Abstract
Changing the abundance of transcripts by regulated RNA degradation is a critical step in the control of various biological pathways. Recently, genome-wide inhibitor-free technologies for determining RNA stabilities in mammalian cells have been developed. In these methods, endogenous RNAs are pulse labeled by uridine analogs [e.g., 4-thiouridine (4sU), 5-etyniluridine (EU) and 5′-bromo-uridine (BrU)], followed by purification of labeled de novo RNAs. These technologies have revealed that the specific half-life of each mRNA is closely related to its physiological function. Genes with short-lived mRNAs are significantly enriched among regulatory genes, while genes with long-lived mRNAs are enriched among housekeeping genes. This review describes the recent progress of experimental procedures for measuring RNA stability.
Keywords: genome-wide technology, RNA degradation, RNA decay, RNA transcription, non-coding RNA, 4sU, BrU
Introduction
All cellular biochemical pathways rely on the regulated expression of genetic information. Hence, genome-wide gene expression profiling by measuring total transcript abundance at the whole transcriptome level is one of the most important issues in the field of molecular biology. Cellular RNA levels are determined by the interplay of tightly regulated processes for RNA transcription and degradation. Transcriptional regulation,1 as well as regulated RNA degradation,2-4 is a critical step in the control of the abundance of transcripts. It has been estimated that the mRNA abundances of 5–10% of human genes are controlled by the regulation of RNA stability.5
In mammals, the rate of RNA degradation is regulated by the interaction between cis-acting elements (specific RNA sequences) and trans-acting elements (RNA binding proteins or microRNAs).6 For example, many short-lived mRNAs that encode proto-oncogenes, nuclear transcription factors and cytokines have AU-rich elements (AREs)5,7 and GU-rich elements (GREs)8 in their 3′ untranslated regions (3′ UTRs). These sequences are targets of many ARE- or GRE-binding proteins, some of which induce degradation, whereas others promote stabilization of the mRNA. Iron responsive elements (IREs)9 are conserved stem-loops, which are bound by iron responsive proteins (IRPs), in the 5′ UTRs and 3′ UTRs of transcripts whose products are involved in iron metabolism. IRPs binding to IREs increase the stability of mRNA. MicroRNAs also regulate mRNA stability by forming imperfect hybrids with the 3′ UTR sequences of target mRNAs during cell growth and development.10,11
RNA degradation pathways also play important roles in mRNA surveillance systems to ensure the fidelity of genetic information flow.12,13 The mRNA surveillance system includes nonsense-mediated mRNA decay (NMD),14,15 nonstop-mediated mRNA decay (NSD),16-19 and no-go decay (NGD).20 The mRNA surveillance systems monitor the integrity of transcripts and sort aberrant transcripts into degradation pathways. Thus, regulation of RNA stability is an important issue in terms of regulation of transcript abundance and RNA quality control. Moreover, changes in RNA stability plays a critical role in shaping the kinetics of gene induction in intricate gene networks in mammalian cells.21-23 In particular, RNA stability significantly influences the induction kinetics of genes encoding inflammatory proteins.24,25
To unravel the underlying processes of the regulated RNA stability, determining RNA stability in genome-wide is indispensable. In this review, we will review chronologically the genome-wide technologies for investigating RNA stabilities in mammalian cells that have been conducted in the past decade and discuss the relationship between RNA stability and physiological function.
