRNAs in wild type through collaboration with other enzymes. For example, it may act cooperatively with SDN1, which truncates small RNAs,32 or another nucleotidyl transferase, which can act on 2’-O-methylated miRNAs. Unmethylated miRNAs that are generated from the activities of these enzymes can then be uridylated by HESO1. Further investigations are needed to determine the relationship between HESO1 and other miRNA-degrading activities.
siRNAs Negatively Influence HEN1-Mediated miRNA Methylation
siRNAs are similar to miRNAs in structure, biogenesis and function. But unlike miRNAs, which come from hairpin pre-miRNAs, siRNAs originate from long double-stranded RNA precursors.33,34 Both miRNAs and siRNAs in plants carry a 2’-O-methyl group on the 3′-terminal nucleotide, a modification introduced by the methyltransferase HEN1.12,19 siRNAs compete with miRNAs for methylation in Arabidopsis when HEN1 function is compromised.35 This conclusion was drawn from studies of mutations in DNA-dependent RNA polymerase IV or RNA-dependent RNA polymerase 2, both of which are essential for the biogenesis of endogenous 24-nt siRNAs.36-38 These mutations can rescue the defects in miRNA methylation of hen1–2, a weak hen1 allele, revealing the negative influence of siRNAs on HEN1-mediated miRNA methylation.35 These mutations do not exhibit large effects on miRNA levels in wild type, suggesting that HEN1 activity is not limiting. However, HEN1 activity may be rendered limiting under conditions when large quantities of siRNAs are produced.
Possible Roles of Argonaute Proteins in miRNA 3′ Truncation or 3′ Uridylation
Based on the well-established framework of miRNA biogenesis, the final step of miRNA maturation, following miRNA methylation, is the formation of RISC that contains an Argonaute protein and a small RNA as the core components. Structures of prokaryotic and eukaryotic Argonaute proteins in complex with a guide strand reveal that the 3′ end of the guide strand is anchored in the Argonaute PAZ domain (reviewed in refs.39 and 40). This raises the question whether the 3′-to-5′ exonuclease SDN1 or the 3′ uridylation enzyme HESO1 can access a miRNA while it is bound by AGO1, the major miRNA effector in Arabidopsis.14
Argonaute association appears to stabilize miRNAs. For example, in Arabidopsis ago1 mutants, many miRNAs are reduced in abundance.41 However, this does not rule out the possibility that AGO1-bound miRNAs are accessible by SDN1 or HESO1. Structural studies suggest that RISC undergoes dynamic changes between conformational states, and in some states, the 3′ end of the miRNA becomes dislodged from the PAZ domain. For example, in the structure of a Thermus thermophilus Argonaute-guide-target ternary complex in which the target strand has a long stretch of sequence complementarity to the guide, the 3′ end of the guide is dislodged from the PAZ domain.42 This suggests that when the RISC targets an mRNA with extensive sequence complementarity to the guide strand, the guide strand’s 3′ end could become vulnerable to 3′ targeting enzymes such as SDN1 or HESO1. In fact, in Drosophila and mammalian cell culture, the introduction of artificial targets with longer stretches of sequence complementarity to the miRNAs than natural targets leads to the 3′ truncation and 3′ uridylation of the miRNAs.30,43
We observed that the 3′ truncated and/or uridylated miRNA species in hen1 and hen1 heso1 mutants are associated with AGO1 in vivo (Fig. 2). The AGO1-bound miRNAs in hen1 or hen1 heso1 resemble their corresponding input RNAs in terms of the presence of the 3′ truncated and 3′ uridylated species. This indicates that most 3′ truncated or uridylated species that are observed are bound by AGO1. The fact that these degradation intermediates are bound by AGO1 implies that the initial 3′ truncation or uridylation occurs on AGO1-bound miRNAs. Perhaps, truncation or uridylation beyond a certain length threshold would cause the miRNA to be released from AGO1. In fact, AGO1 binding may determine the length of the U tails observed in vivo. In vitro activity assays show that HESO1 is capable of adding a long U tail (tens to hundreds of nucleotides), but in vivo, the tail length is at most 7 nt.19-21 It could be that species with longer tails cannot be protected by AGO1 and are degraded. We envision that SDN1 and HESO1 shorten and lengthen, respectively, AGO1-bound miRNAs to trigger their dissociation from AGO1 and their eventual degradation.
