Abstract
The q arm of chromosome 1 is frequently amplified at the gene level in breast cancer. Since the significance of this is unclear we investigated whether 1q genes are overexpressed in this disease. The cDNA levels of 1q-located genes were analysed in a search for overexpressed genes. 26 genes mapping to the 1q arm show highly significant (P≤0.01) overexpression of transcripts in breast cancer compared to normal breast tissue. Amongst those showing the highest levels of overexpression in both expressed sequence tag (EST) and serial analysis of gene expression (SAGE) databases was enzyme quiescin Q6 sulfhydryl oxidase 1 (QSOX1). We investigated QSOX1 cDNA derived from T47D breast carcinoma cells by RT-PCR and 3′-RACE PCR and identified a novel extended form of QSOX1 transcript, containing a long 3′UTR, nearly double the size of the previously reported QSOX1 cDNA, and confirmed its 3′ end nucleotide sequence using RACE-PCR. We also used quantitative real-time PCR to analyse a panel of cDNAs derived from 50 clinically-graded normal and malignant breast tissue samples for the expression of QSOX1 mRNAs. QSOX1 transcription was elevated in an increasing proportion in the grade 2 and grade 3 tumours (graded according to the Nottingham prognostic index), with 10 of the 15 grade 3 tumours (67%) examined exceeding the normal range. There was a significant correlation between relative transcript level and clinical grade (P≤0.01) for all qPCR primer sets tested. QSOX1 mRNA levels, based on SAGE expression data, did not correlate with either Estrogen Receptor (ER) or Epidermal Growth Factor Receptor 2 (ErbB-2 or HER2/neu) expression. Our data indicate that QSOX1 is a potential new prognostic marker which may prove of use in the staging of breast tumours and the stratification of breast cancer patients.
Introduction
It is well established that the chromosome 1q arm is subject to amplification at the DNA level in some 50–60% of breast cancers [1]–[3]. Although this is not the only chromosomal amplification in this disease, it is one of the largest and most common, and has been interpreted as representing an early event in breast carcinogenesis [1]. Estimates of the size and number of individual amplicons involved vary, but more recent high-resolution comparative genomic hybridisation studies have indicated that although the entire q arm can be amplified, the 1q21.1–1q31.1 and 1q32.1–1q44 regions are the most frequently affected [4], [5]. In contrast to 1q, the 1p arm shows little amplification, often having a loss of copy number [1], [3]–[5]. However, the functional significance of 1q gene amplification remains unknown. As might be expected, gene copy number is associated with mRNA overexpression in breast cancer tissue, but the correlation here is less than complete. A cDNA microarray study showed that 44% of genes highly amplified are strongly overexpressed in breast cancer, but so too are 6% of genes at normal copy number [6].
Since the whole 1q arm is subject to gene amplification, we hypothesised that multiple genes mapping in this genomic region are important in breast cancer tumourigenesis and/or progression. However to our present knowledge, only two 1q genes, COX2 [7], [8] and peroxiredoxin-6/PRDX6 [9] have thus far been found to be overexpressed in breast cancer and to have key roles in metastasis and cancer cell survival. A further 1q gene, nectin-4/PVRL4, is known to be overexpressed, but the pathological significance of this is as yet unclear [10]. In order to investigate whether further 1q genes might be overexpressed in breast cancer, we analysed their mRNA levels in normal and cancerous breast tissue using serial analysis of gene expression (SAGE) and EST data collected in the Cancer Genome Anatomy Project (http://cgap.nci.nih.gov). The results of these analyses reveal that several genes show particularly high levels of overexpression including QSOX1 (NM_002826) which encodes the enzyme quiescin Q6 sulfhydryl oxidase 1. This enzyme belongs to a family of flavin adeninedinucleotide (FAD) - dependent sulfhydryl oxidases [11].
We confirmed our bioinformatics findings for QSOX1 experimentally, by conducting RT-PCR, 3′RACE PCR and quantitative real-time PCR. We identified a novel extended 3′UTR form of the QSOX1 transcript and showed that the gene is indeed overexpressed in breast cancers of poor prognosis. Our data identify QSOX1 as a gene worthy of further detailed investigation to define its relevance in the pathogenesis and progression of this disease.
Results
Overexpression and 3′ Extension of QSOX 1 Transcripts in Breast Cancer
In order to investigate whether the frequent gene amplification of the q arm of chromosome 1 might be associated with overexpression of the genes located in this region, we investigated a pair of matching SAGE libraries using the DGED tool. The SAGE data showed that 156 1q arm genes undergo transcriptional upregulation in breast ductal carcinoma compared to normal breast tissue. The upregulation of approximately one third of these genes was considered significant (P≤0.05) and 25 of these genes show highly significant upregulation (P≤0.01). The latter upregulated genes are listed in Table 1 in order of highest degree of their overexpression in breast cancer; genes with (0.01<P≤0.05) are listed in the Supplementary Table S1. The expression of four genes shown was not detectable in the library derived from normal tissue (Table 1), resulting in an ‘infinite-fold increase’ in cancer tissue. All the entries listed were also investigated for their expression in EST libraries. However, because of the limited EST data available no quantitative analysis was possible, EST data were therefore considered as only qualitative. Therefore in Table 1 EST expression results are shown solely as the presence or absence of the relevant cDNA in the two pools of cDNA libraries. Of the four highest ranked genes, which had no detectable expression in normal breast tissue in the SAGE database (with P<0.01), we chose to focus on QSOX1 which encodes quiescin Q6 sulfhydryl oxidase 1, which has not been previously associated with breast cancer. QSOX1 was found to be overexpressed in breast cancer on mRNA level in SAGE expression library, but expressed below the detection level in the matched normal SAGE library (P = 0.01) and was also detected in breast cancer tissue but not normal breast in the EST database.
Table 1. Upregulated expression of 1q genes in breast ductal carcinoma.
| Gene Symbol1 | Gene Name | Chromosome position2 | SAGE3 cancer (A) | SAGE3 normal (B) | Normalised Odds A:B4 | EST5 cancer (A) | EST6 normal (B) |
| UBAP2L | Ubiquitin associated protein 2-like | 1q21.3 | 12 | 0 | ∞ | − | − |
| QSOX1 | Quiescin Q6 sulfhydryl oxidase 1 | 1q24 | 10 | 0 | ∞ | + | − |
| PVRL4 | Poliovirus receptor-related 4 | 1q22–q23.2 | 9 | 0 | ∞ | − | − |
| ACBD3 | Acyl-Coenzyme A binding domain containing 3 | 1q42.12 | 9 | 0 | ∞ | − | − |
| GLUL | Glutamate-ammonia ligase (glutamine synthetase) | 1q31 | 29 | 1 | 16.27 | + | − |
| CCT3 | Chaperonin containing TCP1, subunit 3 (gamma) | 1q23 | 14 | 1 | 7.85 | + | − |
| SLC30A1 | Solute carrier family 30 (zinc transporter), member 1 | 1q32–q41 | 13 | 1 | 7.29 | − | − |
| UCHL5 | Ubiquitin carboxyl-terminal hydrolase L5 | 1q32 | 37 | 3 | 6.92 | − | − |
| PI4KB | Phosphatidylinositol 4-kinase, catalytic, beta | 1q21 | 11 | 1 | 6.17 | − | − |
| GPATCH4 | G patch domain containing 4 | 1q22 | 11 | 1 | 6.17 | + | − |
| TMCO1 | Transmembrane and coiled-coil domains 1 | 1q22–q25 | 16 | 2 | 4.49 | + | − |
| GUK1 | Guanylate kinase 1 | 1q32–q41 | 23 | 3 | 4.30 | + | − |
| MAPKAPK2 | Mitogen-activated protein kinase-activated protein kinase 2 | 1q32 | 28 | 5 | 3.14 | − | − |
| S100A14 | S100 calcium binding protein A14 | 1q21.3 | 89 | 16 | 3.12 | − | − |
| RPS6KC1 | Ribosomal protein S6 kinase, 52 kDa, polypeptide 1 | 1q41 | 20 | 4 | 2.81 | − | − |
| ZNF669 | Zinc finger protein 669 | 1q44 | 44 | 10 | 2.47 | − | − |
| CRABP2 | Cellular retinoic acid binding protein 2 | 1q21.3 | 28 | 7 | 2.24 | + | − |
| HAX1 | HCLS1 associated protein X-1 | 1q21.3 | 21 | 6 | 1.96 | − | − |
| PSMD4 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | 1q21.3 | 17 | 5 | 1.91 | − | − |
| ELF3 | E74-like factor 3 (ets domain transcription factor, epithelial-specific) | 1q32.2 | 10 | 3 | 1.87 | − | − |
| H3F3A | H3 histone, family 3A | 1q41 | 57 | 18 | 1.78 | − | − |
| ENSA | Endosulfine alpha | 1q21.3 | 22 | 7 | 1.76 | + | − |
| LMNA | Lamin A/C | 1q21.2–q21.3 | 132 | 43 | 1.72 | + | − |
| TAGLN2 | Transgelin 2 | 1q21–q25 | 64 | 21 | 1.71 | + | − |
| F11R | F11 receptor) | 1q21.2–q21.3 | 142 | 51 | 1.56 | + | + |
The genes are listed in the order of degree of overexpression in cancer tissue. For all genes listed, the significance factor is P≤0.01.
