Abstract
The GPN proteins are a poorly characterized and deeply evolutionarily conserved family of three paralogous small GTPases, Gpn1, 2, and 3. The founding member, GPN1/NPA3/XAB1, is proposed to function in nuclear import of RNA polymerase II along with a recently described protein called Iwr1. Here we show that the previously uncharacterized protein Gpn2 binds both Gpn3 and Npa3/Gpn1 and that temperature-sensitive alleles of Saccharomyces cerevisiae GPN2 and GPN3 exhibit genetic interactions with RNA polymerase II mutants, hypersensitivity to transcription inhibition, and defects in RNA polymerase II nuclear localization. Importantly, we identify previously unrecognized RNA polymerase III localization defects in GPN2, GPN3, and IWR1 mutant backgrounds but find no localization defects of unrelated nuclear proteins or of RNA polymerase I. Previously, it was unclear whether the GPN proteins and Iwr1 had overlapping function in RNA polymerase II assembly or import. In this study, we show that the nuclear import defect of iwr1Δ, but not the GPN2 or GPN3 mutant defects, is partially suppressed by fusion of a nuclear localization signal to the RNA polymerase II subunit Rpb3. These data, combined with strong genetic interactions between GPN2 and IWR1, suggest that the GPN proteins function upstream of Iwr1 in RNA polymerase II and III biogenesis. We propose that the three GPN proteins execute a common, and likely essential, function in RNA polymerase assembly and transport.
Keywords: RNA polymerase assembly, nuclear localization, small GTPases, genome instability, transcription
CHROMOSOME instability (CIN) refers to an increased rate of aneuploidy, meaning loss or gain of large pieces of DNA (i.e., whole chromosomes, fragments) in daughter cells. CIN is observed in the majority of solid tumors (Weaver and Cleveland 2006) and, because it increases the mutational space in a cell population, it is thought to predispose cells to accumulating the right combination of oncogene, tumor-suppressor, and other mutations that lead to cancer (Stratton et al. 2009; Loeb 2011). While tumor-associated gene variants are being identified at a phenomenal pace (e.g., International Cancer Genome Consortium 2010), the impact of a given gene variant on CIN is typically not known.
Our previous efforts to systematically catalog CIN phenotypes among essential yeast genes have uncovered the cellular pathways required to maintain genome stability (Stirling et al. 2011). This study showed that both predictable and less-predictable pathways emerge as highly enriched for CIN genes. In addition, the CIN gene catalog identified a suite of conserved CIN genes that are poorly characterized (Ben-Aroya et al. 2008; Stirling et al. 2011). In principle, any perturbation of a conserved process or CIN gene could be responsible for modulating genome stability in human cancer. YOR262W (hereafter referred to as GPN2) was identified as a conserved and essential CIN gene in one of our recent efforts (Ben-Aroya et al. 2008). GPN2 belongs to a highly conserved family of small GTPases and exists in yeast and humans with two paralogs, GPN1 (yeast NPA3) and GPN3 (YLR243W, hereafter referred to as GPN3). In archaea, a single GPN gene encodes a protein with a conserved glycine–proline–asparagine insertion in the G domain that gives the family its name. The 3D structure of the archaeal GPN protein (PAB9855) reveals a homodimeric molecule with a canonical GTPase domain and the purified protein exhibits GTPase activity in vitro (Gras et al. 2007)
The first characterized human GPN ortholog (GPN1/XAB1/NPA3) was shown to bind the nucleotide excision repair protein XPA and was named XAB1 (XPA binding protein 1). The XPA–XAB1 interaction was presciently, although indirectly, suggested to play a role in nuclear localization of XPA because deletion of amino acids required for nuclear localization disrupted XAB1 binding (Nitta et al. 2000). Subsequent mass spectrometry studies of partially assembled RNA polymerase II (RNAPII) complexes identified the Gpn1, Gpn2, and Gpn3 proteins and revisited the possibility of a role for GPNs in nuclear transport (Boulon et al. 2010; Forget et al. 2010). These observations initiated directed studies of GPN1 function in human tissue culture and yeast. In yeast, reduction of function of NPA3/GPN1 leads to chromatid cohesion defects, cell-cycle defects, and cytoplasmic mislocalization of the RNAPII subunits Rpb1 and Rpb3 (Alonso et al. 2011; Staresincic et al. 2011). Mutations of NPA3/GPN1 designed to abrogate its GTP binding or hydrolysis activities also cause defects in RNAPII nuclear transport. Work in human cells shows that both GPN1 and GPN3 are required for nuclear import of RNAPII, leaving the function of GPN2 unknown (Calera et al. 2011; Carre and Shiekhattar. 2011). Interestingly, another poorly characterized protein, Iwr1, was recently shown be important in the localization of RNA polymerase II, presumably in cooperation with the GPNs (Czeko et al. 2011). Unlike the GPNs and indeed unlike RNA polymerases themselves, Iwr1 contains a bipartite nuclear localization signal (NLS), which may serve to direct the nascent RNA polymerase to a karyopherin-mediated nuclear import pathway (Czeko et al. 2011). IWR1 function has also been linked to the transcriptional activity of all three nuclear RNA polymerases (Esberg et al. 2011). Importantly, IWR1 is dispensable for cell viability unlike the three GPN genes; therefore, its role in nuclear import of RNA polymerases must be buffered by some other cellular activity.
In this study we characterize mutant alleles of the uncharacterized yeast GPN2 and GPN3 orthologs. Mutations in these genes cause a chromosome-loss phenotype and sensitivity to UV and genome-destabilizing chemicals. The GPNs also exhibit physical and genetic interactions with one another, supporting a common function for this protein family. Remarkably, rather than having independent functions in the nuclear transport of different substrates as might have been predicted, mutants in either GPN2 or GPN3 cause defects in the localization of protein subunits of both RNAPII and III. Finally, we show that fusion of a NLS to the RNAPII subunit Rpb3 does not restore nuclear localization of Rpb1 in GPN mutants, while partially rescuing the defects in iwr1Δ. Our findings, combined with the literature, support a model in which all three GPNs serve independently essential functions in the biogenesis of RNA polymerase II and III that are upstream of the NLS activity contained in Iwr1.
Materials and Methods
Yeast growth, strains, and plasmids
Yeast strains and plasmids used in this study are listed in Supporting Information, Table S1. Temperature-sensitive (ts) alleles were generated as described (Ben-Aroya et al. 2008). Other strains were constructed by PCR-mediated one-step gene replacement using a published set of tagging cassettes and standard PEG/lithium acetate transformation (Longtine et al. 1998). For the RPB3– and RPC40–NLS fusions, the SV40 NLS sequence (PKKKRKV) was incorporated into mutagenic primers designed to fuse the NLS to the C terminus of the protein followed by the KanMX marker from pFA6–KanMX6 (Longtine et al. 1998). The TAP- and GFP-tagged strains were obtained from the proteome-wide TAP (Open Biosystems) and GFP collections (laboratory of Brenda Andrews). Site-directed mutagenesis was performed using the Quickchange lightning kit (Agilent Technologies) and GPN2 and GPN3 clones from the MoBY–ORF collection (Ho et al. 2009). Yeast were grown in rich media (YPD) or synthetic media as indicated and at the temperatures indicated. Hydroxyurea or 6-azauracil (Sigma) was added at the indicated concentrations to YPD or SC–uracil media, respectively. Genetic interactions were assessed by tetrad analysis. Viable double mutants were subjected to spot dilution or growth curve assays to confirm enhanced sickness of double mutants. For spot plating assays, overnight cultures of the indicated strains were normalized to the same OD600 and subsequently spotted in 10-fold serial dilutions onto the indicated media and grown for 24–48 hr before imaging.
