Abstract
Phospholipase D is one of the crucial enzymes involved in lipid mediated signaling, triggered during various developmental and physiological processes. Different members of PLD gene family have been known to be induced under different abiotic stresses and during developmental processes in various plant species. In this report, we are presenting a detailed microarray based expression analysis and expression profiles of entire set of PLD genes in rice genome, under three abiotic stresses (salt, cold and drought) and different developmental stages (3-vegetative stages and 11-reproductive stages). Seven and nine PLD genes were identified, which were expressed differentially under abiotic stresses and during reproductive developmental stages, respectively. PLD genes, which were expressed significantly under abiotic stresses exhibited an overlapping expression pattern and were also differentially expressed during developmental stages. Moreover, expression pattern for a set of stress induced genes was validated by real time PCR and it supported the microarray expression data. These findings emphasize the role of PLDs in abiotic stress signaling and development in rice. In addition, expression profiling for duplicated PLD genes revealed a functional divergence between the duplicated genes and signify the role of gene duplication in the evolution of this gene family in rice. This expressional study will provide an important platform in future for the functional characterization of PLDs in crop plants.
Keywords: abiotic stress, development, gene expression, microarray analysis, phospholipase D
Introduction
Various abiotic stresses impose a set of adverse conditions on plants in a given environment, which hamper their growth and development, longevity and productivity. The major abiotic stresses include high salinity, drought, and temperature fluctuations i.e., cold and heat. To combat these adverse growth conditions plants have devised some adaptive mechanisms, which include the triggering of a number of signaling networks.1-4 Lipid mediated signaling has emerged as one of the major signaling pathways and recently well recognized to be triggered in response to various stresses in plants.5-8 Various environmental cues trigger the hydrolysis of membrane phospholipids leading to the generation of different classes of lipid and lipid-derived signal messengers such as phosphatidic acid (PA), diacylglycerol (DAG), DAG-pyrophosphate (DGPP), lysophospholipids, free fatty acids (FFAs), phosphoinositides, and inositol polyphosphates.9-11 Phospholipase D (PLD) group of enzymes hold a central place in the catalysis of membrane lipid hydrolysis in plants. PLD cleaves the terminal phosphodiesteric bond of phospholipids to release phosphatidic acid (PtdOH) and water-soluble free head group.12 PLD represents a major family of phospholipases in plants and comprised of multiple isoforms. Arabidopsis genome encodes 12 PLD members while 17 members have been reported in the genome of crop plant rice.13,14 In Arabidopsis, PLDs have been grouped into five classes namely; α, β, γ, δ and ζ based on their gene structure, domain organization, sequence similarity and biochemical properties.13 In rice genome; α, β, δ and ζ PLDs have been identified as orthologs of Arabidopsis PLDs, however PLDs of γ class are not encoded.14 In addition, two new classes of PLDs have been predicted in rice and designated as PLDκ and PLDφ, comprised of one member each. Strikingly, PLDφ class is structurally different from any other plant PLDs, as it harbours a signal peptide at N-terminal instead of a C2 domain (found in C2-PLDs) or PX/PH domain (found in PX/PH-PLDs) and therefore belongs to a unique structural class of PLDs known as SP-PLD. This PLD was identified in rice genome for the first time14 and recently also has been reported in grapes and poplar.15 Different PLD isoforms have been associated with abiotic stress (salt, drought and cold) triggered lipid signaling and associated responses in a number of plant species.16-21 Similarly, PLDs have also been implicated in the various stages of plant development.6,22,23 Although, functional role of PLDs have been well dissected in stress and development related processes in various plant species, knowledge about their involvement in similar pathways in crop plant rice has not been investigated adequately. To comprehend the underlying functional mechanism of a gene, expression profiling is a very handy tool as it provides clue to its functional relevance. At the same time, this also leads to development of preliminary platform from where the detailed functional role of respective genes can be unearthed by adopting comprehensive molecular, biochemical and genetic tools. With this view in mind, in this report we are providing a detailed genome-wide expression (transcriptomic) analysis of rice PLD gene family using gene chip microarray under abiotic stress conditions (salt, drought and cold) and during various stages of plant development (vegetative and reproductive). We have identified a set of differentially expressing PLD genes under these conditions and stages, and validated the expression profile for a few selected candidates under abiotic stresses using quantitative real time PCR. Moreover, we have marked the duplicated PLD members and generated an expression profile to show their functional relatedness.
Results
Expression profile of rice PLDs under abiotic stresses
Genome-wide expression profile of rice PLDs was generated using Affymetrix rice genome arrays data for 7 day old rice seedling treated for three abiotic stress conditions (salt, cold and drought). In comparison to control (untreated 7 day old seedling), a total of seven PLD members expressed differentially (either up or downregulated) under three abiotic stress conditions with several fold change in expression and significant P-value (Fig. 1, Table 1). Out of the seven genes, six genes; OsPLDα1, OsPLDα5, OsPLDα4, OsPLDα6, OsPLDδ1 and OsPLDζ1 were found to be significantly upregulated whereas only OsPLDφ was downregulated (gene names are adopted from Li et al. 200714). Analysis for the specific or overlapping expression of genes under stress conditions revealed that OsPLDα5 was commonly expressed under all three stresses. Similarly, OsPLDδ1 and OsPLDζ1 were commonly upregulated under both salt and drought stress conditions whereas OsPLDα4 was upregulated under both cold and drought stresses. Two PLD members; OsPLDα1 and OsPLDα6 were specifically expressed and upregulated under drought condition. None of the PLD members was found to be expressed exclusively either under salt or cold stress conditions (Table 1, Fig. 3A).
