Abstract
Widespread sharing of long, identical-by-descent (IBD) genetic segments is a hallmark of populations that have experienced recent genetic drift. Detection of these IBD segments has recently become feasible, enabling a wide range of applications from phasing and imputation to demographic inference. Here, we study the distribution of IBD sharing in the Wright–Fisher model. Specifically, using coalescent theory, we calculate the variance of the total sharing between random pairs of individuals. We then investigate the cohort-averaged sharing: the average total sharing between one individual and the rest of the cohort. We find that for large cohorts, the cohort-averaged sharing is distributed approximately normally. Surprisingly, the variance of this distribution does not vanish even for large cohorts, implying the existence of “hypersharing” individuals. The presence of such individuals has consequences for the design of sequencing studies, since, if they are selected for whole-genome sequencing, a larger fraction of the cohort can be subsequently imputed. We calculate the expected gain in power of imputation by IBD and subsequently in power to detect an association, when individuals are either randomly selected or specifically chosen to be the hypersharing individuals. Using our framework, we also compute the variance of an estimator of the population size that is based on the mean IBD sharing and the variance in the sharing between inbred siblings. Finally, we study IBD sharing in an admixture pulse model and show that in the Ashkenazi Jewish population the admixture fraction is correlated with the cohort-averaged sharing.
IN isolated populations, even purported unrelated individuals often share genetic material that is identical-by-descent (IBD). Traditionally, the term IBD sharing referred to coancestry at a single site (or autozygosity, in the case of a diploid individual) and was widely investigated as a measure of the degree of inbreeding in a population (Hartl and Clark 2006). Recent years have brought dramatic increases in the quantity and density of available genetic data and, together with new computational tools, these data have enabled the detection of IBD sharing of entire genomic segments (see, e.g., Purcell et al. 2007; Kong et al. 2008; Albrechtsen et al. 2009; Gusev et al. 2009; Browning and Browning 2011; Carr et al. 2011; Brown et al. 2012). The availability of IBD detection tools that are efficient enough to detect shared segments in large cohorts has resulted in numerous applications, from demographic inference (Davison et al. 2009; Palamara et al. 2012) and characterization of populations (Gusev et al. 2012a) to selection detection (Albrechtsen et al. 2010), relatedness detection and pedigree reconstruction (Huff et al. 2011; Kirkpatrick et al. 2011; Stevens et al. 2011; Henn et al. 2012), prioritization of individuals for sequencing (Gusev et al. 2012b), inference of HLA type (Setty et al. 2011), detection of haplotypes associated with a disease or a trait (Akula et al. 2011; Gusev et al. 2011; Browning and Thompson 2012), imputation (Uricchio et al. 2012), and phasing (Palin et al. 2011).
Recently, some of us used coalescent theory to calculate several theoretical quantities of IBD sharing under a number of demographic histories. Then, shared segments were detected in real populations, and their demographic histories were inferred (Palamara et al. 2012). Here, we expand upon Palamara et al. (2012) to investigate additional aspects of the stochastic variation in IBD sharing. Specifically, we provide a precise calculation for the variance of the total sharing in the Wright–Fisher model, either between a random pair of individuals or between one individual and all others in the cohort.
Understanding the variation in IBD sharing is an important theoretical characterization of the Wright–Fisher model, and additionally, it has several practical applications. For example, it can be used to calculate the variance of an estimator of the population size that is based on the sharing between random pairs. In a different domain, the variance in IBD sharing is needed to accurately assess strategies for sequencing study design, specifically, in prioritization of individuals to be sequenced. This is because imputation strategies use IBD sharing between sequenced individuals and genotyped, not-sequenced individuals to increase the number of effective sequences analyzed in the association study (Palin et al. 2011; Gusev et al. 2012b; Uricchio et al. 2012).
In the remainder of this article, we first review the derivation of the mean fraction of the genome shared between two individuals (Palamara et al. 2012). We then calculate the variance of this quantity, using coalescent theory with recombination. We provide a number of approximations, one of which results in a surprisingly simple expression, which is then generalized to a variable population size and to the sharing of segments in a length range. We also numerically investigate the pairwise sharing distribution and provide an approximate fit. We then turn to the average total sharing between each individual and the entire cohort. We show that this quantity, which we term the cohort-averaged sharing, is approximately normally distributed, but is much wider than naively expected, implying the existence of hypersharing individuals. We consider several applications: the number of individuals needed to be sequenced to achieve a certain imputation power and the implications to disease mapping, inference of the population size based on the total sharing, and the variance of the sharing between siblings. We finally calculate the mean and the variance of the sharing in an admixture pulse model and show numerically that admixture results in a broader than expected cohort-averaged sharing. Therefore, large variance of the cohort-averaged sharing can indicate admixture. In the Ashkenazi Jewish population, we show that the cohort-averaged sharing is strongly anticorrelated with the fraction of European ancestry.
Materials and Methods
Coalescent simulations
To simulate IBD sharing in the Wright–Fisher model, we used the Genome haploid coalescent simulator (Liang et al. 2007). Recombination in Genome is discretized to short blocks and mutations (which we ignore in this study) are placed on the simulated branches. In all simulations, we generated one chromosome with recombination rate of 10−8 per generation per base pair and block lengths of 104 bp (corresponding to resolution of 0.01 cM in the lengths of the shared segments).
IBD sharing in simulations
We used an add-on to Genome that returns, for each pair of chromosomes, the locations of all shared segments (Palamara et al. 2012). In that add-on, a segment is shared as long as the two chromosomes share the same ancestor, even if there was a recombination event within the segment. We calculated, for each pair, the total length of shared segments longer than m and divided by the chromosome size. For Figures 2–6, we simulated Npop ≥ 100 populations and n = 100 haploid sequences in each population and calculated all properties of the total sharing among all available pairs. For the cohort-averaged sharing, we averaged, for each of the n chromosomes, their sharing to each of the other n − 1 chromosomes in the cohort and then used the Npopn calculated values to obtain the variance and the distribution. Details on the simulation of an admixture pulse can be found in Supporting Information, File S1, section S4.
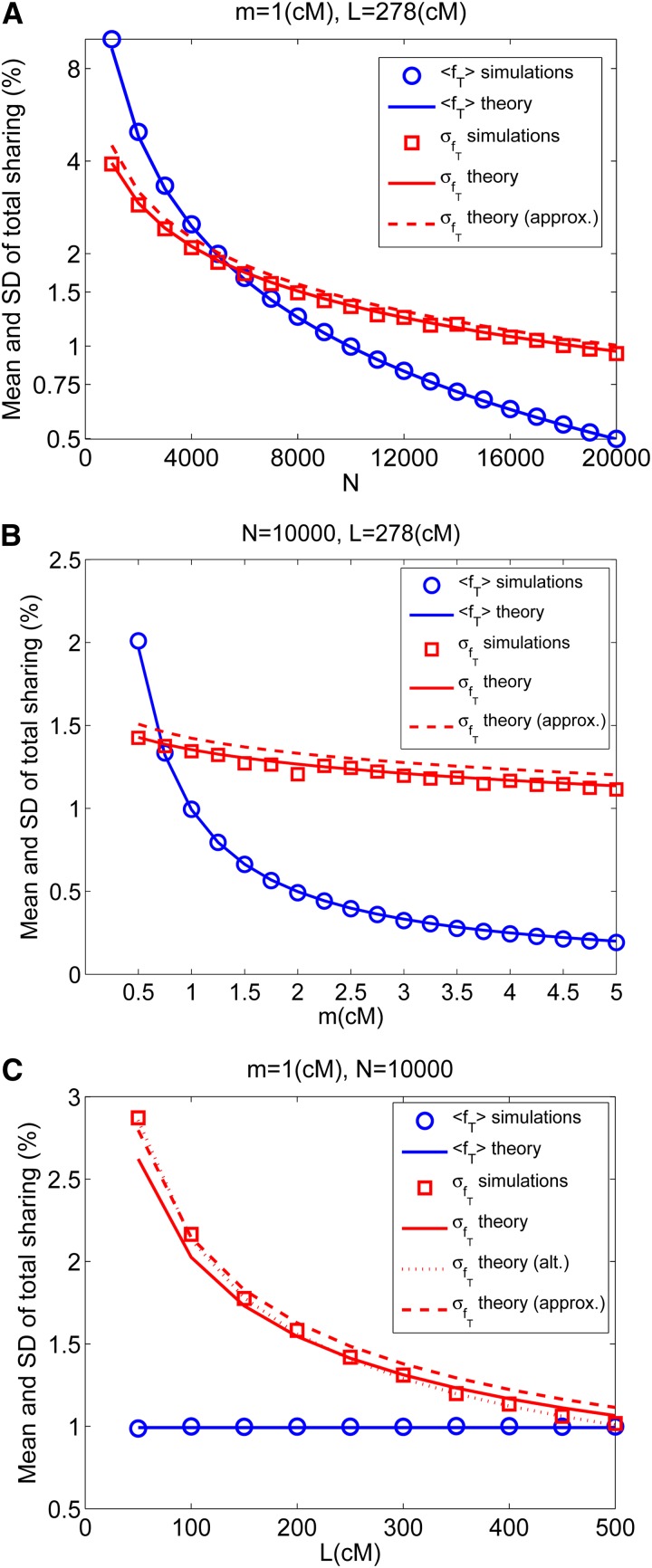
Figure 2.
The mean and standard deviation of the total sharing. For each parameter set, we used the Genome coalescent simulator to generate a number of genealogies (from a population of size N and for one chromosome of size L) and then calculated the lengths of IBD shared segments between random individuals. Each panel presents the results for the mean and standard deviation (SD) of the total sharing, that is, for each pair, the total fraction (in percentages) of the genome that is found in shared segments of length ≥m. Simulation results are represented by symbols and theoretical results by lines (Equation 4 for the mean and Equation 12 for the SD are plotted in solid lines; the approximate form for the SD, Equation 15, is shown in dashed lines). (A) We fixed m = 1 cM and L = 278 cM [the size of the human chromosome 1 (International HapMap Consortium 2007)] and varied N. (B) Same as A, but with fixed N = 10,000 and varying m. (C) Fixed N and m and varying chromosome length L. In C, we also plotted the result of an alternative, more elaborate calculation of the variance (dotted line; see File S1, section S1.3).