GPN mutant allele sequencing
Phenol-chloroform preparations of mutant strain genomic DNA were made and the GPN genes were amplified using flanking primers. PCR products were purified using the ChargeSwitch kit (Invitrogen) and cycle sequencing amplification was conducted using the BigDye kit (Applied Biosystems). Sequencing was performed by the NAPS unit at the Michael Smith laboratories in the University of British Columbia.
Microscopy
Overnight cultures were diluted and allowed to grow for several hours to reenter log phase and subsequently shifted to the indicated temperatures for 3 hr. For live cell imaging, log-phase cells were mounted on concanavalin A coated slides, washed, and imaged in SC media essential as described (Carroll et al. 2009; Stirling et al. 2012b). In some cases 1 μg/ml DAPI was added to live cells immediately prior to imaging to mark the nucleus. Live and fixed cells were imaged with the appropriate filter sets on a Zeiss Axioscope using Metamorph software (Molecular Devices).
Localization scoring and statistics
GFP localization was assessed qualitatively by counting the proportion of cells with strong nuclear signal (i.e., the predominant localization in wild type) and comparing the wild-type control to GPN mutants using Student’s t-test. We also extracted a quantitative measure of mislocalization by defining a pixel area within the DAPI-stained nucleus and another, equally sized area immediately adjacent in the unstained cytosol. The nuclear localization score expresses the total fluorescence pixel intensity within these two areas as a ratio (i.e., nucleus/cytoplasm). Pooling intensity ratio data for three replicates in which at least 30 cells were scored permitted computation of the mean and standard deviation of scores. Where two samples were compared, Student’s t-test was used to determine significance (P-values are reported in the figures). Where more than two samples were compared, a one-way ANOVA followed by Tukey’s post-hoc test was used to determine whether the samples were significantly different (α is reported in the figures).
TAP pulldowns and Western blotting
TAP pulldowns were performed essentially as described (Kobor et al. 2004). Briefly, 100 mL TAP-tagged and control cultures were grown to an OD of 1.0, collected via centrifugation and lysed in TAP-IP buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1.5 mM magnesium acetate, 10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA, protease inhibitors) using glass beads and a Precellys 24 tissue homogenizer. Lysates were cleared via centrifugation at 14,000 rpm for 15 min and then incubated with IgG beads (GE Healthcare) for 90 min with rotation. Beads containing bound proteins were washed four times with TAP–IP buffer followed by elution with 2× SDS–PAGE buffer. TAP pulldowns involving Gpn3–TAP required growth in yeast media containing 2% galactose as the N-terminally tagged Gpn3 was functional only when overexpressed from a GAL1 promoter.
The pulldown fractions and lysates were loaded onto Mini-Protean TGX acrylamide gels (BioRad), transferred to nitrocellulose membranes, and blocked using PBS containing 0.02% Tween-20 and 5% skim milk powder. Blots were probed with mouse monoclonal anti-myc antibodies (Roche Cat no. 11667203001, 1:2500) or rabbit anti-TAP (Thermo Scientific Prod. no. CAB1001, 1:2000) followed by anti-mouse horseradish peroxidase (HRP) or anti-rabbit HRP conjugated antibodies (1:15,000). Blots were imaged using Bio-Rad Chemi-Doc system.
For protein quantitation Western blots, 5 mL log phase cultures were collected by centrifugation, resuspended in TAP–IP lysis buffer, and lysed in the Precellys with acid washed glass beads. Lysates were cleared by centrifugation and quantitated using the Bradford protein assay (Bio-Rad). Total protein, 10 μg, was run on an SDS–PAGE gel, and transferred to a nitrocellulose membrane for blocking. Membranes were washed and incubated with anti-Rpb3 antibody (Neoclone Cat. no. W0012, 1:1000 dilution) followed by goat anti-mouse HRP secondary antibody (1:3000). Blots were imaged, stripped (200 mM glycine, 0.1%SDS, 1% Tween 20, pH 2.2), and reprobed with anti-PGK1 antibody (Invitrogen Cat. no. 6457, 1:10,000) followed by goat anti-mouse HRP (1:15,000) followed by imaging.
Results
Mutational analysis of the GPNs: An evolutionarily conserved protein family
Three distinct GPN GTPases have been identified in humans and yeast and each GPN is more closely related to its interspecies ortholog than its intraspecies paralogs suggesting that three functionally conserved subfamilies may exist (Figure S1). To understand the evolutionary roots of the GPNs, we identified GPN1-, 2-, and 3-like sequences using BLAST-P, from a set of diverse eukaryotes and archaea and built a phylogenetic tree (Figure S1). Our phylogenetic analysis suggests that while GPN-like proteins are also found in diverse archaeal species, they are not universally conserved in archaea and are never found in bacteria (Figure S1).This analysis identified likely members of each GPN1, 2, and 3 group in all the diverse eukaryotes examined, including even one of the most minimal eukaryotic genomes known from the microsporidian Encephalitzoon cuniculi (Figure S1;(Katinka et al. 2001). The remarkable conservation of three distinct GPN proteins across eukaryotes suggests that each GPN serves an important function not served by the paralogous GPN proteins. The importance of each GPN is supported by the fact that NPA3/GPN1, GPN2, and GPN3 are all essential genes in Saccharomyces cerevisiae (Giaever et al. 2002).
NPA3 (GPN1) is the best-characterized yeast GPN, whereas GPN2 and GPN3 are considerably less studied. To generate tools to specifically interrogate the functions of GPN2 and GPN3 we created mutant alleles of each gene using a mutagenic PCR approach described previously (Ben-Aroya et al. 2008). Four gpn2 mutants and three gpn3 mutants that exhibit hypomorphic and, in some cases, ts growth were isolated (Figure 1A and Figure S2). The mutant alleles were sequenced and variants were mapped onto the crystal structure of archaeal GPN PAB0955 to assess the potential functional impact. Each mutant had between one and five amino acid changes resulting from missense mutations (Table S2). In some cases the mutations appeared in the GTPase domain, while in others the putative dimer interface was more directly affected (summarized in Figure 1A). While the specific functional consequence of each mutation remains to be assessed, each allele was a partial loss-of-function mutation that decreased cellular fitness and we selected alleles that showed hypomorphic growth with temperature-dependent phenotypes and low incidence of suppressors, gpn2-2 and gpn3-1 (Figure S2), for further study.
Figure 1.
