Abstract
For decades, cold-adapted, temperature-sensitive (ca/ts) strains of influenza A virus have been used as live attenuated vaccines. Due to their great public health importance it is crucial to understand the molecular mechanism(s) of cold adaptation and temperature sensitivity that are currently unknown. For instance, secondary RNA structures play important roles in influenza biology. Thus, we hypothesized that a relatively minor change in temperature (32–39°C) can lead to perturbations in influenza RNA structures and, that these structural perturbations may be different for mRNAs of the wild type (wt) and ca/ts strains. To test this hypothesis, we developed a novel in silico method that enables assessing whether two related RNA molecules would undergo (dis)similar structural perturbations upon temperature change. The proposed method allows identifying those areas within an RNA chain where dissimilarities of RNA secondary structures at two different temperatures are particularly pronounced, without knowing particular RNA shapes at either temperature. We identified such areas in the NS2, PA, PB2 and NP mRNAs. However, these areas are not identical for the wt and ca/ts mutants. Differences in temperature-induced structural changes of wt and ca/ts mRNA structures may constitute a yet unappreciated molecular mechanism of the cold adaptation/temperature sensitivity phenomena.
Keywords: influenza, RNA, structure, temperature, vaccine
Introduction
Influenza vaccines have been a great public health priority1 and their future is man-made constructs created using molecular biology tools. Compared with other types of influenza vaccines, live attenuated influenza vaccines (LAIV) possess major advantages because of administration convenience and potency of the immune response.2 There are alternative approaches which can lead to viral attenuation and be utilized for LAIV design.3
Since the late 1960s, cold-adapted temperature-sensitive (ca/ts) LAIVs have become an important vaccination instrument in the USSR. The ca/ts phenotype leads to impaired growth at an elevated temperature of approximately 39°C4-9 while permitting viral growth at lower temperatures. Molecular mechanism(s) causing the ca/ts phenotype in influenza A viruses remain unclear. Significant effort was devoted to explaining temperature sensitivity through mutations in the coding regions and amino acid changes. Jin et al. found that certain non-silent mutations in PB1, PB2 and NP might lead to temperature-sensitivity when induced in A/Ann Arbor/6/60.6 According to Song et al., three non-silent mutations in PB1 and one non-silent mutation in PB2 might lead to the ts phenotype.4 Youil et al. investigated several A/Leningrad/134/17/57 subclones and found that the most temperature-sensitive one had amino acid changes in the PB1, PA and NS1 genes.10 Furthermore, Snyder et al. found that it can be sufficient to induce the temperature-sensitive phenotype by replacing the two segments of coding for PA and M1/M2 of a wild type virus with those of A/Ann Arbor/6/60.11 Interestingly, in all these cases at least one subunit of the viral polymerase (PA, PB1 and PB2) is affected.
In addition to the attempts to explain the ca/ts phenotype through mutations in viral proteins, there were also reports implicating RNAs in temperature sensitivity. A promising finding was made by Dalton et al.,12 suggesting that, at an elevated temperature, viral polymerase tends to dissociate from the cRNA-promoter, thereby leading to a decreased vRNA synthesis while the synthesis of cRNA and mRNA remains approximately constant. A decrease in the synthesis of vRNA related to temperature sensitivity, which also maintained mRNA synthesis, was described by Chan et al.9 In a more general vein, RNAs can serve as intracellular thermometers.13 For example, a thermosensitive RNA switch was implicated in the propagation of tick-borne encephalitis virus.14 Recent publications suggest that, apart from RNA abundance, RNA structures may play a comparably important role. The importance of mRNA secondary structures for expression of influenza virus genes was recently demonstrated by Ilyinskii et al.15 Therefore, identification of previously unknown influenza RNA structures16 and the analysis of their functional roles are areas of increasing interest.17-19
We hypothesized that changing temperature causes perturbations in mRNA secondary structures, which contributes to the cold-adapted, temperature-sensitive phenotype. To test this hypothesis, we have developed a new in silico method of analysis to reveal if the structures of two closely related RNA molecules would react differently to temperature elevation. Unfortunately, it is not possible to reliably calculate exact structures of each RNA molecule at two temperatures, compare the differences between the two structures, and then evaluate whether or not these differences are identical for two RNAs. First of all, at each particular temperature an RNA molecule may have different co-existing structures. Furthermore, since the number of possible structures increases rapidly with the length of the input sequence, the precision of RNA structure predictions suffers. Another limitation of RNA secondary structure predictions is that taking pseudoknots into account makes the task non-deterministic polynomial-time hard (NP-hard).20 In this particular case NP-hard means that growth of RNA length elevates time necessary for computation to a restrictive duration. However, in support of our hypothesis, one does not need to know the exact structures before and after perturbation to conclude that the two structures have reacted differently. For example, if two windows are broken into a different number of pieces by soccer balls, we need to know neither the shapes of the windows and nor the exact forms of the pieces to conclude that the perturbations of the two glasses are not identical.
An ensemble of RNA structures can be represented via a partition function,21,22 which is a sum of Boltzmann factors over every possible secondary structure. In using partition functions, one can calculate the probability for each nucleotide to be coupled within a double-stranded conformation.23,24 An advantage of partition functions is that they take into account not just the minimum free energy structure, but rather an ensemble of energetically favorable structures. Thus, if one adenine would be bound to a particular uracil within a single highly likely structure, while another adenine would couple with ten uracils within ten less likely structures, parameters for these two adenines may be the same. Although partition functions are not precisely accurate, they are much more accurate than in silico predictions of the actual RNA structures. Partition functions were used instead of actual structures, for example, by Witwer et al.25 and Thurner et al.26 to investigate secondary structure conservation in Picornaviridae and Flaviviridae, respectively, and by Chursov et al.27 for elucidating sequence-structure relationships in yeast mRNAs. However, so far, partition functions have not been used to assess and compare structural RNA perturbations caused by temperature elevation.