From Unigene (http://www.ncbi.nlm.nih.gov/UniGene).
Total number of short SAGE tags identified for each individual gene.
The sequences Odds ratio is obtained by calculating normalised values of expression of each gene (total number of SAGE tags divided by the total number of tags in each library: 66,128 tags in the breast ductal carcinoma library and 50,512 tags in normal epithelium), and then calculating the ratio of these values (normalised expression in cancer over normalised expression in normal tissue). For the four top entries gene expression was not detected in normal tissues (SAGE only).
Based on ten non-normalised cDNA EST libraries from cancer breast tissues (not cell lines), totalling 11,161 sequences available.
Based on two non-normalised cDNA EST libraries from normal breast tissues (not cell lines), totalling 1,485 sequences available.
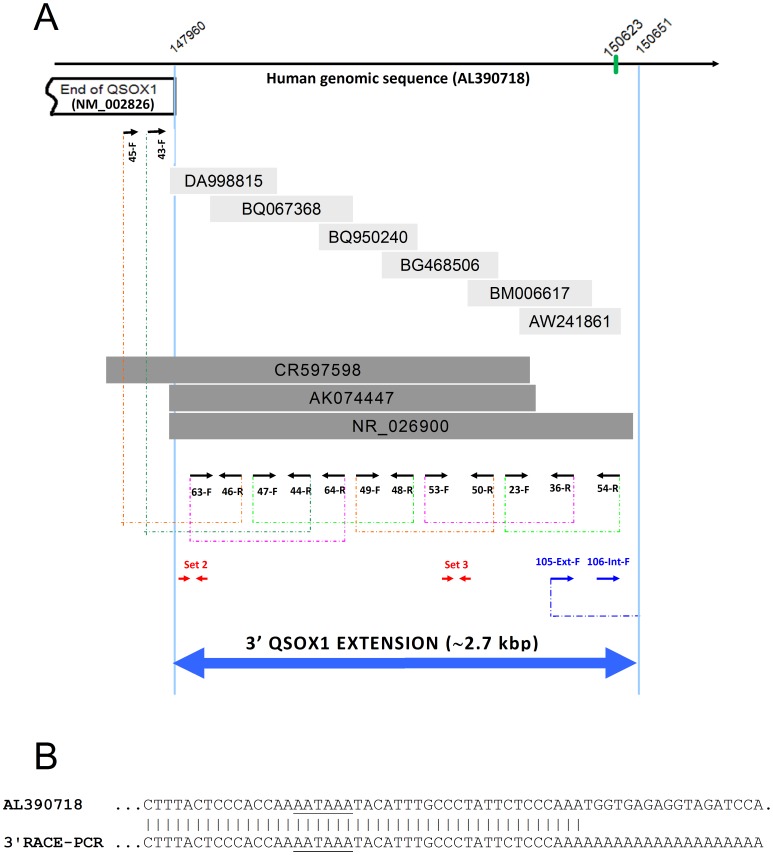
Whilst analysing the extent of EST coverage of QSOX1 cDNA we identified an EST fragment (DA998815) overlapping with the most 3′ end of the reported QSOX1 cDNA (NM_002826) by 14 nucleotides and extending for more than 500 bases beyond the established 3′ end of NM_002826. We therefore used a fragment of genomic sequence immediately beyond the 3′ end of QSOX1 to search human sequence databases for other possible matches. We identified a region of approximately 7,000 bp downstream of QSOX1 containing many ESTs and a few longer cDNA sequences of which some provided a continuous overlapping region of over 2.5 kbp long. A selection of these is shown in Figure 1A, as a series of overlapping sequences indicating a potentially substantial 3′ extension of the QSOX1 cDNA. To prove the existence of the predicted extended version of QSOX1 cDNA experimentally we designed a set of PCR primers (detailed in the Supplementary Table S2) with the aim of obtaining a series of overlapping RT-PCR products at the 3′ end of the QSOX1 gene. Using cDNA from breast cancer derived T47D carcinoma cells we were able to amplify cDNA fragments and to achieve continuous sequence coverage starting from the position 147692 (middle of exon 12 of QSOX1) to position 150620 of the genomic DNA (AL390718) (Figure 1A). Individual amplified cDNA fragments are shown in Figure 2, panels A–H. All of the fragments were of the expected length and all were confirmed by sequencing.
Figure 1. 3′ Extension of QSOX1 cDNA.
Panel A: Overlapping database sequences are aligned with the fragment of human genomic sequence from chromosome 1 (AL390718). The 3′ end of QSOX1 exon 12 (NM_002826) is shown as an open box. Light grey shaded boxes show selected overlapping ESTs (accession numbers are indicated). Dark grey boxes denote the overlapping cDNAs identified in GenBank (accession numbers are indicated). Black arrows indicate the approximate positions and orientation of the PCR primers used to check the expression of the extended QSOX1 transcript. Primer names are shown next to their positions. Dashed lines show the individual overlapping PCR products obtained, which continuously cover the QSOX1 3′ extension. Blue arrows indicate the approximate positions of the sequence-specific RACE-PCR primers used (see Supplementary Table S2 for all the primers’ sequences). The polyadenylation site (AATAAA) was found approximately 30 bases upstream of the poly(A) tail (positions 150623 and 150651 respectively). All positions are numbered relative to the human genomic sequence (AL390718). Red arrow sets indicate the approximate positions of real time qPCR primers sets 2 and 3. The blue double-headed arrow indicates the experimentally confirmed 3′ extension of QSOX1 cDNA. Panel B: Alignment of the experimentally identified 3′ end of the QSOX1 cDNA with the human genomic sequence (AL390718). The polyadenylation signal (AATAAA) is underlined in both sequences.