Inhibitor-Mediated Global Transcriptional Arrest
Historically, the most widely used method for genome-wide analysis of RNA degradation is based on transcriptional inhibitors such as actinomycin D (ActD), 5,6-dichloro-1–D-ribofuranosyl-benzimidazole (DRB) and α-amanitin (α-Am) (Fig. 1).2,26 ActD inhibits transcription initiation broadly by intercalating into DNA, whereas DRB and α-Am specifically inhibit RNA Polymerase II-mediated transcription. Since 2001, genome-wide RNA degradation has been assessed by blocking global transcription with transcriptional inhibitors, and subsequently monitoring ongoing RNA decay over time, using a DNA microarray.21,24,27-33
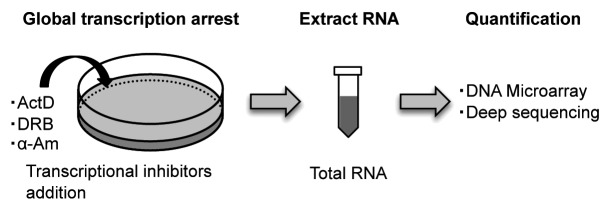
Figure 1. Schematic representation of the genome-wide method for RNA decay using transcriptional inhibitor-mediated global transcriptional arrest. Transcriptional inhibitors such as Actinomycin D (ActD), 5,6-dichloro-1–D-ribofuranosyl-benzimidazole (DRB) or α-amanitin (α-Am) are added to cells, collect the cells at sequential time points after addition of the transcriptional inhibitors, and extract total RNA (blue).
Although transcriptional inhibitors have been widely used for determining RNA stabilities, inhibitor-mediated global transcriptional arrest has a profoundly disruptive impact on cellular physiology, including splicing, polyA addition and other mRNA processing events; moreover, it interferes with the precise determination of the RNA degradation rate.34-38 For instance, some transcripts are rapidly stabilized following ActD or DRB treatment.39 Moreover, ActD alters the localizations and stabilities of a large number of long ncRNAs.34,40 Thus, global inhibition of transcription by inhibitors is not considered suitable for measuring RNA stabilities.
Pulse Labeling by 4-Thiouridine (4sU)
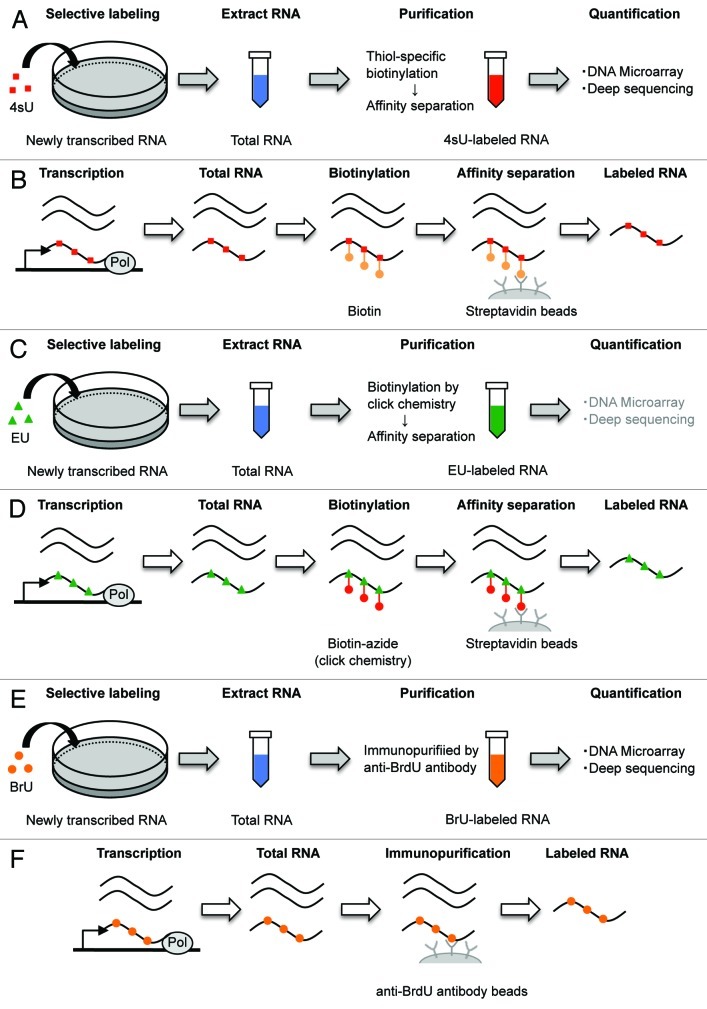
In 1978, 4sU was first used to label endogenous RNAs in mammalian cells as a non-disruptive technology for measuring RNA decay.41 First, similarly to other nucleosides, 4sU is rapidly taken up by cells. After entering a cell, 4sU is phosphorylated by cellular uridine kinases. In consequence, phosphorylated 4sU is continuously accumulated in cells over time. Additional steps, such as electroporation or lipofection, are not necessary for labeling RNA with this compound. This metabolic labeling of newly transcribed RNA with 4sU for short time has minimal adverse effects on gene expression, RNA decay, protein stability and cell viability.41-43 However, prolonged culturing with 4sU causes inhibition of cell growth.34 Although this technique includes the advantages of being able to isolate and analyze nascent RNA, the advantage was not broadly recognized during the next 30 y. In 2007, Kenzelmann et al. reported an integrated approach that combined the advantages of direct incorporation of 4sU following two hours of labeling, with the specific isolation of thiolated RNAs from total RNA using an agarose-based organomercurial affinity matrix.43 Dölken et al. reported an improved approach using thiol-specific