Based on partition functions, we have developed a technique to identify RNA sequence regions where probabilities of nucleotide coupling change the most with temperature elevation. We demonstrate that dense areas of altered nucleotide coupling are not identical for closely related wt and ca/ts RNAs. Thus, although, we cannot predict the exact RNA structures, we know that these structures are changing differently with temperature elevation.
Results
The propensity of nucleotides to appear in double-stranded conformations depends on temperature. As seen in Figure 3, all nucleotides change their base-pairing probabilities upon temperature elevation from 32°C to 39°C, with transitions from a double-stranded to a single-stranded conformation being expectedly more frequent (see Table 2). Between 62.8% and 75.2% of positions in each mRNA change their probability to be coupled to a lower value. Furthermore, between 3.9% and 10.9% of nucleotides in each mRNA change their base-pairing probabilities significantly (more than three standard deviations below or above the mean over all seven temperature increments between 33–39°C and 32°C (see the Materials and Methods section and Table 3). In all but one mRNAs, the majority of significantly changing positions (between 52% and 88.6%) shows a decrease in their base-pairing probability, whereas this percentage is somewhat lower (42.1%) for NS2 Arb/ca.
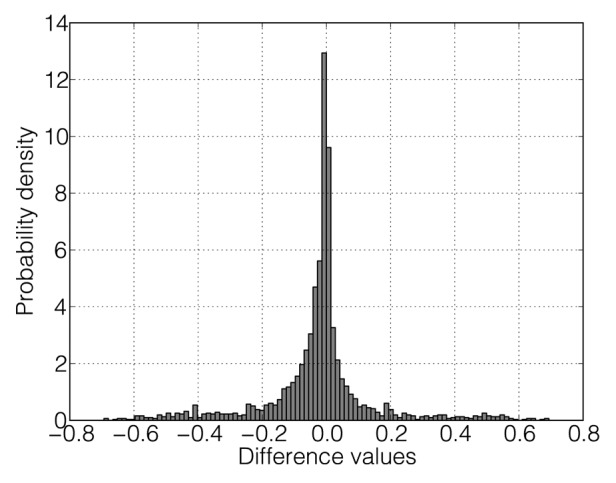
Figure 3. The histogram of differences of the probability values of nucleotides to be in a double-stranded conformation for PB1 Arb/wt upon temperature change between 32°C and 39°C. The vector of probabilities for 32°C was subtracted from the vector for 39°C.
Table 2. The number of positions in each mRNA where the probability of nucleotides to be in a double-stranded conformation decreases (increases) upon temperature elevation from 32°C to 39°C. There are no positions at which the probability to be paired upon temperature change between 32°C and 39°C remains unchanged. Here, and in all subsequent tables for those mRNAs that were not considered in the analysis due to the absence of mutations, values are not shown.
| Strain | M1 | M2 | NP | NS1 | NS2 | PA | PB1 | PB2 |
|---|---|---|---|---|---|---|---|---|
| Arb/wt |
−/− |
207/87 |
1045/452 |
445/209 |
231/135 |
1452/699 |
1433/841 |
1537/743 |
| Arb/ca |
−/− |
211/83 |
1007/490 |
440/214 |
258/108 |
1437/714 |
1510/764 |
1531/749 |
| Len/wt |
534/225 |
221/73 |
900/507 |
−/− |
233/133 |
1467/684 |
1428/846 |
1552/728 |
| Len/17/ca |
525/234 |
219/75 |
−/− |
−/− |
248/118 |
1462/689 |
1525/749 |
1578/702 |
| Len/47/ca | 525/234 | 219/75 | 899/508 | −/− | 248/118 | 1462/689 | 1510/764 | 1622/658 |
Table 3. The number of nucleotides in each mRNA where the base pairing probability decreases (increases) significantly (more than three standard deviations from the mean over all temperature differences between 33°C to 39°C compared with 32°C) upon temperature change between 32°C and 39°C compared with other nucleotides in the same mRNA.
| Strain | M1 | M2 | NP | NS1 | NS2 | PA | PB1 | PB2 |
|---|---|---|---|---|---|---|---|---|
| Arb/wt |
−/− |
19/8 |
88/36 |
42/25 |
31/4 |
58/26 |
130/95 |
133/83 |
| Arb/ca |
−/− |
13/12 |
78/34 |
29/18 |
8/11 |
102/50 |
100/57 |
132/68 |
| Len/wt |
46/17 |
17/8 |
49/34 |
−/− |
15/12 |
84/53 |
129/84 |
130/72 |
| Len/17/ca |
50/16 |
19/13 |
−/− |
−/− |
23/6 |
114/39 |
125/60 |
131/62 |
| Len/47/ca | 50/16 | 19/13 | 48/39 | −/− | 23/6 | 114/39 | 123/60 | 137/52 |
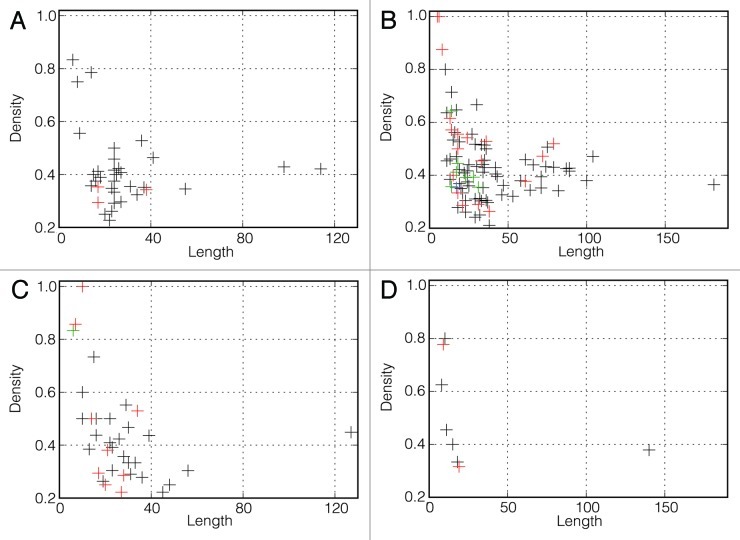
For each mRNA, we computed a density plot of significantly changing positions along the sequence as described in the Materials and Methods section (Fig. 2; Figs. S1–19). From these plots, it becomes immediately apparent that strongly temperature-sensitive positions are not evenly or randomly distributed along the sequence but rather aggregate in clusters. The numbers of clusters defined by the density-based algorithm for each mRNA are presented in Table 4. The only mRNA where no clusters were detected is NS2 Len/wt. The average length of clusters varies between 15.9 and 61.0 positions (Table 5) and the average density of significantly changing positions in the clusters is in the range of 22% to 53% (Table 6). Overall, very short clusters are required by the DBSCAN algorithm to have a very high density while the density in longer clusters can be as low as 21% (Fig. 4).
