Abstract
It has increasingly been recognized that adapting populations of microbes contain not one, but many lineages continually arising and competing at once. This process, termed “clonal interference,” alters the rate and dynamics of adaptation and biases winning mutations toward those with the largest selective effect. Here we uncovered a dramatic example of clonal interference between multiple similar mutations occurring at the same locus within replicate populations of Methylobacterium extorquens AM1. Because these mutational events involved the transposition of an insertion sequence into a narrow window of a single gene, they were both readily detectable at low frequencies and could be distinguished due to differences in insertion sites. This allowed us to detect up to 17 beneficial alleles of this type coexisting in a single population. Despite conferring a large selective benefit, the majority of these alleles rose and then fell in frequency due to other lineages emerging that were more fit. By comparing allele-frequency dynamics to the trajectories of fitness gains by these populations, we estimated the fitness values of the genotypes that contained these mutations. Collectively across all populations, these alleles arose upon backgrounds with a wide range of fitness values. Within any single population, however, multiple alleles tended to rise and fall synchronously during a single wave of multiple genotypes with nearly identical fitness values. These results suggest that alleles of large benefit arose repeatedly in failed “soft sweeps” during narrow windows of adaptation due to the combined effects of epistasis and clonal interference.
Keywords: adaptation, clonal interference, experimental evolution, parallelism, soft sweep
THE classic view of adaptation has been one of periodic selection, whereby the beneficial mutations that escape loss due to drift can rise to fixation unchallenged by additional, independent improvements. Evidence that adaptation consists of a series of discrete events caused by successive selection of individual beneficial mutations came both from the dynamics of rare phage-resistant mutants of Escherichia coli in chemostats (Novick and Szilard 1950) and from the apparent set of punctuated jumps in fitness for long-term populations of E. coli (Lenski et al. 1991). Under this regime, known also as the “strong-selection, weak-mutation” limit (Gillespie 2004), adaptation is directly constrained by the supply rate of rare beneficial mutations into the population.
More recent theory and data have suggested that, under the conditions tested in the laboratory, beneficial mutations occur and escape drift more quickly than the average time to fixation. Since asexual genomes effectively behave as a single locus, beneficial mutations occurring on different backgrounds cannot rise to fixation together and thus interfere with each other. This leads to an extreme version of the Hill–Robertson effect (Hill and Robertson 1966) known as “clonal interference” (Gerrish and Lenski 1998). Relative to the baseline scenario of periodic selection, clonal interference slows the rate of fixation of any particular mutation, skews winning mutations toward larger selective effects, and provides time for competing lineages to continue to accrue additional beneficial mutations before the current sweep is completed (Desai et al. 2007; Desai and Fisher 2007; Rouzine et al. 2008).
Beyond affecting the fate of individual genotypes, at the population level clonal interference changes the dynamics of adaptation relative to the null expectation from periodic selection. First, the rate of adaptation is slower and exhibits a sublinear response to increasing population size because an ever-increasing proportion of the beneficial mutations generated will be outcompeted (de Visser et al. 1999; Miralles et al. 1999; Wilke 2004; Campos and Wahl 2010). Second, unlike the discrete series of fitness steps expected from a periodic process, clonal interference leads to many overlapping rises (and falls) of genotypes with relatively smaller fitness differentials. This leads to a rate of fitness increase that is substantially smoother than the saltation expected from periodic selection (Desai et al. 2007; Desai and Fisher 2007; Rouzine et al. 2008).
Many experimental evolution studies have uncovered multiple evolved lineages that have risen to detectable levels within a population. Some examples have involved adaptive radiations (Rosenzweig et al. 1994; Rainey and Travisano 1998; Rozen and Lenski 2000; Friesen et al. 2004), but as these exhibited stable coexistence due to ecological interactions rather than transient clonal interference, they are not discussed further here. There are several examples of two (or more) evolved alleles at the same locus simultaneously vying for fixation (Notley-McRobb and Ferenci 1999; Wichman et al. 1999; Couñago et al. 2006; Gresham et al. 2008; Barrick and Lenski 2009; Miller et al. 2011; Woods et al. 2011). Although some experiments have persisted long enough to observe a single winning allele at these loci, these alleles are experimental examples of nascent “soft sweeps,” whereby the ancestral allele was being replaced by multiple alleles simultaneously (Pritchard et al. 2010).
To characterize the rate and selective effects of beneficial mutations, as well as the potentially complex dynamics of adaptation due to clonal interference, a number of studies have evolved replicate populations that were started with a mixture of two or three ancestors that differed only by neutral, discernible markers (Rozen et al. 2002; Hegreness et al. 2006; Kao and Sherlock 2008). A more recent elaboration involved constructing an ancestor that would become fluorescently labeled contingent upon a particular class of beneficial mutations occurring (Lang et al. 2011). These experiments have revealed that many populations had substantial deviations from the simple pattern of single beneficial mutations rising alone to fixation, and some adapting lineages can increase, decrease, and then increase again. These more complex dynamics, as long as there are not frequency-dependent, ecological interactions, can be interpreted in light of whether the labeled genotype in question was more (or less) fit than the population average, causing it to rise (or fall) in frequency accordingly (Lang et al. 2011). During a reversal of frequency, the peak abundance (or trough) necessarily represents the time point at which that genotype had fitness equivalent to the population mean (Supporting Information, Figure S1). In most cases, however, there has been no method for tracking the frequencies of many beneficial mutations simultaneously.