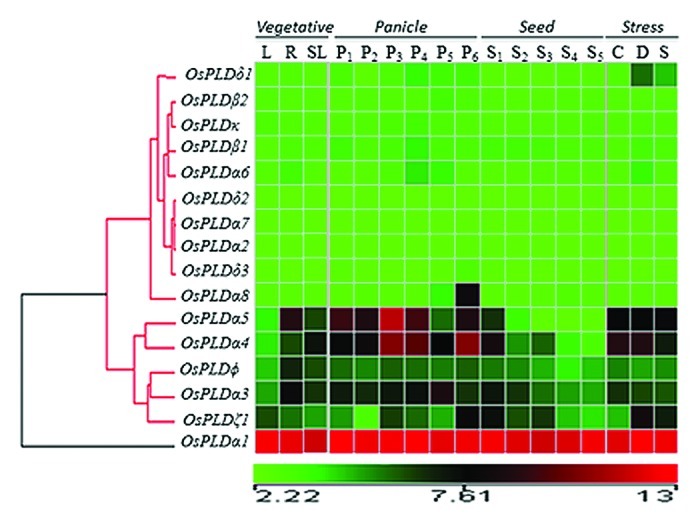
Figure 1. Expression of rice PLD gene family has been represented by a heat map. Developmental stages comprising three vegetative stages (L-leaf, R-root and SL-7dold seedling), six stages of panicle [P1 (0–3 cm), P2 (3–5 cm), P3 (5–10 cm), P4 (10–15 cm), P5 (15–22 cm), and P6 (22–30 cm)] and five stages of seed [S1 (0–2 DAP), S2 (3–4 DAP), S3 (4–10 DAP), S4 (11–20 DAP) and S5 (21–29 DAP)]. Clustering of the expression profile was done with log transformed average values taking mature leaf as base line. Three stress conditions are denoted as C, Cold Stress; D, Drought Stress; S, Salt Stress and SL, control, seven day old unstressed seedling. The scale at the bottom of each heat map is given in log2 intensity value.
Table 1. Microarray expression data for entire set of rice PLD genes under abiotic stresses.
| |
|
|
DROUGHT |
SALT |
COLD |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe Set ID | RGAP Locus | Gene | Fold | Regulation | P value | Fold | Regulation | P value | Fold | Regulation | P value |
| Os.155.1.S1_a_at |
LOC_Os01 g07760 |
OsPLDα1 |
1.76 |
U |
0.0014 |
1.45 |
U |
0.0063 |
1.19 |
U |
0.1379 |
| OsAffx.4272.1.S1_at |
LOC_Os05 g07880 |
OsPLDα2 |
1 |
D |
0.644 |
1.01 |
D |
0.543 |
1 |
U |
0.778 |
| Os.9878.1.S1_at |
LOC_Os06 g40190 |
OsPLDα3 |
1.22 |
D |
0.3375 |
1.39 |
D |
0.1648 |
1.05 |
U |
0.9337 |
| Os.50455.1.S1_at |
LOC_Os06 g40170 |
OsPLDα4 |
3.09 |
U |
0.0031 |
1.2 |
D |
0.55 |
3.16 |
U |
0.0599 |
| Os.50104.1.S1_at |
LOC_Os06 g40180 |
OsPLDα5 |
2.71 |
U |
0.0029 |
3.22 |
U |
0.0297 |
2.93 |
U |
0.0724 |
| Os.54571.2.A1_at |
LOC_Os03 g27370 |
OsPLDα6 |
1.99 |
U |
0.0352 |
1.08 |
U |
0.4458 |
1 |
U |
0.9794 |
| OsAffx.29446.1.S1_at |
LOC_Os08 g31060 |
OsPLDα7 |
1 |
U |
0.644 |
1 |
D |
1 |
1 |
D |
1 |
| Os.50273.1.S1_at |
LOC_Os09 g25390 |
OsPLDα8 |
1 |
D |
1 |
1 |
D |
0.5798 |
1 |
U |
1 |
| Os.9389.2.S1_s_at |
LOC_Os10 g38060 |
OsPLDβ1 |
1.3 |
U |
0.0135 |
1.07 |
U |
0.0754 |
1.06 |
D |
0.8352 |
| Os.34954.1.S1_at |
LOC_Os03 g02740 |
OsPLDβ2 |
1.02 |
U |
0.6604 |
1 |
U |
0.9118 |
1.01 |
U |
0.8858 |
| Os.11408.1.S1_at |
LOC_Os09 g37100 |
OsPLDδ1 |
8.52 |
U |
8.24E-04 |
4.12 |
U |
0.0781 |
1.23 |
U |
0.7385 |
| Os.52881.1.S1_at |
LOC_Os03 g62410 |
OsPLDδ2 |
1.01 |
D |
0.644 |
1.02 |
D |
0.4164 |
1 |
U |
1 |
| Os.5744.1.S1_at |
LOC_Os07 g15680 |
OsPLDδ3 |
1 |
D |
0.6778 |
1.01 |
D |
0.2723 |
1 |
D |
1 |
| OsAffx.14869.1.S1_at |
LOC_Os05 g29050 |
OsPLDζ1 |
9.93 |
U |
0.0019 |
3.79 |
U |
0.0193 |
1.27 |
D |
0.8026 |
| Os.6185.1.S1_at |
LOC_Os06 g44060 |
OsPLDφ |
2.22 |
D |
0.0088 |
1.79 |
D |
0.0094 |
1.65 |
D |
0.0351 |
| Os.56801.1.S1_at | LOC_Os02 g02790 | OsPLDκ | 1.05 | U | 0.027 | 1 | D | 0.9464 | 1 | D | 1 |
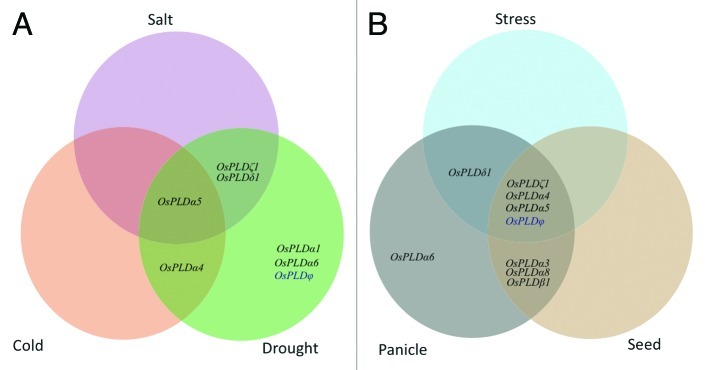
Figure 3. Venn diagram for differentially expressed PLDs. PLD genes up- and downregulated (A) under different abiotic stress conditions and (B) under stresses and developmental stages. Different compartments showing genes specific to either a particular stress/developmental stage or common to more than one stress and/or developmental stage.