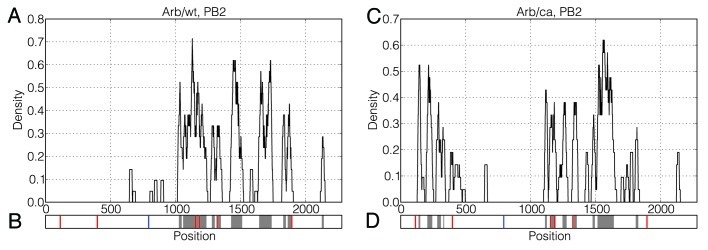
Figure 2. Distributions of significantly changing positions along the PB2 mRNAs of Arb/wt and Arb/ca. A sliding window of size 20 was moved in steps of 1 position over the vector v32–39 and the percentage of significantly changing positions in the window was calculated for each possible starting position. The resulting density plots are depicted in Figure 2A and Figure 2C. Location of clusters of significantly changing positions identified by the DBSCAN algorithm are depicted in Figure 2B and Figure 2D with gray color. Synonymous and non-synonymous mutations are depicted in Figure 2B and Figure 2D with red and blue vertical lines, respectively.
Table 4. The number of clusters in each mRNA as determined by the DBSCAN algorithm.
| Strain | M1 | M2 | NP | NS1 | NS2 | PA | PB1 | PB2 |
|---|---|---|---|---|---|---|---|---|
| Arb/wt |
- |
1 |
7 |
5 |
3 |
5 |
12 |
9 |
| Arb/ca |
- |
1 |
8 |
4 |
1 |
10 |
9 |
11 |
| Len/wt |
4 |
1 |
6 |
- |
0 |
10 |
15 |
11 |
| Len/17/ca |
5 |
1 |
- |
- |
2 |
11 |
9 |
10 |
| Len/47/ca | 5 | 1 | 8 | - | 2 | 11 | 10 | 10 |
Table 5. Average cluster length in each mRNA.
| Strain | M1 | M2 | NP | NS1 | NS2 | PA | PB1 | PB2 |
|---|---|---|---|---|---|---|---|---|
| Arb/wt |
- |
42.0 |
24.9 |
27.0 |
18.3 |
19.4 |
37.8 |
58.1 |
| Arb/ca |
- |
58.0 |
15.9 |
21.5 |
45.0 |
28.6 |
34.9 |
34.9 |
| Len/wt |
22.5 |
42.0 |
26.5 |
- |
- |
25.0 |
34.9 |
39.9 |
| Len/17/ca |
19.8 |
61.0 |
- |
- |
23.0 |
21.5 |
38.9 |
33.3 |
| Len/47/ca | 19.8 | 61.0 | 19.5 | - | 23.0 | 21.5 | 35.0 | 29.0 |
Table 6. Average density of significantly changing positions inside clusters in each mRNA.
| Strain | M1 | M2 | NP | NS1 | NS2 | PA | PB1 | PB2 |
|---|---|---|---|---|---|---|---|---|
| Arb/wt |
- |
0.43 |
0.39 |
0.38 |
0.45 |
0.37 |
0.44 |
0.39 |
| Arb/ca |
- |
0.38 |
0.53 |
0.36 |
0.22 |
0.34 |
0.43 |
0.42 |
| Len/wt |
0.41 |
0.40 |
0.36 |
- |
- |
0.38 |
0.37 |
0.41 |
| Len/17/ca |
0.46 |
0.38 |
- |
- |
0.26 |
0.48 |
0.44 |
0.41 |
| Len/47/ca | 0.46 | 0.38 | 0.46 | - | 0.26 | 0.48 | 0.44 | 0.44 |
Figure 4. Density of significantly changing positions in determined clusters vs. length. (A) Clusters that occur only in wt mRNAs but are not statistically significant (37). (B) Clusters occurring in both wt and ca/ts mRNAs (126). (C) Clusters that occur only in ca/ts mutants but are not statistically significant (45). (D) Statistically significant clusters (9 of them occur only in ca/ts mutants and 1 of them occurs only in wt mRNA). Different colors show different numbers of clusters that have identical values of length and density. Black, one cluster; red, two clusters; green, three clusters; blue, four clusters.
Furthermore, we found that patterns of cluster occurrence exhibit substantial differences between the wild type strains and their cold-adapted, temperature-sensitive mutants, as exemplified in Figure 1 for a subsequence of the PA mRNA. In this case, a cluster of significantly changing positions is observed in Len/17/ca but not in Len/wt. This figure demonstrates that a perturbation of mRNA structure begins at a temperature of approximately 37°C. Out of 218 clusters of temperature-sensitive positions, 126 clusters are present in both wt and ca/ts strains, 38 clusters are present in wt strains but absent in ca/ts mutants, and 54 clusters are present in ca/ts mutants but absent in the wt counterpart (Figure 4 and Supplementary Data).