biotinylation and affinity purification with streptavidin-coated magnetic separation of total RNA into nascent and pre-existing untagged RNA, which achieved high purity following 10 min to 60 min of labeling (Fig. 2A and B).44 Generally, 4sU (Catalog no. T4509, Sigma) and EZ-Link Biotin-HPDP (Catalog no. 21341, Pierce) are used for biotinylation of 4sU-labeled RNA, and the MACS streptavidin kit (Catalog no. 130–074–101, Miltenyi) is used for the specific isolation of biotinylated 4sU-RNA.22,44,45 These methods allow microarray analysis of all three obtained RNA subsets (newly transcribed RNA, total RNA and pre-existing RNA) in parallel. RNA half-lives can then be determined, based on both newly transcribed RNA/total RNA ratios as well as pre-existing RNA/total RNA ratios.44 Based on the newly transcribed RNA/total RNA ratios (R) and the duration of labeling (tL), precise data on mRNA half-life (t1/2) can be calculated according to the following equation:
Figure 2. Schematic representation of the genome-wide method for RNA decay using pulse labeling with uridine analogs. (A) 4sU is added to cells for a pre-defined time. Collecting the cells and extract total RNA (blue). Then, 4sU-labeled RNAs (red) are isolated by thiol-specific biotinylation and affinity separation. (B) 4sU is introduced into newly transcribed RNA in place of uridine. Total RNA extract is biotinylated by covalently linking biotin (orange) to 4sU, followed by binding streptavidin coated beads (gray). Biotinylated RNA is isolated, whereas unlabeled RNA is wash out. Then, cleaving the biotin-4sU disulfide bond releases the 4sU-labeled RNA from beads. (C) EU is added to cells for a pre-defined time. Collecting the cells and extract total RNA (blue). Then, EU-labeled RNAs (green) are isolated by biotinylation of EU in click chemistry and affinity separation. (D) EU is incorporated into newly transcribed RNA in place of uridine. Total RNA extract is biotinylated by copper-catalyzed cycloaddition reaction (click chemistry) (red) to EU, followed by isolation of EU-labeled RNA, similarly to 4sU method. (E) BrU is added to cells for a pre-defined time. Collecting the cells and extract total RNA (blue) at sequential time points after removal of surplus BrU from the culture medium. Then, BrU-labeled RNAs (orange) are isolated by immunopurification. (F) BrU is incorporated into newly transcribed RNA in place of uridine. BrU-labeled RNAs in total RNA extract are immunopurified using an anti-BrdU antibody coated beads (gray), which recognizes both BrdU and BrU.
t1/2 = tL × ln (2) / ln (1 - R)
Removal of newly transcribed RNA from total RNA offers a simple, novel approach for determining RNA decay rates without having to block transcription. Moreover, analysis of newly transcribed RNA allows the quantitative study of regulatory mechanisms governing transcription. This is of particular interest for subsequent promoter analyses. As no cellular stress response is provoked, the regulatory mechanisms that govern mRNA decay, such as the effects exerted by miRNAs, can be studied.