Expression profile of rice PLDs during development
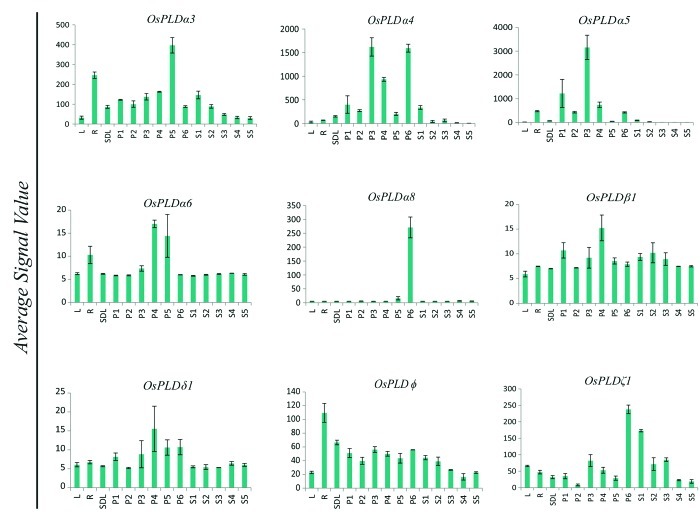
Expression profile for rice developmental stages was generated from the gene chip microarray data. Eleven reproductive developmental stages were analyzed including, six panicle stages (P1-P6) and five seed developmental stages (S1-S5) along with three vegetative stages namely; mature leaf, root and seedling. A detailed analysis of the expression profile revealed that a total of nine PLD genes expressed differentially during reproductive developmental stages when compared with three vegetative developmental stages (Fig. 2, Table S1). Out of the nine genes, eight PLDs; OsPLDα3, OsPLDα4, OsPLDα5, OsPLDα6, OsPLDα8, OsPLDβ1, OsPLDδ1 and OsPLDζ1 were upregulated and a single member OsPLDφ was downregulated. Six PLD members were commonly upregulated in both panicle and seed developmental stages. Two PLD members; OsPLDα6 and OsPLDδ1 were expressed exclusively in the panicle stages whereas none of the PLD gene was exclusively expressed in seed developmental stages (Fig. 3B, Table S1).
Figure 2. Expression profiles of differentially expressed PLD genes during developmental stages. Average signal intensity value of three replicates from microarray for all the developmental stages has been plotted to show the differential expression. Standard error bars have been shown. Y-axis represents signal values from microarray and X-axis shows different developmental stages.
Overlapping expression under abiotic stresses and development
Keeping the fact in mind that abiotic stresses and reproductive development are interconnected processes in the plant life cycle, which has also been proven by overlapping expression of genes in these conditions in prior studies,24-26 we attempted to investigate overlapping expression of PLDs involved in stress and development. Interestingly, all seven PLD genes, which were expressed differentially under abiotic stresses, were also found to be expressed significantly during reproductive developmental stages (Fig. 3A and B). In this subset of seven genes, three genes; OsPLDα4, OsPLDα5 and OsPLDζ1, which were highly upregulated under abiotic stress (mainly drought and high salinity) conditions, also found to be upregulated in panicle and seed developmental stages. OsPLDα4 and OsPLDα5 were mainly upregulated during different stages of panicle development whereas OsPLDζ1 was found to be highly upregulated in the later stages of panicle and early stages of seed development (Fig. 2). A single gene OsPLDδ1 was commonly upregulated under stresses and later stages of panicle development and had no significant expression in seed stages. On the other hand, only OsPLDφ was found to be downregulated in all abiotic stress conditions and developmental stages. Surprisingly, not a single PLD member was found to be commonly expressed in stress and seed developmental stages.