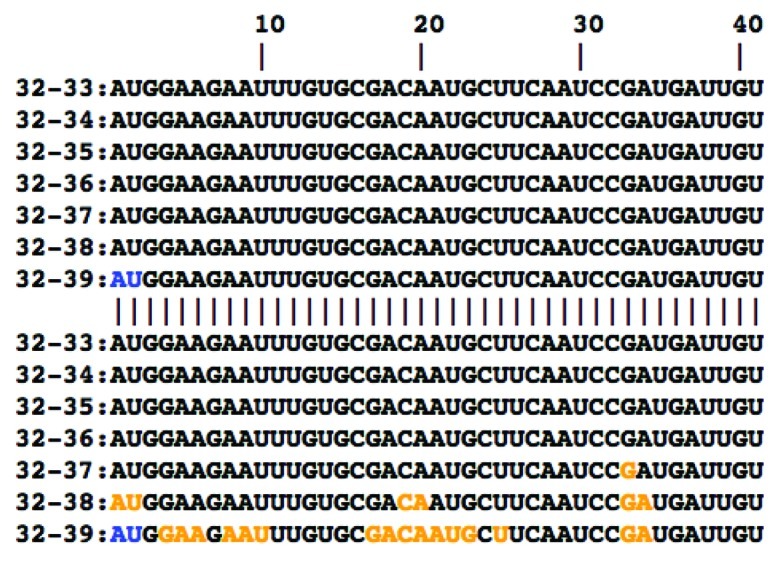
Figure 1. Comparison of significantly changing positions between the PA mRNA of Len/wt (upper 7 rows) and Len/17/ca (lower 7 rows). Each row corresponds to a difference vector v32–33, …, v32–39 containing changes of base pairing probabilities between 32°C and a particular higher temperature. Positions in which base paring probabilities significantly change with temperature elevation in both sequences and those where these changes only affect one of the phenotypes are marked blue and orange, respectively. Only the first 40 bases of each sequence are shown; position numbers of the coding sequence are indicated at the top of the alignment.
The existence of clusters unique for ca/ts strains raises the question whether such clusters are associated with the mutations inducing the ca/ts phenotype or whether random mutations unrelated to the ca/ts phenotype would be as likely to induce these clusters. Likewise, one can ask whether the disappearance of some clusters present in wt strains from ca/ts mutants may be caused by particular ca/ts associated mutations. The best way to approach this problem would be to test whether or not the same pattern of cluster occurrence would be observed while comparing the wt strains investigated here with a high number of naturally occurring influenza virus strains as similar to the wt strains as their ca/ts mutants. However, there are currently not enough naturally occurring strains with the same extent of similarity to the wt as possessed by the ca/ts mutants.
We therefore compared wt sequences with computer-generated mutants possessing random synonymous mutations unrelated to the phenotype of interest. This analysis revealed existence of only one cluster that is present in wt strain (Len/wt) but absent in ca/ts mutant and could be attributed to introducing specific mutations causing the ca/ts phenotype (Table 7). The length of this cluster is 140 nucleotides and the density of significantly changing positions in it equals 38%. At the same time, there are nine clusters (one in Arb/ca, three in Len/17/ca, and five in Len/47/ca) present in ca/ts mutants, and not present in wt, that cannot be observed in the pool of in silico generated random mutants with statistically significant P-values. The length of these ca/ts associated clusters is in the range of 8 to 19 positions and the density of significantly changing positions in them varies between 32% and 80% (Table 7). All the clusters that can be associated with ca/ts phenotype are indicated in Figure 4D. The existence of such clusters suggests that the ca/ts phenotype may be associated with specific perturbations in mRNA secondary structures. Importantly, in all three ca/ts mutants, there are clusters located in the polymerase genes (PA or PB2), in line with previous reports where polymerase genes were consistently associated with the temperature-sensitive phenotype.4,6,10,11
Table 7. Unique clusters potentially associated with the ca/ts phenotype. The P-values of all clusters in one sequence were checked against Bonferroni-corrected significance levels. For each Bonferroni correction, the total number of clusters located in the corresponding sequence was used (11 clusters in Arb/ca PB2, two clusters in Len/17/ca and Len/47/ca NS2, 11 clusters in Len/17/ca and Len/47/ca PA, ten clusters in Len/17/ca and Len/47/ca PB2, eight clusters in Len/47/ca NP, 11 clusters in Len/wt PB2).
| Strain | Sequence | Position | Occurrence in the random data seta | 95% confidence intervalb | P-value |
|---|---|---|---|---|---|
| Arb/ca |
PB2 |
329–336 |
15 |
[0.0, 0.023] |
3.34E-09 |
| Len/17/ca |
NS2 |
290–308 |
16 |
[0.0, 0.024] |
1.11E-08 |
| Len/17/ca |
PA |
93–101 |
32 |
[0.0, 0.043] |
0.0037 |
| Len/17/ca |
PB2 |
1293–1310 |
9 |
[0.0, 0.016] |
5.24E-13 |
| Len/47/ca |
NP |
1017–1026 |
29 |
[0.0, 0.039] |
0.0007 |
| Len/47/ca |
NP |
1178–1192 |
29 |
[0.0, 0.039] |
0.0007 |
| Len/47/ca |
NS2 |
290–308 |
16 |
[0.0, 0.024] |
1.11E-08 |
| Len/47/ca |
PA |
93–101 |
32 |
[0.0, 0.043] |
0.0037 |
| Len/47/ca |
PB2 |
808–818 |
3 |
[0.0, 0.008] |
1.36E-18 |
| Len/wt | PB2 | 1490–1629 | 982 | [0.973, 1.0] | 1.03E-07 |
a The number of times a particular cluster was found in a data set of 1000 sequences with randomly introduced mutations. bEstimated range of values which is likely to include the probability to find a particular cluster with the probability of 95%.
Discussion
Temperature-sensitive mutants were reported for a variety of viruses.39-42 Several studies have demonstrated that thermodynamic stability of certain RNA structures is critical for virus replication.43-45 Temperature-sensitive, anti-viral and anti-bacterial vaccines remain to be promising public health instruments.46-48 So far, cold-adapted temperature-sensitive anti-influenza vaccines have arguably made the largest contribution to the prevention of this infection around the world. Still, molecular mechanism(s) underlining the ca/ts influenza phenotype is poorly understood. Here, we have explored the hypothesis that ca/ts properties of known influenza strains can be (at least partially) explained by temperature-induced perturbations of mRNA structure.