The work described here examines an evolution experiment with Methylobacterium extorquens AM1 that was not explicitly designed to address clonal interference, but which turned out to be uniquely well suited for this purpose. The model system was the adaptation of an engineered M. extorquens AM1 (hereafter “EM”) in which the native pathway for formaldehyde oxidation during growth on methanol was eliminated and then replaced with a foreign pathway from a distant species (Marx et al. 2003). The foreign pathway was expressed upon a plasmid backbone that can replicate, but not conjugate, itself into new lineages (Marx and Lidstrom 2001). These novel enzymes allowed the EM strain to recover the ability to grow on methanol, but it was threefold slower than wild type (Chou et al. 2011). The dependence of these specific defects upon the introduced pathway allowed this system to serve as a model to study adaptation subsequent to the introduction of new metabolic capacities (Chou et al. 2011; Marx 2012). One beneficial mutation that was identified decreased expression of the foreign formaldehyde oxidation pathway itself (Chou et al. 2011). This mutation benefited the host by reducing the costs of overexpressing this pathway in the EM ancestor. Across the eight replicate populations (named F1–F8), all lineages incorporated mutations that reduced expression (a 17–24% fitness increase when tested alone), but these were due to a wide spectrum of mutational types (Chou and Marx 2012).
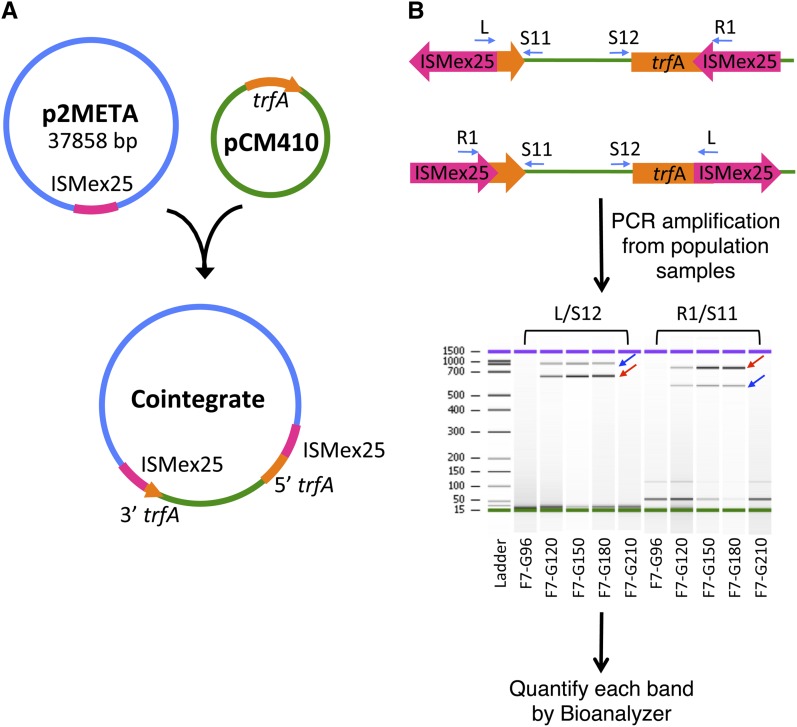
One class of strongly beneficial mutations that repeatedly emerged due to reducing the expression costs of the foreign pathway had unique properties permitting detection at low frequency. Six alleles were discovered that involved transposition of an insertion sequence (IS) element (ISMex25) from a small, single-copy, endogenous plasmid (pMETA2) (Vuilleumier et al. 2009) into the replication initiation gene for this plasmid, trfA (Marx and Lidstrom 2001). This event resulted in a single co-integrate consisting of the two plasmids concatenated together (Chou and Marx 2012) (Figure 1A). Because this event interrupted the catalytic domain of TrfA required to initiate plasmid replication (Lin and Helinski 1992), it rendered the co-integrate dependent upon the replication machinery of pMETA2 and thereby decreased both plasmid copy number and expression of the costly foreign pathway. All six co-integrates involved insertions between positions 835 and 1103 of the trfA-coding sequence. This provided a unique opportunity to study clonal interference because a single PCR amplification technique might be able to detect and distinguish multiple such alleles.
Figure 1.
Formation and quantification of co-integrate alleles in evolving populations. (A) Co-integrates were formed through the incomplete transposition (or subsequent recombination) of ISMex25 from the endogenous p2META plasmid into a small region of the trfA replication gene of the introduced pCM410 plasmid. (B) PCR amplification with pairs of primers internal to ISMex25 and flanking trfA allowed multiple, coexisting trfA::ISMex25 alleles to be quantified.
Here we describe the repeated emergence of the trfA::ISMex25 co-integration alleles and the effects of clonal interference upon their fate in populations. We developed a semiquantitative PCR amplification method that both detected these events at low frequency and simultaneously distinguished each independent insertion. Although only three of the eight populations went on to fix a trfA::ISMex25 allele, we found another 95 co-integrates that transiently rose to detectable frequencies, with up to 17 distinct alleles present simultaneously in a population as a nascent, but usually failed, soft sweep. The dynamics of fitness gains corroborated this picture of abundant clonal interference and allowed us to infer that these alleles usually arose on backgrounds already containing other beneficial mutation(s). Perhaps most remarkably, the trfA::ISMex25 alleles that rose to detection did so in nearly simultaneous waves in any given population, but upon backgrounds of varying fitness values across different populations. We discuss how this synchrony could have emerged due to the joint effects of clonal interference and epistasis.