Expression profile of duplicated PLDs
Analysis for the chromosomal duplication revealed that 6 PLD members (3 pairs); OsPLDα1 (chr. 1): OsPLDα2 (chr. 5), OsPLDβ1 (chr. 10): OsPLDβ2 (chr. 3) and OsPLDζ1 (chr. 5): OsPLDζ2 (chr. 1), were present on the duplicated segments of chromosome and hence, exhibited segmental duplication. On the other hand, three genes; OsPLDα3, OsPLDα4, OsPLDα5 exhibited tandem duplication and were located in a cluster on the chromosome 6 (Table S2). Expression profile was generated for the duplicated PLD genes using microarray data for the three abiotic stresses and entire spectrum of developmental stages. Corresponding probeset was not available for OsPLDζ2 on the microarray gene chip, therefore expression profile could not be generated for the respective gene pair (OsPLDζ1:OsPLDζ2). Average signal values for abiotic stress conditions and developmental stages have been presented as an area-diagram (Fig. 4). Expression profile revealed that one of the segmentally duplicated pair OsPLDα1:OsPLDα2 exhibited pseudo-functionalization as one of the paired partners did not have any expression in tested conditions or developmental stages. On the other hand, another segmentally duplicated pair OsPLDβ1:OsPLDβ2 showed retention of expression as both the partners followed similar expression pattern. In the tandemly duplicated cluster of gene, while OsPLDα3 had negligible expression, pair of OsPLDα4: OsPLDα5 exhibited retention of expression in most of the conditions and stages. However, the magnitude of expression varied between the paired duplicated partners.
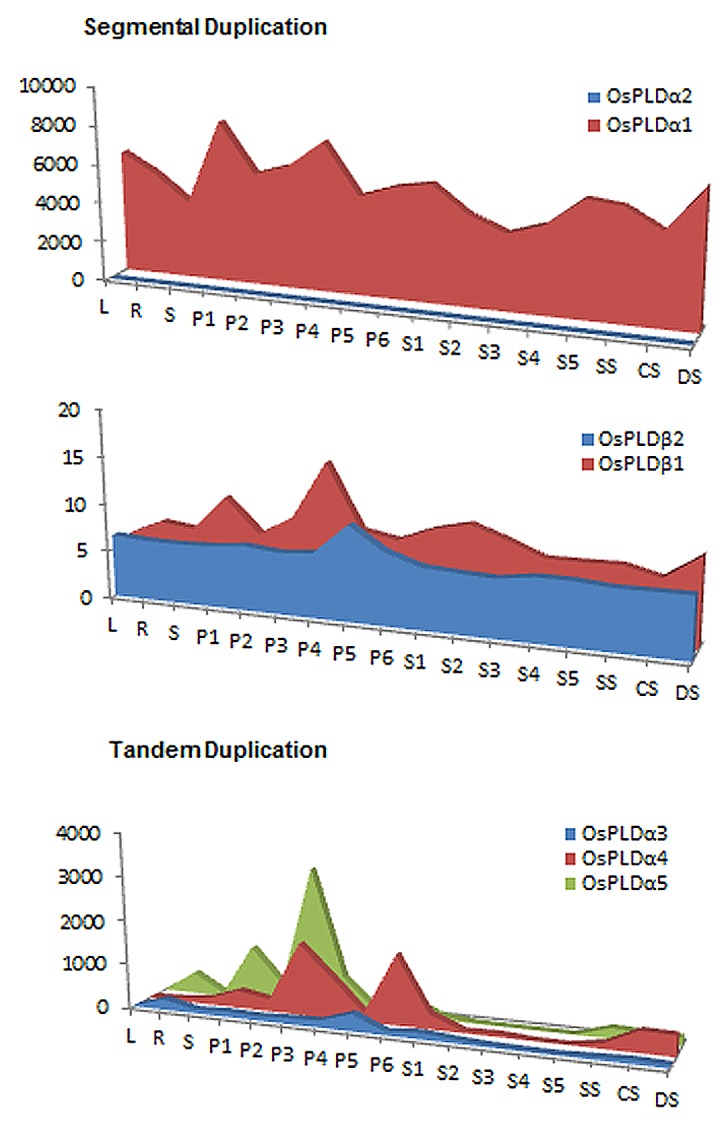
Figure 4. Expression profiles of duplicated PLD genes. Expression profiles of segmentally and tandemly duplicated gene pairs/clusters from microarray data were compared in various developmental stages including leaf (L), root (R) and seven day old seedling (S) tissue, in various stages of panicle development (P1-P6), seed development (S1-S5) and under cold stress (CS), dehydration stress (DS) and salt stress (SS). Each area graph represents compilation of the mean normalized signal intensity values.