It was, therefore, our intention to compare mRNAs at each of the temperatures of interest. However, despite the fact that significant attempts have been made toward theoretical predictions of RNA structure based on energy calculations24,34,49 and co-variation analysis,50,51 it is still not possible to calculate secondary structures of mRNAs accurately using currently available algorithms. At the same time, experimental technologies to determine RNA structures are only beginning to emerge52,53 and are barely available for a broad spectrum of research projects. Thus, we had to develop an indirect computational method aimed to assess if two RNA molecules change their shapes differently in response to temperature elevation.
At each temperature, we calculate probability vectors that contain, for each nucleotide position, the probability to be coupled with another nucleotide within the same RNA, forming a double-helix structure. Apparently, this coupling is temperature-sensitive, with increasing temperature generally leading to a reduced likelihood of “weak” structures. Thus, (1) different structures may constitute an ensemble for the same RNA at different temperatures, and/or (2) at different temperatures the same structures may be present with different abundance. Both of these options are valid and may coexist because, in each given cell, multiple copies of the same RNA molecules may be distributed between alternative shapes.
The fact that the base-paring probability at each position within a probability vector changes with temperature elevation does not necessarily indicate that structural perturbations (or re-distribution of alternative RNA structures) equally involve each nucleotide. Thus, we selected only those nucleotide positions that exhibited the most significant changes of their coupling probabilities. We do not assert that if in two closely related RNA molecules the most temperature-sensitive positions coincide; these two RNA molecules undergo identical temperature-induced structural RNA perturbations. However, it is probably safe to assume that if two RNA variants manifest different nucleotide positions as the most temperature-sensitive ones within the probability vector, temperature elevation influences the structures of these RNA molecules in a different way. Thus, we have proposed here a new technique aimed at identifying mutations that influence temperature-dependent RNA behavior. The central finding upon which our approach is based is that temperature-sensitive positions are not randomly distributed along the length of RNA but rather form distinct clusters. We speculate that such clusters of temperature-sensitive positions may be located within RNA domains that change their shapes particularly strongly with temperature elevation. Although developed for a particular purpose, our method can be applied for studying the role of RNA structure perturbations in a wide range of temperature-related biological phenomena, such as the evolution of warm-bloodedness, thermophilic adaptation of prokaryptic organisms, or susceptibility of parasites and pathogens to increases in host temperature.
Differences in clusters of temperature-sensitive positions are a potential indicator that RNA structures of mutants react differently to temperature change. This raises the question whether these differences can be a causative factor for (or, at least, associated with) the unique ca/ts behavior of the particular influenza virus strains under study. We identified three types of clusters of temperature-sensitive positions that are (1) present in both wt and ca/ts mutants, (2) present in wt, but absent in ca/ts mutants, and (3) absent in wt, but appear in the ca/ts mutants. We, therefore, first tested whether the disappearance of some clusters in the mutants can indicate that they are causative for a rare phenotype, ca/ts. If these clusters would disappear in ca/ts mutants but remain in non-ca/ts RNA variants possessing the same number of mutations, one could conclude that the cluster disappearance and ca/ts behavior are associated. For all such clusters except one, a high number of computer-generated mutants, which are extremely unlikely to be ca/ts, also demonstrate disappearance of the same clusters. Thus, these clusters may simply correspond to temperature-sensitive regions within particular influenza mRNAs unrelated to ca/ts phenotype. Nevertheless, we did observe one cluster, which is associated with the ca/ts phenotype with statistically significant P-value. This cluster is present in the wt strain. It disappears specifically in the ca/ts mutant, but remains in the computer-generated mutants possessing the same number of mutations as the ca/ts one.
Applying the same computational approach, we then tested if appearance of clusters of temperature-sensitive positions in ca/ts mutants, which are lacking in wt, is a phenotype-specific phenomenon. Based on comparisons with computer-generated mutants, we have demonstrated that nine particular clusters are unlikely to appear in mutants other than ca/ts. Thus, we hypothesize that changes in RNA structure caused by raising temperature could be a potential factor contributing to the molecular mechanisms of the temperature-sensitive and/or cold-adapted phenotype in influenza A.
Direct experimental evidence both on secondary structures of mRNAs and their interactions partners will be required to elucidate the exact role of temperature-induced structural changes in the acquisition of the ca/ts phenotype. For example, it is conceivable that conformational changes of influenza mRNA may play a role through altering the RNA ability to associate/dissociate with proteins and other molecules. Also, it cannot be ruled out that temperature-induced structural changes in the untranslated regions, which we have not considered in our analysis, contribute to the ca/ts phenotype. The current scarcity of sequence data for temperature-sensitive strains and their wild type counterparts notwithstanding, we here propose the hypothesis that temperature-induced structural RNA perturbations may be an underlying mechanism of the ca/ts behavior of influenza virus. Further research in this direction might contribute to the rational design of live-attenuated influenza vaccines.
Materials and Methods
Sequences
In our analysis, we have used the cold-adapted, temperature-sensitive mutants A/Ann Arbor/6/60 (Arb/ca) stemming from the wild type (Arb/wt) with the same name and the two mutants A/Leningrad/134/17/57 (Len/17/ca) and A/Leningrad/134/47/57 (Len/47/ca) stemming from the wild type (wt) A/Leningrad/134/57 (Len/wt). Since information on the location of UTRs was not available, only coding regions were used for the analysis. Information on the locations and sequences of coding regions was retrieved from EMBL-ENA (European Nucleotide Archive).28 However, these sequences were adapted according to the publications where they originally were reported29,30 since the mutations annotated in the database were not in agreement with those papers, and no further references were given. The files containing final sequences, used in the current analysis, are presented in the Supplementary Data.
