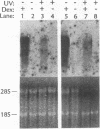
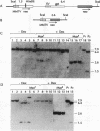
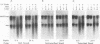Abstract
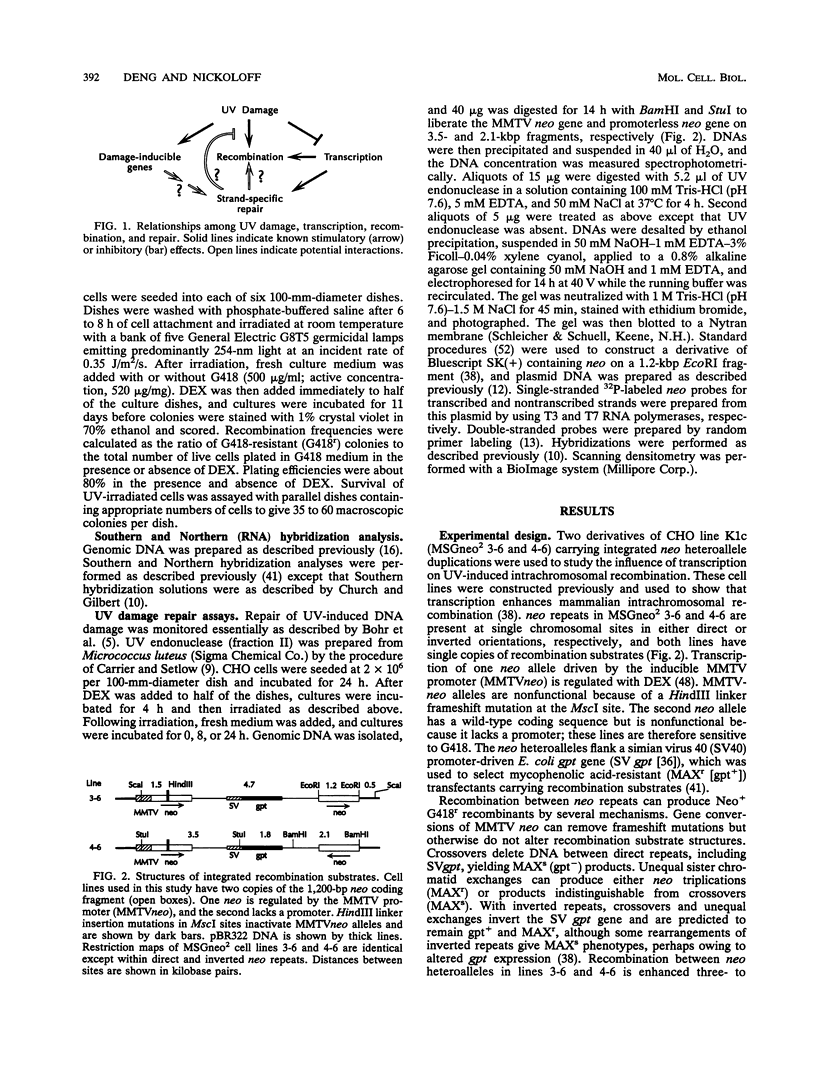
The relationships among transcription, recombination, DNA damage, and repair in mammalian cells were investigated. We monitored the effects of transcription on UV-induced intrachromosomal recombination between neomycin repeats including a promoterless allele and an inducible heteroallele regulated by the mouse mammary tumor virus promoter. Although transcription and UV light separately stimulated recombination, increasing transcription levels reduced UV-induced recombination. Preferential repair of UV damage in transcribed strands was shown in highly transcribed DNA, suggesting that recombination is stimulated by unrepaired UV damage and that increased DNA repair in highly transcribed alleles removes recombinogenic lesions. This study indicates that the genetic consequences of DNA damage depend on transcriptional states and provides a basis for understanding tissue- and gene-specific responses to DNA-damaging agents.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Blackwell T. K., DePinho R. A., Reth M. G., Yancopoulos G. D. Regulation of genome rearrangement events during lymphocyte differentiation. Immunol Rev. 1986 Feb;89:5–30. doi: 10.1111/j.1600-065x.1986.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya N. P., Maher V. M., McCormick J. J. Ability of structurally related polycyclic aromatic carcinogens to induce homologous recombination between duplicated chromosomal sequences in mouse L cells. Mutat Res. 1989 Apr;211(2):205–214. doi: 10.1016/0027-5107(89)90003-1. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya N. P., Maher V. M., McCormick J. J. Effect of nucleotide excision repair in human cells on intrachromosomal homologous recombination induced by UV and 1-nitrosopyrene. Mol Cell Biol. 1990 Aug;10(8):3945–3951. doi: 10.1128/mcb.10.8.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Moore M. W., Yancopoulos G. D., Suh H., Lutzker S., Selsing E., Alt F. W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986 Dec 11;324(6097):585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985 Feb;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Bollag R. J., Waldman A. S., Liskay R. M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- Boothman D. A., Bouvard I., Hughes E. N. Identification and characterization of X-ray-induced proteins in human cells. Cancer Res. 1989 Jun 1;49(11):2871–2878. [PubMed] [Google Scholar]
- Boothman D. A., Wang M., Lee S. W. Induction of tissue-type plasminogen activator by ionizing radiation in human malignant melanoma cells. Cancer Res. 1991 Oct 15;51(20):5587–5595. [PubMed] [Google Scholar]
- Carrier W. L., Setlow R. B. Endonuclease from Micrococcus luteus which has activity toward ultraviolet-irradiated deoxyribonucleic acid: purification and properties. J Bacteriol. 1970 Apr;102(1):178–186. doi: 10.1128/jb.102.1.178-186.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M. L. Oncogenic conversion of transcription factors by chromosomal translocations. Cell. 1991 Aug 23;66(4):619–622. doi: 10.1016/0092-8674(91)90105-8. [DOI] [PubMed] [Google Scholar]
- Deng W. P., Nickoloff J. A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992 Jan;200(1):81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Papathanasiou M. A., Tarone R. E., Wong M., Mitchell J., Hamer D. H. DNA-damage-inducible genes in mammalian cells. Prog Clin Biol Res. 1990;340A:315–325. [PubMed] [Google Scholar]
- Glazer P. M., Greggio N. A., Metherall J. E., Summers W. C. UV-induced DNA-binding proteins in human cells. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1163–1167. doi: 10.1073/pnas.86.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg J., Nawoschik S., Belluscio L., McKee R., Turck A., Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989 Oct 25;17(20):8390–8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Schaer P., Munz P., Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991 Jan;11(1):289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman L., Caron P. R., Mazur S. J., Oh E. Y. Repair of DNA-containing pyrimidine dimers. FASEB J. 1988 Aug;2(11):2696–2701. doi: 10.1096/fasebj.2.11.3294078. [DOI] [PubMed] [Google Scholar]