Generation and characterization of GPN2 and GPN3 ts alleles. (A) Schematic of the 347 amino acid Gpn2 and 272 amino acid Gpn3 proteins with amino acid substitutions in each mutant allele used in this study overlaid. (B) GPN3 mutations cause chromosome integrity defects. Red sectoring is indicative of loss of an artificial chromosome containing a suppressor tRNA that prevents the accumulation of red pigment due to the ade2-101 mutation (Spencer et al. 1990). (C) GPN2 and GPN3 mutants exhibit heightened sensitivity to the DNA-damaging agent hydroxyurea at 30°. (D) GPN mutants exhibit sensitivity to exposure to 25 J/m2 ultraviolet radiation exposure followed by growth at the semipermissive temperature of 34° for 48 hr.
GPN2 was originally described as a chromosomal instability mutant displaying sister chromatid cohesion defects, a phenotype that was also subsequently observed for GPN1 mutants (Ben-Aroya et al. 2008; Alonso et al. 2011). Some hypomorphic alleles of GPN3 created here also show a visible chromosome transmission fidelity (CTF) phenotype (Figure 1B; Spencer et al. 1990; Stirling et al. 2012a). Thus each member of the GPN family appears important for maintaining genome integrity. Some of the GPN2 and GPN3 mutants also exhibited increased sensitivity to hydroxyurea, a nucleotide pool poison that increases stalling of replication forks (Figure 1C). Since human GPN1 has been reported to bind the UV-damage repair protein XPA/Rad14 we also tested UV sensitivity in GPN mutants and found that they were hypersensitive to a UV dose of 25 J/m2 (Figure 1D; Nitta et al. 2000). Importantly mutants in RNAPII show a CTF phenotype and other genome integrity defects (Stirling et al. 2011, 2012b). Together, this suggests that the CIN phenotype of GPN mutants could be directly related to loss of function in RNA polymerase.
GPN2 exhibits genetic and physical interactions with GPN3 and NPA3/GPN1 and IWR1
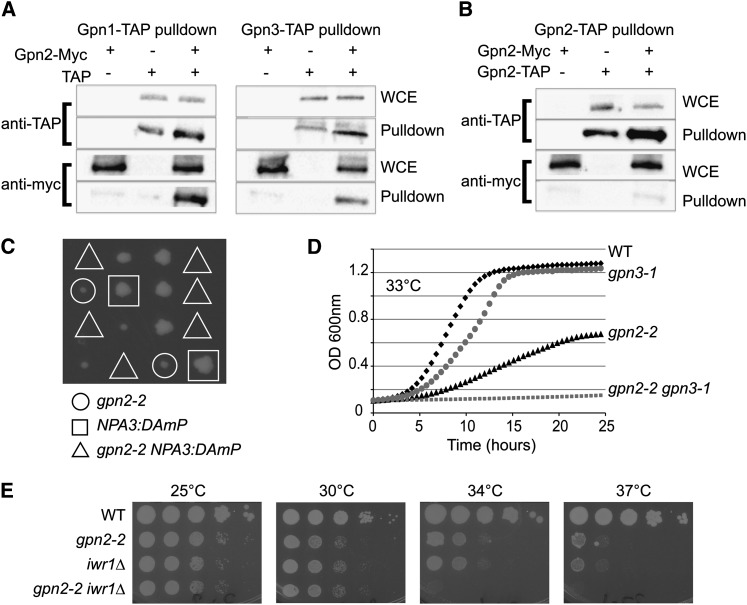
The mutants we describe for GPN2 and GPN3 share genome integrity phenotypes consistent with the possibility that the GPNs cooperate to execute some common cellular function. Indeed, physical interactions between human and yeast GPN family members have previously been shown (Boulon et al. 2010; Staresincic et al. 2011). In yeast, two-hybrid data suggest that Npa3 binds to both Gpn2 and Gpn3 (Uetz et al. 2000). We found that Gpn2 also binds to both Npa3 and Gpn3 by affinity purification and Western blotting of epitope-tagged protein (Figure 2A). It should be noted that while C-terminal Gpn2 and Npa3 epitope fusions (e.g., 13-myc or TAP) supported robust growth as the sole source of the protein, Gpn3 fusions had to be N terminal and were not completely functional, only supporting growth when overexpressed from a GAL promoter (unpublished observation). Gpn3 is the smallest GPN protein (∼9 kDa smaller than Gpn2) and is missing a fungal-specific C-terminal domain found in Gpn2; whether this accounts for its sensitivity to epitope tagging is unknown. Together with our data, interactions between all yeast GPNs have now been observed and the structural and in vitro data suggest that these interactions may involve oligomerization (Figure 2A; (Gras et al. 2007; Staresincic et al. 2011)).
Figure 2.
Functional relationships between GPN proteins and Iwr1. Coprecipitation tests of Gpn2–13-Myc by TAP fusions of Npa3 or Gpn3 (A) or Gpn2–TAP (B) as measured by Western blotting with anti-TAP or anti-Myc antibodies. WCE, whole cell extract. (C, D, and E) Genetic interactions between GPN2 mutants and NPA3/GPN1, GPN3, and IWR1 mutants. (C) Tetrad dissection of gpn2-2 NPA3::DAMP double mutants. Triangles indicate where double mutant colonies should grow. (D) Growth curve assay of gpn2-2, gpn3-1, and double mutants at the indicated temperature. (E) Spot dilution assays of WT, gpn2-2, iwr1Δ, and double mutant cells. Cells were grown for 2 days at the indicated temperature.
Crystallography studies of the archaeal GPN protein PAB0955 suggest that homodimerization occurs between PAB0955 protein (Gras et al. 2007) and, in humans, both homo- and heterodimerization have been observed with purified Gpn1 and Gpn3 (Carre and Shiekhattar 2011). Thus, eukaryotic GPN interactions could occur through homodimerization, heterodimerization, or simply in the context of a large protein complex. To begin to clarify this issue, we determined whether Gpn2 could homodimerize in vivo using a diploid strain containing one 13-myc-tagged copy and one TAP-tagged copy of Gpn2. Pulldown of the TAP-tagged Gpn2 did not result in coprecipitation of 13-myc-tagged Gpn2 beyond background, suggesting that S. cerevisiae Gpn2 may not homodimerize and indeed that the stoichiometry of Gpn2 in stable cellular protein complexes may be 1 (Figure 2B).
To assess a common or partially overlapping cellular function for GPNs we generated double mutants among NPA3, GPN2, GPN3, and IWR1 mutant alleles. Tetrad analysis of double heterozygous mutants of gpn2-2 NPA3::DAmP (decreased abundance by mRNA perturbation; Breslow et al. 2008) showed synthetic lethality between these two genes (Figure 2C). The gpn2-2 gpn3-1 double mutants could be isolated at 25° but growth curve analysis revealed a strong genetic interaction between GPN2 and GPN3 at higher temperatures (Figure 2D). IWR1 plays a role in nuclear localization of RNAPII and would, therefore, be predicted to synergize with other GPN mutants if they play a role similar to NPA3 in the process (Czeko et al. 2011; Staresincic et al. 2011). Consistently, iwr1Δ gpn2-2 double mutants show a severe synergistic temperature-sensitive growth defect, supporting a connection between GPN2 and nuclear transport of RNAPII (Figure 2E). Interestingly, similar to otherwise nonessential core RNAPII mutants (i.e., RPB4 and RPB9; Woychik and Young 1989; Woychik et al. 1991), the function of Iwr1 becomes essential at 37°, as IWR1 mutant cells failed to grow at this temperature (Figure 2E).