Materials and Methods
Growth medium and strains
All strains used are derivatives of M. extorquens AM1. Cultures were routinely grown in Hypho minimal medium containing a modified trace metal mix as described (Chou et al. 2009). The details of the evolution of the F1–F8 populations have been described (Chou et al. 2011). Briefly, the two ancestral EM strains, CM701 or CM702, were derived from introducing the pCM410 plasmid expressing the foreign formaldehyde oxidation pathway into strains CM508 and CM624, which lack the first step of H4MPT biosynthesis (Marx 2008). These two plasmid-free strains differed from each other solely in terms of whether or not they produced pink carotenoids (Van Dien et al. 2003), a trait that has been previously shown to be neutral in this environment (Lee et al. 2009). The odd-numbered F populations were founded with the pink CM701 strain; even populations were founded with the white CM702 version. Every 4 days, 150 μl of the culture was transferred into 9.45 ml of fresh medium containing 15 mM methanol (a 1/64 dilution that permitted six generations per transfer and a final population size of 1–2 × 109). After 300 generations, all cultures had improved sufficiently that the time between transfers was reduced to 2 days.
Four strains with known sites of ISMex25 insertion into trfA were used to test the PCR method for co-integrate detection/quantification. These were strains CM935 (isolate from F1 at generation 120; trfA::ISMex25 inserted “reverse” after nucleotide 1055), CM937 (F7 at generation 120; forward after nucleotide 1103), CM946 (F4 at generation 120; reverse after nucleotide 472), and CM1013 (F6 at generation 300; forward after nucleotide 907) (Chou and Marx 2012).
PCR technique for detection and quantification of co-integration events
PCR products were directly amplified from 1 µl of boiled and prepped frozen population stocks in a volume of 20 µl, using four primers in different pairings, as illustrated in Figure 1B. One of each pair has homology outside of the trfA gene [S11: GACTTCCGGCAAGCTATACG, reverse (to trfA), 173 bp downstream; S12: CCCGCAAGGAGGGTGAATG, forward, starting 16 bp upstream of trfA and overlapping the ATG) and the other within ISMex25 (L: CATGCGGGCTCTATGGTCAAC, reverse, 54 bp downstream of the first nt of ISMex25; R1: CAAAGCGTGAAAGCAGAACGGTAG, forward, 64 bp upstream of the last nt of ISMex25). L/S11 and R1/S12 generate products for a reverse allele and R1/S11 and L/S12 amplify a forward allele. Pools of PCR products from controls or mixed populations were quantified using an Agilent 2100 Bioanalyzer with the DNA1000 kit. The molarity estimated by the Bioanalyzer was used as the intensity of each PCR product. Two bands were considered as a pair if the summation of their size amplified by the primers of forward pair or reverse pair was ∼1500 bp and the pair was detected in at least two generations of the same population. Due to PCR amplification efficiency, some alleles were also identified by just one shorter band amplified from one primer pair, with the condition that this product showed up in consecutive generations. The intensity was determined by the average of the two products for each direction or divided in half for single-band alleles (Figure S2). Integrations were conservatively considered to have two peaks through time if their intensity was >1.0 and changed by fivefold.
Growth rate analysis of evolved populations
Growth rate measurements were conducted as previously described (Lee and Marx 2012). Briefly, 5 μl of cryopreserved mixed populations were inoculated into 640 μl medium supplemented with 13.125 mM methanol and 0.4375 mM succinate (ratio of carbon from methanol:succinate = 7:1) in 48-well microtiter plates (Costar) and grown to saturation to acclimate to growth on methanol. Cultures were then transferred with a 1:64 dilution into fresh Hypho medium supplemented with 20 mM methanol and placed into a humidified plate-shaking tower (Liconic) at 30°. Optical densities were obtained every 2 hr by using a de-lidding station and Twister arm (Caliper) to automatically transfer plates to a Wallac Victor 2 plate reader (Perkin-Elmer) until cultures reached saturation, using the open source laboratory manager software, Clarity (Delaney et al. 2012). Growth rates were determined by fitting a logistic growth model using the custom analysis software, Curve Fitter (N. Delaney, M. Kaczmarek, and C. Marx, unpublished results) with a minimum of four replicates (File S1).
Model fitting for fitness dynamics
For each of the eight populations, the trajectory of mean fitness was fit to three different models that we wanted to compare: exponential, hyperbolic, and step-like models. Exponential and hyperbolic models were performed by using the “nls” function in R (Ihaka and Gentleman 1996), with equations a(1 − e−bx) or (ax)/(b + x), respectively (a and b representing the two parameters to be fitted). As the exponential model performed less well than the hyperbolic for all populations, we focus on the latter. The step-like model fitting was performed by the “pava” function in the “isotone” package (de Leeuw et al. 2009), and a backward elimination procedure was applied to avoid overfitting the model with too many steps (threshold of 0.05) as before (Lenski et al. 1991). Akaike information criterion (AIC) was used to evaluate the model fitting, using an approximate function of AIC = n ln(RSS/n) + 2k with correction for finite sample size: AICc = AIC + 2k(k + 1)/(n − k − 1) (Burnham and Anderson 2002), where n is the sample size and k is the number of parameters.
Results
Semiquantitative PCR method to detect IS-mediated co-integrations
To examine the diversity and frequencies of beneficial, IS-mediated co-integrations that occurred in our eight evolved populations, we developed a semiquantitative PCR method to detect these events. As described above, all co-integrates previously identified involved ISMex25 and occurred within a small region of the IncP plasmid replication gene trfA, yet the exact insertion points were distinct (Chou and Marx 2012). We therefore developed a PCR-based approach to directly detect such events from population samples (Figure 1B).
To first test the approximate limit of detection for this method, we applied this semiquantitative PCR method to mixed cultures of the EM ancestor (CM701) and four trfA::ISMex25-containing strains with known insertion sites (Chou and Marx 2012). We could reliably generate bands with all four primers down to 0.01–0.1% of the population. This compares rather favorably to previous high-throughput methods for quantifying SNPs directly in a mixed population sample (Herring et al. 2006; Gresham et al. 2008), including our recently developed frequency sequencing (FREQ-Seq) method (Chubiz et al. 2012).
