Abstract
Cellulose is synthesized at the plasma membrane by protein complexes known as cellulose synthase complexes (CSCs). The cellulose-microtubule alignment hypothesis states that there is a causal link between the orientation of cortical microtubules and orientation of nascent cellulose microfibrils. The mechanism behind the alignment hypothesis is largely unknown. CESA interactive protein 1 (CSI1) interacts with CSCs and potentially links CSCs to the cytoskeleton. CSI1 not only co-localizes with CSCs but also travels bi-directionally in a speed indistinguishable from CSCs. The linear trajectories of CSI1-RFP coincide with the underlying microtubules labeled by YFP-TUA5. In the absence of CSI1, both the distribution and the motility of CSCs are defective and the alignment of CSCs and microtubules is disrupted. These observations led to the hypothesis that CSI1 directly mediates the interaction between CSCs and microtubules. In support of this hypothesis, CSI1 binds to microtubules directly by an in vitro microtubule-binding assay. In addition to a role in serving as a messenger from microtubule to CSCs, CSI1 labels SmaCCs/MASCs, a compartment that has been proposed to be involved in CESA trafficking and/or delivery to the plasma membrane.
Keywords: cellulose microfibril, cellulose synthase complex, microtubule, plasma membrane
The microtubule cytoskeleton is essential for the cellular organization of eukaryotic cells, playing an important role in cell division and intracellular transport. For plant cells, microtubules provide an additional role in regulating wall polysaccharide deposition. During plant cell division, microtubules form four different organizations including the cortical interphase array, preprophase bands (PPB), mitotic spindles and phragmoplast, which are all involved in cell wall formation.1 The association between cortical microtubules and cellulose microfibril deposition has been well documented in the literature.2-6 The microtubule-microfibrils alignment hypothesis proposes that cortical microtubules, which lie beneath the plasma membrane of elongating cells, provide tracks for cellulose synthase complexes (CSCs) that convert glucose molecules into crystalline cellulose microfibrils.4 Live imaging of cortical microtubules and CSCs in transgenic lines containing fluorescent fusions of both tubulin and CESA proteins supports the alignment model.7 However, the molecular details of how microtubules interact with CSCs remain unknown.
A yeast two-hybrid screen using the entire central cytosolic domain of CESA was used to identify proteins that interact with CESA and potentially link CSCs to the cytoskeleton. CESA interactive protein 1 (CSI1) is among several dozen putative CESA interactive proteins identified.8,9 The sub-cellular localization of CSI1 has been examined in epidermal cells in dark-grown hypocotyls. CSI1 co-localizes with primary cell wall CSCs as demonstrated by simultaneously imaging GFP-CESA3/RFP-CSI1 and YFP-CESA6/RFP-CSI1.8,10 Similar to CSC particles, CSI1 travels bi-directionally with an average speed of 300–350 nm/min.10 Freeze fracture EM has shown CSCs in the Golgi, which has led to the hypothesis that CSCs are assembled in the Golgi. If this hypothesis holds true, CSI1 may not be involved in the assembly of CESA complexes since CSI1 does not show localization in the Golgi.
Functional association between CSI1 and cortical microtubules was recently explored using live imaging of transgenic lines bearing RFP-CSI1 and YFP-TUA5.10 In dark-grown Arabidopsis hypocotyls, RFP-CSI1 and YFP-TUA5 signals overlap extensively, and RFP-CSI1 was observed to move along the underlying microtubule tracks labeled by YFP-TUA5. The quantification of overlapping signals between RFP-CSI1 and YFP-TUA5 revealed that more than 80% of RFP-CSI1 signals co-aligned with microtubules, an extent of overlap that was significantly above random co-localization (45%). Based on the observations that CSI1 containing particles travel bi-directionally in a speed similar to that of CESA along the underlying cortical microtubules, we speculate that the association of CESA with microtubules may be through CSI1. To further determine the spatial and temporal relationship among CSI1, CSCs and cortical microtubules, we examined co-localization patterns in transgenic lines bearing CFP-TUA1, RFP-CSI1 and GFP-CESA6. The percentage of co-localization between pairs of GFP-CESA6/RFP-CSI1, pairs of RFP-CSI1/CFP-TUA1 and pairs of GFP-CESA6/CFP-TUA1 was 80%, 81% and 73%, respectively. For all GFP-CESA6 containing particles that co-localized with CFP-TUA1, only 2 out of 227 GFP-CESA6 particles lacked the accompaniment of RFP-CSI1 (Fig. 1 is a representative experiment). These results led us to hypothesize that the co-localization of GFP-CESA6 and CFP-TUA1 is dependent on RFP-CSI1.