GPN2 and GPN3 mutants are defective in localization and stability of RNAPII subunits
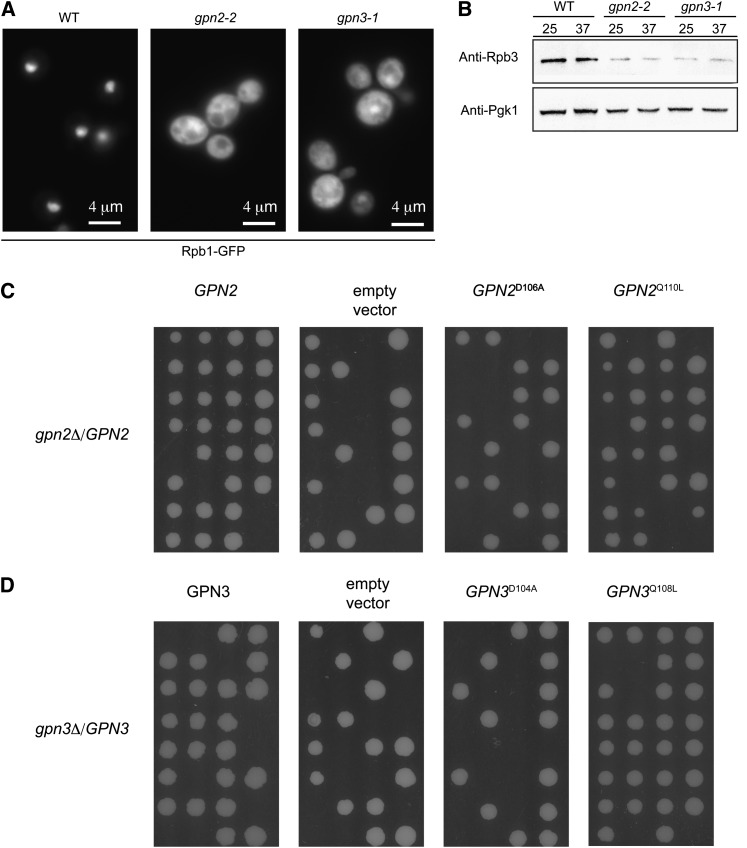
Core subunits of RNAPII were shown to mislocalize from the nucleus to the cytoplasm when GPN1 or GPN3 was disrupted by siRNA in human cells or by mutation of NPA3/GPN1 in yeast (Carre and Shiekhattar 2011; Staresincic et al. 2011). To examine the impact of our GPN2 and GPN3 mutants we expressed a functional GFP-tagged version of Rpb1 in each mutant. We found that both gpn2-2 and gpn3-1 mutants show defects in Rpb1–GFP localization, as there is a strong cytoplasmic accumulation of Rpb1–GFP compared to the wild-type genetic background (Figure 3A). This is not specific to Rpb1 as both Rpb2–GFP and Rbp3–GFP also mislocalized in gpn2-2 mutants (data not shown). To assess the levels of endogenous RNA polymerase subunits we took advantage of a specific antibody to yeast Rpb3. Western blots showed a notable reduction in the levels of Rpb3 in gpn2-2 and gpn3-1 mutants at both 25° and 37°, when compared to wild type (Figure 3B). Together these data demonstrate that each GPN family member plays a role in the localization of RNAPII subunits and that soluble subunit levels are also altered either because of instability or reduced expression. To phenotypically link reduced Rpb3 protein levels to some of the GPN2/3 mutant allele phenotypes, we exploited an RPB3–DAmP allele that expresses reduced levels of Rpb3 (data not shown). Similar to GPN2 and GPN3 mutants, the RPB3–DAmP allele was hypersensitive to both HU and UV treatments, suggesting that reduced RNA Pol II subunit levels could account for at least some of the GPN mutant phenotypes (Figure S3).
Figure 3.
RNA polymerase II mislocalization in GPN mutants. (A) Representative GFP micrographs showing the localization of Rpb1–GFP in WT, gpn2-2, and gpn3-1 mutants. Scale bar indicates 4 μm. (B) Mutation of GPN2 or GPN3 leads to reduced levels of an RNA polymerase II subunit. Rpb3 protein levels in whole-cell lysates of the indicated strains were assessed by Western blot with anti-Rpb3 specific antibodies. Anti-PGK1 blots are shown to indicate equal loading. (C) Tetrad analysis of a heterozygous deletion mutant for GPN2 transformed with wild-type and mutant GPN2 plasmids reveals the requirement for the GTP binding D106 residue for Gpn2 function but not Q110. (D) Mutation of the conserved D104 and Q108 residues in Gpn3 concurs with the observed Gpn2 results.
Previous work has implicated conserved D and Q residues involved in GTP binding and hydrolysis, respectively, as important for the function of NPA3/GPN1 in localization of RNA polymerase II (Staresincic et al. 2011). To assess the importance of these same residues for cell viability in Gpn2 and Gpn3, we engineered point mutations at the orthologous sites via site-directed mutagenesis. Plasmids containing GPN2 with a D106A and Q110L mutation and GPN3 with a D104A and Q108L mutation were transformed into heterozygous deletion mutant strains followed by sporulation and tetrad dissection. As shown in Figure 3C, the gpn2D106A mutation results in a nonfunctional protein that cannot complement the deletion mutant while the gpn2Q110L mutant protein is functional. A similar result was observed for analogous point mutants in GPN3 (Figure 3D). These data suggest that GTP binding is essential for Gpn2 and Gpn3 function but GTP hydrolysis may not be, although at this point it is unclear to what degree the Q110 and Q108 mutations actually impair GTP hydrolysis. Additionally, the previous work on NPA3/GPN1 involved complementing ts-degron alleles with the point mutants, rather than knockouts, which may account for the differences observed here (Staresincic et al. 2011).