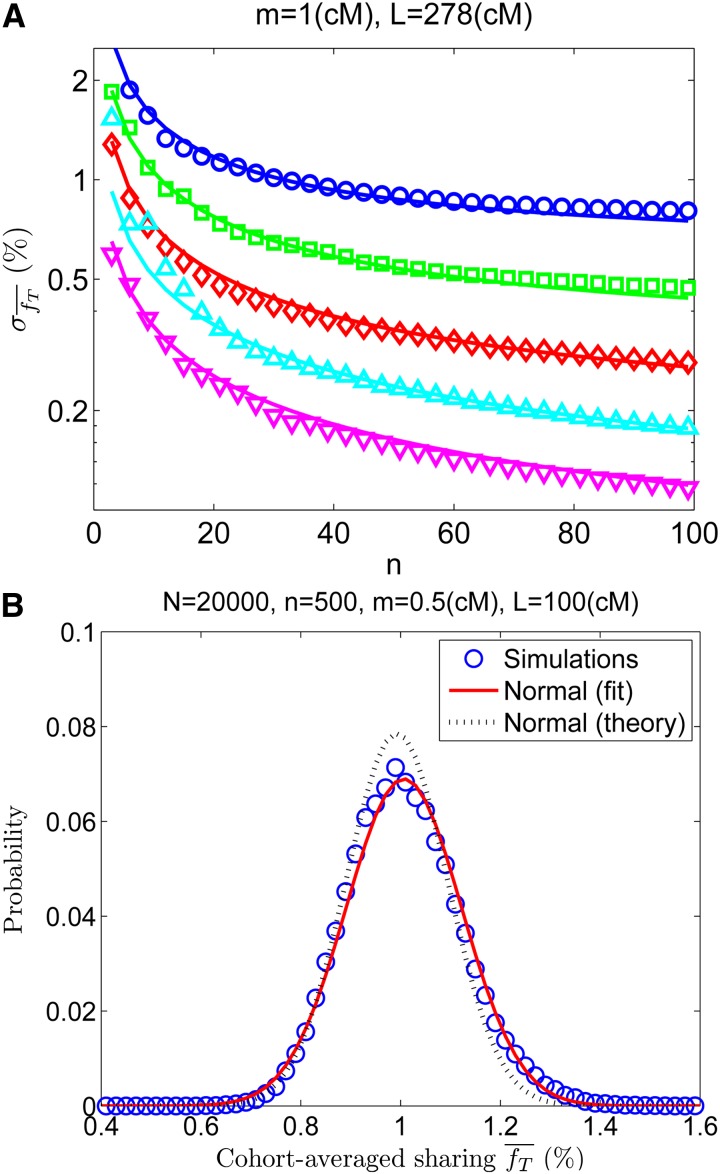
Figure 6.
The cohort-averaged sharing. (A) Simulation results (symbols) for , that is, the standard deviation (SD) of the cohort-averaged sharing (in percentage of the chromosome) vs. the cohort size n. The different curves correspond to different values of N (top to bottom: N = 1000, 2000, 4000, 8000, 16,000). The lines correspond to Equation 28. Details of the simulations are as in Figure 2A. (B) The distribution of the cohort-averaged sharing. The fit is to a normal distribution having the same mean and SD as the real data. Also plotted is a normal distribution with mean given by Equation 4 and SD given by Equation 28.
The Ashkenazi Jewish cohort
The cohort we analyzed was previously described in Guha et al. (2012) and Palamara et al. (2012). Briefly, DNA samples from ≈ 2600 Ashkenazi Jews (AJ) were genotyped on the Illumina-1M SNP array. Genotypes (autosomal only) were subjected to quality control, including removal of close relatives, and phasing [Beagle (Browning and Browning 2009)], leaving finally ≈741,000 SNPs for downstream analysis. IBD sharing was calculated using Germline (Gusev et al. 2009) with the following parameters: bits, 25; err_hom, 0; err_het, 2; min_m, 1; h_extend, 1. The results presented in IBD sharing after an admixture pulse section remained qualitatively the same even when we used a longer length cutoff of m = 5 cM.
Admixture analysis
For the admixture analysis, we merged the HapMap3 CEU population (Utah residents with ancestry from Northern and Western Europe; International HapMap Consortium 2007; release 2) with the AJ data, removed all SNPs with potential strand inconsistency, and pruned SNPs that were in linkage disequilibrium (Purcell et al. 2007). We then ran Admixture (Alexander et al. 2009) with default parameters and K = 2. Admixture consistently classified all individuals according to their population (CEU/AJ). Genome-wide, the AJ ancestry fraction was ≈85%, compared to ≈3% for the CEU population. Principal components analysis [SmartPCA (Patterson et al. 2006)] gave qualitatively similar results.
Simulations of AJ demography
Demographic reconstruction of the AJ population was performed in Palamara et al. (2012), using chromosome 1 of 500 randomly selected individuals and using a novel IBD-based method described therein. Simulations presented here were performed using the final set of inferred demographic parameters: ancestral (diploid) population effective size of ≈2300 individuals, expansion starting 200 generations ago reaching ≈45,000 individuals 33 generations ago, a severe bottleneck of ≈270 individuals, and an expansion to the current size of ≈4.3 million individuals. Simulation of 100 populations was carried out using Genome (Liang et al. 2007).
Results
Variation in IBD sharing in the Wright–Fisher model
Definitions:
The Wright–Fisher model:
We consider the standard Wright–Fisher model for a finite, isolated population, described by 2N haploid chromosomes, where each pair of chromosomes corresponds to one diploid individual. Each chromosome in the current generation descends, with equal probability, from one of the chromosomes in the previous generation, and recombination occurs at rate 0.01/cM per generation. The Wright–Fisher model has been widely investigated both in forward dynamics and under the coalescent (Wakeley 2009). For simplicity of notation, we denote the number of individuals, or the population size, as N, even though we really refer to the number of haploids and not the number of individuals. Throughout most of the analysis, we assume that each individual carries a single chromosome of length L cM.
IBD sharing:
We say that a genomic segment is shared, or is IBD, between two individuals if it is longer than m(cM) and it has been inherited without recombination from a single common ancestor. We do not require the shared segments to be completely identical. That is, if any mutation has occurred since the time of the most recent common ancestor (MRCA), that would not disqualify the segments from being shared IBD according to our definition. The reason is that even in the presence of mutations, an order of magnitude calculation shows that regardless of the segment length, two individuals sharing a segment are expected to differ in just ≈1 site along the segment (see File S1, section S1.1). Therefore, in a long IBD segment, the number of differences should be very small compared to the number of matches. In practice, there are also other sources of error in IBD detection, most notably phase switch errors. We assume, however, that there always exists a large enough length threshold above which segments are detectable without errors (Browning and Browning 2011; Brown et al. 2012), which corresponds to the parameter m introduced above; the precise value of the threshold will depend on the genotyping/sequencing technology. We assume that information is available for M markers, uniformly distributed (in genetic distance) along the chromosome and densely enough that any effect caused by the discreteness of the markers is negligible (say, if m ⋅ (M/L) ≫ 1). We define the total sharing between two individuals as the fraction of their markers that are found in shared segments.
Mean total sharing:
In this subsection, we review the derivation of the mean fraction of the genome found in segments shared between two individuals (Palamara et al. 2012). We assume that the coalescent process along the chromosome can be approximated by the sequentially Markov coalescent (McVean and Cardin 2005) and ignore the different behavior of sites at the ends of the chromosome. Consider first a single site s and assume that its MRCA dates g generations ago. The total length ℓ of the segment in which the site is found is the sum of ℓR and ℓL, where ℓR and ℓL are the segment lengths to the right and left of s, respectively (all lengths are in centimorgans). The distributions of ℓR and ℓL are exponential with rate g/50, since the two individuals were separated by 2g meioses, each of which introduces a recombination event with rate 0.01/cM, and the nearest recombination would terminate the shared segment. The probability π of the total segment length, ℓ, to exceed m is, given g,
| (1) |
According to coalescent theory in the Wright–Fisher model, under the continuous-time scaling g → Nt the times to the MRCA are exponentially distributed with rate 1: Φ(t) = e−t. Therefore,
| (2) |
The total fraction of the genome found in shared segments is
| (3) |
where I(s) is the indicator that site s is in a shared segment, and the sum is over all sites. The mean fraction of the genome shared is
| (4) |
where 〈⋅〉 denotes the average over all ancestral processes. As expected, for mN → ∞, 〈fT〉 → 0 and for mN → 0, 〈fT〉 → 1. For large N, we have 〈fT〉 ≈ 100/(mN).
The variance of the total sharing:
We now turn to calculating the variance of the total sharing. Using Equation 3,
where π2(s1, s2) is the probability that both markers s1 and s2 are on shared segments and π is given by Equation 2. In the rest of the section, we assume that each individual carries one chromosome only, for if we have c chromosomes, each of length Li, then (if the two individuals are not close relatives)
and the variance is
| (5) |
where fT,(i) is the total sharing in chromosome i, assumed independent of the other chromosomes. Rewriting π2(s1, s2) as π2(k), where k is the number of markers separating s1 and s2, we have
| (6) |
All that is left is to evaluate π2(k), for which we provide three approximations. The first is presented below and the second (which is a variation of the first) is presented in File S1, section S1.3. The third approximation, which is the most crude but yields an explicit dependence on the population parameters, is presented in An approximate explicit expression.
In the first approach, we assume that once the times t1, t2 to the MRCA at the two sites are known, the sites are (or are not) in shared segments independently of each other and with probabilities given by Equation 1. Clearly, this assumption is violated when both sites belong to the same shared segment, and in File S1, section S1.3, we show how this assumption can be avoided (but at the cost of significantly complicating the analysis). Nevertheless, it gives a good approximation, as we later see (Figure 2). We can therefore use Equation 1 to write
| (7) |
where Φ(t1, t2) is the joint probability density function (PDF) of t1 and t2 and
is the Laplace transform of Φ(t1, t2). We therefore reduced the problem of finding π2(k) into that of finding .
