Abstract
Aerobactin genes are known to be present in virulent strains and absent from avirulent strains, but contributions of iucC and iucA, which are involved in aerobactin synthesis, to the pathogenicity of avian pathogenic Escherichia coli (APEC) have not been clarified. In this study, effects of double mutants (iucA/iutA or iucC/iutA) compared to those of single mutants (iucA, iucC or iutA) of aerobactin genes on the virulence of APEC strain E058 were examined both in vitro (aerobactin production, ingestion into HD-11 cells, survival in chicken serum) and in vivo (competitive growth against parental strain, colonization and persistence). In competitive co-infection assays, compared to the E058 parental strain, the E058ΔiucA mutant was significantly reduced in the liver, kidney, spleen (all P<0.01), heart and lung (both P<0.001). The E058ΔiutA mutant also was significantly reduced in the liver, lung, kidney (all P<0.01), heart and spleen (both P<0.001). The E058ΔiucC mutant was significantly attenuated in the heart and kidney (both P<0.05) and showed a remarkable reduction in the liver, spleen and lung (P<0.01); meanwhile, both E058ΔiucAΔiutA and E058ΔiucCΔiutA double mutants were sharply reduced as well (P<0.001). In colonization and persistence assays, compared with E058, recovered colonies of E058ΔiucA were significantly reduced from the lung, liver, spleen and kidney (P<0.01) and significantly reduced in the heart (P<0.001). E058ΔiutA was significantly reduced from the heart, lung, liver, spleen and kidney (P<0.01). E058ΔiucC, E058ΔiucAΔiutA and E058ΔiucCΔiutA were significantly decreased in all organs tested (P<0.001). These results suggest that iutA, iucA and iucC play important roles in the pathogenicity of APEC E058.
Introduction
Although iron is an essential element for all living cells, ferric iron is barely soluble at biological pH. Therefore, iron can be taken up in a complexed form via siderophores, a process that has been shown to contribute to the pathogenicity of avian pathogenic Escherichia coli (APEC) strains. The hydroxamate siderophore aerobactin has previously been associated with the pathogenicity of APEC strains [1], [2], [3], [4], [5], [6]. The ability of virulent microorganisms to grow under iron-limiting conditions may promote systemic infections [7], [8], [9]. As determined by the lethality for 1-day-old chicks in avian strains of E. coli [1], this physiological trait is highly correlated with APEC virulence. Lafont et al. stated that aerobactin genes are present in virulent strains but absent from avirulent strains, which has been confirmed by epidemiological studies [3]. Work undertaken by Tivendale et al. found that the iucA gene located on the pVM01 plasmid of APEC is highly correlated with virulence, adding support to the hypothesis that aerobactin is a virulence factor in APEC strains [10]. Dozois et al. revealed that iutA is dedicated to the virulence of APEC strain χ7122 [11]. In the APEC O2 strain, the aerobactin operon is situated on the pAPEC-O2-ColV plasmid [12], which contains genes responsible for the synthesis of the hydroxa siderophore aerobactin (iucABCD) and for ferric aerobactin uptake (iutA) [13], [14], [15]. Among APEC strains, iron acquisition systems can be encoded by plasmid genes [16], [17] or by chromosomal pathogenicity islands [18]. Aerobactin is a hydroxamate siderophore that is encoded by a plasmid operon [19], [20]. Hence, the expression of aerobactin should be associated with virulence of APEC. The aerobactin operon, which includes iucABCD and iutA, is localized to the APEC ColV plasmid [21].
Based on our previous observation that the iucCD gene of APEC E058 is up-regulated in vivo and the iutA transcription was also observed up-regulated in a microarray to detect the transcriptoms of avian pathogenic Escherichia coli (APEC) grown in vivo compared with those in vitro [22], we hypothesized that iutA or iucC and/or iucD is involved in the virulence of APEC E058. To our knowledge, the contribution of iucC to the pathogenesis of APEC and the effects of double mutations in iucA/iutA or iucC/iutA on APEC virulence compared to that of the each single mutation have not been reported. In this study, we chose to delete the single iucC, iucA or iutA gene or the double iucC/iutA or iucA/iutA genes to investigate their pathogenic roles in APEC O2 strain E058. A series of pathogenicity tests, including bactericidal assay in chicken serum, HD-11 cells ingestion assay, colonization and persistence in chickens, and in vivo competition assay, were employed to evaluate the pathogenicity of the APEC E058ΔiucA, E058ΔiucC, E058ΔiutA, E058ΔiucAΔiutA and E058ΔiucCΔiutA mutants compared with the E058 parent strain.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Research Council. All experiments and procedures performed on the animals were approved by the Animal Care and Use Committee of Yangzhou University (approval ID: SYXK (Su) 2007–0005). Chickens were provided with food and water ad libitum. After 24 h of infection, the birds were euthanized by carbon dioxide asphyxiation and then dissected with aseptic surgical techniques.