The influenza A genome is composed of eight segments encoding 12 proteins: three polymerase subunits (PB1, PB2, and PA), a small proapoptotic mitochondrial protein (PB1-F2), hemagglutinin (HA), neuraminidase (NA), the nucleoprotein (NP), the matrix protein M1, an integral membrane protein M2, and the two nonstructural proteins NS1 and NS2.31 Recently, Wise et al. showed that PB1 gene segment also encodes a twelfth gene product, N-terminally truncated version on the polypeptide, N40.32 Sequences for NA and HA were not taken into consideration since these segments do not stem from attenuated viruses in the reassortant live vaccines, and thus cannot be associated with the temperature-sensitive phenotype. For all other genes, the numbers of single nucleotide polymorphisms (SNPs) in the coding sequences of the ca/ts mutants compared with their wild type counterparts are presented in Table 1.
Table 1. The number of SNPs in the coding sequences of the ca/ts mutants compared with their wild type counterparts. The sequences of the ca/ts mutants of the M1, M2, NS2 and PA genes in Len/17/ca and Len/47/ca are identical.
| Strain | M1 | M2 | NP | NS1 | NS2 | PA | PB1 | PB2 |
|---|---|---|---|---|---|---|---|---|
| Arb/ca |
0 |
1 |
2 |
1 |
1 |
3 |
7 |
7 |
| Len/17/ca |
1 |
2 |
0 |
0 |
1 |
3 |
3 |
1 |
| Len/47/ca | 1 | 2 | 1 | 0 | 1 | 3 | 4 | 3 |
Identification of significantly changing positions
For the first step, we wanted to identify those nucleotides within each mRNA that are the most prone to changing their coupling pattern with temperature elevation. These nucleotides would correspond to the most temperature labile positions within RNA chains. To achieve this goal, we proposed and implemented a new technique as discussed here.
At each particular temperature, an RNA sequence consisting of N nucleotides can be presented by a vector of probabilities (hereinafter referred to as “probability vector”) for each nucleotide to be in a double-stranded conformation at this temperature. Thus, we substitute a sequence of N ribonucleotides with a sequence of N real numbers between 0.0 and 1.0. Then, we calculate the probability vectors for each of the influenza mRNAs for the temperatures 32°C up to 39°C (in increments of 1°C) using the RNAfold tool from the Vienna RNA package (v.1.8.5)23,24,33-35 with the command line option –noLP that disallows base pairs that can only occur as helices of length 1. Performing the above described procedure, eight probability vectors were generated for each mRNA. Seven difference vectors v32–33, …, v32–39 were calculated from the probability vectors for 33°C to 39°C for the same RNA and the vector at 32°C, containing the set of differences between the value for each position of the probability vector at higher temperature and the value for the same position at lower temperature. These positions in difference vectors of each mRNA that possess values more than three standard deviations apart from the mean calculated over all values of the seven difference vectors were considered temperature-sensitive. Such “significantly changing” positions are presumed to result from perturbations in secondary RNA structures due to the temperature elevation. Furthermore, to filter out possible calculation artifacts, we considered a position temperature-sensitive only if it appeared at some temperature and remained to be such at all higher temperatures.
Comparison of significantly changing positions between wt and ca/ts strains
To test whether significant temperature-induced structural changes in secondary RNA structures are the same or different for wild type strains and their cold-adapted, temperature-sensitive counterparts, we designed a visualization method allowing simultaneous comparison of temperature-induced changes for two RNAs. For example, Figure 1 depicts a comparison of significantly changing positions between Len/wt and Len/17/ca for a subsequence of the PA mRNA.
Visualization of significantly changing positions demonstrated that such positions are not evenly distributed along the sequences but rather have a tendency to aggregate into clusters, i.e. regions with a high density of significantly changing positions. As a tool to analyze such clusters, we employed density plots obtained by sliding a 20-base long window over the vector v32–39 and calculating the percentage of significantly changing positions in the window for each possible starting position. For example, Figures 2A and 2C depict density plots for the PB2 mRNAs of Arb/wt and Arb/ca, respectively.
Identification of clusters of temperature-sensitive positions
We further sought to provide a definition of clusters of changing positions for each RNA, focusing on the difference vectors v32–39. To these difference vectors, we applied the density-based spatial clustering of applications with noise (DBSCAN) algorithm.36,37 This algorithm needs two parameters as input, a distance threshold r and a density threshold MinPts. For a given set of points D (in our case the set of significantly changing positions in mRNA according to the difference vector v32–39), the density of every point pi from D is calculated as the number of points qi that are within a radius r around pi. If qi > MinPts, then the point pi is classified as a core point. If the distance between two points is less than r, then they are said to be directly-connected. Two points are considered density-connected if they are connected to core points and these core points are, in turn, density-connected. A cluster is constructed as a maximally connected component of the set of points that have a distance of smaller than r to some core point. We used the implementation of DBSCAN from the scikit-learn Python module38 with a distance threshold r equal to 11 and a density threshold MinPts equals 4.
Generation of randomly mutated mRNAs
In order to assess whether the appearance of clusters of temperature-sensitive positions is specific for mutations inducing the ca/ts phenotype or whether random mutations unrelated to ca/ts phenotype would be as likely to induce these clusters, we adopted an approach similar to that employed in our previous paper.27 For each wt mRNA, a data set consisting of 1000 mutant sequences was generated in silico. Each in silico generated variant contained the same number of mutations as the respective ca/ts mutant. All computer-generated mutations were synonymous ones and introduced into the sequences randomly. It is safe to assume that none (or extremely few) of the randomly generated in silico mutants would possess the ca/ts phenotype if tested in vitro and/or in vivo. Significantly changing positions in the sequences from the artificial data sets were determined as described above and used to calculate clusters of changing positions by applying the DBSCAN algorithm. Clusters from computer-generated sequences were compared with the clusters from naturally occurring wt and ca/ts mutants.