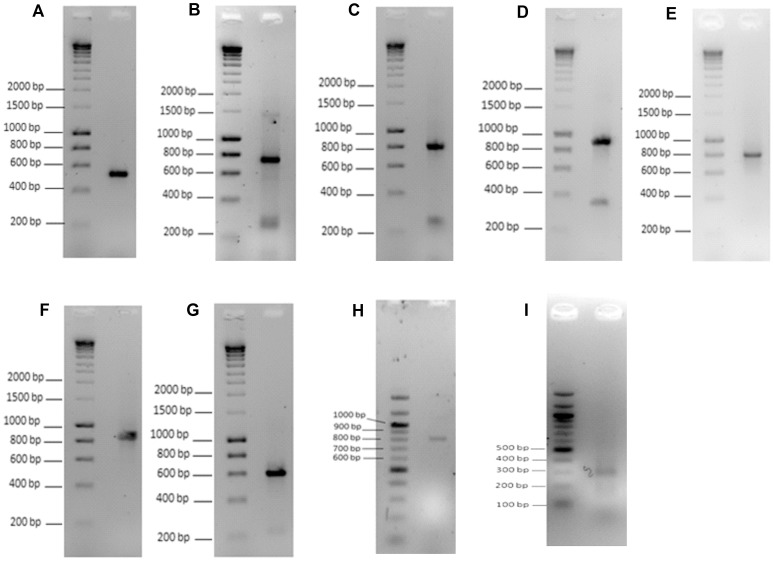
Figure 2. RT-PCR and RACE-PCR amplification of the extended form of the QSOX1 cDNA.
Panel A shows amplified cDNA using primers 45-F and 46-R (expected length 550 bp). Panel B shows amplified cDNA using primers 43-F and 44-R (expected length 735 bp). Other PCR amplifications were using primers: 63-F with 64-R (panel C, expected size 836 bp), 47-F with 48-R (panel D, expected size 922 bp), 49-F with 50-R (panel E, 825 bp), 53-F with 36-R (panel F, 883 bp), 23-F with 54-R (panel G, 620 bp). Panel H shows amplified cDNA of the soluble version of QSOX1 (QSOX1b) using primers 17-F and 18-R (expected length 814 bp). Panel I shows the size of the cDNA fragment amplified with the specific sense primer (106-Int-F) and antisense 3′ RACE adapter primer (Adap-1-R). The specificity of amplification (the identity of all amplified PCR products) was further confirmed by DNA sequencing for all amplified products.
In order to identify the actual 3′ end of the extended QSOX1 cDNA, we carried out 3′-RACE-PCR using a set of nested specific primers and a set of 3′-RACE adapter primers (see Supplementary Table S2 for sequences). The amplified cDNA was detected as a ∼300 bp band, see Figure 2, panel I, and the 3′-end of the QSOX1 cDNA was confirmed by sequencing. The end of the extended QSOX1 cDNA corresponds to position 150651 of the genomic sequence (AL390718), see Figure 1B. We also identified a typical polyadenylation signal AATAAA positioned approximately thirty nucleotides upstream of the poly-A sequence in the cDNA (Figure 1B). The combination of the overlapping RT-PCRs, of the 3′-RACE-PCR and the polyadenylation signal confirmed unequivocally the position of the true 3′ end of the QSOX1 cDNA. Since the translation of QSOX1 stops inside exon12 which contains a stop codon, the newly identified extension is therefore a long non-coding 3′UTR.
Quantitative Real-time PCR of QSOX1 Transcripts in Breast Cancer
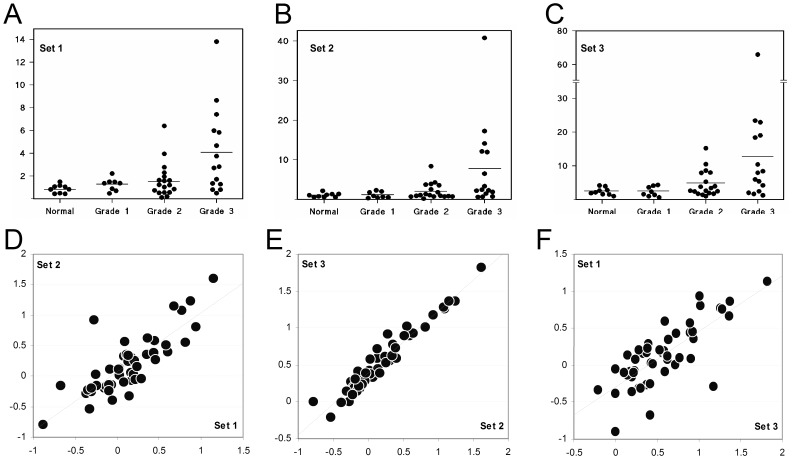
We further sought to confirm experimentally both the existence of this new 3′ QSOX1 mRNA extension and the upregulation of QSOX1 mRNA levels in breast cancer tissue. Hence real-time PCR was conducted using sets of probes based on the QSOX1 coding sequence (CDS) (Set 1) and on the newly identified 3′UTR extension (Sets 2 and 3, see Figure 1A). A panel of cDNA samples prepared from surgically removed breast tissue was screened. All tissues tested revealed the transcription of all the three QSOX1 regions tested, confirming that the QSOX1 mRNA exists as a ∼6 kbp transcript, much longer than previously thought. The expression of all of the three regions was low in normal tissues and in grade 1 tumours, but showed elevated levels in an increasing proportion of the grade 2 and grade 3 tumours, Figure 3A–C. Non-parametric statistical analysis across all four clinical classifications showed a Spearman’s correlation coefficient of 0.53 between relative transcript level and clinical grade, a value significant at the p 0.01 level. Of the 15 grade 3 tumours examined, expression in 10 samples exceeded the upper limit of normal (mean +2 standard deviations) for each primer set.
Figure 3. Quantitative Real-Time PCR of QSOX1 transcripts in a panel of RNA samples from normal and breast cancer tissue samples, clinically graded using the Nottingham prognostic index.
Data for three different TaqMan probe sets are shown (panels A-C). In all panels the vertical axis is relevant abundance normalised to 18s rRNA levels. A: Primer Set 1 (based on the QSOX1 CDS). B: Primer Set 2 (based on the QSOX1 3′UTR). C: Primer Set 3 (based on the QSOX1 3′UTR). The horizontal line in each dot plots shows mean expression data for each probe set/condition D: Scatter plot for Set 2 vs. Set 1 data across all preparations. E: Scatter plot for Set 3 vs. Set 2 data across all preparations. F: Scatter plot for Set 1 vs. Set 3 data across all preparations. For all scatter plots, all axes show relevant abundance values for the relevant probe set in logarithmic scale. Best fit linear regression is shown as a dotted diagonal line.
Of the 41 patients whose cDNA preparations were subject to real time PCR analysis, 25 had tumours classified as ductal (mean relative transcript level, 3.21) and 9 were lobular (mean value, 1.346). The remaining 7 patients were distributed between the mixed (4), mucinous (2) and cribriform (1) pathological classifications. For the results obtained with the Set 1, shown in Fig. 2A, a Two-tailed t tests established a significance difference between the normal mean and the ductal tumour mean (p = 0.00148), and between the ductal and lobular mean values (p = 0.120). There was however no significant differences in the mean values for normal tissue and the lobular tumours (p = 0.112).