Bacterial strains, plasmids, primers, media and growth conditions
The strains, plasmids and cell line used in this study are listed in Table 1. The primers applied in this study are listed in Table 2. Cells were routinely grown at 37°C in Luria-Bertani (LB) broth or on LB agar with/without appropriate antibiotics: kanamycin (Km) (50 μg.ml−1), zeocin (25 μg.ml−1) or ampicillin (Amp) (60 μg.ml−1) unless otherwise specified. The chicken macrophage cell line HD-11 was maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, PAA, Pasching, Australia) at 37°C, 5% CO2.
Table 1. Strains, plasmids and cell line used in this study.
| Description | Source | |
| Strains | ||
| E058 | Wild-type avian E. coli serotype O2 | [44] |
| DH5α | endA1 hsdR17 (rk −mk +)supE44 thi-1 recA1 gyrA (NalR) RelA1Δ(lacIZYA-argF) U169deoR (φ80d lac Δ (lacZ) M15) | Invitrogen |
| LG1522 | Aerobactin indicator strain | [5] |
| E058ΔiutA | iutA mutant of E058, Kanr | This study |
| E058ΔiucA | iucA mutant of E058, Zeor | This study |
| E058ΔiucAΔiutA | iutA and iucA double mutant of E058, Zeor, Kanr | This study |
| E058ΔiucC | iucC mutant of E058, Zeor | This study |
| E058ΔiucCΔiutA | iutA and iucC double mutant of E058, Zeor, Kanr | This study |
| Re-E058ΔiucA | Complementation of E058ΔiucA | This study |
| Re-E058ΔiucC | Complementation of E058ΔiucC | This study |
| Plasmids | ||
| pMD®18-T simple vector | TA cloning vector, ApR | Promega |
| pT-iucA | iucA cloned into pMD®18-T simple vector | This study |
| pT-iucC | iucC cloned into pMD®18-T simple vector | This study |
| pBluescript II SK(-) | Cloning vector | Fermentas |
| pS-iucA | Hind III-BamH I iucA fragment cloned into SK(-) | This study |
| pS-iucC | EcoR I-Xba I iucC fragment cloned into SK(-) | This study |
| pEM7/Zeo | Zeocin-resistant cassette | Invitrogen |
| pUC4K | Kanamycin-resistant cassette | Invitrogen |
| pS-iucA-Zeo | Zeocin-resistant gene inserted into pS-iucA | This study |
| pS-iucC-Zeo | Zeocin-resistant gene inserted into pS-iucC | This study |
| pGEX-6P-1 | Bacterial expression vector | Amersham |
| pGEX-6P-1-iucA | iucA cloned into pGEX-6P-1 vector | This study |
| pGEX-6P-1-iucC | iucC cloned into pGEX-6P-1 vector | This study |
| Cell line | ||
| HD-11 | Chicken macrophage line, chicken myelomonocytic transformed by the myc-encoding MC29 virus | [45] |
Table 2. Primers designed and used in this study.