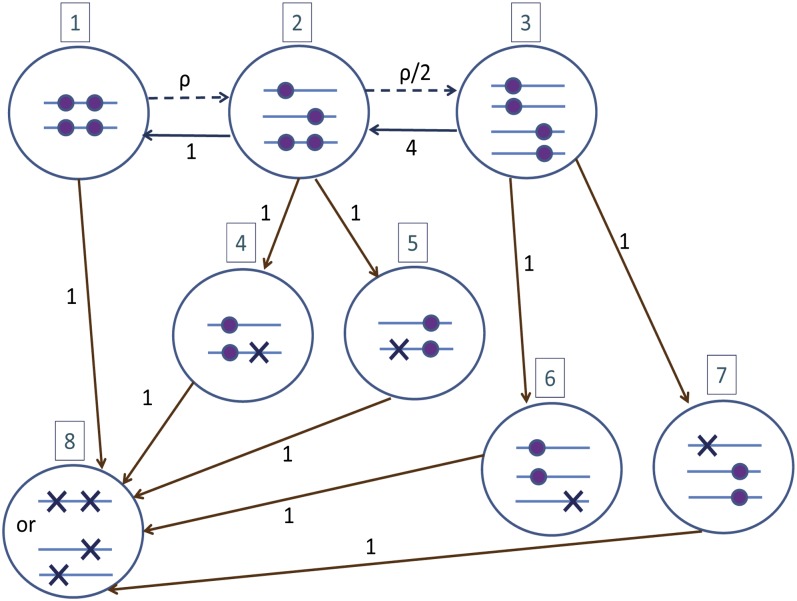
To find Φ(t1, t2) (or rather, its Laplace transform), we use the continuous-time Markov chain representation of the coalescent with recombination (Hudson 1983; Simonsen and Churchill 1997; Wakeley 2009). The chain is illustrated in Figure 1. Initially (present time), the chain is in state 1, corresponding to two chromosomes carrying two sites each. The chain terminates at state 8, when both sites have reached their MRCA. To construct the chain, coalescence events were assumed to occur at rate 1 and recombination events at rate ρ/2, where is the scaled recombination rate (Wakeley 2009).
Figure 1.
An illustration of the continuous-time Markov chain representation of the coalescent with recombination (Simonsen and Churchill 1997; Wakeley 2009). Large circles correspond to states, with the state number in a box on top of each circle. Arrows connecting circles represent transitions (solid lines, coalescence events; dashed lines, recombination events), with their rates indicated. The lines inside each circle represent chromosomes with two sites each. Ancestral sites are indicated as either small circles (as long as there are still two lineages carrying the ancestral material) or crosses (whenever the two lineages coalesced and the site has reached its MRCA). Transitions leading to the MRCA in one or two sites are colored brown. Transitions between states 4 and 6 and between 5 and 7 are not indicated, as they do not affect the final coalescence times. The schematic was adapted from Wakeley (2009).
Denote by Pi(t) the probability that the chain is at state i at time t, given that it started at state 1. The probability that the two sites have reached their MRCA simultaneously in the time range [t, t + dt] is P1(t)dt, since this is the product of the probability that the chain is at state 1 at time t (P1(t)) and the probability of the transition 1 → 8 in the given time interval (dt). The probability that only the left site has reached its MRCA (and the right site has not) in [t, t + dt] is P2(t)dt + P3(t)dt: this corresponds to the transitions 2 → 5 and 3 → 7. This is also the probability that only the right site has reached its MRCA in [t, t + dt] (transitions 2 → 4, 3 → 6). Finally, the probability that the left site has reached its MRCA in [t1, t1, + dt1] and that the right site has reached its MRCA in [t2, t2 + dt2] (t2 > t1) is . This is true, because the exit rate from states 5 and 7 is 1; therefore, the probability that the chain will wait at one of those states for time (t2 − t1) and then leave to the terminal state is . Similar considerations apply for the case t1 > t2 (with the transitions 2 → 4 and 3 → 6). In sum, for t1, t2 > 0,
where δ(t) is the Dirac delta function and Θ(t) = 1 for t > 0 and is otherwise zero. Laplace transforming the last equation,
| (8) |
In the last equation, (i = 1, 2, 3) are the Laplace transforms of Pi(t). The Laplace transforms can be calculated using the general relation
| (9) |
where Q is the transition rate matrix: Qij is the transition rate from i to j ≠ i and ,
| (10) |
Using Equations 8–10 and Mathematica,
| (11) |
where A = (1 + q1)(1 + q2), B = (3 + q1 + q2)(6 + q1 + q2), C = ρ(2 + q1 + q2), D = 13 + 3(q1 + q2), and E = ρ2(2 + q1 + q2). Equation 11 was also derived in Griffiths (1991), using the birth-and-death process equivalent of the coalescent with recombination, and can also be derived using the Feynman–Kac formula (see File S1, section S1.4). Substituting, using Mathematica, Equation 11 in Equation 7, and then using Equation 6 gives an expression for the variance,
| (12) |
The function ℱ is too long to reproduce here, but can be found in the Matlab code (File S2). For further discussion on the approximations made, see File S1, section S1.2. The standard deviation (SD) of the total sharing is defined as usual as .
To evaluate the accuracy of our expressions for the mean and SD of the total sharing, we used the Genome coalescent simulator (Liang et al. 2007), along with an add-on that returns, for each generated genealogy, the locations of the segments that are IBD between each pair of individuals (Palamara et al. 2012). The simulation results (see also Methods) are presented and compared to the theory in Figure 2. In each panel, we varied one of N, m, and L, keeping the two others fixed (as long as the marker density is large enough, the number of markers M has no effect on the variance). Across most of the parameter space, our expressions agree well with simulations. Notable deviations, however, arise for the SD in particularly short or long chromosomes. For these cases, the second, more complicated approximation, which we mentioned above and appears in File S1, section S1.3, is more accurate (Figure 2).
An approximate explicit expression:
In this subsection, we derive another, simpler approximation of the variance, one that is less accurate but that has an explicit dependence on the population and genetic parameters. The gist of this approximation is that the main contribution to the variance comes from the long-distance probability of pairs of sites to reside on the same segment. Denote the distance between two given sites by d, and assume that d > m. For a given pair of individuals, if there was no recombination event between the two sites in the history of the two lineages, then both sites lie on a shared segment of length ≥d > m. Of course, even if there was a recombination event, the two sites could still be each on a different shared segment. However, this occurs with probability very close to π2, the probability that the two sites are on shared segments given that they are independent.
In terms of Equation 6, the above approximation translates to, for d > m,
| (13) |
where pnr is the probability of no recombination,
| (14) |
for distant sites where Nd ≫ 50. This is true because in the ancestral process (Figure 1), no recombination corresponds to a coalescence event taking place before any recombination event. Since the coalescence rate is 1 and the recombination rate is ρ, Equation 14 follows. We can then further simplify and neglect the contribution to the variance from sites separated by short distance d < m. Finally, we can also neglect the single-site term of the variance, since it scales as 1/M and therefore vanishes when the markers are dense. Overall, the simplified Equation 6 gives
| (15) |
for L ≫ m. Nicely, Equation 15 provides an explicit (and rather simple) dependence on N, L, and m, and as expected, the expression does not depend on the marker density. Equation 15 is also plotted in Figure 2, showing that it fits quite well to the simulation results, although it is usually less accurate than Equation 12.
For the entire (autosomal) human genome, we use Equation 5,
| . |
For m ≈ 1(cM), the last equation gives
| (16) |
A variable population size:
The framework presented above can be extended to calculate the variance for a generalization of the Wright–Fisher model in which the population size is allowed to change in time. Denote the population size as N(t) = N0λ(t), where t is the time (scaled by N0) before present. The PDF of the (scaled) coalescence time for two lineages is (see, e.g., Li and Durbin 2011)
| . |
As shown in Palamara et al. (2012), the mean of the total sharing is obtained by substituting the above Φ(t) in Equation 2, giving
| (17) |
For the variance, following Equation 13, we need to calculate the probability of no recombination, pnr. For sites distance d apart,
| (18) |
since for coalescence time t the sites are separated by 2N0t meioses, in each of which the probability of no recombination is e−d/100. Equation 18 reduces to Equation 14 for λ(t) = 1 [where Φ(t) = e−t]. Equation 18 can then be substituted into Equation 15, giving
| (19) |
In File S1, section S1.5 (Figure S1), we work out an example of a linearly expanding population, where Equation 18 was solvable and the integral of Equation 19 was evaluated numerically.
The total sharing in a length range:
Consider the quantity , defined as the total fraction of the genome found in shared segments of length in the range [ℓ1, ℓ2]. Clearly, , that is, the difference between the usual total sharing when m = ℓ1 and when m = ℓ2. The average is simply
an equation that was derived in Palamara et al. (2012) and then used for demographic inference. Here, we calculate the variance, , as follows:
| (20) |
The covariance term can be expanded as
where I(s; m = ℓ) is the indicator that site s is in a shared segment of length at least ℓ, πm=ℓ is the probability associated with the indicator, and π2(s1, ℓ1; s2, ℓ2) is the probability that site s1 is in a shared segment of length at least ℓ1 and site s2 is in a shared segment of length at least ℓ2. The key approximation, similar to the one made in An approximate explicit expression section (Equation 15), is that is nonzero only when the two sites lie on the same segment and the segment is longer than ℓ2. Defining pnr, as before, as the probability of no recombination between s1 and s2 in the history of the two individuals, we have
| (21) |
where the last step follows from Equation 15. Substituting Equation 21 into Equation 20, we obtain
For a constant population size, using Equation 15 (taking all terms in that equation) gives
| (22) |
Equation 22 is compared to simulations in Figure 3, showing good agreement. Note that as long as ℓ1, ℓ2 ≪ L, the variance depends only on the ratio ℓ2/ℓ1.
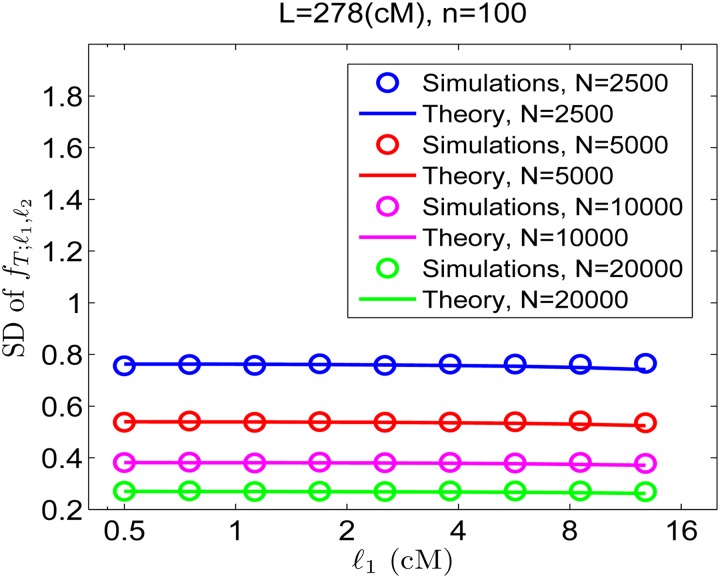
Figure 3.
