Abstract
The photorespiratory enzyme glycolate oxidase (GOX) was found to be involved in nonhost resistance by regulating plant defense responses through the production of H2O2. Silencing of a gene encoding NADPH oxidase (AtRBOHD) in the gox mutants did not further increase susceptibility to a nonhost pathogen, P. syringae pv tabaci, although it caused an increase in bacterial growth in the Atgox1 and Atgox3 mutant backgrounds. In order to confirm this finding, we created double homozygous knockouts AtrbohD x Atgox1 and AtrbohD x Atgox3 to evaluate symptom development and bacterial growth. Here we show that there is no additive effect of disease symptoms or bacterial growth in the AtrbohD x Atgox1 and AtrbohD x Atgox3 double mutants when compared with individual mutants. Slight additive effect observed previously upon silencing of AtRBOHD in Atgox1 and Atgox3 mutants was most likely due to cross-silencing of AtRBOHF. These results further prove that GOX plays a role in nonhost resistance independent of NADPH oxidase.
Keywords: NADPH oxidase, glycolate oxidase, nonhost resistance
Plants are able to detect the threat of pathogens by mechanisms that include a plethora of preformed compounds as well as complex induced responses.1 In most cases these mechanisms are enough to stop the pathogen before it is able to cause significant damage to the plant. In nature, the most common form of disease resistance is attributed to nonhost resistance defined as the resistance exhibited by entire plant species to all isolates of a microbial species.”2,3 A typical feature of nonhost resistance is a hypersensitive response (HR), a form of programmed cell death that occurs around the site where pathogens or elicitors interact with the plant.3 This HR involves accumulation of the reactive oxygen species (ROS) like superoxide (O2-), and the most stable form hydrogen peroxide (H2O2).4
The source of ROS during plant defense has been controversial and for many years it has been proposed that upon pathogen recognition, ROS production occurs through the action of NADPH oxidases, also called respiratory burst oxidases (RBOH).5 NADPH oxidase reaction uses O2 to generate superoxide (O2-) which is converted into H2O2 by superoxide dismutase (SOD).5 In Arabidopsis, 10 genes encode NADPH oxidases but only two, AtRBOHD and AtRBOHF, have been implicated in plant defense.6,7 AtrbohD mutant was shown to be defective in the production of H2O2 after inoculation with the avirulent bacterium P. syringae pv tomato DC3000 (AvrRpm1) although it was not compromised in HR6 and exhibited distinct behavior depending on the pathogen used for inoculation. Thus, the growth of P. syringae pv tomato DC3000 (AvrRpm1) was not affected in this mutant and bacteria grew to the same level as wild-type plants8 However, the accumulation of the oomycete Peronospora parasitica6 and the fungus Alternaria brassicicola9 was reduced in the AtrbohD mutant when compared with wild-type plants. Conversely, AtrbohF mutant which was not defective in H2O2 accumulation,6 allowed higher accumulation of the virulent bacteria P. syringae pv tomato DC3000 while the accumulation of the avirulent P. syringae pv tomato DC3000 (AvrRpm1) was not different than in wild-type plants.8 Similar to AtrbohD mutant, there was less sporangiophore formation due to Peronospora parasitica infection in AtrbohF mutant plants.6
We recently showed that another source of H2O2 involved in defense responses against the nonhost pathogen P. syringae pv tabaci is the photorespiratory enzyme glycolate oxidase (GOX).10 The generation of H2O2 by GOX occurs when it catalyzes the oxidation of glycolate to glyoxylate.11 Similarly to NADPH oxidase, there are multiple genes encoding GOX in Arabidopsis: AtGOX1, AtGOX2, AtGOX3, AtHAOX1 and AtHAOX2.12 Two of these genes AtGOX3 and AtHAOX2 were induced by P. syringae pv tabaci in wild-type plants and their corresponding mutants showed significant reduction in the expression of defense genes. These results suggested that the H2O2 generated by GOX is used as a signal to activate several defense signal transduction pathways.10