Validation of microarray expression under abiotic stress conditions
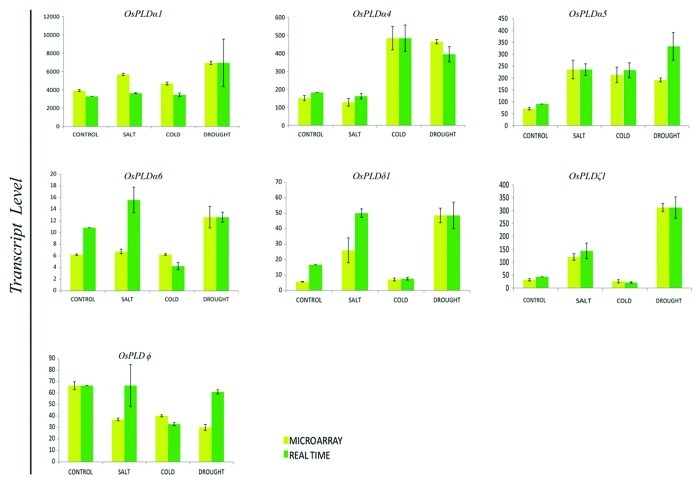
Expression pattern and profile of the seven PLD genes, which were expressing differentially and significantly either exclusively or overlapping under abiotic stress conditions were validated employing quantitative real time PCR. Interestingly, all the candidate genes exhibited anticipated expression pattern and clearly revealed the differential regulation of transcript level under different abiotic stresses (Fig. 5). For a few genes, the level of transcript was found to be much higher than estimated by microarray profile in conditions such as for OsPLDα6 and OsPLDδ1 in salt stress and for OsPLDφ in salt and drought stresses.
Figure 5. Validation of microarray expression profiles for differentially expressing PLD genes by quantitative RT-PCR. Two and three biological replicates were analyzed by real time PCR and microarray, respectively. Standard error bars have been shown for both microarray and real time PCR data. Y-axis represents raw expression values from microarray and normalized expression value from real time PCR and X-axis shows different experimental conditions.
Discussion
Abiotic stresses are the major factors, which hamper the normal plant growth and productivity. After extensive research, it has been appreciated that plants acquire tolerance or adaptability against these stresses by a plethora of signal transduction network. One of the important signaling networks is mediated by lipid molecules generated in response to multiple stimuli in plants. Therefore, it is imperative to understand the role of major lipid modifying enzymes such as phospholipase A, C and D. Phospholipase D (PLD) class has been implicated in multiple stress responses in Arabidopsis. In crop plant such as rice, not enough has been investigated for the role of PLD, which enticed us to look for the functional role of this family by focusing on gene expression analysis during different stages of development and abiotic stresses.
Expression profile of a gene provides an important clue regarding its functional role and molecular action; therefore, we performed a whole-genome transcript profiling for the rice PLD genes in three different abiotic stresses and during various developmental stages. Microarray data revealed that a subset of PLD genes expressed differentially and significantly under specific or multiple abiotic stress conditions (Table 1). In this subset of genes, out of six upregulated PLDs, four genes belong to α subfamily (OsPLDα1, OsPLDα4, OsPLDα5 and OsPLDα6), whereas one member each from δ and ζ subfamilies. This observation suggests that α subfamily of PLDs, which have evolved together in due course of time in rice and Arabidopsis, exhibited functional conservation and is majorly responsive to abiotic stresses. A single member of φ subfamily (OsPLD φ) was found to be repressed in all the three abiotic stresses. This deviation in expression for OsPLDφ suggests that this might act as a negative regulator of stress signaling or it has functional role in some processes other than stress signaling, which might be attributed to its novel domain structure (N-terminal signal peptide), because C2 domain and PX/PH domains in other PLDs are known to mediate the targeting of these enzymes to plasma membrane where the lipid hydrolysis occur in response to various stresses.27,28 These results were further supported by quantitative transcript analysis by real time PCR, which showed that all the tested PLD members had expression pattern and transcript level similar to microarray expression pattern under different abiotic stress conditions (Fig. 5). However, in case of OsPLDφ, the transcript abundance was not as low in case of salt and drought stresses as revealed by the microarray data. This type of variation in expression level from these two techniques (microarray and real time PCR) has been observed in previous studies also.24-26 Gene induction and transcript accumulation for PLDs in response to similar abiotic stresses has been observed previously, in different plant species such as Arabidopsis and tomato.21,29AtPLDα1, Arabidopsis ortholog of rice PLDα1 has been studied most extensively and was found to regulate ABA and drought mediated responses.30-33 Overexpression of AtPLDα1 made plants highly sensitive to ABA, which lead to enhanced stomatal closure and resulted in reduced transpirational water loss and drought tolerance, whereas the knockout plants showed opposite response.30,31 Similarly, AtPLDα1 has also been implicated in salinity and freezing stress mediated signaling pathways.21,34 Tobacco and tomato PLDα1 were also reported to mediate drought and salt stress responses.21,35 Another study showed that, AtPLDα3 knockout mutant plants exhibited high sensitivity towards salinity, dehydration and ABA, while gain-of-function of AtPLDα3 led to reduced sensitivity in transgenic plants.5 These observations are supportive to our findings of involvement of various PLDα subfamily members in different abiotic stresses in rice. Other PLD member, AtPLDδ has also been found to be involved in salinity and freezing responses and tolerance.17,21 With similar kind of expression profiles, sequence conservation and close evolutionary relationship,14 a functional conservation might also be predicted and significant role can be corroborated in abiotic stress signaling for rice PLD gene family members. Involvement of PLDs in various stress responses has been further supported by the presence of various cis-regulatory elements in their promoter, including ABRE (abscissic acid responsive element), LTR (low temperature responsive), MBS (myb binding site), Skn-1, GCN4, RY-element and motif IIb (Table S3). These motifs have been previously known to regulate various stress responses and plant development.36-40 Promoter of several PLD genes, which were significantly and differentially regulated in stress and developmental stages, and found to contain multiple cis-regulatory elements especially ABRE, MBS and Skn-1 known to control ABA, drought and development mediated induction of genes. Notably, OsPLDα1, OsPLDα5, OsPLDβ1, OsPLDδ1 and OsPLDζ1 were commonly induced in abiotic stresses and development, and found to contain most of these motifs. This observation indicated regulation of PLD genes by the cis-regulatory promoter elements and also accounted for their overlapping expression in abiotic stresses and development. Expression profiling for various developmental stages revealed that nine PLD genes showed significant variation in their expression level through a spectrum of