The standard deviation (SD) of the total sharing in a length range. Simulation results (symbols) are shown for the SD of the fraction of the genome found in shared segments of specific length ranges. The total sharing for each range was calculated for random pairs of individuals in Wright–Fisher populations of the sizes indicated in the inset. The SD is plotted vs. the starting point of each length range, ℓ1 (where for each ℓ1, the successive data point is ℓ2). Note the logarithmic scale in the x-axis and hence that ℓ2/ℓ1 is fixed (equal to 1.5). Theory (lines) corresponds to Equation 22.
The total sharing distribution and an error model:
Having the first two moments of the total sharing, we sought to find its distribution, P(fT). While we could not find an exact expression, we could find, inspired by the numerical results of Huff et al. (2011), a reasonable fit. Huff et al. (2011) showed empirically that for HapMap’s Europeans (International HapMap Consortium 2007), the number of segments shared between random individuals was distributed as a Poisson and that the length of each segment was distributed exponentially with a lower cutoff at m, independently of the number of segments. If this is true also for the Wright–Fisher model, then the total length of the shared segments, defined as LT = LfT, is distributed as a sum of a Poisson-distributed number of these exponentials. In equations,
| (23) |
where n0 is the mean number of segments, the density of the ℓi’s is exp[−(ℓi − m)/ℓ0]/ℓ0 (ℓ0 + m is the mean segment length), and ℓi ≥ m. Such an expression is easier to handle in Laplace space, where the Laplace transform of P(LT), , is
| (24) |
and we used the convolution theorem. For given n0 and ℓ0, P(LT) [and then P(fT)] was uniquely determined from by numerical inversion (de Hoog et al. 1982; Hollenbeck 1998). For specific values of (N, L, m), we fitted the parameters n0 and ℓ0 by minimizing the squared error between the simulated distribution and P(fT) (from Equation 24) in a grid search. The results are plotted in Figure 4, with the fitted n0 and ℓ0 plotted in Figure S2. It can be seen that Equation 24 captures quite well the unique features of P(fT) (except in the tail; see Figure S2).
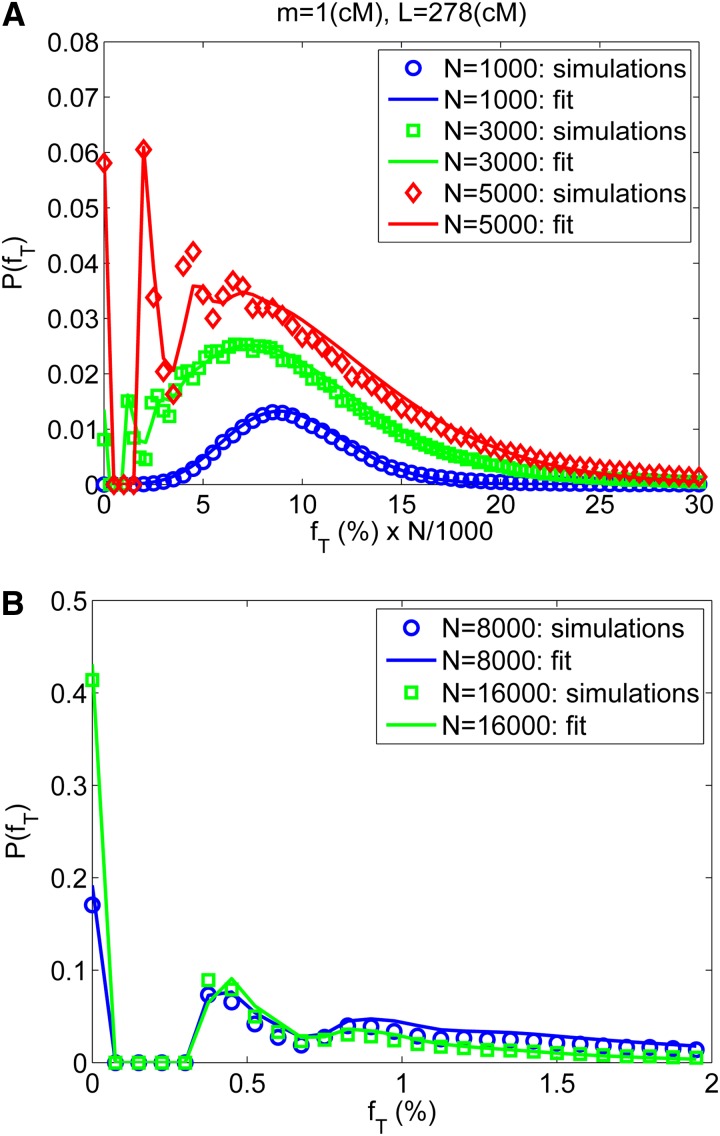
Figure 4.
The distribution of the total sharing. Simulation results (symbols) are shown for the distribution of the total sharing between random pairs of individuals in the Wright–Fisher model. Details of the simulation method are as in Figure 2A. (A) The distribution of the total sharing for N = 1000, 3000, and 5000. For better readability, the x-axis (the total sharing fT) is given in percentages and scaled by N/1000, shifting the distributions for N = 3000 and N = 5000 to the right. (B) The distribution of the total sharing for N = 8000 and 16,000. Here the x-axis is not scaled. In A and B, lines represent the fit to a sum of a Poisson number of shifted exponentials, Equation 24.
Inspection of the distributions (Figure 4) for several values of N leads to some interesting observations. For small N (e.g., N ≈ 1000 and for m = 1 cM and L = 278 cM), where the typical amount of sharing is large (〈fT〉 ≈ 5−10%, n0 ≈ 10, ℓ0 ≈ 1 cM), the distribution is unimodal (but not normal), centered around 〈fT〉. As N increases (e.g., N ≈ 3000), a discontinuous peak appears at fT = 0, with P(fT) = 0 for 0 < fT < m/L (≈0.4%). This is of course due to the restriction on the minimal segment length: a pair of individuals can share either nothing or at least one segment of length m. For fT > m/L the distribution is continuous, still centered around 〈fT〉, but with small, yet notable peaks at fT = m/L, 2m/L, 3m/L, … corresponding to pairs of individuals sharing a small number of minimal length segments. For even larger N (e.g., N ≈ 10,000 and beyond), 〈fT〉 drops below 1%, n0 ≈ 1 (ℓ0 still ≈1 cM), and the peaks at fT = 0 and fT = m/L increase such that the distribution decreases almost monotonically beyond m/L. An analytical bound on the fraction of pairs not sharing any segment is given in File S1, section S2.1 (Figure S3).
An error model:
To model errors during IBD detection, suppose that we set m large enough to avoid any false positives (i.e., detected segments that are not truly IBD). We model false negatives as true IBD segments being missed with probability ε (independent of the segment length). It is possible to extend the above formulation (Equation 23) to the case with errors, as follows. Summing over the true number of segments, n′, the distribution of the number of detected segments, n, is
that is, a Poisson with parameter n0(1 − ε). Then, as a sum of a random number of independent variables, the mean and variance of LT are 〈LT〉 = 〈n〉〈ℓ〉 and Var[LT] = 〈n〉 Var[ℓ] + 〈ℓ〉2 Var[n], where n is the number of segments and ℓ is the segment length. In our case,
| (25) |
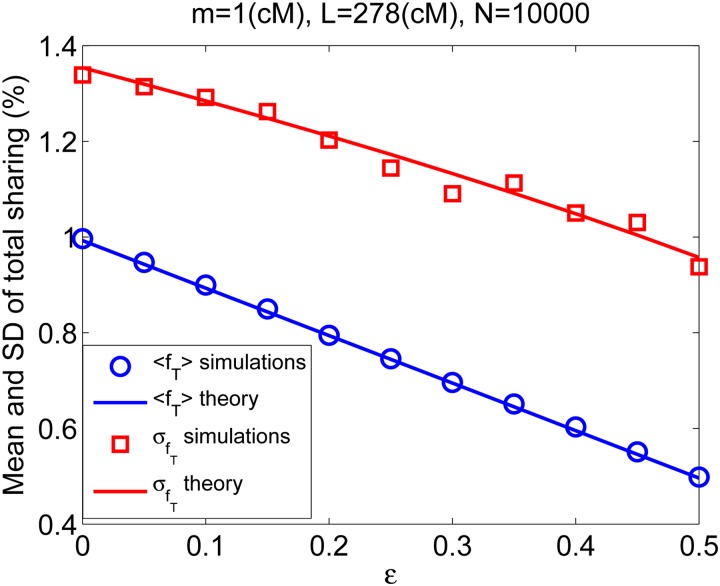
demonstrating that in the presence of detection errors, both the mean and the variance of the total sharing are (1 − ε) times their noise-free values. This is confirmed by simulations in Figure 5.
Figure 5.
The mean and standard deviation (SD) of the total sharing in the presence of detection errors. Simulation results (symbols) are plotted for mean and SD of the total sharing in the Wright–Fisher model. Simulation details are as in Figure 2, except that each segment was dropped with probability ε. Theory (lines) is from Equation 4 for the mean and Equation 12 for the SD, but where the mean is multiplied by (1 − ε) and the SD by , as in Equation 25.
Other approaches:
We note that a similar approach dates back to R. A. Fisher (Fisher 1954) and others (Bennet 1954; Stam 1980; Chapman and Thompson 2003) in their work on IBD sharing in a model where the population has been recently founded by a number of unrelated individuals. Briefly, those authors also assumed a Poisson number of IBD segments, each of which is exponentially distributed. They then matched the Poisson and exponential parameters to the average IBD sharing and the average number of segments, which they calculated using their population model. Here, we used a different population model (the coalescent; see also File S1, section S2.2) and assumed the exponentials have a cutoff at m. In principle, the parameters n0 and ℓ0 can also be directly calculated, by matching the mean and variance of the total sharing; see File S1, section S2.3. In practice, however, this does not give a good fit. In Palamara et al. (2012), a similar compound Poisson approach was developed but with a different, coalescent theory-based approximation of the segment length PDF, leading to an improved fit of the remaining parameter n0.