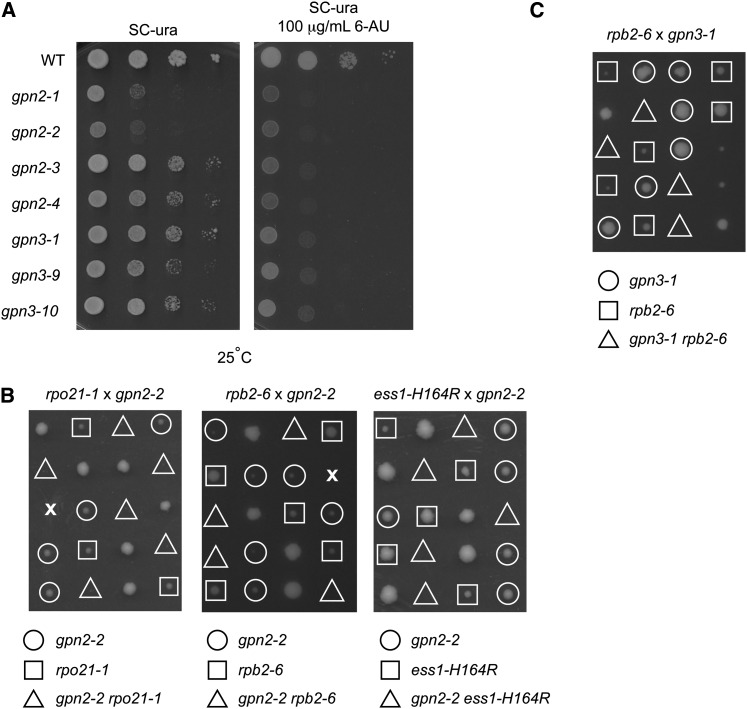
Given the disrupted localization and levels of some RNAPII subunits it would be expected that GPN mutants would be sensitized to additional perturbation of transcription. Consistently, the GPN mutants showed increased sensitivity to the transcription elongation inhibitor 6-azauracil (Figure 4A). Moreover, gpn2-2 showed synthetic lethality or synthetic growth defects when combined with mutants in the core RNAPII subunits RPB1 (rpo21-1), RPB2 (rpb2-6) or the transcriptional regulator ESS1 (ess1-H164R) (Figure 4B), and gpn3-1 also showed synthetic lethality with rpb2-6 (Figure 4C). These data, together with published data (Staresincic et al. 2011), show that loss-of-function mutations in any GPN family member lead to incomplete RNAPII localization and function (Staresincic et al. 2011).
Figure 4.
Functional impact of GPN mutations on RNAPII. (A) Spot-dilution assay for 6-azauracil (6-AU) sensitivity in the indicated GPN2 and 3 mutants. Genetic interactions were tested by (B) tetrad analysis of gpn2-2 crossed with RNA polymerase II transcription mutants and (C) tetrad analysis of gpn3-1 crossed with rpb2-6.
GPN2, GPN3, and IWR1 mutants are defective in localization of RNAPIII
Our data and the literature demonstrate a role for GPNs in nuclear import of RNA polymerase II; however, it is unclear whether the GPNs have a broader role in the nuclear import of other proteins. To address this we tested for mislocalization of other nuclear proteins with known roles in maintenance of chromosomal stability (i.e., the replisome component Ctf4, the cohesin subunit Smc1, and Swr-complex member Swc4) and a control NLS–GFP fusion. We did not detect strong differences in the localization of Ctf4, Smc1, Swc4, or NLS–GFP when GPN2 is mutated (Figure S4). This result indicates that the canonical nuclear import pathways are likely functioning normally at least in the GPN2 mutant background.
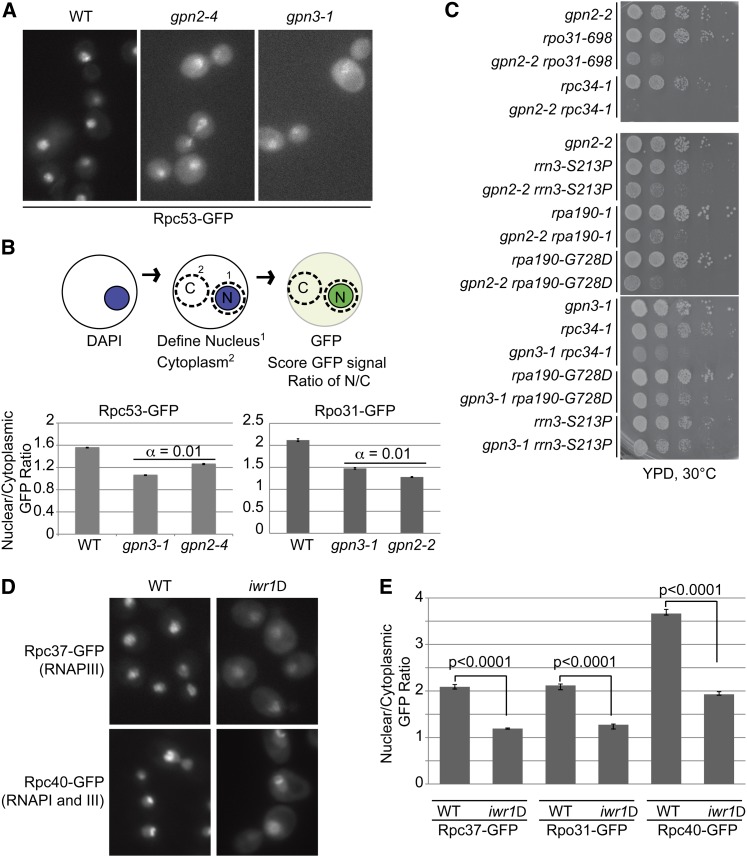
While the transport of the four model nuclear proteins mentioned above was not obviously affected by GPN mutation, it remains possible that transport of an unknown subset of the yeast nuclear proteome is affected by GPNs. Among the best candidates for additional substrates of the GPNs are the RNA polymerase I and III complexes. While comparatively little is known about the assembly of RNAPI and III, they have paralogous subunit architectures to RNAPII and share a number of subunits. Therefore, GFP fusions to representative RNA polymerase I and III subunits were crossed to GPN2 and GPN3 mutant strains and visualized. Fluorescence microscopy revealed that the RNAPIII subunit Rpc53–GFP is mislocalized in both gpn2-4 and gpn3-1 mutant backgrounds, whereas the RNAPI subunit Rpa135–GFP appeared unaffected by either GPN mutation (Figure 5A and Figure S5). It should be noted that introduction of the GFP-tagged versions of several RNA polymerase I and III subunits into the GPN mutant background resulted in lethality, suggesting genetic interactions of GPN mutants with both RNAPI and III; indeed, this is why the gpn2-4 allele was used instead of gpn2-2 (data not shown). While the localization of Rpc53–GFP in GPN mutants was qualitatively different than wild type, a clear nuclear signal usually remained (Figure 5A). While this might be expected given the hypomorphic nature of the GPN alleles, it prompted us to develop a quantitative measure of nuclear localization to which we could assign statistical significance. Figure 5B reports the ratio of GFP signal in the nucleus vs. the cytoplasm for the data in Figure 5A. Importantly, we confirmed our observations using an independent RNA polymerase III subunit, Rpo31–GFP, in gpn2-2 and gpn3-1 mutants using the same quantification scheme (Figure 5B, bottom). This method recapitulates the qualitative observations and shows a significant reduction in nuclear Rpc53-GFP levels in gpn2-4 and gpn3-1 cells and Rpo31-GFP levels in gpn2-2 and gpn3-1 (α = 0.01).
Figure 5.