| Primer name | Sequence 5′–3′ | Position (bp) | Source |
| iutA-F | CGAAGCTTTCTCAACCCACTGCTTCTT (iutA sense; Hind III site underlined) | 33–51 | This study |
| iutA-R | TAGGATCCTGGTATAGCCATCGACCTT (iutA antisense; BamH I site underlined) | 1981–1999 | This study |
| iucA-F | CTCAAGCTTAGTGCTTCCTGAATGCCT (iucA sense; Hind III site underlined) | 44–61 | This study |
| iucA-R | CTCGGATCCTATGAGTCACCTGGTCAC (iucA antisense; BamH I site underlined) | 1556–1573 | This study |
| pS-iucA-F | CTCGATATCTCAGGCAGGTATCGTTCA (pS-iucA sense; EcoR V site underlined) | 492–509 | This study |
| pS-iucA-R | CTCGAATTCCGATGCTTACTGTCAGCA (pS-iucA antisense; EcoR I site underlined) | 1200–1217 | This study |
| iucC-F | CTCGAATTCACTGGGATTTGGTCAACC (iucC sense; EcoR I site underlined) | 14–31 | This study |
| iucC-R | CTCTCTAGAATTCCTGAGTTACCAGCC (iucC antisense; Xba I site underlined) | 1715–1732 | This study |
| Zeo-BamH I-F | CTCGGATCCCACGTGTTGACAATTAAT (Inserting zeocin gene; BamH I site underlined) | 1938–1955 | This study |
| Zeo-BamH I-R | CTCGGATCCTCAGTCCTGCTCCTCGGC (Inserting zeocin gene; BamH I site underlined) | 2366–2383 | This study |
| GF | CACAGTCTTACTGCCAGT (shiG sense) | 114–131 | This study |
| GR | TTCTCGGTATCGGACAGA (shiG antisense) | 266–283 | This study |
| BF | TTGGTGAACAGCAATGGC (iucB sense) | 701–718 | This study |
| BR | ACCTCCGTGAAGAAGTGA (iucB antisense) | 921–938 | This study |
| DF | ATGGCATCACTGCCGATT (iucD sense) | 782–799 | This study |
| DR | TACGTGCAGATCTCCATG (iucD antisense) | 1181–1198 | This study |
| O2F | ATGTCGTGTTCCGTGCTCA (O2ColV sense) | 1–19 | This study |
| O2R | TCAGTAAGTTGGCAGCATC (O2ColV antisense) | 204–222 | This study |
| Re-iucA-F | TCAGGATCCATGATCCTGCCCTCTGAA (iucA sense; BamH I site underlined) | 1–18 | This study |
| Re-iucA-R | TCAGAATTCTCAGACCTCCTGAGCCTG (iucA antisense; EcoR I site underlined) | 1708–1725 | This study |
| Re-iucC-F | CGCGAATTCATGAATCACAAAGACTGG (iucC sense; EcoR I site underlined) | 1–18 | This study |
| Re-iucC-R | GCGGTCGACTCATGATTCATATTCCTG (iucC antisense; Sal I site underlined) | 1726–1743 | This study |
DNA and genetic manipulations
All restriction and DNA-modifying enzymes and a 200 bp DNA marker purchased from Takara (Dalian, Liaoning, China) were used according to the supplier's recommendations. The Lamda DNA/EcoR I-plus-Hind III molecular size standard was purchased from Fermentas (Shenzhen, Guangdong, China). Purification of PCR products and DNA fragments was performed using kits manufactured by Promega (Madison, WI, USA). The DIG High Prime DNA Labeling and Detection kit was purchased from Roche (Indianapolis, IN, USA). Transformation of E. coli strains was routinely carried out using electroporation. DNA and deduced amino acid sequence analyses were performed using DNASTAR Lasergene 8 software to predict conserved domains and using the search engine at http://blast.ncbi.nlm.nih.gov/Blast.cgi. DNA nucleotide sequences were determined by Sangon Co. LTD. (Shanghai, China). Reactions were carried out using Taq DNA polymerase (Fermentas) under the following conditions: 94°C for 4 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min and finally an extension at 72°C for 10 min.
Construction of iutA, iucA, iucC, iucA/iutA and iucC/iutA mutant strains
The iucA, iucC and iutA genes were amplified from template which was a boiled preparation of the overnight culture of E058 using primer pairs iucA-F/iucA-R, iucC-F/iucC-R and iutA-F/iutA-R respectively (Table 2), and the PCR products were cloned into pBluescript SK (–) to form pS-iucA, pS-iucC and pS-iutA respectively. To form pS-iucA-Zeo, the iucA PCR product, which was amplified from pS-iucA using primers pS-iucA-F/R (Table 2) and digested with EcoR I-EcoR V, was ligated with the Zeor gene cassette originating from pEM7/Zeo digested with the same enzymes. To form pS-iucC-Zeo, the pS-iucC fragment digested with BamH I was ligated with the Zeor gene cassette amplified from pEM7/Zeo using primers Zeo-BamH I-F/R and digested with BamH I. To form pS-iutA-Kan, the pS-iutA plasmid and the pUC4K plasmid (containing the Kanr gene) were both digested with Pst I and ligated together to form pS-iutA-Kan. Deletions of target genes iucA, iucC, iutA, iucA/iutA and iucC/iutA in the APEC O2 E058 strain essentially were performed as described by Xiong et al. [23].