4sU is efficiently incorporated into RNA by broad range of cell types of human and murine origin, including fibroblasts, endothelial cells, dendritic cells, macrophages, B-cells and T-cells.44 The median mRNA half-life in murine NIH3T3 fibroblasts and human B-cells were 4.9 and 5.3 h, respectively, as estimated by the 4sU–Microarray method.38,44 Using the 4sU–seq method (massively parallel sequencing analysis of 4sU-containing mRNAs), the median mRNA half-life in NIH3T3 was estimated as 9.0 h (Table 1).45 This discrepancy may be caused by inherent limitations in indirect estimation of RNA degradation rates when comparing purified 4sU-containing mRNAs with preexisting mRNAs. Indeed, the yield of purification of 4sU-containing mRNAs would dramatically alter the estimation.22,38,45 In this regard, directly chasing decreasing levels of labeled RNA would be more suitable for determining RNA decay.
Table 1. Median mRNA half-lives in various studies.
| Reagent | Platform | Species | Cell line | Median half-life (h) | Reference |
|---|---|---|---|---|---|
| Actinomycin D |
Microarray |
Human |
Hepatocellular carcinoma cell (HepG2) /Primary fibroblast cell (Bud8) |
10.0 |
30 |
| Actinomycin D |
Microarray |
Mouse |
Embryonic stem (ES) cell (MC1/MC2-B6) |
7.1 |
24 |
| Actinomycin D |
Microarray |
Mouse |
Neuro-2a neuroblastoma cell (N2A) |
5.1 |
27 |
| Actinomycin D |
Microarray |
Mouse |
Fibroblast cell (NIH3T3) |
4.9 |
45 |
| 4-Thio-Uridine |
Microarray |
Human |
B-cell (BL41) |
5.3 |
39 |
| 4-Thio-Uridine |
RNA-seq |
Mouse |
Fibroblast cell (NIH3T3) |
9.0 |
25 |
| 5′-Bromo-Uridine | RNA-seq | Human | Cervical cancer cell (HeLa) | 3.4 | 26 |
Pulse Labeling by 5-Ethynyluridine (EU)
In 2008, EU was first used to label endogenous RNAs in mammalian cells.46 EU is efficiently incorporated into the nascent RNA in living cells, similarly to 4sU. Newly transcribed EU-labeled cellular RNAs are separated from total RNA by biotinylation of EU in a copper-catalyzed cycloaddition reaction (often referred to as click chemistry), followed by purification on streptavidin magnetic beads. The isolated RNAs are used as templates for reverse transcriptase-mediated cDNA synthesis for subsequent quantification of RNA (Fig. 2C and D).
This method is commercially available as the Click-iT Nascent RNA Capture Kit (Catalog no. C10365, Invitrogen).47 The manufacturer’s instruction of this kit recommends that the pulse labeling time is 30 min to 60 min for a 0.5mM EU dose, or 1 to 24 h for a 0.1 or 0.2 mM EU dose. This instructions state that Jurkat cells were pulsed with EU for 24 h, after which the medium was replaced with growth medium without EU. Then, total RNA was isolated and subjected to nascent RNA capture using the Click-iT Nascent RNA Capture Kit. RT-qPCR analysis was performed and the RNA stabilities of several RNAs were determined. Ideue et al. also reported that several histone mRNAs stabilities were determined by measuring the half-life of EU pulse-labeled histone mRNAs.48 HeLa cells were incubated with 0.5 mM EU for 30 min, and total RNAs were isolated from cells at sequential time points after removal of surplus EU from the culture medium. EU-labeled RNAs were biotinylated and captured using the Click-iT Nascent RNA Capture Kit, followed by RT-qPCR analysis.
EU has been shown to be non-toxic to the cells, as evidenced by propidium iodide/Annexin staining and it does not affect the global transcriptome of the cell.47 However, prolonged culture with EU causes inhibition of cell growth.34 Moreover, the EU-labeled RNA can be used for DNA microarrays or deep sequencing on a genome-wide scale; however, this method has been mainly used for RT-qPCR, so far.