- Hanawalt P. C. Heterogeneity of DNA repair at the gene level. Mutat Res. 1991 Apr;247(2):203–211. doi: 10.1016/0027-5107(91)90016-h. [DOI] [PubMed] [Google Scholar]
- Hanawalt P. C. Preferential repair of damage in actively transcribed DNA sequences in vivo. Genome. 1989;31(2):605–611. doi: 10.1139/g89-113. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988 Dec;52(4):536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E. N., Boothman D. A. Effect of caffeine on the expression of a major X-ray induced protein in human tumor cells. Radiat Res. 1991 Mar;125(3):313–317. [PubMed] [Google Scholar]
- Husain I., Griffith J., Sancar A. Thymine dimers bend DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2558–2562. doi: 10.1073/pnas.85.8.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R. L., Roeder G. S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984 Dec;39(2 Pt 1):377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Kim R. A., Wang J. C. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989 Jun 16;57(6):975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Hicks J. B. A position-effect control for gene transposition: state of expression of yeast mating-type genes affects their ability to switch. Cell. 1981 Aug;25(2):517–524. doi: 10.1016/0092-8674(81)90070-2. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadon S. A., Lawrence D. A. Preferential repair of DNA damage on the transcribed strand of the human metallothionein genes requires RNA polymerase II. Mutat Res. 1991 Jul;255(1):67–78. doi: 10.1016/0921-8777(91)90019-l. [DOI] [PubMed] [Google Scholar]
- Leadon S. A., Snowden M. M. Differential repair of DNA damage in the human metallothionein gene family. Mol Cell Biol. 1988 Dec;8(12):5331–5338. doi: 10.1128/mcb.8.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H., Maizels N. Transcriptional regulatory elements stimulate recombination in extrachromosomal substrates carrying immunoglobulin switch-region sequences. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4154–4158. doi: 10.1073/pnas.89.9.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990 Jan;10(1):113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989 Nov 2;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Selection for animal cells that express the Escherichia coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2072–2076. doi: 10.1073/pnas.78.4.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn R. S., Adair G. M., Christmann C. B., Humphrey R. M. Ultraviolet stimulation of intermolecular homologous recombination in Chinese hamster ovary cells. Mol Carcinog. 1991;4(6):519–526. doi: 10.1002/mc.2940040616. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Chen E. Y., Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989 Jun 5;207(3):527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol Cell Biol. 1992 Dec;12(12):5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto D. S., Bohr V. A. DNA repair in the metallothionein gene increases with transcriptional activation. Nucleic Acids Res. 1987 Dec 10;15(23):10021–10030. doi: 10.1093/nar/15.23.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T., Wrange O. Inhibition of chromatin assembly in Xenopus oocytes correlates with derepression of the mouse mammary tumor virus promoter. Mol Cell Biol. 1991 Oct;11(10):5259–5265. doi: 10.1128/mcb.11.10.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Machin N., Stahl F. W. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Ringold G. M., Yamamoto K. R., Bishop J. M., Varmus H. E. Glucocorticoid-stimulated accumulation of mouse mammary tumor virus RNA: increased rate of synthesis of viral RNA. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2879–2883. doi: 10.1073/pnas.74.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Shulman M., Landy A. Biochemical analysis of att-defective mutants of the phage lambda site-specific recombination system. J Mol Biol. 1982 Apr 15;156(3):505–522. doi: 10.1016/0022-2836(82)90263-7. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985 Jan;5(1):75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W. D., Wilde C. E., 3rd, Reno D. L., Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971 Oct 14;61(1):25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- Sauerbier W., Hercules K. Gene and transcription unit mapping by radiation effects. Annu Rev Genet. 1978;12:329–363. doi: 10.1146/annurev.ge.12.120178.001553. [DOI] [PubMed] [Google Scholar]
- Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J. H., Chambon P., Egly J. M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993 Apr 2;260(5104):58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993 Apr 2;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Smith A. J., Berg P. Homologous recombination between defective neo genes in mouse 3T6 cells. Cold Spring Harb Symp Quant Biol. 1984;49:171–181. doi: 10.1101/sqb.1984.049.01.020. [DOI] [PubMed] [Google Scholar]
- Snyder U. K., Thompson J. F., Landy A. Phasing of protein-induced DNA bends in a recombination complex. Nature. 1989 Sep 21;341(6239):255–257. doi: 10.1038/341255a0. [DOI] [PubMed] [Google Scholar]
- Song K. Y., Chekuri L., Rauth S., Ehrlich S., Kucherlapati R. Effect of double-strand breaks on homologous recombination in mammalian cells and extracts. Mol Cell Biol. 1985 Dec;5(12):3331–3336. doi: 10.1128/mcb.5.12.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S. E., Roeder G. S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Aug;9(8):3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S., Rubnitz J. Recombination events after transient infection and stable integration of DNA into mouse cells. Mol Cell Biol. 1985 Apr;5(4):659–666. doi: 10.1128/mcb.5.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Garrett D. S., Brockie I. R., Svoboda D. L., Telser J. 1H NMR assignment and melting temperature study of cis-syn and trans-syn thymine dimer containing duplexes of d(CGTATTATGC).d(GCATAATACG). Biochemistry. 1990 Sep 18;29(37):8858–8866. doi: 10.1021/bi00489a049. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl F. W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Tsaneva I. R., Müller B., West S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992 Jun 26;69(7):1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Tsujimura T., Maher V. M., Godwin A. R., Liskay R. M., McCormick J. J. Frequency of intrachromosomal homologous recombination induced by UV radiation in normally repairing and excision repair-deficient human cells. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1566–1570. doi: 10.1073/pnas.87.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Roeder G. S. A chromosome containing HOT1 preferentially receives information during mitotic interchromosomal gene conversion. Genetics. 1990 Mar;124(3):561–572. doi: 10.1093/genetics/124.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Roeder G. S. Gene conversion tracts stimulated by HOT1-promoted transcription are long and continuous. Genetics. 1990 Dec;126(4):851–867. doi: 10.1093/genetics/126.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Y., Maher V. M., Liskay R. M., McCormick J. J. Carcinogens can induce homologous recombination between duplicated chromosomal sequences in mouse L cells. Mol Cell Biol. 1988 Jan;8(1):196–202. doi: 10.1128/mcb.8.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Young H. A., Shih T. Y., Scolnick E. M., Parks W. P. Steroid induction of mouse mammary tumor virus: effect upon synthesis and degradation of viral RNA. J Virol. 1977 Jan;21(1):139–146. doi: 10.1128/jvi.21.1.139-146.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]