We further tested the degree to which our PCR technique produces quantitative data. For this, we varied the ratio of strains with two different insertions (Figure S2). Although PCR amplification favors the smaller amplified products, fortunately, these effects will be well-balanced by two pairs of primers since an insertion allele with a shorter upstream amplicon will have a longer downstream one. Correspondingly, by taking the average of the two control curves, this bias became a sigmoidal shape centered around the expected template ratios (Figure S2). It should be noted, however, that, because different insertion events will amplify with varying efficiency, we do not aim to interpret the absolute values between amplified products for different insertions, but simply the temporal dynamics of the detected alleles.
Individual populations demonstrated waves of synchronous co-integration
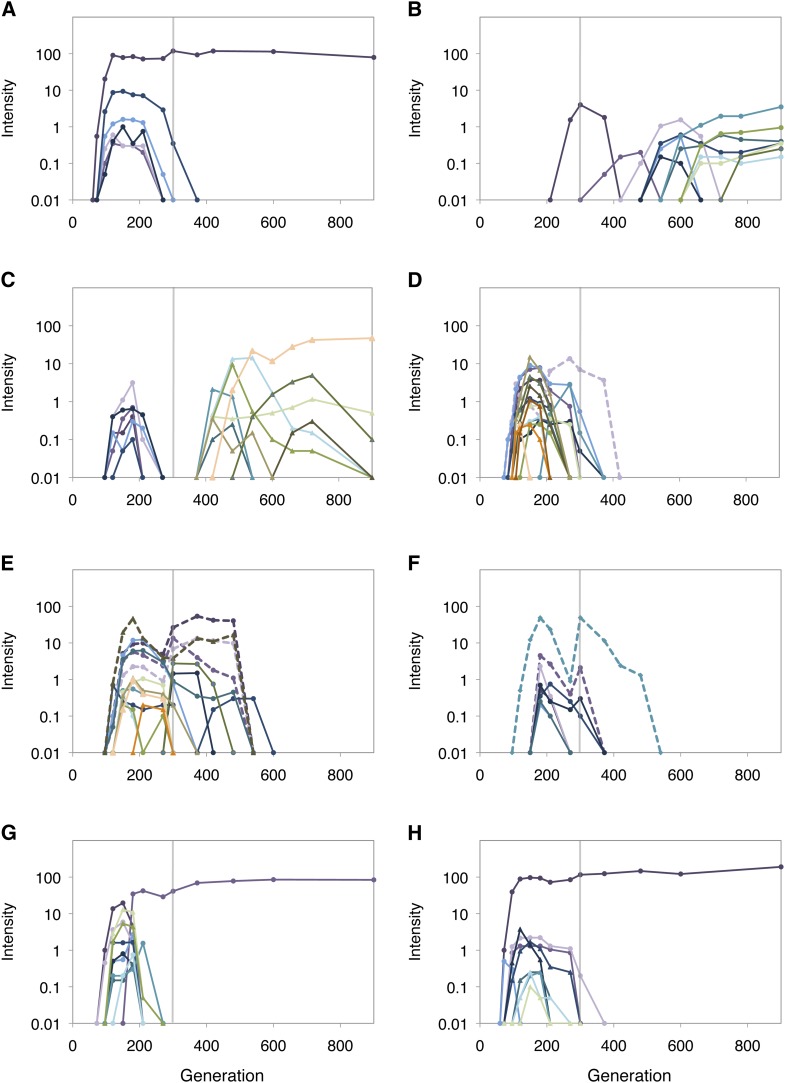
We applied the above PCR method directly to frozen samples for 23 time points from each of the eight replicate populations to quantify co-integration events and track their dynamics (Figure 2 and File S1). First, we found that co-integration events occurred in all populations despite only rising to fixation in three (F1, F7, F8). Second, even when just considering this particular class of beneficial mutation, there was a tremendous degree of repeatability and clonal interference. We detected between 8 and 17 different events for each population, with 98 co-integrations in total. Third, the typical dynamics for transient alleles was a rapid rise to peak intensity, followed by a rapid decrease. This pattern indicated that the genotype of the strain containing that allele was initially more fit than the population average, peaked in frequency when its fitness matched that of the population, and then fell when outcompeted by more fit lineages (see Figure S1). Fourth, for several alleles, mainly in populations F5 and F6, we detected a more complicated trajectory whereby that allele reached a maximum, fell, and then rose to a second peak value before falling again. There are three possible interpretations for this dynamic: a second, independent genotype with an identical (or nearly so) trfA::ISMex25 allele; the acquisition of an additional beneficial mutation within the co-integrate lineage; or a change in the selective regime. Two lines of evidence support this last hypothesis: most of these second peaks rose in synchrony in F5 and F6 at precisely 300 generations, coincident with the shift in the transfer regime from 4 to 2 days. This suggests that the fitness of these genotypes increased at that juncture, but they still subsequently lost out as other lineages later moved on to fixation.
Figure 2.
Dynamics of trfA::ISMex25 co-integrate alleles in evolved populations. (A–H) Intensities of diagnostic PCR products resulting from co-integrate quantification in populations F1–F8, respectively. Vertical guideline on each panel indicates the change in transfer scheme from 4 to 2 days at generation 300. Populations F1, F7, and F8 (A, G, H) had a co-integrate allele rise to fixation. Double-peak alleles in F4, F5, and F6 are shown by dashed lines.