Pulse-Labeling by 5′-Bromo-Uridine (BrU)
In 1959, BrU was first used to label endogenous RNAs in mammalian cells.49 During the next 50 y, BrU was widely used for immunocytochemical detection of nascent RNA in a broad range of cell types to determine the number of RNA polymerases that are active at any moment, the number of transcription sites and the number of polymerases associated with one transcription unit.50-58 An anti-bromodeoxyuridine (BrdU) antibody was used to immunoprecipitate this BrU-labeled nascent RNA because the antibody recognizes both BrdU and BrU. In 2008, Ohtsu et al. reported an integrated method that combined immunopurification of anti-BrdU antibody-coated magnetically separated BrU-RNAs from total RNA with a DNA microarray method for transcriptional profiling of nascent mRNAs.59 They applied this method to mouse FM3A cells. Moreover, Core et al. reported an improved approach that combined nuclear run-on assays (NRO) using BrU-labeling of nascent RNA during the run-on step, with deep sequencing for mapping the position, amount and orientation of transcriptionally engaged RNA polymerases on a genome-wide scale.60 They used this method, which is called global run-on sequencing (GRO-seq), on human IMR90 cells.
Recently, BrU was used for pulse labeling endogenous transcripts by adding it to cell culture media as non-disruptive technology for directly measuring RNA decay (Fig. 2E and F).34 BrU is incorporated into cells, which converted it to BrUTP, similarly to 4sU. BrUTP is recognized as the same substrate as UTP; therefore, nascent RNA is labeled by BrU. Total RNAs are isolated from cells at sequential time points after removal of surplus BrU from the culture medium, and BrU-labeled RNAs (BrU-RNAs) are recovered by immunopurification. Generally, BrU (Catalog no. 320–34741, Wako), anti-Bromodeoxyuridine (Clone. 2B1, Catalog no. MI-11–3, MBL), and Protein G Sepharose [In our experience, Protein G Immobilied Protein G (Catalog no. 20399, Thermo Scientific) is better than Protein G Sepharose 4 FF (GE Healthcare)] are used for immunopurification of BrU-labeled RNA.34 The ongoing decrease in BrU-RNA levels over time is measured directly using deep sequencing. The half-lives of RNAs are determined by calculating the time when the RNA-seq value reached half of the initial RNA-seq value. This method is called 5′-bromo-uridine immunoprecipitation chase–deep sequencing analysis (BRIC-seq). The isolated BrU-RNAs can be used as templates for reverse transcriptase–mediated cDNA synthesis for subsequent quantification of RNA, because the BrU-RNA template does not cause misincorporation by reverse transcriptase.60 By contrast, 4sU permits base-paring with guanine instead of adenine, which causes base-changes in the RNA sequence. This is a serious disadvantage for determination of RNA abundance based on RNA-sequencing. Moreover, unlike ActD treatment, BrU does not alter RNA localization of long ncRNAs.34 Thus, BrU is also a suitable uridine analog for labeling RNAs in the monitoring of RNA degradation under physiological conditions. BrU does not cause harmful effects compared with other uridine analogs, such as 4sU and EU; however, BrU-labeling takes 24 h for effective incorporation,34 as compared with 4sU-labeling, which takes about 1 h.22,44
The median mRNA and ncRNA half-lives in HeLa cells were 3.4 and 3.4 h, respectively, as estimated by BRIC-seq.34 We found that the relative distribution of ncRNA half-lives was similar to that of mRNA half-lives, suggesting that the stabilities of ncRNAs are regulated, as is the case with mRNAs.
Relationship Between RNA Stability and Physiological Function
Genome-wide technologies for investigating mRNA stability have revealed that the specific half-life of each mRNA is closely related to its physiological function.22,27,34,45 Genes with short-lived mRNAs are significantly enriched among regulatory genes, which encode proteins that are required for only a limited time in the cell, such as transcription factors, signaling genes, chromatin modifying enzymes and cell cycle regulators. Conversely, genes with long-lived mRNAs are enriched among housekeeping genes, such as those involved in translation, respiration, energy metabolism and protein degradation. Recently, two independent research groups reported that non-coding RNA (ncRNA) half-lives vary over a wide range that is comparable with that of mRNAs. ncRNAs with short half-lives included known regulatory ncRNAs, while those with long half-lives contain a significant proportion of ncRNAs involved in housekeeping functions.28,34 These results suggest that the half-lives of ncRNAs is indicative of functionality. The median mRNA half-lives in E. coli and S. cerevisiae are in the range of 5–21 min;61,62 however, the median mRNA half-lives in mouse and human cells are reported to be 5–10 h (Table 1).27-29 The half-life of the mRNA pool of a cell is roughly in proportion to the length of its cell cycle.