vegetative and reproductive developmental stages (Table S1). Out of the nine PLDs, eight genes were upregulated in one or more panicle and seed development stage w.r.t three vegetative stages, whereas a single gene OsPLDφ was downregulated in the entire spectrum of reproductive development, similar to abiotic stresses. Among the upregulated genes, five PLD members belong to α subfamily and all of them were significantly expressed in different panicle stages and interestingly none of them is expressing during seed developmental stages (Fig. 2). Transcripts of OsPLDα3 were accumulated at P5 stage (vacuolated pollen), OsPLDα6 at P5-P6 and OsPLDα8 specifically at P6 (mature pollen), whereas OsPLDα5 expressed at P3 (meiosis) and OsPLDα4 in almost all the panicle stages.This has suggested the significant role of this subfamily of PLDs in the floral organ development in rice. OsPLDβ1 significantly expressed in panicle and seed developmental stages whereas OsPLDζ1 had significant expression in the late panicle stage (P6) and early seed developmental stages S1-S3 (early globular embryo to embryo morphogenesis stages) (Fig. 2). Previously, OsPLDβ1 expression was found to be upregulated in immature seeds and it was implicated in seed germination process in rice where it activated ABA signaling by SAPK leading to inhibition of seed germination.14 Some other studies also supported our finding regarding PLD genes in various developmental stages. In soybeans, the expression of a PLD gene varied during seed developmental and seed germination stages.22 Similarly, the high expression for the two Arabidopsis PLD members; PLDζ1 and PLDζ2 was observed in the roots and both of these genes were implicated in root elongation and patterning.41 Phosphatidic acid (PA) generated by the activity of PLD has been found to regulate ABA mediated seed germination in Arabidopsis.42 Apart from PLDs, the differential expression pattern has been observed for the members of phospholipase A (PLA) class under similar developmental stages in rice.26 Analysis for overlapping expression under abiotic stresses and reproductive development provided interesting results and revealed that all the seven PLD members, which expressed significantly and differentially under abiotic stresses were also differentially expressed during reproductive developmental stages (Fig. 3A and B). This kind of overlapping expression has been observed earlier for various gene families in plants,24-26,43,44 which further strengthen the fact that abiotic stresses and reproductive development are interconnected phenomena in plants, as dehydration conditions set in during the later stages of seed maturation and leads to seed dormancy.43,45,46 The overlapping expression as discussed earlier, can be attributed to the presence of cis-regulatory element such as ABRE in the promoters (especially for the genes such as OsPLDβ1), which controls both stress response and development through regulation of phytohormone ABA responses.25,49-51 Therefore, signaling pathways triggered by abiotic stresses and during development can be hypothesized to be commonly controlled by ABA involving PLDs. Additionally, expression profile of duplicated PLD genes revealed variable expression pattern for the paired partners. In the segmentally duplicated PLDs one pair (OsPLDα1: OsPLDα2) exhibited pseudo-functionalization while other pair (OsPLDβ1: OsPLDβ2) showed retention of expression (Fig. 4, Table S2). This observation is consistent with previous reports, which mention that segmentally duplicated genes exhibit a functional divergence.50 Among the three tandemly duplicated genes, one gene (OsPLDα3) had almost negligible expression exhibiting pseudo-functionalization while other two had similar expression pattern hence showed retention of expression. However, the amplitude of expression was variable, which could be attributed to the variability in the cis-regulatory elements.25,51 This kind of variable expression pattern among the duplicated genes could be due to the lack of intense selection pressure during the course of evolution.50,52-54 Gene duplication, therefore, might have played a significant role in the functional diversification of this gene family in rice.
In conclusion, this study provides a comprehensive expression profile for the entire set of rice PLD genes under abiotic stress conditions and during developmental stages, which has not been performed until now. Subsets of PLD genes with differential expression profile during abiotic stresses and reproductive development have been identified, which signify the role of PLDs in abiotic stress signaling and during development in rice. Moreover, most of the differentially expressed PLD genes exhibited overlapping expression under stress conditions and during development, indicating the crosstalk of these pathways involving PLDs possibly through a common component such as ABA. Duplicated PLD genes showed variable expression pattern indicating the role of gene duplication in the functional diversification of PLD gene family in rice.
Materials and Methods
Plant material, growth conditions and abiotic stress treatment
The tissues samples were collected from field grown rice plants (Oryza sativa ssp. Indica var IR64), at different panicle and seed development stages. To avoid wounding, collected panicles were instantly frozen in liquid nitrogen. Stress treatment was given to IR64 rice seeds, which were first sterilized with 0.1% HgCl2 and grown in culture room conditions at 28 ± 1°C with a daily photoperiodic cycle of 14 h light and 10 h dark. After seven days growth, seedlings were subjected to different stress treatments according to Ray et al. 2007.51
Microarray expression analysis
To perform the microarray based expression analysis, the total RNA was isolated from the three replicates of rice tissues representing different stages of plant development, which included three vegetative stages (mature leaf, 7 days old seedling and their roots), 11 reproductive stages (P1–P6 and S1–S5; representing panicle and seed developmental stages, respectively) and three abiotic stress conditions, i.e., cold, salt, and drought. Microarray experiments were then performed using 51 Affymetrix GeneChip Rice Genome Arrays (Gene Expression Omnibus, GEO, platform accession number GPL2025). The raw data (*.cel) files generated from all the chips were imported to Array Assist 5.0 software (Stratagene, USA) for detailed analysis according to Arora et al. 2007.24 The microarray expression data have been deposited in the gene expression omnibus (GEO) database at NCBI under the series accession numbers GSE6893 and GSE6901.