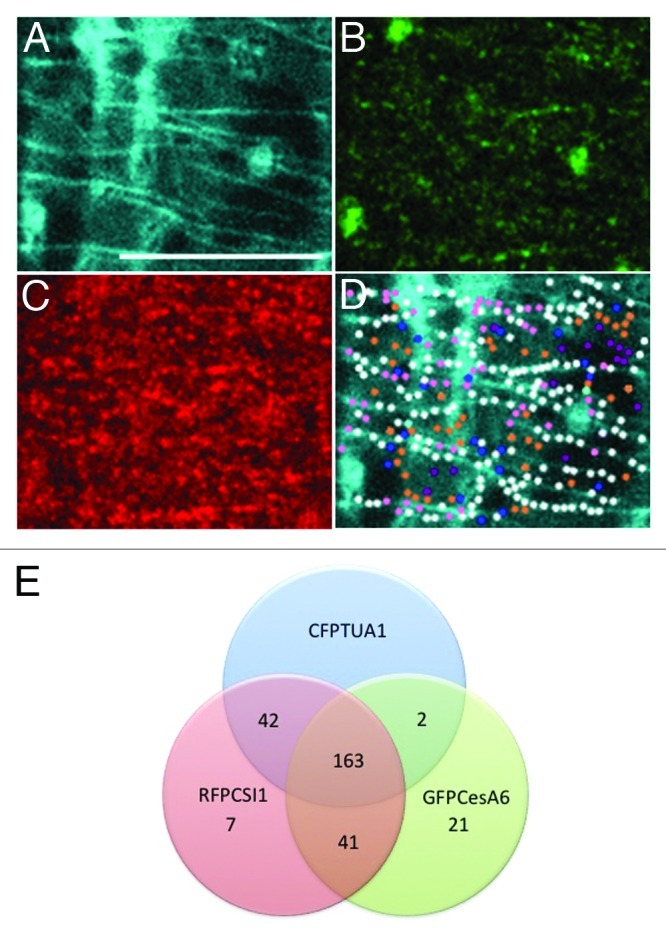
Figure 1. Co-localization of CESA complexes, CSI1 and cortical microtubules. (A-C) Three-channel imaging of epidermal cells expressing CFP-TUA1 (A), GFP-CESA6 (B) and RFP-CSI1 (C). (D) Co-localization analysis of A, B and C. Venn diagram of co-localization pattern showing in (D). White dots, 163; Orange dots, 41; Pink dots, 42; Cyan dots, 2; Blue dots, 21; purple dots, 7. Scale bar = 10 μm.