Effect of the GPN-IWR1 system on RNA polymerase III localization. (A) Nuclear localization of Rpc53-GFP in gpn2-4 and gpn3-1 mutants. (B) Schematic of nuclear localization scoring system and quantification of Rpc53-GFP localization from A. α indicates the results of Tukey’s posthoc analysis of a one-way ANOVA for the three data sets. (C) Spot dilution assays of GPN2 (left) and GPN3 (right) mutants in combination with RNAPI and III subunit mutant alleles. (D) Localization of Rpc37–GFP and Rpc40–GFP in iwr1Δ. (E) Quantification of Rpc37–Rpc40– and Rpo31–GFP fusions using the scoring system from B. P-values indicate the results of Student’s t-test.
When we directly tested for genetic interactions with RNAPI and III subunit ts alleles, we identified synthetic slow growth interactions between gpn2-2 and alleles in all three RNA polymerases (RPA190, RPO31, and RPC34) and a RNA polymerase I transcription factor (RRN3) and between gpn3-1 and RPC34 and potentially RPA190 (Figure 5C). Since transcription by all three RNA polymerases is required to coordinate cellular activities (for example, ribosome assembly), it is not surprising that the GPN mutants could exhibit genetic interactions with all three RNAPs while affecting only localization of RNAPII and III. Alternatively, it is possible that in different GPN mutant backgrounds or with different GFP fusions, RNAPI mislocalization would become visible.
Since the GPNs may cooperate with IWR1 in RNAPII localization we also sought to explore the effects of IWR1 mutations on RNAPI and III localization. For this experiment we introduced the IWR1 deletion into four GFP-fusion strains: Rpo31–GFP and Rpc37–GFP, subunits of only RNAPIII; Rpc40–GFP, a subunit of both RNAPI and RNAPIII; and Rpa135, a subunit of only RNAPI. We observed clear mislocalization of Rpc37, Rpo31, and Rpc40 in the iwr1Δ strains but did not see any mislocalization of Rpa135–GFP, indicating that Iwr1 and the GPNs are affecting RNAPIII but not obviously affecting RNAPI (Figure 5D and supporting Figure S5). Quantification of the nuclear/cytoplasmic GFP ratios from Figure 5D showed that the mislocalization of RNAPIII subunits in iwr1Δ was statistically significant (Figure 5E; P < 0.0001).
Fusing a nuclear localization signal to Rpb3 partially bypasses IWR1 but not GPN mutants
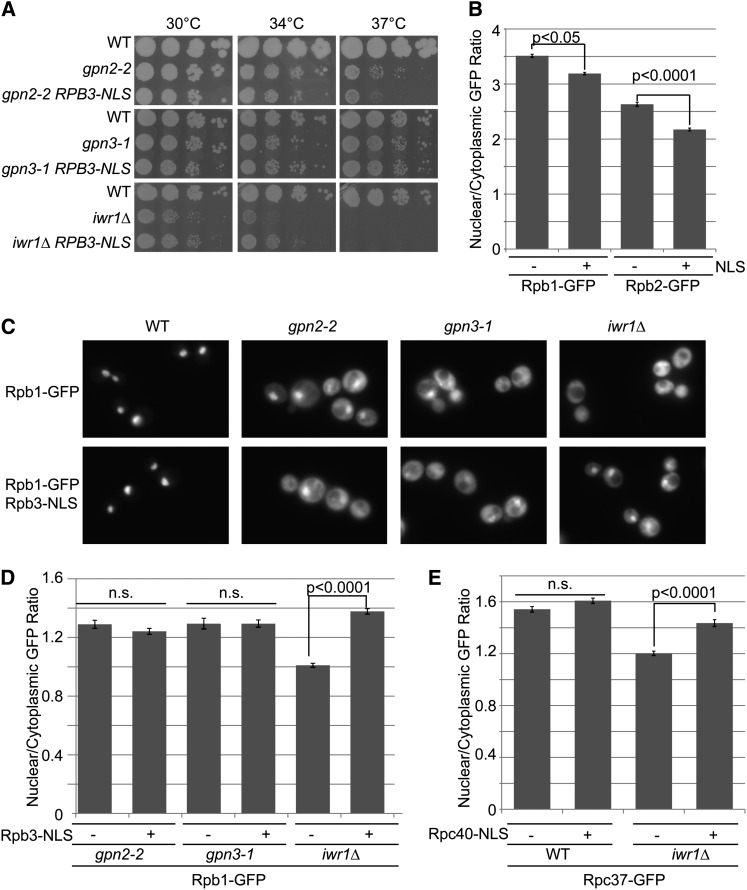
In light of the data presented in Figures 2 and 5 and in recent publications on the GPN proteins and the NLS-containing protein Iwr1 (Di Croce 2011), we propose a cooperative role for these proteins in RNAPII biogenesis and nuclear import. However, it is unclear whether the GPN proteins function primarily in assembly or nuclear import. To address this, and to begin to dissect the functional contributions of GPNs and IWR1, we hypothesized that fusion of a strong NLS directly to Rpb3 could bypass the nuclear localization defect in iwr1Δ and potentially GPN mutants. To test this hypothesis in vivo, we generated a fusion of the SV40 NLS (PKKKRKV) to RPB3. As RPB3 is an essential gene and the fusion strain grew normally, we inferred that the fusion protein was functional. Remarkably, the Rpb3–NLS fusion protein exacerbated the growth defect of gpn2-2 mutants at 37° while partially rescuing the growth defect of iwr1Δ mutants at 34° (Figure 6A).
Figure 6.
Cellular effects of Rpb3–NLS fusion on RNAPII localization. (A) Fitness of gpn2-2, gpn3-1, and iwr1Δ mutants with or without Rpb3–NLS fusion. Spot dilution assays of the indicated strains were performed as in Figure 2. (B) Effect of NLS fusion on nuclear localization of Rpb1–GFP and Rpb2–GFP in wild-type cells. (C) Qualitative localization defects of Rpb1–GFP in the indicated mutant background with (bottom) and without (top) the RPB3–NLS fusion construct. (D) Quantification of Rpb1–GFP localization in the strains from C as in Figure 5B. (E) Quantification of Rpc37–GFP localization in iwr1Δ cells with or without the Rpc40–NLS fusion. P-values indicate the results of Student’s t-tests.