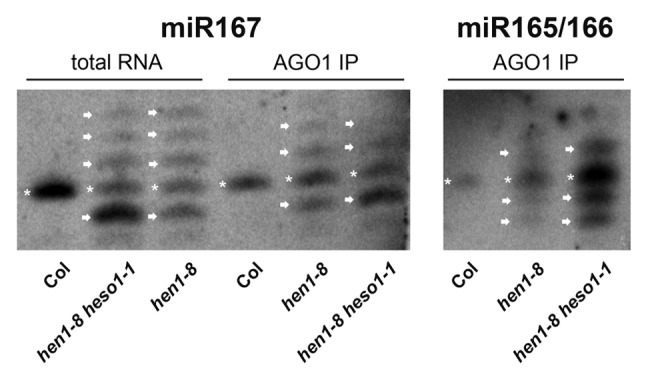
Figure 2. northern blots of miR167 and miR165/166 from total RNA or AGO1 immunoprecipitates from hen1–8 and hen1–8 heso1–1 inflorescences. Note that the Col (wild type) samples only serve as a size reference; RNA from seedlings was used and was not in the equivalent quantity as the hen1–8 or hen1–8 heso1–1 samples. The bands representing miRNAs of the wild-type size are indicated by asterisks. The arrows above and below the asterisks mark bands that represent 3′ uridylated and 3′ truncated species, respectively.
Materials and Methods
Phylogenetic analysis
For ten putative Arabidopsis nucleotidyl transferases and selected homologs from other eukaryotes, sequences corresponding to the NT_PAP_TUTase (cd05402) domain44 were aligned using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default parameters.45 The phylogenetic tree in Figure 1 was generated using TreeView.46
Immunoprecipitation and northern blotting
AGO1 was immunoprecipitated as described47 from hen1 and hen1 heso1 inflorescences and wild-type seedlings with mouse-anti-AGO1 antibodies generated by Dr. Xiaofeng Cao’s group at Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. RNAs were isolated from AGO1 immunoprecipitates as well as input samples (total RNA) using Tri-reagent (Molecular Research Center, Inc.). Northern blotting to detect miRNAs was performed as described.48 The DNA oligonucleotide probe for detection of miR167a is 5′-TAGATCATGTTGGCAGTTTCA-3′; that for detecting miR165/166 is 5′-AATGAAGCCTGGTCCGA-3′.
Conclusion
miRNA degradation is a major factor affecting the steady-state levels of miRNAs. Factors that participate in or control miRNA degradation are just beginning to be understood. Many questions await elucidation. Does HESO1 play a role in miRNA degradation in wild type, in which miRNAs are 2’-O-methylated? What is the enzyme(s) that degrades uridylated miRNAs? Does SDN1 or HESO1 act on AGO1-bound miRNAs? What factors influence the accessibility of the 3′ end of a miRNA to SDN1 or HESO1 when it is AGO-bound? Is there temporal or spatial regulation of miRNA degradation? Are there mechanisms that impart specificity to miRNA degradation (such that a particular miRNA is made unstable under certain conditions)? Answers to these questions will help establish a framework for understanding miRNA turnover.
Acknowledgments
We thank Dr. Xiaofeng Cao at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for providing anti-AGO1 antibodies. This work was supported by grants from the National Science Foundation (MCB-1021465) and the National Institutes of Health (GM061146) to X.C., and Chinese National Science Foundation (30970265) and Guangdong Natural Science Foundation (9151806001000017) to B.M. We thank the anonymous reviewers for their valuable suggestions that helped improve the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22034
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 3.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 4.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 7.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 8.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–26. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–8. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–95. doi: 10.1016/S0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–5. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma JB. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009;461:823–7. doi: 10.1038/nature08433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–33. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–90. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 17.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 18.Abe M, Yoshikawa T, Nosaka M, Sakakibara H, Sato Y, Nagato Y, et al. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining MicroRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol. 2010;154:1335–46. doi: 10.1104/pp.110.160234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–7. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, et al. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012;22:689–94. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren G, Chen X, Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, et al. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29:3688–700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, et al. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 2012;8:e1002617. doi: 10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, et al. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 2012;8:e1002616. doi: 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–85. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–8. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–72. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 29.Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, et al. Differential Impact of the HEN1 Homolog HENN-1 on 21U and 26G RNAs in the Germline of Caenorhabditis elegans. PLoS Genet. 2012;8:e1002702. doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–9. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–9. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 32.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–2. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 34.Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–31. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu B, Bi L, Zhai J, Agarwal M, Li S, Wu Q, et al. siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 2010;38:5844–50. doi: 10.1093/nar/gkq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–22. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–20. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki HM, Tomari Y. The true core of RNA silencing revealed. Nat Struct Mol Biol. 2012;19:657–60. doi: 10.1038/nsmb.2302. [DOI] [PubMed] [Google Scholar]
- 40.Parker JS. How to slice: snapshots of Argonaute in action. Silence. 2010;1:3. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–97. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–61. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, et al. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012;8:e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 46.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 47.Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105:10073–8. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]