Complementation of mutants with the native iucA or iucC gene
For the complementation study, the native genes iucA (1725 bp) and iucC (1743 bp) were handled similarly by amplification using primers Re-iucA-F/R and Re-iucC-F/R (Table 2), with the introduced restriction enzyme recognition sites BamH I-EcoR I and EcoR I-Sal I, respectively. To determine that the sequences were in frame, the iucA and iucC fragments were inserted into the pMD®18-T simple vector and sequenced by Sangon Co. LTD. (Shanghai, China). The iucA and iucC PCR products and expression vector pGEX-6P-1 were digested with BamH I-EcoR I and EcoR I-Sal I, respectively, and ligated using T4 DNA ligase for 4 h at 22°C. The ligation mix was then transformed into DH5α and plated on LB agar plates containing ampicillin. Colonies were tested for the presence of iucA and iucC using standard primers iucA-F/iucA-R and iucC-F/iucC-R (Table 2), respectively. The modified plasmid pGEX-6p-1 with the iucA or iucC insert was isolated from DH5α and electroporated into E058ΔiucA or E058ΔiucC to complement the gene deleted.
RT-PCR analysis
To extract total RNA from each strain, 1 ml of bacteria culture was collected by centrifugation. RNA was extracted by using the RNeasy Mini kit purchased from QIAGEN (Qiagen, Dusseldorf, Germany) and treated with an on-column Rnase-Free Dnase set. The first-strand synthesis of cDNA was primed with random primers using a high capacity cDNA archive kit (Applied Biosystems, Foster City, CA, USA). Primer sets for PCR amplification of target genes shiG, iucA, iucB, iucC, iucD, iutA and O2-ColV16 in cDNA samples are listed in Table 2. In parallel, PCRs were performed with pAPEC-O2-ColV-like plasmid DNA as a positive control and cDNA samples without activation of the reverse-transcription (RT) as a negative control. The PCR products were resolved on 0.8% agarose gels and visualized by ethidium bromide staining.
Growth in LB and under iron limitation
Each strain was cultured in LB medium with the appropriate antibiotic at 37°C overnight. The next day the cell density was estimated by spectrophotometry, and cultures were diluted in PBS prior to inoculation in LB medium with or without 200 µM 2,2′-dipyridyl (DIP) to achieve an approximate starting concentration of 103 to 104 colony-forming units (CFUs) per ml, which was confirmed by viable counts. Mixtures were incubated at 37°C with shaking, and aliquots of these cultures were removed at set time intervals for use in determining viable counts. The data represent averages of three independent assays.
Bactericidal activity of specific-pathogen-free (SPF) chicken serum
The serum bactericidal assay was performed in a 96-well plate essentially as described previously [24] but with the following modifications. Briefly, complement-sufficient serum was prepared and pooled from SPF chickens (White Leghorn, Jinan SPAFAS poultry Co. LTD., Jinan, Shandong, China). The chicken serum was diluted to 0.5, 2.5, 5, 12.5 and 25% in pH 7.2 phosphate-buffered saline (PBS). Bacteria (10 μl containing 106 CFU) were inoculated into reaction wells containing 190 μl of the diluted (25%) heat-inactivated SPF chicken serum or PBS alone and incubated at 37°C for 30 min. Serial dilutions (1∶10) of each well were plated onto LB agar plates. The resulting colonies were counted after 24 h of incubation.
Animal infection models
A competitive co-infection model and a comparative single-strain infection model were used to investigate the contribution of the iucA, iucC, iutA, iucA/iutA and iucC/iutA genes to the virulence of APEC E058. Animals used in these studies were White Leghorn SPF chickens obtained from Jinan SPAFAS Poultry Co. LTD. The birds were treated in the experiments in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (Approved by the State Council on October 31, 1988).
Briefly, for competitive co-infection assays, fifteen 3-week-old chickens were infected intratracheally with equal doses (1×107 CFU each) of the wild-type strain E058 or the mutant strains E058ΔiucA, E058ΔiucC, E058ΔiutA, E058ΔiucAΔiutA or E058ΔiucCΔiutA. At 24 h post-infection, the spleen, heart, liver, lung and kidney of inoculated chickens were collected, weighed and homogenized, and serial dilutions were plated on LB medium with or without kanamycin or zeocin for selection of the mutant or total bacteria [25].