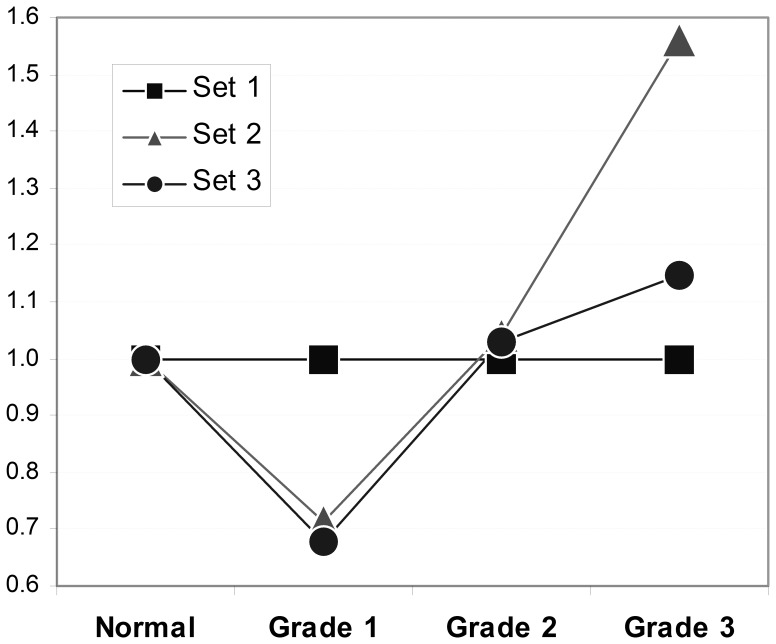
Correlation of the expression levels of individual regions (probe sets 1, 2 and 3) in individual tissue samples, showed some differences in degree of scattering between signals measured with the Set 1 probe (QSOX1 CDS) and either of the Set 2 or 3 probes, both of which are within the newly identified extended QSOX1 3′UTR, (see Figure 3D–F). This raise further the possibility that alternative splicing events might occur to account for a degree of variability in the expression of distinct regions of the transcript. When normalised against expression in normal breast tissue the data revealed that the QSOX1 CDS expresses at relatively higher levels in Grade 1 cancers, compared to the expression level of the QSOX1 long 3′UTR (both Set 2 and Set 3 probes), but the trend is reversed in Grade 2 and 3 cancers (Figure 4).
Figure 4. Differential overexpression of the CDS and 3′UTR of QSOX1.
Mean expression data (as in Figure 2A–C) for the three different TaqMan probe sets normalised per mean expression data for probe Set 1 are shown. Sets 2 and 3 (3′UTR region of QSOX1) show a lower degree of expression in Grade 1 tumours and a higher degree of expression in Grade 3 tumours relative to Set 1 (QSOX1 CDS).
We attempted to identify experimentally any alternatively spliced QSOX1 transcripts which could have contributed to the above observations by PCR amplification of cDNA derived from T47D carcinoma cells using primer pairs designed around all known and predicted splice variants. We did not detect any further alternatively spliced variants beyond the previously reported QSOX1a and QSOX1b forms (the latter encoding the soluble form of the protein [12] and the extended 3′-UTR reported above.
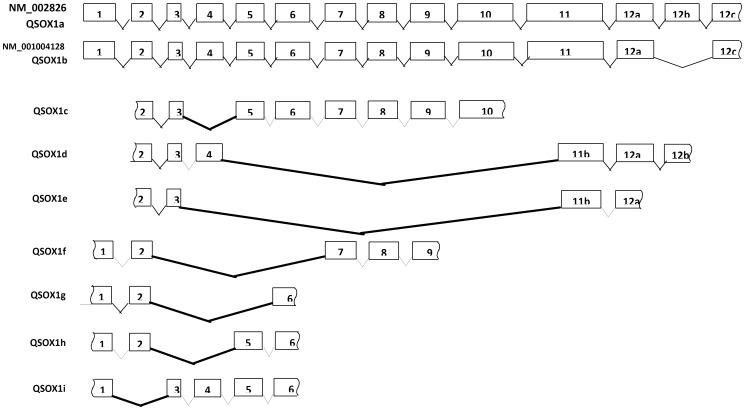
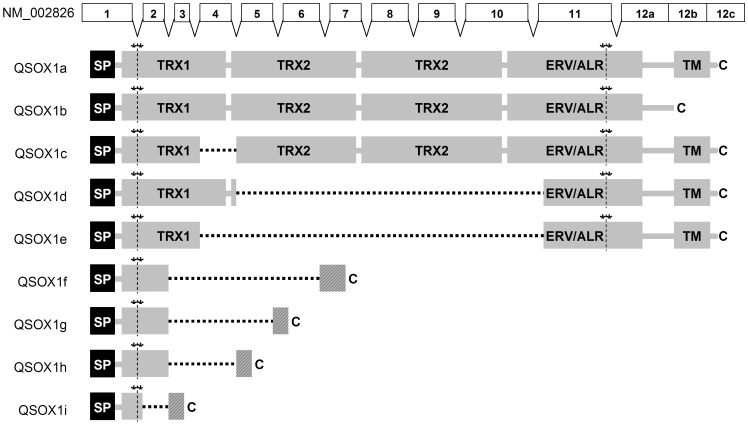
We therefore searched EST databases for any evidence of additional alternative splicing. We identified eight ESTs showing novel putative alternative splicing patterns for the QSOX1 mRNA. These are summarised in Figure 5 and Table 2. Since these sequences show alternative splicing, all of which occurs at the existing intron-exon boundaries, and they do not have any unspliced intronic sequences, it is likely that these may indeed represent alternative splicing rather than random cloning artefacts, despite them being undetectable in T47D carcinoma cells. The two putative alternative splicing variants which could have contributed to the differences in expression level between exon 7 and the 3′UTR would be QSOX1d and QSOX1e (Figure 5). It is interesting that in the protein products of both of these splice variants all three CxxC disulfide motifs present in the full length QSOX1 and essential for the activity of the protein [12] remain intact. In QSOX1d the Thioredoxin TRX1 domain remains intact, whilst the TRX2 and HRR domains are removed completely, making these isoforms similar to plant QSOXs which lack the second Trx domain but have the same three redox sites. TRX1 is truncated at its C terminus in QSOX1e and both of the splicing isoforms encode slightly N-terminally shortened ERV/ALR domains. Both splicing variants preserve the QSOX1 reading frame. Protein domain structures for the products of these and the other putative QSOX1 alternative splice variants are summarised in Figure 6.
Figure 5. Alternative splicing of the QSOX1 mRNA.
QSOX1 exons are shown. Alternatively spliced variants QSOX1a and QSOX1b are based on NM_002826 and NM_001004128 respectively. Other alternative splicing variants are based on the EST mapping data (see Table 2) and are named QSOX1c – QSOX1i for consistency with the previously used nomenclature. The identified splicing sites are shown with bold connecting lines. Exon 12b denotes the sequence fragment missing in the shorter alternatively spliced version of QSOX1b.
Table 2. Putative new QSOX1 splicing variants and their corresponding ESTs.
Figure 6. QSOX1 internal splicing variants and their translation.
QSOX1 protein domains Trx1, Trx2, HRR and ARV/ALR are shown as light grey shaded boxes. Asterisks and vertical yellow lines indicate the position of cysteine-pairs in the product of the QSOX1 transcript. SP (black boxes) denotes a signal peptide, TM indicates a transmembrane domain and C indicates a C terminus. Square dotted horizontal lines indicate the joining of truncated domains. Hatched boxes show amino acid sequences produced where such joining results in reading frame-shifts.