In order to determine whether NADPH oxidase was involved in the phenotypes observed, we silenced AtRBOHD in the wild-type (Col-0) and the gox mutant backgrounds and evaluated disease development and bacterial growth.10 Silencing of AtRBOHD in wild-type background slightly increased symptoms after inoculation with the nonhost pathogen P. syringae pv tabaci when compared with non-silenced control plants. However, AtRBOHD silencing in gox mutant backgrounds did not have an additive effect on disease symptoms after inoculation with P. syringae pv tabaci.10 Interestingly, when bacterial growth was quantified, we observed a slight increase in the number of bacteria after silencing of AtRBOHD in Atgox1 and Atgox3 backgrounds that warrants further investigation.10 In addition, AtRBOHD silencing in both wild-type and gox mutant backgrounds caused cross silencing of AtRBOHF and therefore the exact role of AtRBOHD in gox mutants was not clear. We therefore decided to create double mutants by crossing AtrbohD mutant with Atgox1 and Atgox3 mutants. Double knockouts of AtrbohD x Atgox1 and AtrbohD x Atgox3 were made and double homozygous plants were identified. For inoculations, double homozygous mutant plants were grown simultaneously with single mutant parents and with the wild type, Col-0. Six-week old plants were syringe-inoculated with the nonhost pathogen P. syringae pv tabaci and the host pathogen P. syringae pv maculicola to observe symptom development (Fig. 1). Inoculation with the nonhost pathogen P. syringae pv tabaci did not cause disease symptoms in wild-type Col-0 nor in the AtrbohD mutant indicating that AtRBOHD is not involved in nonhost resistance. The single mutants Atgox1 and Atgox3 showed enhanced disease susceptibility to the nonhost pathogen exhibiting significant chlorosis as previously shown.10 Similar to the results of silencing AtRBOHD in the Atgox1 mutant background, the double mutant AtrbohD x Atgox1 has symptoms similar to that of single Atgox1 mutant. Interestingly, the double mutant AtrbohD x Atgox3 did not have any disease symptoms. Inoculation with the host pathogen P. syringae pv maculicola showed dramatic disease symptoms in all genotypes without any significant difference among them (Fig. 1).
Figure 1. Effect of AtrbohD mutation in Atgox1 and Atgox3 mutants on symptom development associated with the nonhost pathogen P. syringae pv tabaci. Wild-type Col-0, single mutants AtrbohD, Atgox1, Atgox3 and double mutants AtrbohD x Atgox1 and AtrbohD x Atgox3 were inoculated with the nonhost pathogen P. syringae pv tabaci and P. syringae pv maculicola at 1 × 104 CFU/ml. Symptoms were evaluated at 4 d post-inoculation.
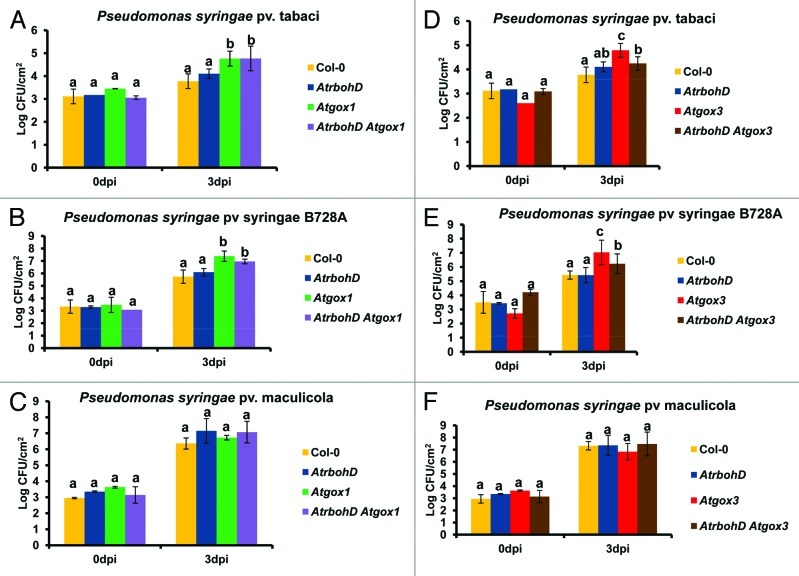
In order to quantify the effects of AtrbohD x Atgox double mutants on bacterial growth, we syringe-inoculated the plants with two nonhost pathogens, P. syringae pv tabaci and P. syringae pv syringae B728A, and a host pathogen, P. syringae pv maculicola (Fig. 2). As reported previously, single mutants of Atgox1and Atgox3 inoculated with nonhost pathogens (Fig. 2A, B, D and E) showed ~10-fold increase in bacterial population at 3 dpi when compared with the wild type, while the growth of the host pathogen P. syringae pv maculicola was not different in these mutants in comparison with the wild type.10 In contrast to the Atgox1 and Atgox3 mutants, the AtrbohD mutant did not support a significant increase in bacterial growth after inoculation with the nonhost pathogens similar to what was observed in wild type (Fig. 2A and B). These results are in line with previous studies on the AtrbohD mutant inoculated with the avirulent pathogen P. syringae pv tomato DC3000 (AvrRpm1).8 In the double mutant, AtrbohD x Atgox1, both nonhost pathogens tested grew ~10-fold more than the wild type (Fig. 2A and B) and was in agreement with the symptoms observed (Fig. 1). This finding indicates that AtrbohD x Atgox1 double mutant behaved similar to Atgox1 single mutant, in response to nonhost pathogen inoculation, and therefore we determined that AtGOX1 functions independently of AtRBOHD.