Quantitative expression analysis by real time PCR
Real time PCR was performed to verify the microarray data for a few selected genes, which showed significant differential expression pattern under abiotic stress conditions. The primers were designed for all the selected genes preferentially, from 3′ end, using PRIMER EXPRESS (PE Applied Biosystems, USA), with default parameters. Primers were further checked for their specificity, using BLAST tool of NCBI and dissociation curve analysis after the PCR reaction (Table S4). First strand cDNA was prepared from 4 µg of DNase treated total RNA, in 100 µl of reaction volume using high-capacity cDNA Archive kit (Applied Biosystems, USA). SYBRGreen PCR Master Mix (Applied Biosystems, USA) was used to determine the transcript levels in ABI Prism 7000 Sequence detection System (Applied Biosystems, USA). Biological duplicates of each sample were taken for the analysis. The average Ct values were calculated by taking the average of three technical replicates for each sample. The cDNA variance among samples was normalized using ACTIN as the endogenous control. Relative expression values were calculated by ΔΔCt method and normalized the data against the maximum average expression value from microarray.
Gene duplication
The RGAP (rice genome annotation project) version 6.1 (http://rice.plantbiology.msu.edu/) segmental duplication database was explored to find out the segmentally duplicated genes. Genes separated by five or fewer genes were considered as tandemly duplicated and amino acid sequence homology of these gene products was computed using MegAlign software 5.07©.
Promoter analysis
To find out various cis-acting regulatory elements in the promoter of all the PLD genes, 1 kb upstream region from translation start site was extracted from RGAP ver 6.1. One kb upstream region was subsequently scanned in PlantCARE database55 for the presence of various regulatory motifs and elements. Various motifs involved in stress responses and plant development were identified.
Supplementary Material
Acknowledgment
This work is partially supported by internal grants of University Grant Commission (UGC), Department of Biotechnology (DBT), Department of Science and Technology (DST) and Council for Scientific and Industrial Research (CSIR), India to G.K.P., A.S. and V.B. acknowledge CSIR for their research fellowship.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20385
References
- 1.Munnik T, Meijer HJ. Osmotic stress activates distinct lipid and MAPK signalling pathways in plants. FEBS Lett. 2001;498:172–8. doi: 10.1016/S0014-5793(01)02492-9. [DOI] [PubMed] [Google Scholar]
- 2.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–73. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–83. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 5.Hong Y, Pan X, Welti R, Wang X. Phospholipase Dalpha3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell. 2008;20:803–16. doi: 10.1105/tpc.107.056390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong Y, Devaiah SP, Bahn SC, Thamasandra BN, Li M, Welti R, et al. Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 2009;58:376–87. doi: 10.1111/j.1365-313X.2009.03788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Hong Y, Wang X. Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta 2009; 1791:927-35. [DOI] [PubMed]
- 8.Munnik T, Testerink C. Plant phospholipid signaling: “in a nutshell”. J Lipid Res. 2009;50(Suppl):S260–5. doi: 10.1194/jlr.R800098-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bargmann BO, Munnik T. The role of phospholipase D in plant stress responses. Curr Opin Plant Biol. 2006;9:515–22. doi: 10.1016/j.pbi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Boss W, Lynch D, Wang X. Lipid-mediated signaling. Annu Plant Rev. 2008;33:202–43. [Google Scholar]
- 11.Tuteja N, Sopory SK. Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal Behav. 2008;3:79–86. doi: 10.4161/psb.3.2.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijer HJ, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- 13.Qin C, Wang X. The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol. 2002;128:1057–68. doi: 10.1104/pp.010928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, Lin F, Xue HW. Genome-wide analysis of the phospholipase D family in Oryza sativa and functional characterization of PLD beta 1 in seed germination. Cell Res. 2007;17:881–94. doi: 10.1038/cr.2007.77. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Zhang C, Yang Y, Hu X. Genome-wide and molecular evolution analyses of the phospholipase D gene family in Poplar and Grape. BMC Plant Biol. 2010;10:117. doi: 10.1186/1471-2229-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sang Y, Zheng S, Li W, Huang B, Wang X. Regulation of plant water loss by manipulating the expression of phospholipase Dalpha. Plant J. 2001;28:135–44. doi: 10.1046/j.1365-313X.2001.01138.x. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Li M, Zhang W, Welti R, Wang X. The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat Biotechnol. 2004;22:427–33. doi: 10.1038/nbt949. [DOI] [PubMed] [Google Scholar]
- 18.Testerink C, Munnik T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005;10:368–75. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Rajashekar CB, Zhou HE, Zhang Y, Li W, Wang X. Suppression of phospholipase Dalpha1 induces freezing tolerance in Arabidopsis: response of cold-responsive genes and osmolyte accumulation. J Plant Physiol. 2006;163:916–26. doi: 10.1016/j.jplph.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Mane SP, Vasquez-Robinet C, Sioson AA, Heath LS, Grene R. Early PLDalpha-mediated events in response to progressive drought stress in Arabidopsis: a transcriptome analysis. J Exp Bot. 2007;58:241–52. doi: 10.1093/jxb/erl262. [DOI] [PubMed] [Google Scholar]