High-resolution analysis of fitness improvement revealed relatively smooth dynamics through time and increasing variance between populations
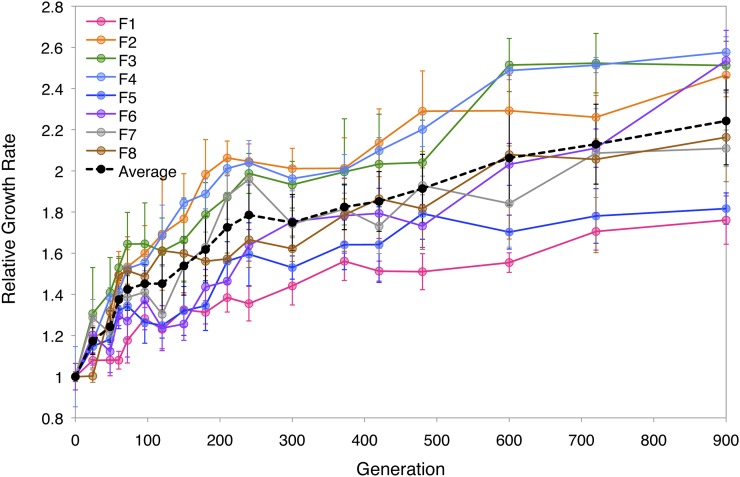
How do the complex allele dynamics of repeated co-integrations map onto the pattern of fitness increase? To obtain high-resolution phenotypic data, we used the growth rate of mixed populations as a proxy for fitness (Figure 3 and File S1). These growth rates were determined via an automated robotic system (Figure S3; see Materials and Methods for details). Previous work with these populations has shown that growth rates are very highly correlated with fitness values obtained from competition experiments (Chou et al. 2011). Growth rate improvements were quite rapid, with an average increase of ∼50% by 150 generations and ∼125% by 900 generations. Consistent with the almost universal trend observed in experimental evolution, the rate of fitness improvement decelerated through time. For example, the average rate of fitness increase between generations 48 and 72 was 151% per 100 generations, whereas from generations 600 to 900, it fell to 11.5% per 100 generations.
Figure 3.
Relative growth rates through 900 generations. Average across populations is indicated by the black dashed line.
Putting aside the direct evidence already amassed for the existence of clonal interference, we can ask in an approximate way how well the dynamics of fitness gains reflected underlying genotypic complexity compared to the null expectation for periodic selection. The adaptation dynamics of individual populations did not reveal a dramatic series of punctuated jumps, but were rather smooth through time (Figure 3). To characterize where the dynamics fell on the spectrum from highly periodic to continuous, we compared the dynamics of fitness relative to simplistic models representing these extreme behaviors (Figure S4). Periodic selection has previously been modeled using an isotonic, step-wise fitting model with backward elimination procedure (stepwise F test) to choose an appropriate number of such steps (Lenski et al. 1991). We compared the quality of fit and complexity of the resultant model by AIC to a hyperbolic model that uses two parameters to describe a continuous smooth rise in fitness with declining slope. Despite its simplicity, a hyperbola was the better model to describe the data than an optimal step model for all but two of the eight populations.
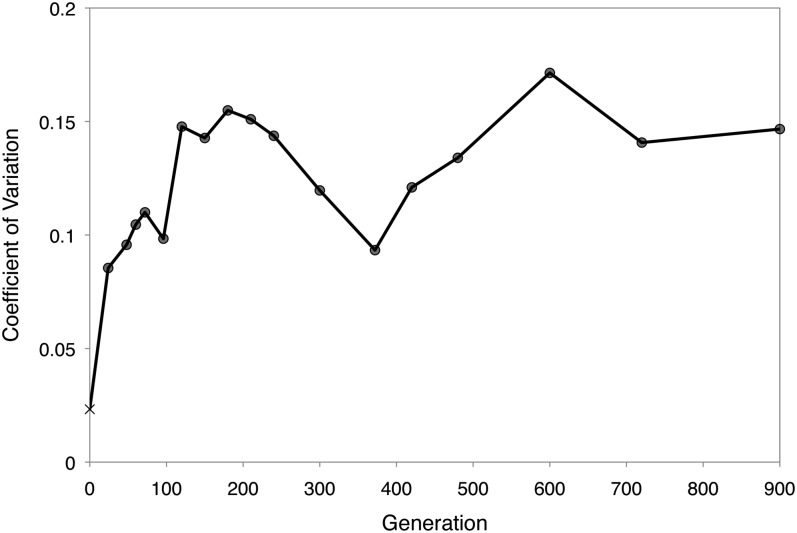
Turning to differences between populations, there was substantial sustained variation in growth rate between populations that suggested that different populations had found distinct adaptive solutions (Figure 4). Early in the experiment the variance grew rapidly; the coefficient of variation at generation 120 was indistinguishable from that at generation 900 (F test; P = 0.28), despite a transient decrease in between these times.
Figure 4.
Maintenance of among-population variance in growth rate. After a rapid rise in the first 120 generations, the coefficient of variation was sustained between 10 and 17% despite continued adaptation. The first point shows the experimental variation for the ancestor.