Figure 2. Differential response of AtrbohD x Atgox1 and AtrbohD x Atgox3 after inoculation with the nonhost pathogens P. syringae pv tabaci and P. syringae pv syringae B728A. Wild-type Col-0, single mutants AtrbohD, Atgox1, Atgox3 and double mutants AtrbohD x Atgox1 and AtrbohD x Atgox3 were inoculated with the nonhost pathogen P. syringae pv tabaci (A and D), P. syringae pv syringae (B and E) and P. syringae pv maculicola (C and F) at 1 × 104 CFU/ml. At 0 and 3 dpi, two leaf samples (0.5 cm2) from four biological replicates were collected, grounded, serially diluted and plated. Bacterial colonies were counted after two days. Bars represent means and standard deviation. One way ANOVA was used to determine statistical significance among treatments. After significance was found, LSD (least significant difference) test was used to determine differences between genotypes. Means with the same letter for a given time point are not significantly different at p < 0.05.
Interestingly, the growth of nonhost pathogens in the double mutant AtrbohD x Atgox3 was slightly compromised when compared with Atgox3 single mutant (Fig. 2D and E). This result suggests that the AtrbohD mutation is epistatic to the Atgox3 mutation and the former causes a reversion of the phenotypes observed in Atgox3 upon inoculation with nonhost pathogens. The mechanism behind this reversion is unknown but perhaps is related to the accumulation of the phytoalexin camalexin as observed in the cat2 AtrbohD double mutant or involves accumulation of defense metabolites by the AtrbohD mutant.8 The discrepancy between these and our previous results10 wherein silencing of AtRBOHD in Atgox1 and Atgox3 mutant did not show any effect on disease symptoms and the apparent additive effect in the bacterial growth is probably due to the cross-silencing of AtRBOHF. None of the mutants tested were hyper-susceptible to the host pathogen P. syringae pv maculicola (Fig. 2C and F), indicating that neither AtGOX genes nor AtRBOHD are involved in basal resistance responses.
In conclusion, we show that there is no additive effect of disease symptoms or bacterial growth in the AtrbohD x Atgox1 and AtrbohD x Atgox3 double mutants when compared with individual mutants. Slight additive effect observed previously10 upon silencing of AtRBOHD in Atgox1 and Atgox3 mutants was most likely due to cross-silencing of AtRBOHF. These results further prove that GOX plays a role in nonhost resistance independent of NADPH oxidase.
Acknowledgments
We thank Conner Boatright and Ziying Lei for assistance in the screening of double homozygous and Colleen Elles for plant care. This work was supported by The Samuel Roberts Noble Foundation and in part by a grant from the Oklahoma Center for the Advancement of Science and Technology (OCAST; PSB09-020).
Glossary
Abbreviations:
- HR
hypersensitive response
- ROS
reactive oxygen species
- Atrboh
respiratory burst oxidase
- GOX
glycolate oxidase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20429
References
- 1.Hammond-Kosack KE, Parker JE. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol. 2003;14:177–93. doi: 10.1016/S0958-1669(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 2.Heath MC. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol. 2000;3:315–9. doi: 10.1016/S1369-5266(00)00087-X. [DOI] [PubMed] [Google Scholar]
- 3.Heath MC. Hypersensitive response-related death. Plant Mol Biol. 2000;44:321–34. doi: 10.1023/A:1026592509060. [DOI] [PubMed] [Google Scholar]
- 4.Van Breusegem F, Dat JF. Reactive oxygen species in plant cell death. Plant Physiol. 2006;141:384–90. doi: 10.1104/pp.106.078295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–75. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 6.Torres MA, Dangl JL, Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A. 2002;99:517–22. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres MA, Jones JD, Dangl JL. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet. 2005;37:1130–4. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- 8.Chaouch S, Queval G, Noctor G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 2012;69:613–27. doi: 10.1111/j.1365-313X.2011.04816.x. [DOI] [PubMed] [Google Scholar]
- 9.Pogány M, von Rad U, Grün S, Dongó A, Pintye A, Simoneau P, et al. Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 2009;151:1459–75. doi: 10.1104/pp.109.141994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojas CM, Senthil-Kumar M, Wang K, Ryu CM, Kaundal A, Mysore KS. Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell. 2012;24:336–52. doi: 10.1105/tpc.111.093245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foyer CH, Bloom AJ, Queval G, Noctor G. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol. 2009;60:455–84. doi: 10.1146/annurev.arplant.043008.091948. [DOI] [PubMed] [Google Scholar]
- 12.Reumann S, Ma C, Lemke S, Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 2004;136:2587–608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]