Discussion
Our bioinformatics analyses clearly indicate that a number of the genes located in sub-regions of the q arm of chromosome 1 which are commonly amplified at the DNA level in breast cancer [1]–[5] are also highly significantly upregulated at the mRNA level compared to normal breast tissue (Table 1). For example the PVRL-4 gene product, Nectin-4 was found to be undetectable in normal breast epithelium, but expressed in some 60% of ductal breast carcinomas, correlating strongly with basal-like markers [10]. Another gene, GLUL encodes the enzyme glutamine synthetase which is highly expressed both in normal breast luminal epithelial cells and in luminal breast cancer [13]. Both mitogen-activated protein kinase-activated protein kinase 2 protein levels and enzyme activity are elevated in breast cancer [14]. S100A14 overexpression is also well established in breast cancer. Thus Leth-Larsen et al. [15] derived a single cell clone of a human primary ductal breast carcinoma cell line HMT309 with an epithelial-line cancer stem phenotype and established that it expressed higher levels of S100A14 protein than the parental cell line. Elevated levels of the ELF3 protein have been detected in some but not all breast cancer cell lines [16]. Transgelin 2 was amongst a number of proteins found by LC-MS analysis to be overexpressed in microvessels isolated from invasive ductal breast carcinoma compared to those from adjacent non-malignant tissue [17].
Of particular interest was the identification among the found genes, of QSOX1, which encodes quiescin Q6 sulfhydryl oxidase 1, because no information existed at the time of its association with breast cancer. We confirmed our findings independently using bioinformatics approach and experimentally by RT-PCR, 3′RACE PCR and quantitative real-time PCR. We demonstrate here that the transcript for QSOX1 is highly overexpressed in some breast cancers, with a strong correlation between expression level and prognostic index. Indeed, out of 15 cases with the worst prognostic index, 10 (67%) showed expression levels above the normal range. This high rate of overexpression is at least comparable with that of established prognostic markers such as ESR1 or HER2 [18]. However, the expression pattern of QSOX1 correlated with neither ESR1 nor HER2 (Supplementary Figure S1), indicating that quiescin Q6 might be a new prognostic factor suitable for cancer staging and for the further stratification of patients with breast cancer. In this respect overexpression of QSOX1 may have good prognostic potential and would be technically more straightforward to test than, for example, the reduction in the expression level of stromal caveolin Cav-1, reported recently to be indicative of advanced breast cancer, metastases, early disease recurrence and poor outcome [19], [20]. Our results show that the QSOX1 transcript was significantly elevated in ductal tumours, but since this classification also included all tumours of advanced grade, the patient sample set we studied does not enable us to disentangle the effects of tumour classification versus tumour grade.
Overexpression of QSOX1 in breast cancers has not previously been reported, however in a recent proteomics study, quiescin Q6-derived peptides were found to be secreted at elevated levels by two breast cancer derived cell lines, BT474 and MDA-MB-468, compared to the normal breast epithelial cell line, MCF-10A [21]. Transcription of QSOX1 in oestrogen receptor-expressing breast cancer cell lines has also been documented in two further studies, although in one of these expression was found to be repressed by 17β-oestradiol [22] whereas in the other such treatment resulted in a modest induction [23]. In other cancers, peptides derived from a C-terminal part of the secreted form of QSOX1 have been identified by liquid-chromatography-tandem mass spectrometry (LC-MS/MS) in the plasma of patients with ductal adenocarcinoma of the pancreas, but not in normal healthy donors, leading to the suggestion of QSOX1 being a biomarker of cancer of the pancreas [24]. Moreover in a rodent model of prostate cancer, QSOX1 is highly overexpressed in prostatic hyperplasia and intraepithelial neoplasia (PIN) lesions of mice lacking the transcriptional regulator and tumour suppressor gene Nkx3.1 [25]. The overexpression of QSOX1 in different types of tumours, and the ability to detect the protein or its fragments in body fluids including plasma [24], and urine [26], [27] makes QSOX1 a promising target for studying as a candidate universal prognostic cancer marker gene.
Previously, only two alternative splicing-derived variants of the enzyme had been reported in human, mouse and rat tissues (NM_002826 and NM_001004128 human sequences encoding short and long mRNAs respectively). The latter encodes a full length membrane anchored protein, found in the Golgi, secretory granules, and in the endoplasmic reticulum [28], [29]. The shorter version of the QSOX1 mRNA is produced by an alternative splicing event, whereby a 733 base long fragment is deleted from the middle of exon 12 resulting in a truncated QSOX1 protein missing the transmembrane domain, and this encodes the secreted isoform [12], [30]. The extended 3′UTR reported here does not seem to encode an alternative protein sequence. Most likely it is involved in the post-transcriptional regulation of QSOX1 mRNA translation and mRNA nuclear export, cleavage and polyadenylation. 3′UTRs are considered vital for transcript stability and gene expression regulations such as translational repression mediated by RNA-binding proteins and micro RNAs [31]–[33]. The QSOX1 3′UTR extension reported here could be involved in the regulation of QSOX1 transcript stability and may explain the observed variability in the apparent levels of QSOX1 CDS vs. 3′UTR regions.
Our systematic analysis of EST databases revealed eight putative new QSOX1 splice variants. Of these three sequence variants, QSOX1c, d and e, have an internal in-frame deletion which preserves fully or partially the primary structure of the two main known functional domains of the protein: the PDI-like oxidoreductase thioredoxin domain Trx1 and the FAD - dependent sulfhydryl oxidase domain ERV/ALR [34]–[37]. The thioredoxin domain, Trx2, and the helix-rich-helix region HRR are deleted from all but the QSOX1c variant. The retaining of the main functional domains of the protein and all of the redox-active disulfides in the isoforms QSOX1c,d,e may preserve the main function of the enzyme - the oxidation of sulfhydryl groups to disulfides by reducing oxygen to hydrogen peroxide. This assumption is in agreement with the recently published functional studies of QSOX1 fragments which indicated the importance of interaction of the Trx and ERV/ALR domains for the thiol-oxidation activity of the QSOX1 protein [37], [38]. The products of the remaining splice variants (QSOX1f-i) lack all but the N-terminal fragment of the Trx1 domain and are most likely dysfunctional proteins.
This enzyme belongs to a family of flavin adeninedinucleotide (FAD) - dependent sulfhydryl oxidases [11]. It is a multi-domain protein formed by fusion of two ancient genes. The N-terminal region has a tandem pair of thioredoxin (TRX) domains, related to protein disulfide isomerase (PDI), and there is a C-terminal sulfhydryl oxidase–like endogenous retroviral element (ERV1). It also has a helix-rich-helix region (HRR) domain and a transmembrane domain. This enzyme catalyses the oxidation of protein thiol groups to disulphides with attendant reduction of oxygen to hydrogen peroxide [12], but its cellular role is currently unclear. Although initially identified as being strongly up-regulated in fibroblasts reaching confluence, it has also been since associated with growth factor activity, and it is highly expressed in cells with a large secretory load [12]. In different studies on QSOX1 a variety of functions have been ascribed to this enzyme including protection from apoptosis [11], [12], [39]–[42] and facilitation of tumour cells invasion [38]. Thus, the forced overexpression of quiescin 6 in human MCF-7 breast cancer cells rendered them more resistant to apoptosis arising from oxidative stress compared to control transfected cells, thereby implicating QSOX1 in cell survival [11] whilst the suppressed expression of quiescin in pancreatic cancer cell lines BxPC-3 and Panc-1 inhibited cancer cell invasion by activation of MMP-2 and MMP-9 Matrix Metalloproteinases [43]. The consequences of elevated levels of quiescin Q6 mRNA and its splicing variants in breast cancer are currently far from certain.