While the RPB3–NLS fusion was competent to support robust growth, it did slightly but significantly reduce the nuclear/cytoplasmic GFP ratio in Rpb1– and Rpb2–GFP-bearing strains (Figure 6B). This could indicate that aberrant nuclear targeting of RNAPII subunits may not permit efficient RNAP assembly. When we examined the localization of Rpb1–GFP in the GPN and IWR1 mutants containing the Rpb3–NLS fusion, we observed qualitatively similar mislocalization in the gpn2-2 and gpn3-1 mutants but observed partial rescue in the iwr1Δ strain (Figure 6C). Quantification of these data confirmed a significantly higher nuclear/cytoplasmic GFP ratio in the iwr1Δ strains bearing the NLS fusion than the strain without the NLS fusion (Figure 6D). To prove that this function was also important for RNA polymerase III, we fused the same NLS sequence to RPC40 and found that this could partially restore localization of Rpo31–GFP in iwr1Δ strains (Figure 6E). Together these data show that addition of an alternative nuclear import signal can partially rescue IWR1 but not GPN2 or GPN3 mutants. In support of the literature on Gpn1 and Iwr1, these data imply that the Gpn2 and Gpn3 proteins are working upstream of import to assemble functional RNAPII complexes while at least part of the function of Iwr1 is specifically tied to nuclear import. Moreover, these data suggest that the NLS contained within IWR1 is critical for its role in RNA polymerase III localization. Indeed, we found that, while expression of full-length IWR1 from a plasmid rescues RNAPII and III localization in iwr1Δ, expression of partial or complete deletions of the NLS sequence failed to improve localization of Rpb1–, Rpo31–,or Rpc37–GFP fusion (Figure S6).
Discussion
The GPNs represent a highly conserved family of small GTPases that evolved in the common ancestor of eukaryotes and archaea (Figure S1). They appear to be active as GTPases and experiments with Npa3 suggest the potential for the nucleotide switch-like behavior seen in better characterized GTPases like Ras or the nuclear import regulator Ran (Staresincic et al. 2011). The precise role of the GTPase activity of the GPNs is not clear, although there is evidence that the affinity of Npa3/Gpn1 for RNAPII is regulated by its GTP-binding status (Staresincic et al. 2011). Our mutational analysis suggests that at least GTP binding by Gpn2 and Gpn3 is an absolute requirement of cell viability; thus all three GPN proteins require GTP. Based on this study (Figures 3–6) and the literature, it appears that GPN1, GPN2, and GPN3 activities are all required for normal RNA polymerase II nuclear localization (Carre and Shiekhattar 2011; Staresincic et al. 2011). While GPNs are also conserved in many archaea, a conserved function cannot be related to nuclear transport but could feasibly relate to RNAP assembly. Consistent with a distinct function for Iwr1, no significant BLAST-P hits for Iwr1 are observed in archaea (data not shown), supporting the notion that Iwr1 is most important for nuclear import and is accordingly not present in archaea. Prior to this study, GPN2 and GPN3 were virtually uncharacterized in yeast and thus, our focus was accordingly on understanding their function, particularly that of GPN2.
The genome instability phenotypes of the GPN mutants (Figure 1) could be ascribed to their role in RNA polymerase biogenesis, as defects in RNAPII subunits are known to elicit many of the same phenotypes (e.g., (Stirling et al. 2011, 2012b). The causes of transcription-associated CIN are potentially diverse including RNA:DNA hybrid formation, loss of specific transcripts, or defects in DNA repair (Herrero and Moreno 2011; Lagerwerf et al. 2011; Aguilera and Garcia-Muse 2012). In this study, reducing the levels of RNAPII subunits by using a DAmP allele was sufficient to recapitulate some of the phenotypes of the GPN2 and GPN3 mutants, suggesting that one important role for GPNs is maintaining sufficient levels of RNA polymerase subunits (Figure S3). The reported interaction between GPN1/XAB1 and XPA/Rad14p has not so far been observed in high-throughput yeast interaction studies or explored thoroughly in mammalian cells, although this could be another possible connection to genome integrity (Nitta et al. 2000).
The mechanism by which the GPN proteins might cooperate is not clear. Mass spectrometry studies have identified all three proteins in complex, and direct tests have validated Gpn1–Gpn2 and Gpn2–Gpn3 interactions (Figure 2; (Boulon et al. 2010; Forget et al. 2010; Carre and Shiekhattar 2011; Staresincic et al. 2011)). In this study we observed genetic interactions between gpn2-2, gpn3-1, and NPA3::DAmP alleles, also supporting a common function. Archaeal GPNs homodimerize in vitro and, while in vitro studies have suggested the capability of human Gpn1 and Gpn3 to homodimerize, we could not find evidence of a Gpn2 homodimer by coprecipitation from cell lysates (Figure 2; Gras et al. 2007; Carre and Shiekhattar 2011). Indeed, the question of why virtually all eukaryotes have retained three GPN genes remains unclear. While an appealing model would be that each GPN is responsible for a different RNA polymerase (i.e., three essential GPNs for three essential RNAPs), our results do not support this idea. GPN2 and GPN3 mutants had defects in both RNAPII and III localization but did not mislocalize RNAPI subunits (Figure 3–5 and Figure S5). While this could suggest that GPNs truly have no role in RNAPI assembly, it is possible that our hypomorphic alleles simply did not perturb some specific aspect of GPN function required for RNAPI assembly. Moreover, the role of NPA3/GPN1 in localization of RNAPI and III has not been thoroughly assessed and could affect RNAPI. Finally, the assembly of RNAPI appears to be fundamentally different from RNAPII as individual subunits are brought to the rDNA prior to assembly (Dundr et al. 2002). Therefore, GPNs may play a role in assembly within the nucleolus itself or be responsible for the localization of single subunits or subcomplexes not assessed in the present study. The case may be similar for IWR1 mutants, as we showed defects in RNAPIII localization but not RNAPI in the absence of Iwr1 (Figure 5). In the literature, single GFP markers of RNAPI and III used previously did not mislocalize in IWR1 mutants (Czeko et al. 2011), whereas other data suggests that Iwr1 functions in initiation of transcription for all three nuclear RNA polymerases (Esberg et al. 2011). Some of these discrepancies could be due to the choice of specific assay, which may be important for detecting defects in RNA polymerases with different assembly pathways.
Our data together with the literature support a model in which all three GPNs act upstream of Iwr1 to assemble RNA polymerase II and III (Figure 7). The inability of Rpb3–NLS fusions to rescue GPN mutants, while partially rescuing the fitness and localization defects in iwr1Δ suggests that the role of GPNs is predominantly upstream of import at the level of RNA polymerase assembly, which supports and extends the previous model for RNA polymerase II biogenesis (Wild and Cramer 2012). We consistently found that Rpb3 was unstable, suggesting that it was not undergoing normal biogenesis in GPN mutants; however, based on the data collected we cannot rule out that this is due to reduced transcription (Figure 3). To perform an assembly and stability function, the GPNs are likely acting as part of a network of molecular chaperones known to interact with RNA polymerases. In particular, Hsp90 and the R2TP complex have been shown to play a role, in complex with the GPNs, in assembling RNA polymerases (Boulon et al. 2010). Interestingly, there are also potential physical connections to the CCT (chaperonin-containing tailless complex polypeptide 1) complex, which is primarily involved in folding actin and tubulin subunits. However, many other less abundant proteins have been implicated as CCT interactors including RNAPII and RNAPIII subunits (Dekker et al. 2008; Yam et al. 2008). Notably, tubulin seems to be important for RNA polymerase assembly and the CCT complex is known to cooperate with the hexameric prefoldin chaperone, subunits of which are found in R2TP, which has been proposed to play a role in RNAP assembly (Vainberg et al. 1998; Forget et al. 2010). The potentially elaborate chaperone pathway for RNA polymerase assembly is not unprecedented; for example, tubulin heterodimer assembly requires at least eight chaperones and cofactors (reviewed in (Lundin et al. 2010).