Statistical tests
For each particular cluster of interest identified in wt and/or the ca/ts mutants, the frequency of its occurrence in the in silico generated mutants was calculated. Using these frequencies we conducted a statistical analysis to test if occurrence/disappearance of a particular cluster is associated with the ca/ts phenotype. For each cluster, which we observed in a ca/ts mutant but not in the wt, we tested the null hypothesis (H0) that the probability to observe this cluster among the computer generated sequences was 5% or higher. Conversely, for each cluster, which was observed in the wt but not in ca/ts strain, the null hypothesis (H0) was that the probability to observe this cluster was less than 95%. In other words, a low frequency means that a cluster, which we observe in naturally occurring ca/ts strain although it is absent in the wt, is unlikely to occur by chance. Thus, the appearance of this cluster is likely to be associated with the ca/ts phenotype. Similarly, the fact that a cluster was present in the wt but disappeared in the ca/ts mutant can only be explained by the ca/ts phenotype if the probability to observe this cluster in the random mutants is 95% or higher.
To that end, we used one-sided binomial tests. The significance level for the test was Bonferroni-corrected by dividing the significance level of 5% by the total number of clusters in that sequence. H0 was rejected for P-values lower than the adjusted significance level. For these calculations, a cluster was considered to be ‘present’ in an artificial sequence if that sequence contained a cluster overlapping, by at least one position, with the cluster from the real sequence.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the support of the TUM Graduate School’s Thematic Graduate Center Regulation and Evolution of Cellular Systems (RECESS) at the Technische Universität München
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The DFG International Research Training Group ‘Regulation and Evolution of Cellular Systems’ (GRK 1563) and the Russian Foundation for Basic Research (RFBR 09–04–92742).
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/cc/article/22081
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22081
References
- 1.Stöhr K, Kieny MP, Wood D. Influenza pandemic vaccines: how to ensure a low-cost, low-dose option. Nat Rev Microbiol. 2006;4:565–6. doi: 10.1038/nrmicro1482. [DOI] [PubMed] [Google Scholar]
- 2.Belsey MJ, de Lima B, Pavlou AK, Savopoulos JW. Influenza vaccines. Nat Rev Drug Discov. 2006;5:183–4. doi: 10.1038/nrd1988. [DOI] [PubMed] [Google Scholar]
- 3.Ilyinskii PO, Thoidis G, Shneider AM. Development of a vaccine against pandemic influenza viruses: current status and perspectives. Int Rev Immunol. 2008;27:392–426. doi: 10.1080/08830180802295765. [DOI] [PubMed] [Google Scholar]
- 4.Song H, Nieto GR, Perez DR. A new generation of modified live-attenuated avian influenza viruses using a two-strategy combination as potential vaccine candidates. J Virol. 2007;81:9238–48. doi: 10.1128/JVI.00893-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falcón AM, Marión RM, Zürcher T, Gómez P, Portela A, Nieto A, et al. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J Virol. 2004;78:3880–8. doi: 10.1128/JVI.78.8.3880-3888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin H, Lu B, Zhou H, Ma C, Zhao J, Yang CF, et al. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (FluMist) derived from cold-adapted A/Ann Arbor/6/60. Virology. 2003;306:18–24. doi: 10.1016/S0042-6822(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 7.Jin H, Zhou H, Lu B, Kemble G. Imparting temperature sensitivity and attenuation in ferrets to A/Puerto Rico/8/34 influenza virus by transferring the genetic signature for temperature sensitivity from cold-adapted A/Ann Arbor/6/60. J Virol. 2004;78:995–8. doi: 10.1128/JVI.78.2.995-998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsfasman TM, Markushin SG, Akopova II, Ghendon YZ. Molecular mechanisms of reversion to the ts+ (non-temperature-sensitive) phenotype of influenza A cold-adapted (ca) virus strains. J Gen Virol. 2007;88:2724–9. doi: 10.1099/vir.0.83014-0. [DOI] [PubMed] [Google Scholar]
- 9.Chan W, Zhou H, Kemble G, Jin H. The cold adapted and temperature sensitive influenza A/Ann Arbor/6/60 virus, the master donor virus for live attenuated influenza vaccines, has multiple defects in replication at the restrictive temperature. Virology. 2008;380:304–11. doi: 10.1016/j.virol.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 10.Youil R, Kiseleva I, Kwan WS, Szymkowiak C, Toner TJ, Su Q, et al. Phenotypic and genetic analyses of the heterogeneous population present in the cold-adapted master donor strain: A/Leningrad/134/17/57 (H2N2) Virus Res. 2004;102:165–76. doi: 10.1016/j.virusres.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Snyder MH, Clements ML, De Borde D, Maassab HF, Murphy BR. Attenuation of wild-type human influenza A virus by acquisition of the PA polymerase and matrix protein genes of influenza A/Ann Arbor/6/60 cold-adapted donor virus. J Clin Microbiol. 1985;22:719–25. doi: 10.1128/jcm.22.5.719-725.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalton RM, Mullin AE, Amorim MJ, Medcalf E, Tiley LS, Digard P. Temperature sensitive influenza A virus genome replication results from low thermal stability of polymerase-cRNA complexes. Virol J. 2006;3:58. doi: 10.1186/1743-422X-3-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–60. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 14.Elväng A, Melik W, Bertrand Y, Lönn M, Johansson M. Sequencing of a tick-borne encephalitis virus from Ixodes ricinus reveals a thermosensitive RNA switch significant for virus propagation in ectothermic arthropods. Vector Borne Zoonotic Dis. 2011;11:649–58. doi: 10.1089/vbz.2010.0105. [DOI] [PubMed] [Google Scholar]
- 15.Ilyinskii PO, Schmidt T, Lukashev D, Meriin AB, Thoidis G, Frishman D, et al. Importance of mRNA secondary structural elements for the expression of influenza virus genes. OMICS. 2009;13:421–30. doi: 10.1089/omi.2009.0036. [DOI] [PubMed] [Google Scholar]