Quiescin Q6 is found both within the endoplasmic reticulum/Golgi apparatus and as a secreted protein [12]. This dual location, together with its activity towards unfolded polypeptides implies functions both in the initial folding of secreted proteins and their remodelling once they have reached the cell surface and extracellular matrix. The latter activity might be related to cancer cell signalling, migration and metastasis. In this context it is pertinent that in a recent genetic screen of cDNAs searching for transcripts able to promote the metastasis of the breast cancer cell line 168FARN, the thiol isomerase ERp5 was identified as promoting tumour cell migration and invasion, and as being up-regulated in invasive clinical breast cancer samples [44]. Taken together with our findings, it is emerging that the cellular mechanisms for the formation of appropriate disulphide linkages in secreted proteins is a fertile area of investigation in breast cancer of poor prognosis.
Methods
SAGE and EST Expression Data Analysis
cDNA and SAGE databases accessible on-line through the Cancer Genome Anatomy Project (CGAP) portal (http://cgap.nci.nih.gov) were used for the initial analysis of the expression of chromosome 1 genes in normal and cancerous breast tissues. The SAGE experimental viewer (SEV) and digital gene expression displayer (DGED) (http://cgap.nci.nih.gov/Tissues/SAGE) were used to identify genes potentially overexpressed in breast cancer. We used a matched pair of libraries derived from breast ductal carcinoma (BEREP4+_AP_DCIS_2, containing 66128 short SAGE tags) and normal epithelium (BEREP4+_AP_N2, containing 50512 short SAGE tags), both libraries were derived from tissues obtained from the same patient. Both under- and over- expressed genes were selected in SAGE DGED (F = 1) and no limitations were put on the values of the significance factor (P = 1). DGED analysis yielded 1915 SAGE tags (data not shown), Because some of the genes were represented by more than one short SAGE sequence tag, all such entries were combined. Unique gene entries having the significance factor P<0.2 were then annotated to their respective chromosome arms and cytogenetic bands and those genes located on the “q” arm of chromosome 1 were selected for analysis.
To confirm the results of the SAGE DGED search independently, we used expressed sequence tag (EST) expression data and the DGED tool (http://cgap.nci.nih.gov/Tissues/GXS). The following criteria were used to select libraries. For pool A (“cancer”): tissue selection - mammary gland/breast; minimum number of sequences per library - 10; library protocol - non-normalised, tissue histology - cancer. The same settings were used for pool B (“normal”), except for the tissue histology, which was set to normal. For the both pools, only human EST libraries were selected, all types were allowed (CGAP, MGC and ORESTES); in neither case were libraries from cell lines used. These settings yielded 10 cancer libraries (NCI_CGAP_Br1.1, NCI_CGAP_Br12, NCI_CGAP_Br22, NCI_CGAP_Br16, NCI_CGAP_Br18, NCI_CGAP_Br15, NCI_CGAP_Br3, NCI_CGAP_Br13, NCI_CGAP_Br17 and NIH_MGC_151) and two normal libraries (NCI_CGAP_Br14 and NCI_CGAP_Br7). These libraries contained 7985 “cancer” and “942” normal EST sequences.
EST Mapping and QSOX1 Gene Extension
To find out the extent of EST coverage of the of QSOX1 mRNA, the long form of the reported QSOX1 sequence (NM_002826) was searched against human EST (Expressed sequence tags) databases using BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Following the identification of an EST fragment (DA998815) having a short 14 nucleotides overlap with the very 3′ end of the QSOX1 cDNA and extending for over 500 bases downstream, another search for any ESTs downstream of the QSOX1 gene was conducted using a fragment of human genomic DNA (AL390718, positions 147940 to 154450). A similar search was then performed using the non-redundant DNA database “nr/nt” to identify any cDNAs covering the region downstream of the reported exon 12 of the QSOX1 gene. To identify putative alternatively spliced variants of QSOX1, its cDNA sequence (NM_002826) was searched against EST databases. Default BLAST search settings were used in all cases except the word size was set to 16, and no sequence filters were applied.
To identify possible correlations between QSOX1 expression and the levels of two established breast cancer prognostic markers ESR1 or HER2, 67 breast cancer SAGE expression libraries were analysed (last accessed 30/01/2011). Tag counts for short SAGE tag “AGCAGGTGCC” were used as a measure of ESR1 expression, counts for short SAGE tag “AGGAAGGAAC” were used as a measure of HER2 expression, and the counts for short SAGE tag “CTTGATTCCC” were used as a measure of QSOX1 expression. The total number of tags were 63 (ESR1), 574 (HER2) and 355 for QSOX1 across all 67 breast SAGE libraries (including 18 normal tissue derived, 10 cell line derived and 39 cancer tissue derived libraries).
cDNA Preparation
T47D breast carcinoma cells were from European Collection of Cell Cultures (ECACC), Health Protection Agency Porton Down, Salisbury, SP4 0JG, UK. Total RNA was purified from cultured T47D cells using an Aurum Total RNA purification kit (Bio-Rad) and following the manufacturer’s protocol. Approximately 2x106 cells were used and the RNA was finally eluted in 80 µl of elution buffer. cDNA was synthesized from 3 µl of the total RNA preparation, 100 pmol of Oligo-d(T)21 primer and using either iScript (Biorad) or BioScript (Bioline) kits and following the manufacturers’ recommendations. The synthesized cDNAs were diluted 10-fold with deionised water and stored at −20°C. Both cDNAs were tested by RT-PCR amplification of a short fragment of GAPDH cDNA using forward and reverse GAPDH gene specific primers, as detailed in Supplementary Table S2. cDNA obtained with the BioScript kit was used for RT-PCRs and cDNA produced using the iScript kit was used for 3′-RACE-PCR.
RT-PCR
A Mastercycler gradient thermal cycler (Eppendorf) was used for RT-PCR amplifications. All the reactions were assembled using a REDTaq ReadyMix kit (Sigma-Aldrich). The amplification conditions included an initial denaturing step of 1 min at 96°C, followed by 30 cycles, each consisting of a 30 s denaturing step at 96°C, 30 s annealing step at (Tm−7°C) and 1 min extension step at 72°C, and a final incubation of 5 min at 72°C. Primer sequences are listed in the Supplementary Table S2. All amplified cDNAs were analysed by electrophoresis in 1.5% agarose gels. All cDNAs were purified using a QIAquick PCR purification kit (QIAGEN) and their identity was confirmed by sequencing (GATC Biotech).
3′-RACE PCR
Two nested adapter oligonucleotides were devised to have identical annealing temperatures with the QSXO1-specific forward nested primers “105-Ext-F” and “106-Int-F”. One additional long adapter primer “Oligo-dT-21-Adap-0-R” containing Oligo(dT)21 was made based on the designed adapters “Adap-1-R” and “Adap-2-R”. Adapter primers were designed to have no significant sequence similarity with known human DNA sequences from the “nr/nt” database. All primer sequences are listed in Supplementary Table S2. Two rounds of 3′ RACE PCR amplification were performed using a Mastercycler gradient thermal cycler (Eppendorf). In the first round 2 µl of the Oligo-dT(21)- primed cDNA was amplified using a mixture of adapter primers “Oligo-dT-21-Adap-0-R” and “Adap-1-R” (at 1∶10 molar ratio) and one sequence specific primer “105-Ext-F”; and a REDTaq ReadyMix PCR kit (Sigma-Aldrich). The amplification procedure included one initial denaturing step for 1 min at 96°C, followed by 3 cycles, each consisting of a 30 s denaturing step at 96°C, 30 s annealing step at 65°C and 3 min extension step at 72°C, following by another 3 cycles, each consisting of a 30 s denaturing step at 96°C, 30 s annealing step at 50°C and 3 min extension step at 72°C, followed by 25 cycles of a 30 s denaturing step at 96°C, 30 s annealing step at 65°C and 3 min extension step at 72°C. Final extension was at 72°C for 5 min. The second round of 3′-RACE-PCR used primers “106-Int-F” and “Adap-2-R” and 0.2 µl of the first round of 3′-RACE-PCR reaction. The amplification conditions were: initial denaturing for 1 min at 96°C, followed by 30 cycles of a 30 s denaturing step at 96°C, 30 s annealing step at 56°C and 3 min extension step at 72°C. Final extension was at 72°C for 5 min. The amplified cDNA was analysed by electrophoresis in 2% agarose gels, purified using a QIAquick PCR purification kit (QIAGEN) and sequenced (GATC Biotech).