- 21.Bargmann BO, Laxalt AM, ter Riet B, van Schooten B, Merquiol E, Testerink C, et al. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 2009;50:78–89. doi: 10.1093/pcp/pcn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu SB, Zheng L, Wang X. Changes in phospholipase D expression in soybeans during seed development and germination. J Am Oil Chem Soc. 1995;•••:73. [Google Scholar]
- 23.Chen G, Snyder CL, Greer MS, Weselake RJ. Biology and biochemistry of plant phospholipases. Crit Rev Plant Sci. 2011;30:239–58. doi: 10.1080/07352689.2011.572033. [DOI] [Google Scholar]
- 24.Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics. 2010;11:435. doi: 10.1186/1471-2164-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh A, Baranwal V, Shankar A, Kanwar P, Ranjan R, Yadav S, et al. Rice phospholipase A superfamily: organization, phylogenetic and expression analysis during abiotic stresses and development. PLoS One. 2012;7:e30947. doi: 10.1371/journal.pone.0030947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopka J, Pical C, Hetherington AM, Müller-Röber B. Ca2+/phospholipid-binding (C2) domain in multiple plant proteins: novel components of the calcium-sensing apparatus. Plant Mol Biol. 1998;36:627–37. doi: 10.1023/A:1005915020760. [DOI] [PubMed] [Google Scholar]
- 28.van Leeuwen W, Okrész L, Bögre L, Munnik T. Learning the lipid language of plant signalling. Trends Plant Sci. 2004;9:378–84. doi: 10.1016/j.tplants.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Wang C, Sang Y, Zheng L, Qin C. Determining functions of multiple phospholipase Ds in stress response of Arabidopsis. Biochem Soc Trans. 2000;28:813–6. doi: 10.1042/BST0280813. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Qin C, Zhao J, Wang X. Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci U S A. 2004;101:9508–13. doi: 10.1073/pnas.0402112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra G, Zhang W, Deng F, Zhao J, Wang X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312:264–6. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- 32.Zhang TT, Song YZ, Liu YD, Guo XQ, Zhu CX, Wen FJ. Overexpression of phospholipase D alpha gene enhances drought and salt tolerance of Populus tomentosa. Chin Sci Bull. 2008;53:3656–65. doi: 10.1007/s11434-008-0476-1. [DOI] [Google Scholar]
- 33.Peng Y, Zhang J, Cao G, Xie Y, Liu X, Lu M, et al. Overexpression of a PLDα1 gene from Setaria italica enhances the sensitivity of Arabidopsis to abscisic acid and improves its drought tolerance. Plant Cell Rep. 2010;29:793–802. doi: 10.1007/s00299-010-0865-1. [DOI] [PubMed] [Google Scholar]
- 34.Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, et al. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–2002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 35.Hong Y, Zheng S, Wang X. Dual functions of phospholipase Dalpha1 in plant response to drought. Mol Plant. 2008;1:262–9. doi: 10.1093/mp/ssm025. [DOI] [PubMed] [Google Scholar]
- 36.Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, et al. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–48. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 37.Jiang C, Iu B, Singh J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol. 1996;30:679–84. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- 38.Takaiwa F, Oono K, Kato A. Analysis of the 5′ flanking region responsible for the endosperm-specific expression of a rice glutelin chimeric gene in transgenic tobacco. Plant Mol Biol. 1991;16:49–58. doi: 10.1007/BF00017916. [DOI] [PubMed] [Google Scholar]
- 39.Kim SY, Wu R. Multiple protein factors bind to a rice glutelin promoter region. Nucleic Acids Res. 1990;18:6845–52. doi: 10.1093/nar/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Qin C, Welti R, Wang X. Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 2006;140:761–70. doi: 10.1104/pp.105.070995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katagiri T, Ishiyama K, Kato T, Tabata S, Kobayashi M, Shinozaki K. An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. Plant J. 2005;43:107–17. doi: 10.1111/j.1365-313X.2005.02431.x. [DOI] [PubMed] [Google Scholar]
- 43.Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol. 2007;65:467–85. doi: 10.1007/s11103-007-9199-y. [DOI] [PubMed] [Google Scholar]
- 44.Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant. 2008;1:732–50. doi: 10.1093/mp/ssn047. [DOI] [PubMed] [Google Scholar]
- 45.Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410:327–30. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- 46.Hetherington AM. Guard cell signaling. Cell. 2001;107:711–4. doi: 10.1016/S0092-8674(01)00606-7. [DOI] [PubMed] [Google Scholar]
- 47.Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–63. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 48.Sirichandra C, Wasilewska A, Vlad F, Valon C, Leung J. The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J Exp Bot. 2009;60:1439–63. doi: 10.1093/jxb/ern340. [DOI] [PubMed] [Google Scholar]
- 49.Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–37. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 51.Ray S, Agarwal P, Arora R, Kapoor S, Tyagi AK. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica) Mol Genet Genomics. 2007;278:493–505. doi: 10.1007/s00438-007-0267-4. [DOI] [PubMed] [Google Scholar]
- 52.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 53.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–64. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cusack BP, Wolfe KH. When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet. 2007;23:270–2. doi: 10.1016/j.tig.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–7. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.