- 16.Moss WN, Priore SF, Turner DH. Identification of potential conserved RNA secondary structure throughout influenza A coding regions. RNA. 2011;17:991–1011. doi: 10.1261/rna.2619511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gultyaev AP, Fouchier RA, Olsthoorn RC. Influenza virus RNA structure: unique and common features. Int Rev Immunol. 2010;29:533–56. doi: 10.3109/08830185.2010.507828. [DOI] [PubMed] [Google Scholar]
- 18.Priore SF, Moss WN, Turner DH. Influenza A virus coding regions exhibit host-specific global ordered RNA structure. PLoS ONE. 2012;7:e35989. doi: 10.1371/journal.pone.0035989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motard J, Rouxel R, Paun A, von Messling V, Bisaillon M, Perreault JP. A novel ribozyme-based prophylaxis inhibits influenza A virus replication and protects from severe disease. PLoS ONE. 2011;6:e27327. doi: 10.1371/journal.pone.0027327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyngso RB. Complexity of pseudoknot prediction in simple models. Lect Notes Comput Sci. 2004;3142:919–31. doi: 10.1007/978-3-540-27836-8_77. [DOI] [Google Scholar]
- 21.Pipas JM, McMahon JE. Method for predicting RNA secondary structure. Proc Natl Acad Sci USA. 1975;72:2017–21. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M, Sankoff D. Rna Secondary Structures and Their Prediction. Bull Math Biol. 1984;46:591–621. [Google Scholar]
- 23.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–19. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 24.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–90. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witwer C, Rauscher S, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 2001;29:5079–89. doi: 10.1093/nar/29.24.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurner C, Witwer C, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol. 2004;85:1113–24. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- 27.Chursov A, Walter MC, Schmidt T, Mironov A, Shneider A, Frishman D. Sequence-structure relationships in yeast mRNAs. Nucleic Acids Res. 2012;40:956–62. doi: 10.1093/nar/gkr790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanz C, Aldebert P, Althorpe N, Baker W, Baldwin A, Bates K, et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2005;33(Database issue):D29–33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox NJ, Kitame F, Kendal AP, Maassab HF, Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2) Virology. 1988;167:554–67. [PubMed] [Google Scholar]
- 30.Klimov AI, Cox NJ, Yotov WV, Rocha E, Alexandrova GI, Kendal AP. Sequence changes in the live attenuated, cold-adapted variants of influenza A/Leningrad/134/57 (H2N2) virus. Virology. 1992;186:795–7. doi: 10.1016/0042-6822(92)90050-Y. [DOI] [PubMed] [Google Scholar]
- 31.Nelson MI, Holmes EC. The evolution of epidemic influenza. Nat Rev Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- 32.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83:8021–31. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–48. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast Folding and Comparison of Rna Secondary Structures. Monatsh Chem. 1994;125:167–88. doi: 10.1007/BF00818163. [DOI] [Google Scholar]
- 35.Bompfünewerer AF, Backofen R, Bernhart SH, Hertel J, Hofacker IL, Stadler PF, et al. Variations on RNA folding and alignment: lessons from Benasque. J Math Biol. 2008;56:129–44. doi: 10.1007/s00285-007-0107-5. [DOI] [PubMed] [Google Scholar]
- 36.Ester M, Kriegel H-P, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the 2nd International Conference on Knowledge Discovery and Data MIning 1996:226-31. [Google Scholar]
- 37.Kriegel H-P, Kröger P, Sander J, Zimek A. Density-based clustering. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery. 2011;1:231–40. doi: 10.1002/widm.30. [DOI] [Google Scholar]
- 38.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 39.Kung YH, Huang SW, Kuo PH, Kiang D, Ho MS, Liu CC, et al. Introduction of a strong temperature-sensitive phenotype into enterovirus 71 by altering an amino acid of virus 3D polymerase. Virology. 2010;396:1–9. doi: 10.1016/j.virol.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Lulla V, Sawicki DL, Sawicki SG, Lulla A, Merits A, Ahola T. Molecular defects caused by temperature-sensitive mutations in Semliki Forest virus nsP1. J Virol. 2008;82:9236–44. doi: 10.1128/JVI.00711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y, Zhao Q, Zhao Y, Wang Q, Ning Y, Zhang Z. Complete genome sequence of attenuated low-temperature Thiverval strain of classical swine fever virus. Virus Genes. 2008;36:531–8. doi: 10.1007/s11262-008-0229-x. [DOI] [PubMed] [Google Scholar]
- 42.Sparks JS, Donaldson EF, Lu X, Baric RS, Denison MR. A novel mutation in murine hepatitis virus nsp5, the viral 3C-like proteinase, causes temperature-sensitive defects in viral growth and protein processing. J Virol. 2008;82:5999–6008. doi: 10.1128/JVI.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berkhout B, Klaver B, Das AT. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 1997;25:940–7. doi: 10.1093/nar/25.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirmomeni MH, Hughes PJ, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–70. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe A, Ferguson GL, Minor PD, Macadam AJ. Coding changes in the poliovirus protease 2A compensate for 5’NCR domain V disruptions in a cell-specific manner. Virology. 2000;269:284–93. doi: 10.1006/viro.2000.0244. [DOI] [PubMed] [Google Scholar]
- 46.Sugimoto M, Yamanouchi K. Characteristics of an attenuated vaccinia virus strain, LC16m0, and its recombinant virus vaccines. Vaccine. 1994;12:675–81. doi: 10.1016/0264-410X(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 47.White MD, Bosio CM, Duplantis BN, Nano FE. Human body temperature and new approaches to constructing temperature-sensitive bacterial vaccines. Cell Mol Life Sci. 2011;68:3019–31. doi: 10.1007/s00018-011-0734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins PL, Murphy BR. New generation live vaccines against human respiratory syncytial virus designed by reverse genetics. Proc Am Thorac Soc. 2005;2:166–73. doi: 10.1513/pats.200501-011AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–8. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–7. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr., Swanstrom R, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–6. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.