Animal experiments were also carried out to determine the colonization ability of the E058ΔiucA, E058ΔiucC, E058ΔiutA, E058ΔiucAΔiutA and E058ΔiucCΔiutA mutant strains in comparison with the E058 parent strain. For this single-strain infection model, 3-week-old White Leghorn chickens (15 chickens per group) were inoculated in the left thoracic air sac with 0.1 ml (107 CFU) of a suspension containing the wild-type E058 strain or mutant derivatives. After 24 h, all chickens were euthanized and examined for macroscopic lesions. The heart, liver, spleen, lung and kidney of the chickens were collected, weighed, suspended in PBS and homogenized. Bacterial loads were determined by plating serial dilutions of the homogenates on selective LB agar medium [26].
HD-11 ingestion assay
For ingestion assays, avian macrophage HD-11 cells were grown in DMEM with 10% FBS at 37°C with 5% CO2 at 2×105 cells per well in 24-well cell culture plates and incubated for 24 h prior to ingestion assays. Bacteria were inoculated into cells at a multiplicity of infection (MOI) of 100. Inoculated cells were incubated at 37°C with 5% CO2 for 1 h under to allow the bacteria to be ingested by the cells. Thereafter, the cells were washed with PBS, and the extracellular bacteria were eliminated by culturing in DMEM medium containing gentamicin (100 µg/ml) at 37°C for 1.5 h prior to washing the cells again using PBS. The intracellular bacteria were treated with 1 ml 0.1% Triton X-100, and a 100 µl aliquot of this suspension was inoculated into 900 µl PBS. Serial dilutions (1∶10) of each sample were plated onto LB agar plates. The resulting colonies were counted after 24 h of incubation. Wells containing only HD-11 cells were used as negative controls. The assay was performed in triplicate. The ingestion ratio was determined by dividing the number of ingested bacteria by the number of bacteria in the initial inoculation.
Aerobactin production
Mutants were also assessed for aerobactin production as described by Vidotto et al. [5]. Low-iron agar assay plates, composed of M-9 minimum salts, containing 200 µM 2,2′-dipyridyl and 0.2% glucose, were seeded with 1 ml/liter of an overnight culture of the indicator organism, E. coli LG1522, which is incapable of producing aerobactin but can use exogenously produced aerobactin. The E058 strain and its mutants were stab inoculated into the agar, and the plates were incubated at 37°C for 24 h. Following incubation, plates were observed for growth of the indicator organism around the stabs in a halo as evidence of aerobactin elaboration by the test mutants.
Statistical analyses
Statistical analyses for in vivo tests were performed using GraphPad Prism v5.0 software package (GraphPad Software). The Wilcoxon matched-pair test was used to analyze data from the competition assay. The Mann Whitney U test was performed for analysis of results from the colonization and persistence assay.
Results
Construction of the iutA, iucA and iucC knockout mutants
The iucA and iucC genes situated on the pAPEC-O2-ColV-like plasmid from APEC E058 were sequenced and confirmed to be 1725 bp and 1743 bp in length, respectively. The putative iutA gene in the wild-type strain E058 was identified by BLAST searches of the E. coli plasmid pAPEC-O2-ColV complete sequence. Nucleotide sequence analysis showed that the iutA DNA fragment was 1966 bp in length. The APEC E058 mutants were created by the method described by Xiong et al. [23]. Sequence analysis revealed that the shiG gene encoding a conserved hypothetical protein is upstream of iucA. The iucB gene encoding the aerobactin biosynthesis protein IucB is downstream of iucA and upstream gene of iucC. The iucD gene, encoding the aerobactin IucD protein, is downstream of iucC. The iutA gene is downstream of iucD and upstream of the O2-ColV16 gene.
The iucA and iucC knockout mutants were constructed by allelic exchange with an insertion of a Zeor cassette, while the iutA knockout mutant was made with an insertion of a Kanr cassette. By sequence analysis of the PCR products, the iucA and iucC genes were confirmed to have been replaced by the Zeor gene in the pAPEC-O2-ColV-like plasmid of APEC E058 at the predicted position, and the generated mutants were named E058ΔiucA and E058ΔiucC, respectively. The iutA gene was also confirmed to have been replaced by the Kanr gene in the pAPEC-O2-ColV-like plasmid of APEC E058 at the predicted position, and the generated mutant was named E058ΔiutA.