The cohort-averaged sharing
We have so far considered the total sharing between any two random individuals in a population. In practice, we usually collect genetic information on a cohort of n individuals. In this context, we can attribute each individual with the amount of genetic material it shares with the rest of the cohort. Define, for each individual, the cohort-averaged sharing as the average total sharing between the given individual and the other n − 1 individuals in the cohort. Naively, one may anticipate that the width of the distribution of will approach zero for large n, because the averaging will tend to eliminate any randomly arising differences between the individuals. We show that in fact, the width of the distribution approaches a nonzero limit. The individuals at the right tail of the cohort-averaged sharing distribution can be seen as “hypersharing”, meaning they are, on average, more genetically similar to members of the cohort than are others. Similarly, individuals at the left tail are “hyposharing”. The existence of hypersharing individuals is important for prioritizing individuals for sequencing, as we show in Implications to sequencing study design section.
Define the fraction of the genome shared by individuals i and j as . The cohort-averaged sharing of i, , is
The variance of is
| (26) |
where we assumed n ≫ 1 and used the fact that the covariance term is identical for all (i, j1, j2) combinations and therefore, for simplicity of notation, we set i = 1, j1 = 2, and j2 = 3. Recall that (Equation 3), where I(s) is the indicator that site s is on a shared segment. Thus, the covariance can be written as
where I(i,j)(s) is the indicator that site s is on a segment shared between individuals i and j, and is the probability that a given site is on a segment shared between 1 and 2 and that another site, k markers away from the first, is on a segment shared between 1 and 3. As in An approximate explicit expression section (e.g., Equation 15), we will keep only the most dominant term in the sum. Consider the coalescent tree relating the three individuals 1, 2, and 3 and assume that the distance between the sites is d > m. If there was no recombination event in the entire tree between the two sites, then immediately . Otherwise, we assume that each of the two sites belongs to a shared segment (or not) independently of the other; that is, . The probability of no recombination, pnr, depends on T3, the total size of the tree of three lineages. Since the PDF of T3 is (Wiuf and Hein 1999; Wakeley 2009),
or, for dN ≫ 100,
| . |
The covariance becomes
Substituting Equations 15 and 27 in Equation 26, the standard deviation of the cohort-averaged sharing is
| (28) |
For , , while for (but <N, as the cohort size cannot exceed the population size), , which is independent of n. This implies that even for very large cohorts, the distribution of the cohort-averaged sharing retains a minimal width. Equation 28 is in good agreement with simulations, as shown in Figure 6A (although some deviations are seen for larger n). We note that the variance was computed for a given individual over all ancestral processes of a cohort of size n. Therefore, the variance within the cohort, for a given ancestral process might actually be smaller. Simulations results (Figure S4), however, show that unless n is very small, Equation (28) is a good approximation also for the variance within the cohort.
For a genome with c chromosomes,
| , |
where is the cohort-averaged sharing of chromosome i. For the human genome and for small n and m ≈ 1 cM, Equation 16 gives
| (29) |
whereas for large n, Equation 27 gives
| , | (30) |
which is, as explained above, independent of n.
The fact that the width of the cohort-averaged sharing distribution does not approach zero for large n results from the “long-range” correlations between the averaged (n − 1) variables or, in other words, the fact that for all i, j1, j2. In Hilhorst and Schehr (2007), it was found that when all pairs of random variables are weakly correlated, the PDF of their average converges to a normal distribution. In our case, the covariance is (Equation 27), much smaller, for typical values of N, L, and m, than (Equation 15). The variables we average are therefore weakly dependent, as we also observe in simulations (Figure S5). We thus conjectured that the distribution of the cohort-averaged sharing is normal or is close to one. This is confirmed by simulation results, as shown in Figure 6B. We note, however, that a small but systematic deviation from a normal distribution exists in all parameter configurations we tested, in the form of a broader right tail and a narrower left tail than expected (Figure S5). This seems to be due to the small probability (≈1/N) of random pairs of individuals to be close relatives.
Implications to sequencing study design
Suppose we have sparse genotype information for a large cohort, as well as whole-genome sequences for a subset of it. If the genotype data allow detection of IBD shared segments, then alleles not typed can be directly imputed if they lie on haplotypes shared with sequenced individuals (see, e.g., Uricchio et al. 2012). In fact, such a strategy is expected to be quite successful; as we mentioned in the Definitions section, only about one recent mutation is expected on each shared segment. Since some individuals share more than others, their sequencing should be prioritized if imputation power is to be maximized. Recently, Gusev et al. (2012b) developed an algorithm (Infostip) for sample selection based on the observed IBD sharing. Here, using our results in The cohort-averaged sharing section, we calculate the theoretical maximal imputation power.
Assume first that individuals are haploids; the case of diploids is treated later. Assume a cohort of size n, a budget that enables the sequencing of ns individuals, and two selection strategies: either of random ns individuals or of the ns individuals with the largest cohort-averaged sharing. Define an imputation metric, , as the average fraction of the genome of i, an individual not sequenced, that is shared IBD with at least one sequenced individual. Let the selected individuals be , and denote the fraction of the genome that i shares with mj as . To calculate , we assume that for all j1, j2, = 1, … , ns (), is independent of (which is justified, as we showed in The cohort-averaged sharing section). We also assume that the locations of the shared segments are independent and uniformly distributed along the genome. Under these assumptions, the probability of a locus to be covered by at least one sequenced individual is
| (31) |
and this is also the average covered fraction of the genome. We note, however, that assuming that the locations of shared segments are independent and uniformly distributed is mostly for mathematical convenience. Simulation results (Figure S6) show that sharing tends to concentrate at specific loci, implying that Equation 31 can be thought of as an upper bound (see Figure 7). When fT ≪ 1,
| , |
and for random selection of individuals for sequencing,
| (32) |
where 〈fT〉 is given by Equation 4. When selecting the highest-sharing individuals, values of come from the right tail of the cohort-averaged sharing distribution, ,
where and are the minimum and average, respectively, of the cohort-averaged sharing among the sequenced individuals . Since we argued in The cohort-averaged sharing section that is approximately normal with parameters 〈fT〉 and (Equation 28), satisfies
| (33) |
We can thus finally write
| (34) |
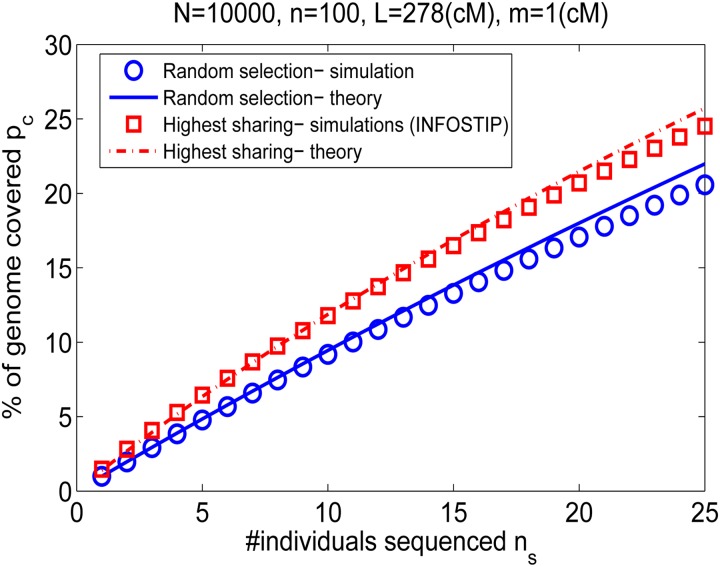
Before getting to simulations, we note that in practice, selection of exactly those individuals with the largest cohort-averaged sharing will not achieve the imputation power of Equation 34. This is because the top sharing individuals usually share many segments with each other and thus sequencing of all of them will be redundant (e.g., in the extreme case of siblings, both will appear as top sharing, but sequencing of both will add little power beyond sequencing just one). To avoid such redundancies, we selected the high-sharing (simulated) individuals using Infostip (Gusev et al. 2012b), which proceeds in a greedy manner, each time selecting the individual who shares the most with the rest of the cohort in regions that are not yet covered by the already selected individuals. We then compared the imputation power when individuals were selected either randomly or using Infostip. The results, shown in Figure 7, suggest good agreement between the theoretical Equations 32 and 34 and the simulations, at least as long as ns is not large (relative to n). For large ns, the coverage is lower than predicted, likely due to the nonuniform concentration of the shared segments.
Figure 7.
Coverage of genomes not selected for sequencing by IBD shared segments. We simulated 500 Wright–Fisher populations with N = 10,000, n = 100, and L = 278 cM and searched for IBD segments with length ≥m = 1 cM. For each plotted data point, we selected ns individuals either randomly or using Infostip. Then, for each of the n − ns individuals not selected, we calculated the fraction of their genomes shared with at least one selected individual. We plotted (symbols) the average coverage over all individuals in all populations. Lines correspond to theory: Equation 32 for random selection and Equation 34 for Infostip selection.
For a cohort of n diploid individuals (assuming phase can be resolved) we redefine the cohort-averaged sharing as
(where, e.g., is the cohort-averaged sharing of the first chromosome of individual i) and assume that the individuals selected for sequencing have the largest diploid cohort-averaged sharing. Since the two terms in are weakly dependent,
where is given by Equation 28. The coverage metric pc is interpreted, as before, as the probability of a locus on a given chromosome to be in a segment shared with at least one sequenced chromosome. The theory developed above is still valid, provided that in Equations 32 and 34 ns is replaced by 2ns and that in Equation 33 is replaced by .
Increase in association power:
Using our results for the power of imputation by IBD, we calculate below the expected subsequent increase in power to detect rare variant association. We use the simple model of Shen et al. (2011), in which we consider rare variants that appear in cases but not in any control, and assume that the causal variant is dominant.