Concluding Remarks
Genome-wide technology for determining RNA stability has been employed to examine the response of mouse fibroblasts to type I and II interferons (IFN)44 or the response of mouse dendritic cells to lipopolysaccharide (LPS).22 Dölken et al. identified a previously undisclosed, highly connected network of short-lived transcripts that are selectively downregulated by IFN, between 30 and 60 min after IFN treatment, which showed strong associations with cell cycle and apoptosis. Rabani et al. found that changes in transcription rates determine the majority of temporal changes in RNA levels, but that changes in degradation rates are important for shaping sharp ‘peaked’ responses to LPS.22
We believe that the genome-wide technology for determining RNA stability, which is becoming more sensitive, represents an increasingly important facet in the characterization of the regulatory circuitry of biological systems in mammalian cells.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22036
References
- 1.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–45. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 2.Raghavan A, Bohjanen PR. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief Funct Genomic Proteomic. 2004;3:112–24. doi: 10.1093/bfgp/3.2.112. [DOI] [PubMed] [Google Scholar]
- 3.Friedel CC, Dölken L. Metabolic tagging and purification of nascent RNA: implications for transcriptomics. Mol Biosyst. 2009;5:1271–8. doi: 10.1039/b911233b. [DOI] [PubMed] [Google Scholar]
- 4.Keene JD. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology. 2010;151:1391–7. doi: 10.1210/en.2009-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–9. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 6.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–59. doi: 10.1038/nrg3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL. AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res. 2011;39(Database issue):D66–9. doi: 10.1093/nar/gkq990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlasova-St Louis I, Bohjanen PR. Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1. Curr Opin Genet Dev. 2011;21:444–51. doi: 10.1016/j.gde.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434:365–81. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 12.Akimitsu N. Messenger RNA surveillance systems monitoring proper translation termination. J Biochem. 2008;143:1–8. doi: 10.1093/jb/mvm204. [DOI] [PubMed] [Google Scholar]
- 13.Imamachi N, Tani H, Akimitsu N. Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay. Drug Discov Ther. 2012;6:55–61. [PubMed] [Google Scholar]
- 14.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–7. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27:543–53. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 16.Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–61. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 17.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–4. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 18.Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–95. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akimitsu N, Tanaka J, Pelletier J. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 2007;26:2327–38. doi: 10.1038/sj.emboj.7601679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–4. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elkon R, Zlotorynski E, Zeller KI, Agami R. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics. 2010;11:259. doi: 10.1186/1471-2164-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–42. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonso CR. A complex ‘mRNA degradation code’ controls gene expression during animal development. Trends Genet. 2012;28:78–88. doi: 10.1016/j.tig.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–8. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–90. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 26.Chen CY, Ezzeddine N, Shyu AB. Messenger RNA half-life measurements in mammalian cells. Methods Enzymol. 2008;448:335–57. doi: 10.1016/S0076-6879(08)02617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, et al. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–72. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2:H0041. doi: 10.1186/gb-2001-2-10-research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, et al. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–38. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, et al. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barenco M, Brewer D, Papouli E, Tomescu D, Callard R, Stark J, et al. Dissection of a complex transcriptional response using genome-wide transcriptional modelling. Mol Syst Biol. 2009;5:327. doi: 10.1038/msb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–56. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorospe M, Wang X, Holbrook NJ. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol. 1998;18:1400–7. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blattner C, Kannouche P, Litfin M, Bender K, Rahmsdorf HJ, Angulo JF, et al. UV-Induced stabilization of c-fos and other short-lived mRNAs. Mol Cell Biol. 2000;20:3616–25. doi: 10.1128/MCB.20.10.3616-3625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 38.Friedel CC, Dölken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shyu AB, Greenberg ME, Belasco JG. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 40.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melvin WT, Milne HB, Slater AA, Allen HJ, Keir HM. Incorporation of 6-thioguanosine and 4-thiouridine into RNA. Application to isolation of newly synthesised RNA by affinity chromatography. Eur J Biochem. 1978;92:373–9. doi: 10.1111/j.1432-1033.1978.tb12756.x. [DOI] [PubMed] [Google Scholar]
- 42.Woodford TA, Schlegel R, Pardee AB. Selective isolation of newly synthesized mammalian mRNA after in vivo labeling with 4-thiouridine or 6-thioguanosine. Anal Biochem. 1988;171:166–72. doi: 10.1016/0003-2697(88)90138-8. [DOI] [PubMed] [Google Scholar]
- 43.Kenzelmann M, Maertens S, Hergenhahn M, Kueffer S, Hotz-Wagenblatt A, Li L, et al. Microarray analysis of newly synthesized RNA in cells and animals. Proc Natl Acad Sci USA. 2007;104:6164–9. doi: 10.1073/pnas.0610439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–72. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 46.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA. 2008;105:15779–84. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Invitrogen. Click-iT® Nascent RNA Capture Kit. http://www.invitrogen.jp/mp/pdf/clickit_nascent/mp10365.pdf
- 48.Ideue T, Adachi S, Naganuma T, Tanigawa A, Natsume T, Hirose T. U7 small nuclear ribonucleoprotein represses histone gene transcription in cell cycle-arrested cells. Proc Natl Acad Sci USA. 2012;109:5693–8. doi: 10.1073/pnas.1200523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eidinoff ML, Cheong L, Rich MA. Incorporation of unnatural pyrimidine bases into deoxyribonucleic acid of mammalian cells. Science. 1959;129:1550–1. doi: 10.1126/science.129.3362.1550. [DOI] [PubMed] [Google Scholar]
- 50.Vanderlaan M, Thomas CB. Characterization of monoclonal antibodies to bromodeoxyuridine. Cytometry. 1985;6:501–5. doi: 10.1002/cyto.990060603. [DOI] [PubMed] [Google Scholar]
- 51.Schutte B, Reynders MM, van Assche CL, Hupperets PS, Bosman FT, Blijham GH. An improved method for the immunocytochemical detection of bromodeoxyuridine labeled nuclei using flow cytometry. Cytometry. 1987;8:372–6. doi: 10.1002/cyto.990080405. [DOI] [PubMed] [Google Scholar]
- 52.Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–93. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci. 1996;109:1427–36. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 54.Grande MA, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–91. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- 55.Haider SR, Juan G, Traganos F, Darzynkiewicz Z. Immunoseparation and immunodetection of nucleic acids labeled with halogenated nucleotides. Exp Cell Res. 1997;234:498–506. doi: 10.1006/excr.1997.3644. [DOI] [PubMed] [Google Scholar]
- 56.Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell. 1998;9:1523–36. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoshino M, Tagawa K, Okuda T, Okazawa H. General transcriptional repression by polyglutamine disease proteins is not directly linked to the presence of inclusion bodies. Biochem Biophys Res Commun. 2004;313:110–6. doi: 10.1016/j.bbrc.2003.11.086. [DOI] [PubMed] [Google Scholar]
- 58.Kageyama S, Nagata M, Aoki F. Isolation of nascent messenger RNA from mouse preimplantation embryos. Biol Reprod. 2004;71:1948–55. doi: 10.1095/biolreprod.104.031906. [DOI] [PubMed] [Google Scholar]
- 59.Ohtsu M, Kawate M, Fukuoka M, Gunji W, Hanaoka F, Utsugi T, et al. Novel DNA microarray system for analysis of nascent mRNAs. DNA Res. 2008;15:241–51. doi: 10.1093/dnares/dsn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99:9697–702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA. 2002;99:5860–5. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]