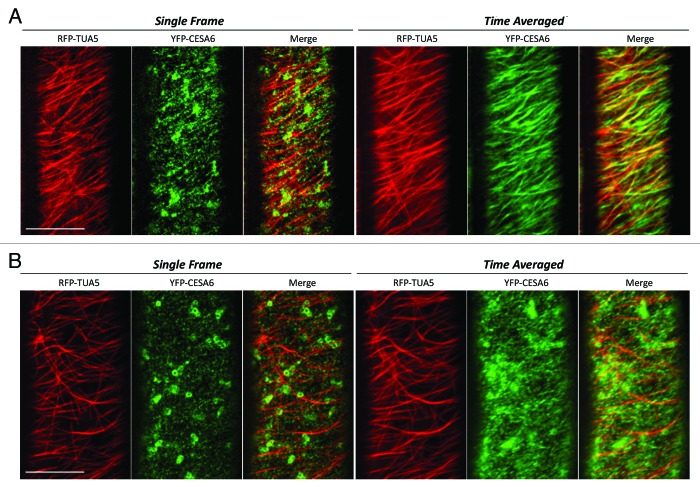
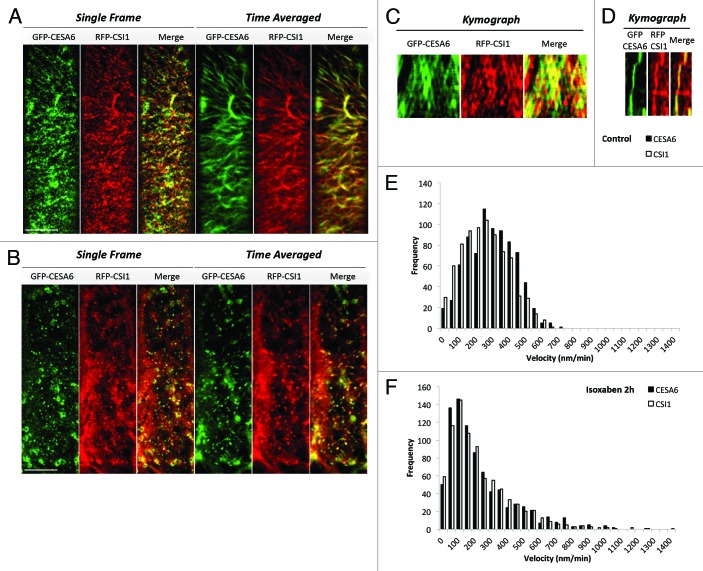
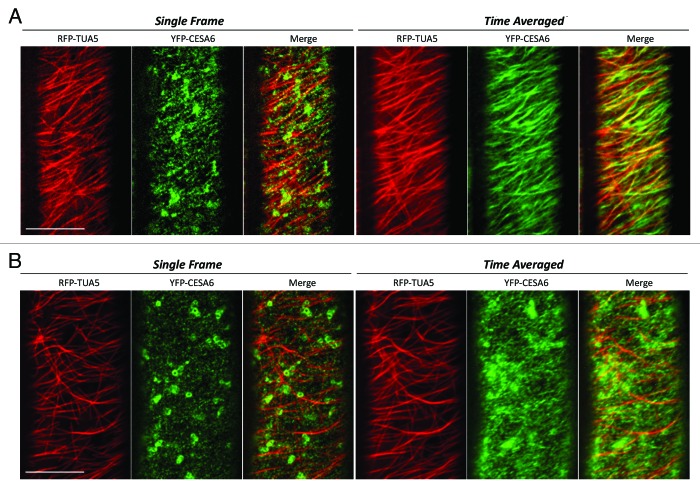
We next examined the co-localization patterns between microtubule and CSCs in the csi1 mutant. The co-alignment between YFP-CESA6 and RFP-TUA5 was diminished to a level that was not significantly above random colocalization in the csi1 mutant (Fig. 2). The mis-alignment between YFP-CESA6 and RFP-TUA5 is evident in time projection images. These data uphold the role of CSI1 in bridging CSCs and cortical microtubules. Recently, evidence suggests that microtubules position the delivery of CSCs through the interaction with small CESA-containing compartments (SmaCCs) or microtubule-associated cellulose synthase compartment (MASCs).11,12 In untreated cells, SmaCCs/MASCs are present in fully elongated cells 10 mm below the apical hook where primary CSCs are nearly absent at the plasma membrane. SmaCCs/MASCs accumulate in the cell cortex upon osmotic stress, protein synthesis inhibition, or cellulose synthesis inhibition.11,12 Upon 100 nM isoxaben treatment for 2 h, GFP-CESA6 and RFP-CSI1 accumulated in small CESA-containing compartments that resembled SmaCCs/MASCs (Fig. 3). SmaCCs/MASCs and CESA complexes were distinguished by their velocities. Similar to SmaCCs/MASCs in YFP-CESA6 cells, RFP-CSI1 punctae traveled with a variable speed from 10 to 3000 nm/min after isoxaben treatment (Fig. 3). RFP-CSI1 particles in isoxaben treated seedlings were also observed to track with microtubule minus ends (data not shown). The role of SmaCCs/MASCs has been proposed to be involved in delivery, internalization, and/or recycling.13 The association of CSI1 with SmaCCs/MASCs indicates CSI1 may have a role in CESA trafficking in addition to the guidance of CSCs along the microtubules.