Assume that we have genotyped and detected IBD segments in a cohort of nc (diploid) cases and nt controls and that we sequenced a subset of ns individuals, of which nc,s are cases and nt,s are controls (ns = nc,s + nt,s). After imputation by IBD, a locus in a (diploid) individual not sequenced has probability to be successfully imputed, where pc is given by Equation 32 or Equation 34. For a given locus, we define the effective number of cases (controls), as the number of cases (controls) for which genotypes are known either directly from sequencing or from imputation. Since there are nc − nc,s cases not sequenced and nt − nt,s controls not sequenced,
| (35) |
In the last equation we assumed, without loss of generality, that cases can only be imputed using other cases and vice versa. The probability of a variant to appear in exactly b cases but in no controls, under the null hypothesis that the variant assorts independently of the disease, is given by Fisher’s exact test,
Define Q as the threshold P-value and denote by b* the smallest integer above which P(cases only) < Q. When the causal variant carrier frequency in cases is β, the probability of the variant to appear in b cases is binomial, and the power is, for a given Q,
| (36) |
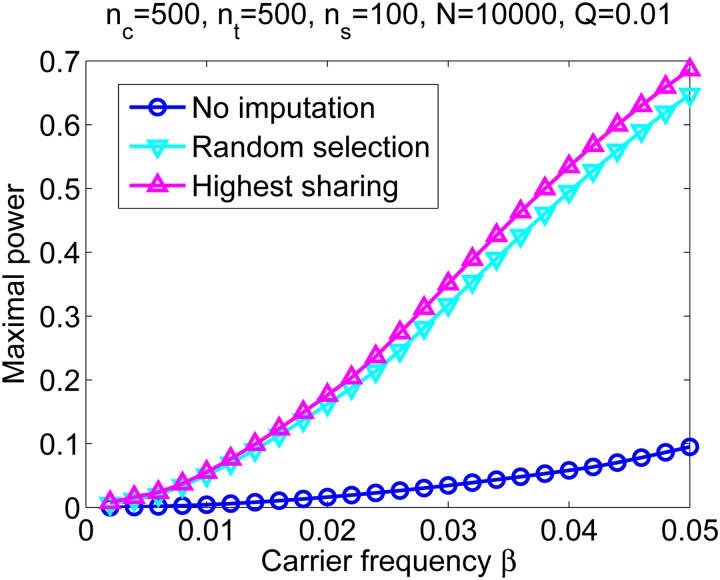
In Figure S7, we plot the power vs. nc,s, when the sequencing budget ns = nc,s + nt,s is fixed and for representative parameter values. In Figure 8, we plot the power vs. the carrier frequency for the optimal value of nc,s. Figure 8 demonstrates that the power increases by severalfold when imputation by IBD is used. This is, however, an expected consequence of increasing the effective sample size and would likely be achieved with any imputation algorithm (e.g., Howie et al. 2012). Figure 8 also shows an additional, slight increase when the highest-sharing individuals are selected for sequencing. Thus, while it should be easy to identify the highest-sharing individuals given a genotyped cohort [e.g., using Infostip (Gusev et al. 2012b)], and doing so will increase the association power, our results suggest that the gain in power over a random selection will be minor.
Figure 8.
Power to detect an association after imputation by IBD. The maximal power to detect an association is shown, with and without imputation by IBD and with sequenced individuals selected either randomly or according to their total sharing. The parameters we used were N = 10,000, L = 278 cM (one chromosome), m = 1 cM, cohort size of 500 cases and 500 controls, a total sequencing budget of ns = 100 individuals, and a threshold P-value of Q = 0.01. For each carrier frequency β, we computed the power for each pair of nc,s and nt,s (number of sequenced cases and controls, respectively), such that nc,s + nt,s = ns, and recorded and plotted the maximal power. The power was calculated using Equations 35 and 36, where in Equation 35, pc was set to zero for the case of no imputation, or calculated using Equations 32 and 34 (random selection and selection by total sharing, respectively, and adjusted for diploid individuals). For the studied parameter set, imputation by IBD leads to a major increase in power. Proper selection of individuals for sequencing also contributes to the power but only slightly.
Other applications of the variance of IBD sharing
An estimator of the population size:
Assume that we have genotyped or sequenced a diploid chromosome of one individual and calculated fT, the fraction of the chromosome shared between the individual’s paternal and maternal chromosomes. Can we estimate the effective population size?
According to Equation 4, . Solving for N gives (see also Palamara et al. 2012)
for 〈fT〉 ≪ 1. This suggests the following estimator,
| (37) |
Below, we investigate the properties of the simple estimator of Equation 37. Using Jensen’s inequality, it is easy to see that the estimator is biased,
The variance of is proportional to Var[1/fT], which we could not calculate, but could approximate as follows. Let us write as
where we applied the Taylor expansion , assuming |fT − 〈fT〉| ≪ 〈fT〉 (in which regime clearly ). Since additive constants do not contribute to the variance, the standard deviation is
| , |
where we used 〈fT〉 ≈ 100/(mN) (Equation 4) and Equation 15 for . The effective population size can also be inferred using Watterson’s estimator, which is , where S2 is the number of heterozygous sites and μ is the mutation rate (per chromosome per generation). Watterson’s estimator is unbiased, , and has variance (assuming no recombination) . Therefore, , compared to for the IBD estimator.
Note that in practice, the proposed estimator is not very useful, as it diverges whenever fT = 0 (which is common for large N). Suppose, however, that we have sequences for n (haploid) chromosomes and that we have computed the total sharing between all pairs. Define . The estimator now takes the form
| (38) |
This is again an overestimate, . In File S1, section S3, we show that is approximately
| . | (39) |
For comparison, in Watterson’s estimator for n (haploid) chromosomes, (for large N and n), which decays to zero with increasing n slower than the IBD estimator. Simulation results, shown in Figure S8 and Figure S9, confirm the accuracy of Equation 39 and show that the bias is limited to very small values of n.
In the context of the error model in The total sharing distribution and an error model section, introducing a probability ε to miss a true IBD segment will decrease the average total sharing by (1 − ε) (Equation 25). Consequently, Equation 38 will estimate a population size ∼1/(1 − ε) [≈ (1 + ε) for small ε] larger than the true one.
IBD sharing between siblings:
The total IBD sharing between relatives can usually be decomposed into sharing due to the recent coancestry and “background” sharing due to population inbreeding (Huff et al. 2011; Henn et al. 2012). While much is known about the distribution of sharing in pedigrees (e.g., Hill and Weir 2011), less is known about the population-level sharing, and relatedness detection algorithms (e.g., Huff et al. 2011; Henn et al. 2012) estimate it empirically. In a different domain, the variance in sharing between relatives appears in theoretical calculations of the variance of heritability estimators (Visscher et al. 2006). Our results for the variance of the total sharing in the Wright–Fisher model (Variation in IBD sharing in the Wright–Fisher model section) can thus have practical applications if modified to account for recent coancestry.
Here, we calculate the variance of the sharing between siblings by combining the approach of Visscher et al. (2006) with that of our An approximate explicit expression section. Assume that two individuals are siblings, either half or full: we calculate, without loss of generality, only the sharing between the two chromosomes that descended from the same parent and denote the fraction of sharing as fS. Assume as before a population of size N and one chromosome of length L. For a given marker to be on a shared segment, it can either be on a segment directly coinherited from the same grandparent (probability 1/2) or otherwise be on a segment shared between the grandparents (probability π/2, Equation 2). We ignore boundary effects near the sites of recombination at the parent. The mean fraction of the genome shared is therefore just 〈fS〉 = (1 + π)/2. The variance can be written as in Equation 6,
where π2,S(k) is the probability of two sites separated by k markers [or genetic distance ] to be on segments shared between the siblings. The probability that the two sites are both coinherited from the same grandparent is
where r is the recombination fraction and we used Haldane’s map function (Visscher et al. 2006). Also with probability psame, the sites are both inherited from different grandparents, and we use the expressions developed in An approximate explicit expression section for the probability of the sites to be in shared segments: π2,S(k) = π2 + Θ(d − m)pnr [where pnr ≈ 50/(Nd) and Θ(x) = 1 for x > 0 and is zero otherwise]. With probability (1 − 2psame), one site is coinherited and the other is not; in that case π2,S(k) = π. Approximating the sum as an integral and simplifying, we finally have
| (40) |
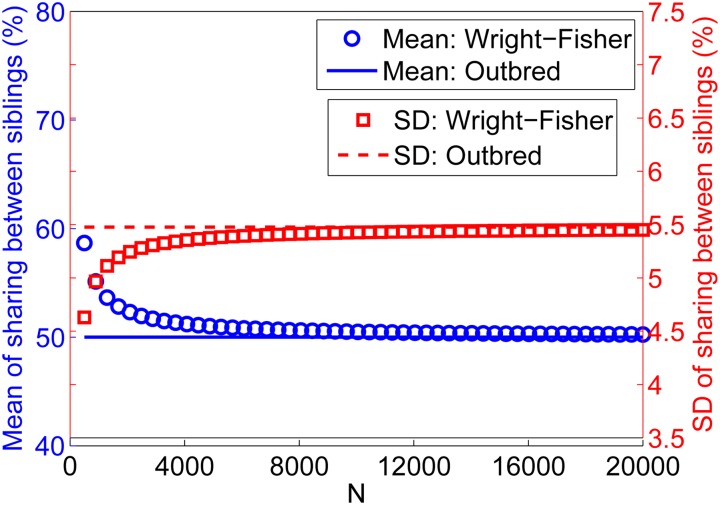
We solved Equation 40 using Mathematica and summed over all chromosomes as in Equation 5. The results for the mean and SD of the total sharing between siblings are plotted in Figure 9 and compared to an outbred population where the grandparents are unrelated. The SD in the outbred population overestimates the Wright–Fisher SD, up to ≈18% for N as small as 500.
Figure 9.
IBD sharing between siblings in the Wright–Fisher model. We plot the theoretical mean and standard deviation (SD) of the IBD sharing between the (maternal only or paternal only haploid) genomes of siblings. Lines correspond to an outbred population (unrelated grandparents): the mean sharing is 50% and the SD is taken from Visscher et al. (2006). Symbols correspond to the theory for the Wright–Fisher model: the mean sharing is (1 + π)/2 (where π is given by Equation 2), and the SD is given by Equation 40. We used m = 1 cM and the chromosome lengths of the autosomal human genome. Note that the y-axis is on the left side for the mean and on the right side for the SD.
IBD sharing after an admixture pulse:
In this final subsection, we study the IBD sharing in a simple admixture model. In our model, a single population A of constant size N has received gene flow from population B, Ga generations ago. We assume that gene flow took place for one generation only (hence, an admixture pulse) and, further, that population B is sufficiently large that the chromosomes it donated to A share no detectable IBD segments. Denote the fraction of the lineages coming from population A at the admixture event as α (fraction 1 − α coming from B), and let Ta = Ga/N be the scaled admixture time. We are interested in IBD sharing between extant chromosomes in population A.