Synchronous waves of co-integration events occurred upon backgrounds of distinct fitness values in different populations
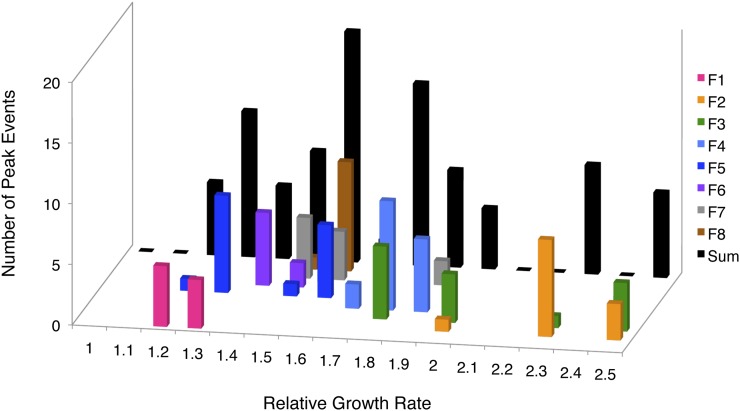
Obtaining fine-scale data through time for both co-integration allele frequencies and population growth allowed us to compare the context in which co-integrations occurred in different populations. By identifying the time point at which a given trfA::ISMex25 allele hit its apex, we estimated that genotype’s fitness value (i.e., the population mean fitness at that moment; see Figure S1). The inferred fitness values for each of the 98 co-integrate genotypes exhibited great variety across populations (Figure 5). In population F1, for example, co-integrates peaked in frequency when the growth rate was 1.2–1.3× the EM ancestor. Given that this is close to the benefit of the integration alone (Chou and Marx 2012), these peaks likely represent strains with just this event or perhaps additional mutation(s) of relatively small effect. On the other hand, detectable co-integrate genotypes peaked at much higher growth rates in the other seven populations, indicating that these trfA::ISMex25 alleles occurred on backgrounds with other mutations of substantial net benefit. Perhaps most remarkably, although co-integrations were present in genotypes across a broad range of fitness levels when summed across all eight populations, in any particular population they occurred in a single wave. We found that these data were very highly clustered compared to a null model whereby the co-integrate genotype fitness values observed for a single population could have been drawn from anywhere within the distribution of observed alleles in the aggregate distribution across populations (Kruskal–Wallis rank-sum test; P = 2.26 × 10−15).
Figure 5.
Distribution of inferred growth rates for genotypes containing co-integrate alleles. The growth rates for each of these genotypes were estimated by determining the population average at the time point for which the allele peaked in frequency and were binned (±0.05 relative growth rate). In the foreground are color-coded distributions for the genotype fitness values observed for each individual population; in the rear (in black) is the summed distribution across the eight populations.
Discussion
Experimental evolution has been instrumental in demonstrating that the simple dynamics of periodic selection are rarely, if ever, observed under the regimes employed in the laboratory. Most studies have either identified segregating evolved alleles or examined the dynamics of marked subpopulations, but few have combined these perspectives (Lang et al. 2011). Here we have taken advantage of the unique molecular details of a particular class of frequent, large-benefit mutations that arose in evolving populations of the EM engineered Methylobacterium strain to directly detect their dynamics from cryopreserved mixed-population samples. Our analysis revealed that multiple co-integration events occurred and rose in nearly simultaneous waves in a given population and thus clonally interfered with each other, as well as with other adapting lineages, as they vied for fixation.
Co-integration of an introduced plasmid was remarkably common
When we first recognized that trfA::ISMex25 alleles from a couple of evolved isolates arose from co-integration of the introduced plasmid into the host genome, it appeared to be a rather unlikely type of mutation (Chou and Marx 2012). After observing nearly identical events in isolates from other populations, we decided it was worth investigating this class of mutations further. That said, we were quite surprised to see up to 17 different types of the same event co-occurring in the same population. These represent attempted soft sweeps of distinct mutational events rising toward fixation. In most populations, the whole soft sweep went extinct; however, in three of eight replicates, one such allele did rise to fixation and thus became a “hard sweep.” It should be kept in mind that the numbers that we report represent a lower bound of the true number of co-integrations that occurred, as many such events will have been lost to drift, never rose beyond our limit of detection, or were indistinguishable from each other due to integration into the same (or nearly so) site within trfA. Furthermore, there were likely many more successful alleles on the introduced plasmid with similar phenotypic effects, given that the trfA::ISMex25 alleles constituted less than half of the mutations reported from isolates common enough to be sampled from three colonies per population (Chou and Marx 2012).
Why did we observe so many coexisting beneficial alleles at a single locus relative to most previous reports? One reason may have been the great sensitivity and ability to distinguish similar events afforded by our PCR method. Thus far, no method for observing rare SNPs from this many mixed-population samples would have unveiled dynamics of variants as rare as our PCR technique could detect. On the other hand, it is likely that the physiological and genetic context of our ancestral strain evolving in methanol medium was key to our repeated observation of their rise in populations. The rate of occurrence of IS-mediated events is often quite high and has been shown to contribute to the observation of parallelism across replicates (Cooper et al. 2001; Chou et al. 2009; Stoebel et al. 2009). Furthermore, the selective effect of co-integrations was quite large, which would have helped co-integrations both escape drift and rapidly rise in frequency.
Rampant clonal interference was revealed in both the complex dynamics of co-integration frequencies and the smooth dynamics of fitness increases
Although the trfA::ISMex25 alleles are interesting in their own right, the ease of detecting them at low frequencies—rather like phage-resistance mutants in early chemostat work (Novick and Szilard 1950)—allowed them to also serve as sentinels for the dynamics of the bulk of the population. Similar to other studies (Lang et al. 2011), we observed examples of selective sweeps, clonal interference, and multiple mutations. However, due to the high sensitivity for detecting distinct co-integrations of our method, we detected many similar co-integrations present simultaneously in the populations.
The availability of fine-scale data on the dynamics of phenotypic improvement provided a second window into the underlying population dynamics during adaptation. Indeed, despite the massive gains in fitness that we observed in our system relative to most previous evolution experiments with bacteria, the observed that dynamics were quite smooth compared to a punctuated model of periodic selection. The first 2000 generations of the 12 replicate populations of the long-term experimental evolution of E. coli were all better fit as a series of steps with an isotonic model (Lenski et al. 1991). In contrast, although the adaptation of EM in methanol was certainly more “bumpy” than that captured by a two-parameter function, the AIC criteria of a step-like isotonic model indicated an improved fit in only two of eight cases. This relatively continuous improvement emphasizes the interesting divergent effects of clonal interference upon adaptation: allele trajectories become much more complicated than under periodic selection; fitness gains become much smoother.