Figure 7.
Model for GPN1, 2, and 3 function in biogenesis of RNA polymerases II and III. After synthesis RNA polymerase subcomplexes begin to assemble in the cytoplasm. The three GPN proteins bind with the assembly intermediates and aid the formation of the RNA polymerase complex, possibly cooperating with the Hsp90/CCT chaperone machinery. Iwr1 association with the RNA polymerases provides the NLS required for import through the nuclear pore complex (NPC). Once in the nucleus, the RNA polymerase is able to associate with transcription factors (TF) and carry out transcription activity while the GPN proteins and Iwr1 are recycled to the cytoplasm via a nuclear export pathway, potentially driven by a nuclear export sequence in Gpn1 (Reyes-Pardo et al. 2012).
There are several outstanding questions regarding the cellular role of the GPN proteins: Given their common phenotypes, what specialized roles do each GPN play that make all three genes essential? Is the substrate repertoire of the GPNs for protein complex assembly limited to nuclear RNA polymerases? More broadly, the existence of the GPN and Iwr1 system suggests that regulated assembly and transport of RNAPs is important for proper functioning or regulation of transcription since RNAPs do not encode NLSs in their primary sequence.
Supplementary Material
Acknowledgments
We thank Brenda Andrews, Richard Wozniak, Patrick Cramer, and Elizabeth Conibear for providing strains and plasmids. P.H. acknowledges support from the National Institutes of Health and Canadian Institutes of Health Research (CIHR). S.W.M. acknowledges scholarship support from CIHR and University of British Columbia. P.C.S. is a fellow of the Terry Fox Foundation (no. 700044) and the Michael Smith Foundation for Health Research.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Aguilera A., Garcia-Muse T., 2012. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46: 115–124. [DOI] [PubMed] [Google Scholar]
- Alonso B., Chaussinand G., Armengaud J., Godon C., 2011. A role for GPN-loop GTPase yGPN1 in sister chromatid cohesion. Cell Cycle 10: 1828–1837. [DOI] [PubMed] [Google Scholar]
- Ben-Aroya S., Coombes C., Kwok T., O’Donnell K. A., Boeke J. D., et al. , 2008. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell 30: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., et al. , 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39: 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., et al. , 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calera M. R., Zamora-Ramos C., Araiza-Villanueva M. G., Moreno-Aguilar C. A., Pena-Gomez S. G., et al. , 2011. Parcs/Gpn3 is required for the nuclear accumulation of RNA polymerase II. Biochim. Biophys. Acta 1813: 1708–1716. [DOI] [PubMed] [Google Scholar]
- Carre C., Shiekhattar R., 2011. Human GTPases associate with RNA polymerase II to mediate its nuclear import. Mol. Cell. Biol. 31: 3953–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. Y., Stirling P. C., Stimpson H. E., Giesselmann E., Schmitt M. J., et al. , 2009. A yeast killer toxin screen provides insights into a/b toxin entry, trafficking, and killing mechanisms. Dev. Cell 17: 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeko E., Seizl M., Augsberger C., Mielke T., Cramer P., 2011. Iwr1 directs RNA polymerase II nuclear import. Mol. Cell 42: 261–266. [DOI] [PubMed] [Google Scholar]
- Di Croce L., 2011. Regulating the shuttling of eukaryotic RNA polymerase II. Mol. Cell Biol. 31: 3918–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C., Stirling P. C., McCormack E. A., Filmore H., Paul A., et al. , 2008. The interaction network of the chaperonin CCT. EMBO J. 27: 1827–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L. I., et al. , 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298: 1623–1626. [DOI] [PubMed] [Google Scholar]
- Esberg A., Moqtaderi Z., Fan X., Lu J., Struhl K., et al. , 2011. Iwr1 protein is important for preinitiation complex formation by all three nuclear RNA polymerases in Saccharomyces cerevisiae. PLoS ONE 6: e20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget D., Lacombe A. A., Cloutier P., Al-Khoury R., Bouchard A., et al. , 2010. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteomics 9: 2827–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Gras S., Chaumont V., Fernandez B., Carpentier P., Charrier-Savournin F., et al. , 2007. Structural insights into a new homodimeric self-activated GTPase family. EMBO Rep. 8: 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A. B., Moreno S., 2011. Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J. 30: 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. H., Magtanong L., Barker S. L., Gresham D., Nishimura S., et al. , 2009. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 27: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Cancer Genome Consortium, 2010. International network of cancer genome projects. Nature 464: 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka M. D., Duprat S., Cornillot E., Metenier G., Thomarat F., et al. , 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453. [DOI] [PubMed] [Google Scholar]
- Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., et al. , 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2: E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H., 2011. DNA damage response and transcription. DNA Repair 10: 743–750. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., 2011. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer 11: 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lundin V. F., Leroux M. R., Stirling P. C., 2010. Quality control of cytoskeletal proteins and human disease. Trends Biochem. Sci. 35: 288–297. [DOI] [PubMed] [Google Scholar]
- Nitta M., Saijo M., Kodo N., Matsuda T., Nakatsu Y., et al. , 2000. A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res. 28: 4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Pardo H., Barbosa-Camacho A. A., Perez-Mejia A. E., Lara-Chacon B., Salas-Estrada L. A., et al. , 2012. A nuclear export sequence in GPN-loop GTPase 1, an essential protein for nuclear targeting of RNA polymerase II, is necessary and sufficient for nuclear export. Biochim. Biophys. Acta 1823: 1756–1766. [DOI] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P., 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staresincic L., Walker J., Dirac-Svejstrup A. B., Mitter R., Svejstrup J. Q., 2011. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J. Biol. Chem. 286: 35553–35561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Bloom M. S., Solanki-Patil T., Smith S., Sipahimalani P., et al. , 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7: e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Crisp M. J., Basrai M. A., Tucker C. M., Dunham M. J., et al. , 2012a Mutability and mutational spectrum of chromosome transmission fidelity genes. Chromosoma 121: 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Chan Y. A., Minaker S. W., Aristizabal M. J., Barrett I., et al. , 2012b R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton M. R., Campbell P. J., Futreal P. A., 2009. The cancer genome. Nature 458: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., et al. , 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Vainberg I. E., Lewis S. A., Rommelaere H., Ampe C., Vandekerckhove J., et al. , 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93: 863–873. [DOI] [PubMed] [Google Scholar]
- Weaver B. A., Cleveland D. W., 2006. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18: 658–667. [DOI] [PubMed] [Google Scholar]
- Wild T., Cramer P., 2012. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 37: 99–105. [DOI] [PubMed] [Google Scholar]
- Woychik N. A., Young R. A., 1989. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol. 9: 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Lane W. S., Young R. A., 1991. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J. Biol. Chem. 266: 19053–19055. [PubMed] [Google Scholar]
- Yam A. Y., Xia Y., Lin H. T., Burlingame A., Gerstein M., et al. , 2008. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.