Figure 2.
CESA distribution and motility is affected in csi1 seedlings. Epidermal cells in 3-d-old dark grown hypocotyls co-expressing YFP-CESA6 and RFP-TUA5 in wild type and csi1 seedlings. (A) In wild type, the co-alignment of CESA and microtubules was evident in merge image in both single frame and time projection images. (B) In csi1 seedlings, CESA were randomly distributed. CESA linear tracks were apparently shorter and did not follow the underlying microtubule tracks. Time average image (duration 5 min, 5 sec interval) Scale bar = 10 μm.
Figure 3. CSI1 associates with SmaCCs/MASCs. Epidermal cells in 3-d-old dark grown hypocotyls co-expressing GFP-CESA6 and RFP-CSI1 were incubated in Murashige and Skoog liquid solution containing diluted DMSO control (0.01%) (A) or 100 nM isoxaben for 2 h (B). (C) Kymographs display CESA6 and CSI1 movement in control cells. (D) Kymographs display CESA6 and CSI1 movement in isoxaben-treated cells. (E) Histogram of GFP-CESA6 and RFP-CSI1 in control cells. (F) Histogram of GFP-CESA6 and RFP-CSI1 in isoxaben-treated cells. Scale bar = 10 μm.
CSI1 encodes a large protein (2,151 amino acids) with multiple Armadillo (ARM) repeats. ARM repeat containing proteins have been shown to be involved in various processes including cytoskeleton function.14-17 To test whether CSI1 directly binds to microtubules, we purified full-length CSI1 protein from E. coli and performed an in vitro microtubule-binding assay. CSI1 pelleted with tubulin polymers, similar to the positive control, MAP2.10 The binding of CSI1 to microtubules is saturated at: 2 μM. Scatchard plots revealed a dissociation constant of 1.07 ± 0.33 μM, n=3 (Fig. 4 is a representative experiment). Based on the observations that CSI1 binds to microtubules in vitro and CSI1 associates with microtubules in vivo, CSI1 can be classified as a bona fide microtubule associated protein (MAP). We did not observe CSI1 decorating other forms of microtubules during cell division in our preliminary analysis of root cells during cell division. Moreover, CSI1 does not share sequence homology with known structural MAPs therefore CSI1 may belong to a loose group of microtubule-interacting proteins rather than structural MAPs. Further analysis is currently underway in our lab to determine the microtubule-binding domain in CSI1.
Figure 4. Saturation binding assay of CSI1/tubulin. The increasing amount of tubulin was added to a constant amount of CSI1 in the saturation-binding assay. The dissociation constant for CSI1 binding to tubulin polymers was determined by the best fit to the saturation-binding curve. The Scatchard plot was derived from a representative saturation-binding curve.
Here we present evidence that CSI1 is required for guidance of CSCs along cortical microtubules. CSI1 does so by directly mediating the interaction between cortical microtubules and CSCs, as demonstrated by its direct binding to microtubules in vitro10 and to CESA proteins in yeast two hybrid assay.8 As a bona fide MAP, CSI1 also decorates microtubules in vivo. However, the dynamics of CSI1 more closely resembles the dynamics of CSCs than microtubules as CSI1 moves bi-directionally at an average speed of 300–350 nm/min.8,10 CSI1 does not co-localize with CSCs in the Golgi, suggesting that the association of CSI1 with CSCs might occur at the membrane or before the insertion to the membrane. It is not known how CSI1 is delivered to the plasma membrane or how CSI1 is associated with CSCs and cortical microtubules. One possible scenario could be that CSI1 associates with CSCs before insertion at the plasma membrane and then CSI1 positions CSCs along cortical microtubules. Alternatively, CSI1 could be delivered to cortical microtubules before it associates with CSCs. CSI1 labels SmaCCs/MASCs, a compartment that has been proposed to be involved in CESA trafficking and/or delivery to the plasma membrane. Pinpointing the exact role of SmaCCs/MASCs will be crucial to dissecting the mechanism by which CSI1 associates with CSCs and cortical microtubules.