Adaptation of the engineered strain of Methylobacterium was more variable than typical evolution experiments
One goal of experimental evolution has been to permit inferences to be made about the underlying adaptive landscape based upon the dynamics of improvement, notably the extent to which replicates converge phenotypically to what appears to be a single fitness peak vs. diverge to find very different solutions (Lenski et al. 1991). Over 10,000 generations of long-term evolution of E. coli populations on glucose, for example, the fitness of the 12 replicate populations increased to an average of ∼1.4, with a coefficient of variation of ∼3% (Lenski and Travisano 1994). Earlier experiments evolving wild-type M. extorquens AM1 on methanol or on the multi-carbon compound succinate also revealed relatively little variation (3.6% and 2.0%, respectively) (Lee et al. 2009). In comparison, the coefficient of variation of the populations founded with the EM strain was sustained at ∼15% from generation 120 to 900, a value five- to eightfold higher than the above examples. This is perhaps surprising, given that these populations also witnessed multiple examples of parallelism, including all populations reducing expression costs of the foreign formaldehyde oxidation pathway (Chou and Marx 2012) and six of eight having identical insertions of ISMex4 into the promoter of a novel cobalt transporter encoded by icuAB (Chou et al. 2009).
Why might the adaptive outcomes for the EM strain have been so much more variable than usual? Compared to most experiments, the rewiring of EM’s central metabolic pathway for growth on methanol made this strain particularly unfit. This starting situation is unusual for experimental evolution, but is analogous to the series of events that occur following horizontal gene transfer. Indeed, the exchange of formaldehyde oxidation pathways across natural bacterial lineages has been well documented (Chistoserdova et al. 2004). Regardless of starting genotype, for evolving populations to scale alternative fitness peaks, there must be reciprocal sign epistasis to guarantee that beneficial mutations that would bridge from one apparent optimum to another do not exist (Weinreich et al. 2005). In this regard, beneficial mutations within a single adaptive trajectory from one of the populations studied here exhibited solely positive effects (Chou et al. 2011). A better test for distinct fitness optima, however, would be to test interactions between beneficial mutations that occurred independently in separate trajectories, as has been done in a few cases (Cooper et al. 2003; Kvitek and Sherlock 2011; Woods et al. 2011).
Parallel co-integration events occurred in distinct waves at different times and fitness levels across populations
The most surprising result regarding the repeated emergence of co-integrations was their synchronous dynamics within single populations and distinct timings across populations. One possible explanation for this synchronicity could be that the occurrence of co-integrations itself was episodic due to a factor such as environmental stress leading to increased transposition rates. Although this is possible, the timing of maximal frequency was more synchronous than the first detection of alleles, and replicate populations had waves of detectable co-integrations at distinct times, but were cultured together. Furthermore, we have previously shown the direct ability to select for co-integrates via antibiotic resistance on a plasmid (Chou and Marx 2012). We therefore found this hypothesis less likely than just a simple form of epistasis: once a lineage has one of the possible mutations that decreased expression of the foreign formaldehyde oxidation pathway, there may be little benefit (or perhaps even detriment) to obtain another. Thus, once one expression-decreasing mutation occurs in a lineage, it likely shuts the door on all others.
But what prevented the highly beneficial trfA::ISMex25 from rising in frequency early in populations, at least transiently? There is always a lag in adaptation for experiments that begin from a single genotype; however, in all populations except perhaps F1 there were substantial fitness gains before the first co-integrate peaked in frequency. This demonstrates that other mutations had occurred and rose in frequency well before these alleles became detectable. Either clonal interference and/or epistasis could have prevented the early success of co-integrates. At the start of the experiment there may have simply been mutations even more beneficial than the 17–24% conferred by co-integration alleles. Indeed, the three populations where trfA::ISMex25 peaked at the highest fitness values (F2, F3, F4) were also the three that were most consistently among the most fit populations through time. Using our recently developed allele-frequency sequencing method (FREQ-Seq), we demonstrated that the first beneficial mutation in the F4 population was in gshA (encodes γ-glutamylcysteine synthetase). The selective benefit of gshAEvo is approximately threefold higher than that of trfA::ISMex25 (Chubiz et al. 2012). Furthermore, the gshAEvo mutation from F4 reduces the benefit of turning down the foreign pathway by half (Chou et al. 2011). These data suggest that the complex interaction of clonal interference and epistasis may have collaborated to radically alter the relative likelihood of this class of beneficial mutations rising to high frequency, thereby producing surprisingly synchronous waves of success and failure of independent beneficial mutations occurring at a single locus.
Acknowledgments
We thank David Robinson for expert assistance with statistical analyses; David Chou for initial information about the populations and strains examined; and Michael Desai, members of the Marx lab, and anonymous reviewers for helpful suggestions on the manuscript. This work was supported by National Science Foundation grant DEB-0845893 (to C.J.M.) and National Institutes of Health grant R01 GM078209 (to C.J.M.).