To approximate the mean IBD sharing in the sample, note that if admixture was very recent, then two chromosomes will be potentially shared only if both descend from population A, which occurs with probability α2. Therefore, the mean sharing is α2 times its value without admixture. While this is a good approximation (Figure S10), it does not account for two chromosomes, one or two of which are from the external population B, having their common ancestor more recently than the admixture event. We therefore calculate the mean IBD sharing using Equation 17, using the following (nonnormalized) PDF for the coalescence times,
| (41) |
which gives
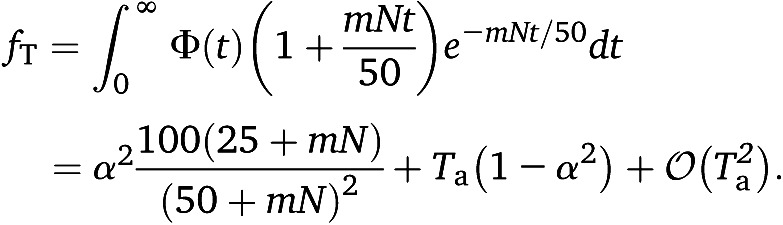 |
(42) |
Note that this is just 〈fT〉admix ≈ α2〈fT〉no admix + (1 − α2)Ta. The first term corresponds to lineages descending from population A; the second term corresponds to at least one of the lineages descending from population B but where the lineages have coalesced already in the hybrid population. The variance can be similarly calculated, by substituting Equation 41 into Equation 19,
| (43) |
where γ is the Euler–Mascheroni constant, and we solved the integrals in Mathematica and later simplified under specific assumptions (see File S1, section S4). Equation 43 usually predicts a variance slightly smaller than the case of no admixture. Simulation results are shown in Figure S10 for the mean and variance. While agreement is not perfect (as Equation 19 is itself approximate), Equations 42 and 43 capture the main effects of changing α and Ta. Note that the result of Equation 42 implies that, for small Ta and large N, the observed mean IBD sharing is as if the population is of size ≈N/α2.
A test for admixture:
For recent admixture (small Ta), the fractions of ancestry vary among individuals (Verdu and Rosenberg 2011; Gravel 2012). In our model, since a pair of segments is shared mostly when both descend from population A, some individuals will share more than others merely due to having a larger fraction of A ancestry. In turn, this will increase the variance of the cohort-averaged sharing. This observation suggests the following test for a recent gene flow into a population: (i) extract IBD segments and calculate the mean fraction of total sharing over all pairs, , as well as the SD of the cohort-averaged sharing, ; (ii) use Equation 38 to infer the population size, ; (iii) simulate Npop populations of size , extract IBD sharing, and calculate the SD of the cohort-averaged sharing in each population; and (iv) the P-value for rejecting the null hypothesis of no admixture is the fraction of the Npop populations where the SD of the cohort-averaged sharing was larger than the observed one. Note that the identity of the external population need not be known, nor are the admixture fraction and time; the test relies on admixture creating a gradient of ancestry fractions and hence an increased variability in the similarity between individuals. Simulation results are plotted in Figure S10, showing that for a P-value of 0.05 and Ga = 5, gene flow with α ≈ 0.9 or lower can be detected (α ≈ 0.8 or lower for Ga = 10). We stress that a broader than expected distribution of cohort-averaged sharing does not necessarily indicate admixture, and there might be other factors responsible for the effect (see also the Discussion). We validated, however, that IBD detection errors alone (as in the model in The total sharing distribution and an error model section) as well as variable population size (in a simple two-size model) do not lead to significant P-values in the admixture test (Figure S11).
IBD sharing and admixture in the Ashkenazi Jewish population:
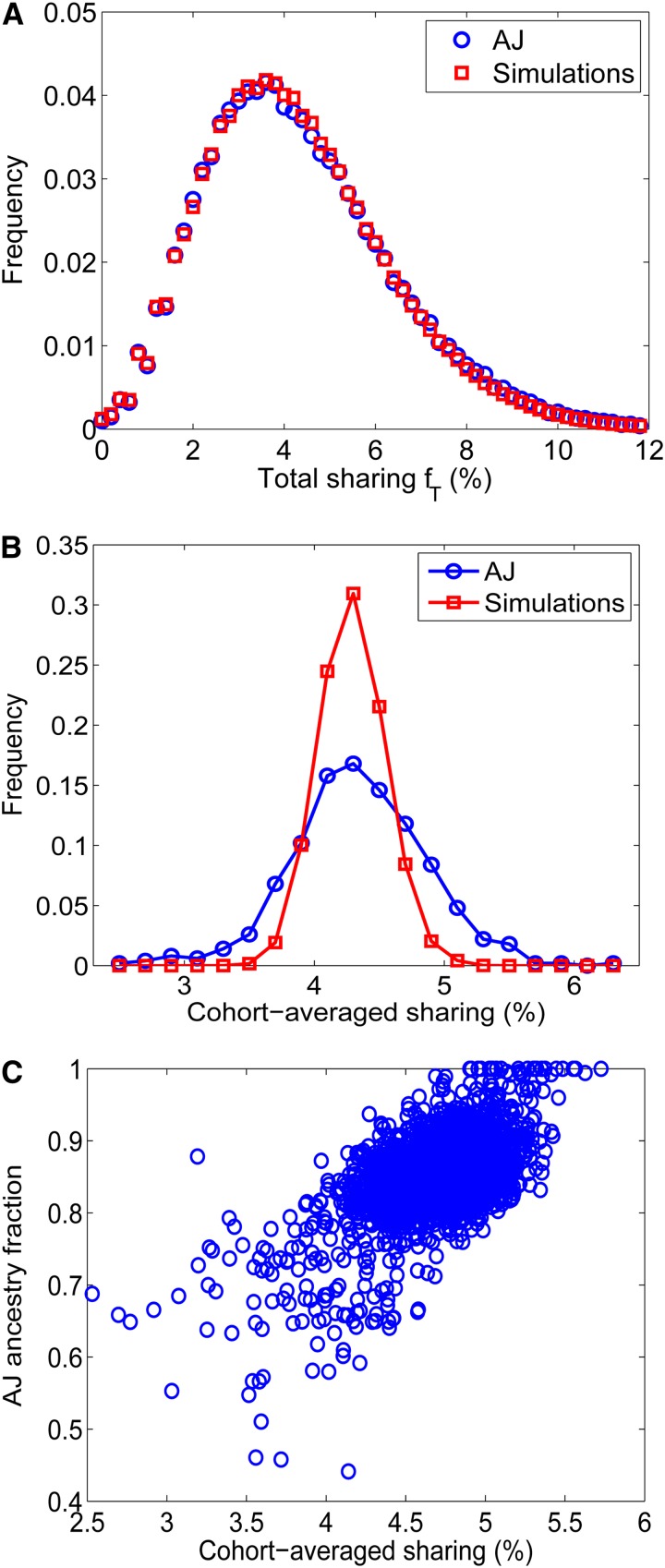
As our final result, we apply the admixture test to the real population of Ashkenazi Jews (AJ). Historical records, and recently also genetic studies, suggest that AJ form a genetically distinct group of likely Middle-Eastern origin. However, the AJ population was also shown to receive a significant amount of gene flow from neighboring European populations (Ostrer 2001; Atzmon et al. 2010; Behar et al. 2010; Bray et al. 2010; Guha et al. 2012). We analyzed a data set of ≈ 2600 AJ, details of which have been published elsewhere (Guha et al. 2012; Palamara et al. 2012) and are summarized in the Methods section. To detect IBD shared segments in the AJ population, we used Germline (Gusev et al. 2009). For 500 individuals on chromosome 1, and with m = 1 cM, the average fraction of sharing over all pairs is ≈4.4%, leading to an estimated population size of . The SD of the cohort-averaged sharing is 0.52%, higher than the SD in all 500 populations we simulated with a constant size (typically 0.34%, maximum 0.41%). The recent history of Ashkenazi Jews, however, has likely involved bottlenecks and expansions, different from the constant size assumption. In Palamara et al. (2012), a population model was inferred based on the fraction of the genome shared at different segment lengths. The model’s best estimate of AJ history is a slow expansion until ∼35 generations ago and then a severe bottleneck (effective population size of just 270) followed a by rapid expansion to a current size of a few millions. As can be seen in Figure 10, A and B, the model agrees well with the distribution of the fraction of total sharing over all pairs, but predicts a much narrower distribution of cohort-averaged sharing than the true one. Here too, in none of 100 simulated populations with the inferred demography was the SD of the cohort-averaged sharing as large as in the real data. These results, therefore, suggest (based on the AJ population alone) that the AJ population was the target of a recent gene flow. To confirm that the increase in the variance of the cohort-averaged sharing is due (at least partly) to admixture, we ran an admixture analysis [Admixture (Alexander et al. 2009)] comparing AJ to HapMap’s CEU (International HapMap Consortium 2007). As can be seen in Figure 10C, the fraction of “AJ ancestry” is indeed highly correlated with the cohort-averaged sharing (Pearson’s r = 0.59).
Figure 10.
IBD sharing and admixture in the Ashkenazi Jewish (AJ) population. We detected IBD shared segments using Germline in chromosome 1 of n = 500 AJ individuals and compared them to simulations of the demographic history inferred in Palamara et al. (2012). (A) The distribution of the total sharing over all pairs. (B) The distribution of the cohort-averaged sharing. While the demographic model fits well the sharing distribution over all pairs, the distribution of the real cohort-averaged sharing is broader than in the model. (C) We used Admixture to calculate the admixture fraction of AJ individuals compared to the CEU population. The “AJ ancestry fraction” of each individual is plotted against its cohort-averaged sharing. C shows results for the full data set (≈2600 individuals).
Discussion
The recent availability of dense genotypes, together with sophisticated detection tools, has transformed IBD sharing into an increasingly important tool in population genetics. Here, we used coalescent theory to compute the variance and other properties of the total sharing in the Wright–Fisher model. For the variance, we suggested three derivations, one of which was more coarse but had a simple closed form that was later extended to populations of variable size. Investigating the cohort-averaged sharing, we discovered the curious phenomenon of hypersharing. We showed how this can be exploited to improve power in imputation and association studies. We also calculated the variance of the total sharing between siblings and briefly considered some implications to the accuracy of demographic inference. We finally investigated IBD sharing in a hybrid population and suggested a test for admixture based on the cohort-averaged sharing, which we then applied to the Ashkenazi Jewish population. We provide Matlab routines for the main results (File S2).