Disclosure of Potential Conflicts of Interest
No portential conflicts of interest were disclosed.
Acknowledgments
L.L., S.L., and Y.G. are in part supported by the Center for LignoCellulose Structure and Formation, an Energy Frontier Research Center funded by the US. Department of Energy, Office of Science under Award Number DE-SC0001090. Y.G. is supported by grants from National Science Foundation (1121375).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20338
References
- 1.Lloyd C, Chan J, Hussey P. Microtubules and microtubule-associated proteins. In: Hussey, P, ed. The Plant Cytoskeleton in Cell Differentiation and Development. Oxford: Blackwell Publishing Ltd., 2004; 3-31. [Google Scholar]
- 2.Heath IB. A unified hypothesis for the role of membrane bound enzyme complexes and microtubules in plant cell wall synthesis. J Theor Biol. 1974;48:445–9. doi: 10.1016/S0022-5193(74)80011-1. [DOI] [PubMed] [Google Scholar]
- 3.Robinson DG. The microtubule-microfibril syndrome. In: CW, L, ed. The Cytoskeleton in Plant Growth and Development. Waltham: Academic Press, 1982; 109-126. [Google Scholar]
- 4.Baskin TI. On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma. 2001;215:150–71. doi: 10.1007/BF01280311. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd C, Chan J. The parallel lives of microtubules and cellulose microfibrils. Curr Opin Plant Biol. 2008;11:641–6. doi: 10.1016/j.pbi.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 7.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–5. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 8.Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, et al. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci U S A. 2010;107:12866–71. doi: 10.1073/pnas.1007092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu Y, Somerville C. Cellulose synthase interacting protein: a new factor in cellulose synthesis. Plant Signal Behav. 2010;5:1571–4. doi: 10.4161/psb.5.12.13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Lei L, Somerville CR, Gu Y. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci U S A. 2012;109:185–90. doi: 10.1073/pnas.1118560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 12.Vernhettes S, Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Hofte H. Pausing of Golgi Bodies on Microtubules Regulates Secretion of Cellulose Synthase Complexes in Arabidopsis. Plant Cell. 2009;21:1141–54. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashline L, DU J, Gu Y. The trafficking and behavior of cellulose synthase and a glimpse of potential cellulose synthesis regulators. Front Biol. 2011;6:377–83. doi: 10.1007/s11515-011-1161-3. [DOI] [Google Scholar]
- 14.Neilson LI, Schneider PA, Van Deerlin PG, Kiriakidou M, Driscoll DA, Pellegrini MC, et al. cDNA cloning and characterization of a human sperm antigen (SPAG6) with homology to the product of the Chlamydomonas PF16 locus. Genomics. 1999;60:272–80. doi: 10.1006/geno.1999.5914. [DOI] [PubMed] [Google Scholar]
- 15.Smith EF, Lefebvre PA. PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J Cell Biol. 1996;132:359–70. doi: 10.1083/jcb.132.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith HM, Raikhel NV. Nuclear localization signal receptor importin alpha associates with the cytoskeleton. Plant Cell. 1998;10:1791–9. doi: 10.1105/tpc.10.11.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sapiro R, Tarantino LM, Velazquez F, Kiriakidou M, Hecht NB, Bucan M, et al. Sperm antigen 6 is the murine homologue of the Chlamydomonas reinhardtii central apparatus protein encoded by the PF16 locus. Biol Reprod. 2000;62:511–8. doi: 10.1095/biolreprod62.3.511. [DOI] [PubMed] [Google Scholar]