Real Time PCR
Quantitative PCR was performed on an AB 7500 Fast Real Time system (Applied Biosystems, Warrington, UK) as previously described [45]. Cycle-cycle fluorescence changes in each sample were measured to generate a kinetic profile of DNA amplification over a 40-cycle PCR reaction. The cycle threshold number (C T) at which amplification entered the exponential phase was determined to indicate the amount of target RNA in each tissue sample, thus a lower C T value indicates a larger quantity of starting RNA. The total RNA amounts in each sample were normalised relative to the endogenous level of 18S rRNA transcripts. Three primer sets were used (Supplementary Table S2). The primer and probe sequences were based on the database sequence of Homo sapiens QSOX1 (Set 1) and on the newly identified 3′UTR extension of QSOX1 transcript (Sets 2 and 3). All primers and probes were designed using Primer Express 1.0 Software (PE Applied Biosystems). A collection of RNA samples derived from normal and malignant mammary tissue was analysed [45]. These samples, obtained under informed consent, comprised 9 samples of normal mammary tissue acquired from breast reduction surgery and 41 surgically removed breast cancer samples graded according to the Nottingham prognostic index [46]. The patient group was aged between 42 and 88 at diagnosis, with 7 being pre-menopausal. 25 of the tumours were classified as ductal, 9 were lobular, and the remainder were various other pathological types.
Supporting Information
Scatter plot showing lack of correlation between QSOX1 and ESR1 or HER2 expression. Horizontal axis - SAGE expression data for ESR1 or HER2 expressed as Log of the reported SAGE counts. Vertical axis - QSOX1 SAGE counts, Log scale. QSOX1 vs. HER2 (filled circles), QSOX1 vs ESR1 (open circles). The expression data show no significant correlation. The Pearson correlation coefficient calculated for libraries where both ESR1 and QSOX1 were detected was −0.06, and −0.03 for HER2 and QSOX1.
(TIF)
Upregulated expression of 1q genes in breast ductal carcinoma (having significance factor 0.01<P≤0.05). The upregulated genes with the significance factor P≤0.01 are listed in Table 1.
(DOC)
Nucleotide sequences of the primers used for all PCR amplifications.
(DOC)
Acknowledgments
We thank Caroline J. Pennington and Dylan R. Edwards, School of Biological Sciences, University of East Anglia, UK, for conducting the qPCR studies.
Funding Statement
The authors have no support or funding to report.
References
- 1. Tirrkonen M, Tanner M, Karbhu R, Kallioniemi A, Isola J, at al (1998) Molecular cytogenetics of primary breast cancer by CGH. Genes, Chromosomes Cancer 21(3): 177–184. [PubMed] [Google Scholar]
- 2. Nessling M, Richter K, Schwaenen C, Roerig P, et al. (2005) Candidate genes in breast cancer revealed by microarray-based comparative genomic hybridisation of archived tissue. Cancer Res. 65(2): 439–447. [PubMed] [Google Scholar]
- 3. Fridlyand J, Snijders A, Ylstra B, Li H, Olshen A, et al. (2006) Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer 6: 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stange D, Radlwimmer B, Schubert F, Traub F, Pich A, et al. (2006) High-resolution genomic profiling reveals association of chromosomal aberrations on 1q and 16p with histologic and genetic subgroups of invasive breast cancer. Clin. Cancer Res. 12(2): 345–352. [DOI] [PubMed] [Google Scholar]
- 5. Orsetti B, Nugoli M, Cervera N, Lasora L, Chuchana P, et al. (2006) Genetic profiling of chromosome 1 in breast cancer: mapping of regions of gains and losses and identification of candidate genes on 1q. Brit. J. Cancer, 95 (10): 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res., 62 (21): 6240–6245. [PubMed] [Google Scholar]
- 7. Li Z, Schem C, Shi Y, Medina D, Zhang M (2008) Increased COX-2 expression enhances tumor-induced osteoclastic lesions in breast cancer. Clin. Exp. Metastasis 25(4): 389–400. [DOI] [PubMed] [Google Scholar]
- 8. Lucci A, Krishnamurthy S, Singh B, Bedrosian I, Merc-Bernstam F, et al. (2009) Cyclogenase-2 expression in primary breast cancer predicts dissemination of cancer cells to the bone marrow. Breast Cancer Res. Treat., 117 (1): 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang X-Z, Li D-Q, Hou Y-F, Wu J, Lu J-S, et al. (2007) Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res. 9(6): R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabre-Lafay S, Monville F, Garrido-Urbani S, Berruyer-Pouyet C, Ginestier C, et al. (2007) Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer 7: 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morel C, Adami P, Muscard JF, Duval D, Radom J, et al. (2007) Involvement of sulfhydryl oxidase QSOX1 in the protection of cells against oxidative stress-induced apoptosis. Exp. Cell. Res. 313(19): 3971–3982. [DOI] [PubMed] [Google Scholar]
- 12. Heckler E, Rancy P, Kodali V, Thorpe C (2008) Generating disulfides with the quiescin-sulfhydryl oxidases. Biochim Biophys Acta 1783(4): 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kung HN, Marks JR, Chi JT (2011) Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 7(8): e1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salh B, Marotta A, Wagey R, Sayed M, Pelech S (2002) Dysregulation of phosphatidylinositol 3-kinase and downstream effectors in human breast cancer. Int J Cancer 98(1): 148–154. [DOI] [PubMed] [Google Scholar]
- 15. Leth-Larsen R, Terp MG, Christensen AG, Elias D, Kühlwein T, Jensen ON, Petersen OW, Ditzel HJ (2012) Functional Heterogeneity within the CD44 High Human Breast Cancer Stem Cell-Like Compartment Reveals a Gene Signature Predictive of Distant Metastasis. Molec. Medicine. 18(1): 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He J, Pan Y, Hu J, Albarracin C, Wu Y, Dai JL (2007) Profile of Ets gene expression in human breast carcinoma. Cancer Biol Ther. 6(1): 76–82. [DOI] [PubMed] [Google Scholar]
- 17. Hill JJ, Tremblay TL, Pen A, Li J, Robotham AC, Lenferink AE, Wang E, O’Connor-McCourt M, Kelly JF (2011) Identification of vascular breast tumor markers by laser capture microdissection and label-free LC-MS. J Proteome Res. 10(5): 2479–2493. [DOI] [PubMed] [Google Scholar]
- 18. Allred D, Clark G, Molina R, Tandon A, Schnitt S, et al. (1992) Overexpression of HER-2/neu and its relationship with other prognostic factors change during the progression of in situ to invasive breast cancer. Human Pathol. 23(9): 974–979. [DOI] [PubMed] [Google Scholar]
- 19. Witkiewicz AK, Dasgupta A, Nguyen KH, Liu C, Kovatich AJ, et al. (2009) Stromal caveolin-1 levels predict early DCIS progression to invasive breast cancer. Cancer Biol. Ther. 8(11): 1071–1079. [DOI] [PubMed] [Google Scholar]