Footnotes
Communicating editor: J. Lawrence
Literature Cited
- Barrick J. E., Lenski R. E., 2009. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 74: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham K. P., Anderson D. R., 2002. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, Ed. 2. Springer-Verlag, Berlin; Heidelberg, Germany; New York. [Google Scholar]
- Campos P. R., Wahl L. M., 2010. The adaptation rate of asexuals: deleterious mutations, clonal interference and population bottlenecks. Evolution 64: 1973–1983. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L., Jenkins C., Kalyuzhnaya M. G., Marx C. J., Lapidus A., et al. , 2004. The enigmatic Planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 21: 1234–1241. [DOI] [PubMed] [Google Scholar]
- Chou H.-H., Marx C. J., 2012. Optimization of gene expression through divergent mutational paths. Cell Rep. 1: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.-H., Berthet J., Marx C. J., 2009. Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet. 5: e1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.-H., Chiu H.-C., Delaney N. F., Segrè D., Marx C. J., 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubiz L. M., Lee M.-C., Delaney N. F., Marx C. J., 2012. FREQ-Seq: a rapid, cost-effective, sequencing-based method to determine allele frequencies directly from mixed populations. PLoS ONE 7: e47959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. F., Rozen D. E., Lenski R. E., 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100: 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper V. S., Schneider D., Blot M., Lenski R. E., 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183: 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couñago R., Chen S., Shamoo Y., 2006. In vivo molecular evolution reveals biophysical origins of organismal fitness. Mol. Cell 22: 441–449. [DOI] [PubMed] [Google Scholar]
- Delaney N. F., Rojas Echenique J. I., Marx C. J., 2012. Clarity: an open source manager for laboratory automation. J. Lab. Autom. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw J., Hornik K., Mair P., 2009. Isotone optimization in R: pool-adjacent-violators algorithm (PAVA) and active set methods. J. Stat. Softw. 32: 1–24. [Google Scholar]
- Desai M. M., Fisher D. S., 2007. Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics 176: 1759–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., Murray A. W., 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17: 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E., 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283: 404–406. [DOI] [PubMed] [Google Scholar]
- Friesen M. L., Saxer G., Travisano M., Doebeli M., 2004. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58: 245–260. [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R. E., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103: 127–144. [PubMed] [Google Scholar]
- Gillespie J. H., 2004. Population Genetics: A Concise Guide, Ed. 2 Johns Hopkins Press, Baltimore. [Google Scholar]
- Gresham D., Desai M. M., Tucker C. M., Jenq H. T., Pai D. A., et al. , 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M., Shoresh N., Hartl D. L., Kishony R., 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617. [DOI] [PubMed] [Google Scholar]
- Herring C. D., Raghunathan A., Honisch C., Patel T., Kenyon Applebee M., et al. , 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 38: 1406–1412. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Ihaka R., Gentleman R., 1996. R: a language for data analysis and graphics. J. Comput. Graph. Statist. 5: 299–314. [Google Scholar]
- Kao K. C., Sherlock G., 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek D. J., Sherlock G., 2011. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 7: e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Botstein D., Desai M. M., 2011. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-C., Marx C. J., 2012. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 8: e1002651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-C., Chou H.-H., Marx C. J., 2009. Asymmetric, bi-modal tradeoffs during adaptation of Methylobacterium to distinct substrates. Evolution 63: 2816–2830. [DOI] [PubMed] [Google Scholar]
- Lenski R. E., Travisano M., 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 91: 6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C., 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- Lin J., Helinski D. R., 1992. Analysis of mutations in trfA, the replication initiation gene of the broad-host-range plasmid RK2. J. Bacteriol. 174: 4110–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C. J., 2008. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res. Notes 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C. J., 2012. Recovering from a bad start: rapid adaptation and tradeoffs to growth below a threshold density. BMC Evol. Biol. 12: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C. J., Lidstrom M. E., 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147: 2065–2075. [DOI] [PubMed] [Google Scholar]
- Marx C. J., Chistoserdova L., Lidstrom M. E., 2003. The formaldehyde detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J. Bacteriol. 185: 7160–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. R., Joyce P., Wichman H. A., 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187: 185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles R., Gerrish P. J., Moya A., Elena S. F., 1999. Clonal interference and the evolution of RNA viruses. Science 285: 1745–1747. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L., Ferenci T., 1999. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ. Microbiol. 1: 33–43. [DOI] [PubMed] [Google Scholar]
- Novick A., Szilard L., 1950. Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. USA 36: 708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey P. B., Travisano M., 1998. Adaptive radiation in a heterogeneous environment. Nature 394: 69–72. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R. F., Sharp R. R., Treves R. S., Adams J., 1994. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137: 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I., Brunet E., Wilke C., 2008. The traveling-wave approach to asexual evolution: Muller’s ratchet and the speed of adaptation. Theor. Popul. Biol. 73: 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., Lenski R. E., 2000. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am. Nat. 155: 24–35. [DOI] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J. A. G. M., Gerrish P. J., 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Stoebel D. M., Hokamp K., Last M. S., Dorman C. J., 2009Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 5: e1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dien S. J., Marx C. J., O’Brien B. N., Lidstrom M. E., 2003. Genetic characterization of the carotenoid biosynthetic pathway in Methylobacterium extorquens AM1 and isolation of a colorless mutant. Appl. Environ. Microbiol. 69: 7563–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier S., Chistoserdova L., Lee M.-C., Bringel F., Lajus A., et al. , 2009. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS ONE 4: e5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D. M., Watson R. A., Chao L., 2005. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59: 1165–1174. [PubMed] [Google Scholar]
- Wichman H. A., Badgett M. R., Scott L. A., Boulianne C. M., Bull J. J., 1999. Different trajectories of parallel evolution during viral adaptation. Science 285: 422–424. [DOI] [PubMed] [Google Scholar]
- Wilke C. O., 2004. The speed of adaptation in large asexual populations. Genetics 167: 2045–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R. J., Barrick J. E., Cooper T. F., Shrestha U., Kauth M. R., et al. , 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331: 1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]