Most of our analytical results depend on certain assumptions and simplifications, as specified in the individual sections and in File S1, section S1.2. Additionally, in reality, the Wright–Fisher model and the coalescent are only approximations of the true ancestral process, and procedures such as phasing, IBD inference, and imputation are also prone to error. IBD detection errors will particularly affect our results for imputation and association studies (Implications to sequencing study design section), and these results should therefore be considered as idealized upper bounds. The error model we introduced, where each IBD segment is missed with a certain probability, gives a sense of the effect of errors. Investigation of more detailed models, e.g., length-dependent error rate for segment misdetection or more realistic models for imputation and association studies, is challenging and left for future work.
Prospects of our work are in a few fields. First, as shown in Palamara et al. (2012), theoretical characterization of IBD sharing can lead to new methods for demographic inference, which are expected to perform particularly well when investigating the recent history of genetic isolates. Here, we expanded the theory of IBD sharing to compute the variance of the total sharing and the cohort-average sharing. This turned useful, for example, when we provided in An estimator of the population size section expressions for the variance of an estimator of the population size based on the average sharing over all pairs of chromosomes and in IBD sharing after an admixture pulse section a test for recent admixture. In another domain, understanding the distribution of sharing between relatives can improve the accuracy of relatedness detection (IBD sharing between siblings section). Other potential applications are in the detection of regions either positively selected or associated with a disease based on excess sharing, although more work is needed for these. Finally, our results provide the first estimate for the potential success of imputation by IBD strategies (Implications to sequencing study design section). We note that of course, once a given cohort has been genotyped, IBD can be calculated directly to estimate the expected success of imputation. However, in many cases, study design takes place before the actual recruiting and genotyping, and then, if a rough estimate of the population size is available, our results can be invoked to estimate the amount of resources needed.
One of our interesting findings was the presence of hypersharing individuals. While we did not define the term precisely, we referred to the fact that even for large cohorts, the variance of the cohort-averaged sharing does not decrease below a certain value. This result, while somewhat counterintuitive, follows naturally from the population model. In the real population of AJ, we showed that the distribution of the cohort-averaged sharing is even broader, indicating possible admixture, and indeed, we found that the cohort-averaged sharing is highly correlated with the Ashkenazi ancestry fraction. This is not to say that admixture was the only factor shaping the distribution of IBD sharing; other factors such as selection or population substructure could have been playing a role as well. Our results, however, emphasize the importance of reconstructing the AJ demography simultaneously with that of their neighboring populations.
Acknowledgments
We thank the reviewers for insightful comments and Omer Bobrowski for discussions. S.C. thanks the Human Frontier Science Program for financial support. I.P. acknowledges support from National Science Foundation grant CCF 0845677 and National Institutes of Health grant U54 CA121852.
Footnotes
Communicating editor: Y. S. Song
Literature Cited
- Akula N., Detera-Wadleigh S., Shugart Y. Y., Nalls M., Steele J., et al. , 2011. Identity-by-descent filtering as a tool for the identification of disease alleles in exome sequence data from distant relatives. BMC Proc. 5: S76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen A., Korneliussen T. S., Moltke I., van Overseem Hansen T., Nielsen F. C., et al. , 2009. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet. Epidemiol. 33: 266–274. [DOI] [PubMed] [Google Scholar]
- Albrechtsen A., Moltke I., Nielsen R., 2010. Natural selection and the distribution of identity-by-descent in the human genome. Genetics 186: 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G., Hao L., Pe’er I., Velez C., Pearlman A., et al. , 2010. Abraham’s children in the genome era: Major Jewish diaspora populations comprise distinct genetic clusters with shared middle eastern ancestry. Am. J. Hum. Genet. 86: 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar D. M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., et al. , 2010. The genome-wide structure of the Jewish people. Nature 466: 238–242. [DOI] [PubMed] [Google Scholar]
- Bennet J. H., 1954. The distribution of heterogeneity upon inbreeding. J. Roy. Stat. Soc. B 16: 88–99. [Google Scholar]
- Bray S. M., Mulle J. G., Dodd A. F., Pulver A. E., Wooding S., et al. , 2010. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc. Natl. Acad. Sci. USA 107: 16222–16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. D., Glazner C. G., Zheng C., Thompson E. A., 2012. Inferring coancestry in population samples in the presence of linkage disequilibrium. Genetics 190: 1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2011. A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 88: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Thompson E. A., 2012. Detecting rare variant associations by identity-by-descent mapping in case-control studies. Genetics 190: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr I. M., Markham A. F., Pena S. D. J., 2011. Estimating the degree of identity by descent in consanguineous couples. Hum. Mutat. 32: 1350–1358. [DOI] [PubMed] [Google Scholar]
- Chapman N. H., Thompson E. A., 2003. A model for the length of tracts of identity by descent in finite random mating populations. Theor. Popul. Biol. 64: 141–150. [DOI] [PubMed] [Google Scholar]
- Davison D., Pritchard J., Coop G., 2009. An approximate likelihood for genetic data under a model with recombination and population splitting. Theor. Popul. Biol. 75: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog F. R., Knight J. H., Stokes A. N., 1982. An improved method for numerical inversion of Laplace transforms. SIAM J. Sci. Stat. Comput. 3: 357–366. [Google Scholar]
- Fisher R. A., 1954. A fuller theory of “junctions” in inbreeding. Heredity 8: 187–197. [Google Scholar]
- Gravel S., 2012. Population genetics models of local ancestry. Genetics 191: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, R. C., 1991 The two-locus ancestral graph, pp. 100–117 in Selected Proceedings of the Symposium on Applied Probability, Sheffield, 1989, Vol. 18 (IMS Lecture Notes—Monograph Series). Institute of Mathematical Statistics, Hayward, CA. [Google Scholar]
- Guha S., Rosenfeld J. A., Malhotra A. K., Lee A. T., Gregersen P. K., et al. , 2012. Implications for health and disease in the genetic signature of the Ashkenazi Jewish population. Genome Biol. 13: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Lowe J. K., Stoffel M., Daly M. J., Altshuler D., et al. , 2009. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 19: 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Kenny E. E., Lowe J. K., Salit J., Saxena R., et al. , 2011. DASH: a method for identical-by-descent haplotype mapping uncovers association with recent variation. Am. J. Hum. Genet. 88: 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Palamara P. F., Aponte G., Zhuang Z., Darvasi A., et al. , 2012a The architecture of long-range haplotypes shared within and across populations. Mol. Biol. Evol. 29: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Shah M. J., Kenny E. E., Ramachandran A., Lowe J. K., et al. , 2012b Low-pass genome-wide sequencing and variant inference using identity-by-descent in an isolated human population. Genetics 190: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., 2006. Principles of Population Genetics, Ed. 4 Sinauer Associates, Sunderland, MA. [Google Scholar]
- Henn B. M., Hon L., Macpherson J. M., Eriksson N., Saxonov S., et al. , 2012. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS One 7: e34267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst H. J., Schehr G., 2007. A note on q-Gaussians and non-Gaussians in statistical mechanics. J. Stat. Mech., P06003. [Google Scholar]
- Hill W. G., Weir B. S., 2011. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. 93: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck K. J., 1998. INVLAP.M: a matlab function for numerical inversion of Laplace transforms by the de Hoog algorithm. J. Sci. Stat. Comp. 3: 357–366. [Google Scholar]
- Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G. R., 2012. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 1983. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23: 183–201. [DOI] [PubMed] [Google Scholar]
- Huff C. D., Witherspoon D. J., Simonson T. S., Xing J., Watkins W. S., et al. , 2011. Maximum-likelihood estimation of recent shared ancestry (ERSA). Genome Res. 21: 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick B., Li S. C., Karp R. M., Halperin E., 2011. Pedigree reconstruction using identity by descent. J. Comput. Biol. 18: 1481–1493. [DOI] [PubMed] [Google Scholar]
- Kong A., Masson G., Frigge M. L., Gylfason A., Zusmanovich P., et al. , 2008. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 9: 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Zöllner S., Abecasis G. R., 2007. GENOME: a rapid coalescent-based whole genome simulator. Bioinformatics 23: 1565–1567. [DOI] [PubMed] [Google Scholar]
- McVean G. A. T., Cardin N. J., 2005. Approximating the coalescent with recombination. Philos. Trans. R. Soc. B 360: 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrer H., 2001. A genetic profile of contemporary Jewish populations. Nat. Rev. Genet. 2: 891–898. [DOI] [PubMed] [Google Scholar]
- Palamara P. F., Lencz T., Darvasi A., Pe’er I., 2012. Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet. 91: 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palin K., Campbell H., Wright A. F., Wilson J. F., Durbin R., 2011. Identity-by-descent-based phasing and imputation in founder populations using graphical models. Genet. Epidemiol. 35: 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale N., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M. N., Gusev A., Pe’er I., 2011. HLA type inference via haplotypes identical by descent. J. Comput. Biol. 18: 483–493. [DOI] [PubMed] [Google Scholar]
- Shen Y., Song R., Pe’er I., 2011. Coverage tradeoffs and power estimation in the design of whole-genome sequencing experiments for detecting association. Bioinformatics 27: 1995–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen K. T., Churchill G. A., 1997. A Markov chain model of coalescence with recombination. Theor. Popul. Biol. 52: 43–59. [DOI] [PubMed] [Google Scholar]
- Stam P., 1980. The distribution of the fraction of the genome identical by descent in finite random mating populations. Genet. Res. 35: 131–155. [Google Scholar]
- Stevens E. L., Heckenberg G., Roberson E. D. O., Baugher J. D., Downey T. J., et al. , 2011. Inference of relationships in population data using identity-by-descent and identity-by-state. PLoS Genet. 7: e1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uricchio L. H., Chong J. X., Ross K. D., Ober C., Nicolae D. L., 2012. Accurate imputation of rare and common variants in a founder population from a small number of sequenced individuals. Genet. Epidemiol. 36: 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P., Rosenberg N. A., 2011. A general mechanistic model for admixture histories of hybrid populations. Genetics 189: 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Medland S. E., Ferreira M. A. R., Morley K. I., Zhu G., et al. , 2006. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2009. Coalescent Theory: An Introduction. Roberts & Co. Greenwood Village, Colorado. [Google Scholar]
- Wiuf C., Hein J., 1999. Recombination as a point process along sequences. Theor. Popul. Biol. 55: 248–259. [DOI] [PubMed] [Google Scholar]