- 20. Witkiewicz AK, Dasgupta A, Sotgia F, Mercier I, Pestell RG, et al. (2009) An absence of stromal caveolin-1 expression predicts early tumor recurrence and poor clinical outcome in human breast cancers. Am. J. Pathol. 174(6): 2023–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kulasingam V, Diamandis EP (2007) Proteomics analysis of conditioned media from three breast cancer cell lines. Molec. and Cellular Proteomics 6(11): 1997–2011. [DOI] [PubMed] [Google Scholar]
- 22. Inoue A, Yoshida N, Omoto Y, Oguchi S, Yamori T, et al. (2002) Development of cDNA microarray for expression profiling of estrogen-responsive genes. J. Molec. Endocrinology 29(2): 175–192. [DOI] [PubMed] [Google Scholar]
- 23. Moggs JG, Murphy TC, Lim FL, Moore DJ, Stuckey R, et al. (2005) Anti-proliferative effect of estrogen in breast cancer cells that re-express ERα is mediated by aberrant regulation of cell cycle genes. J. Molec. Enodcrinology 34(2): 535–551. [DOI] [PubMed] [Google Scholar]
- 24. Antwi K, Hostetter G, Demeure M, Katchman B, Decker G, et al. (2009) Analysis of the plasma peptidome from pancreas cancer patients connects a peptide in plasma to overexpression of the parent protein in tumors. J. Proteome Res. 8(10): 4722–4731. [DOI] [PubMed] [Google Scholar]
- 25. Song H, Zhang B, Watson MA, Humphrey PA, Lim H, et al. (2009) Loss of Nkx3.1 leads to activation of discrete downstream target genes during prostate tumorigenesis. Oncogene, 28 (37): 3307–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castagna A, Cecconi D, Sennels L, Rappsilber J, Guerrier L, et al. (2005) Exploring the hidden human urinary proteome via ligand library beads. J. Proteome Res. 4(6): 1917–1930. [DOI] [PubMed] [Google Scholar]
- 27. Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M (2006) The human urinary proteome contains more than 1500 proteins including a large proportion of membranes proteins. Genome Biology 7(9): R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mairet-Coello G, Tury A, Esnard-Feve A, Fellmann D, Risold P, et al. (2004) FAD-linked sulfhydryl oxidase QSOX: topographic, cellular, and subcellular immunolocalization in adult rat central nervous system. J. Comp. Neurol. 473(3): 334–363. [DOI] [PubMed] [Google Scholar]
- 29. Tury A, Mairet-Coello G, Poncet F, Jacquenard C, Risold P, et al. (2004) QSOX sulfhydryl oxidase in rat adenohypophysis: localization and regulation by estrogens. J. Endocrinol 183(2): 353–363. [DOI] [PubMed] [Google Scholar]
- 30. Radom J, Colin D, Thiebault F, Dognin-Bergeret M, Mairet-Coello G, et al. (2006) Identification and expression of a new splice variant of FAD-sulfhydryl oxidase in adult rat brain. Biochim. Biophys. Acta 1759(5): 225–233. [DOI] [PubMed] [Google Scholar]
- 31. Grzybowska E, Wilczynska A, Siedlecki J (2001) Regulatory functions of 3′UTRs. Biochem. Biophys. Res. Commun. 288(2): 291–295. [DOI] [PubMed] [Google Scholar]
- 32. Lopez de Silanes IL, Quesada MP, Esteller M (2007) Aberrant regulation of messenger RNA 3′-untranslated region in human cancer. Cell. Oncol., 29 (1): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andreassi C, Riccio A (2009) To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 19(9): 465–474. [DOI] [PubMed] [Google Scholar]
- 34. Raje S, Thorpe C (2003) Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: role of a thioredoxin domain in disulfide bond formation. Biochemistry 42(15): 4560–4568. [DOI] [PubMed] [Google Scholar]
- 35. Heckler E, Alon A, Fass D, Thorpe C (2008) Human quiescin-sulfhydryl oxidase, QSOX1: probing internal redox steps by mutagenesis. Biochemistry 47(17): 4955–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kodali VK, Thorpe C (2010) Quiescin sulfhydryl oxidase from Trypanosoma brucei: catalytic activity and mechanism of a QSOX family member with a single thioredoxin domain. Biochemistry 49(9): 2075–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng W, Chu Y, Yin Q, Xu L, Yang C, et al. (2011) Crucial effect of the first CXXC motif of human QSOX 1b on the activity to different substrates. J. Biochem. 149(3): 293–300. [DOI] [PubMed] [Google Scholar]
- 38. Zheng W, Zhang W, Hu W, Zhang C, Yang Y (2012) Exploring the Smallest Active Fragment of HsQSOX1b and Finding a Highly Efficient Oxidative Engine. PLoS One. 7(7): e40935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coppock DL, Cina-Poppe D, Gilleran S (1998) The quiescin Q6 gene (QSCN6) is a fusion of two ancient gene families: thioredoxin and ERV1. Genomics 54(3): 460–468. [DOI] [PubMed] [Google Scholar]
- 40. Thorpe C, Hoober K, Raje S, Glynn N, Burnside J, et al. (2002) Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch. Biochem. Biophys. 405(1): 1–12. [DOI] [PubMed] [Google Scholar]
- 41. Wittke I, Wiedemeyer R, Pillmann A, Savelyeva L, Westermann F, et al. (2003) Neuroblastoma-derived sulfhydryl oxidase, a new member of the sulfhydryl oxidase/Quiescin6 family, regulates sensitization to interferon gamma-induced cell death in human neuroblastoma cells. Cancer Res. 63(22): 7742–7752. [PubMed] [Google Scholar]
- 42. de Andrade CR, Stolf BS, Debbas V, Rosa DS, Kalil J, et al. (2011) Quiescin sulfhydryl oxidase (QSOX) is expressed in the human atheroma core: possible role in apoptosis. In Vitro Cell Dev Biol Anim. 47(10): 716–727. [DOI] [PubMed] [Google Scholar]
- 43. Katchman BA, Antwi K, Hostetter G, Demeure MJ, Watanabe A, et al. (2011) Quiescin sulfhydryl oxidase 1 promotes invasion of pancreatic tumor cells mediated by matrix metalloproteinases. Mol Cancer Res. 9(12): 1621–1631. [DOI] [PubMed] [Google Scholar]
- 44. Gumireddy K, Sun F, Klein-Szanto AJ, Gibbins J, Gimotty P, et al. (2007) In vivo selection for metastasis promoting genes in the mouse. Proc. Natl. Acad. Sci. USA 104(16): 6696–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, et al. (2004) Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin. Cancer. Res. 10(7): 2429–2440. [DOI] [PubMed] [Google Scholar]
- 46. Galea MH, Blamey RW, Elston C, Ellis I (1992) The Nottingham Prognostic Index in primary breast cancer Breast Cancer Res. Treat. 22(3): 207–219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter plot showing lack of correlation between QSOX1 and ESR1 or HER2 expression. Horizontal axis - SAGE expression data for ESR1 or HER2 expressed as Log of the reported SAGE counts. Vertical axis - QSOX1 SAGE counts, Log scale. QSOX1 vs. HER2 (filled circles), QSOX1 vs ESR1 (open circles). The expression data show no significant correlation. The Pearson correlation coefficient calculated for libraries where both ESR1 and QSOX1 were detected was −0.06, and −0.03 for HER2 and QSOX1.
(TIF)
Upregulated expression of 1q genes in breast ductal carcinoma (having significance factor 0.01<P≤0.05). The upregulated genes with the significance factor P≤0.01 are listed in Table 1.
(DOC)
Nucleotide sequences of the primers used for all PCR amplifications.
(DOC)