The iucA gene mutant plasmid containing the replacement Zeor gene was PCR amplified with primers iucA-F and iucA-R (Table 2) and purified for electroporation to E058ΔiutA competent cells as described previously [23]. After 18 h of incubation, the resulting Zeor Kanr colonies were selected for PCR identification using primers iucA-F and iucA-R or iutA-F and iutA-R (Table 2) and analyzed by sequencing. The iucA and iutA double mutant was named E058ΔiucAΔiutA. The iucC and iutA double mutant E058ΔiucCΔiutA was generated using the same method.
RT-PCR analysis
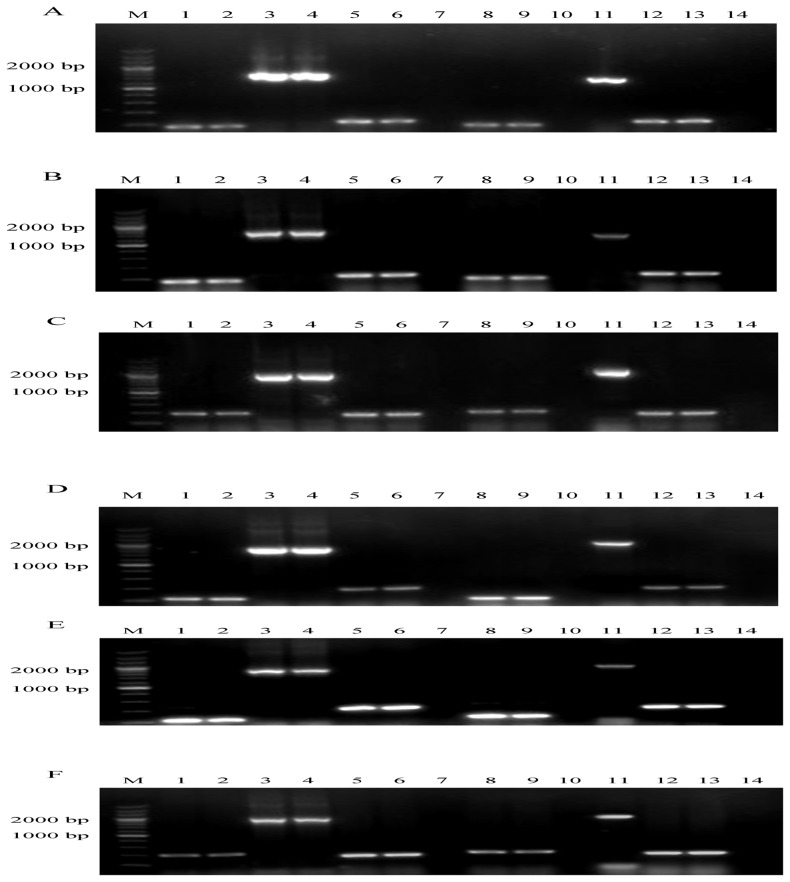
To determine whether the insertion had a polar effect on the upstream or downstream genes, total RNA samples extracted from the parental E058 and mutants E058ΔiucA, E058ΔiucC, E058ΔiucAΔiutA and E058ΔiucCΔiutA were analyzed by RT-PCR using primer sets designed for shiG [GF/GR], iucA [iucA-F/iucA-R], iucB [BF/BR], iucC [iucC-F/iucC-R], iucD [DF/DR], iutA [iutA-F/iutA-R] and O2-ColV16 [O2F/O2R] (Table 2). When compared to the parental E058, the insertion of the Zeor gene in the E058ΔiucA, E058ΔiucAΔiutA, E058ΔiucC or E058ΔiucCΔiutA mutant only disrupted the transcription of the iucA or iucC gene (Figure 1A, 1B, lane10; 1D, 1E, lane 10). Meanwhile, insertion of the Kanr gene in the E058ΔiucAΔiutA or E058ΔiucCΔiutA mutant only disrupted transcription of the iutA gene (Figure 1C, 1F, lane 10), and the upstream and downstream genes of all three target genes (iucA, iucC, iutA) were not influenced.
Figure 1. Detection of iucA (A, B), iucC (D, E) and iutA (C, F) gene expression by RT-PCR.
(A). Detection of iucA gene transcription in E058 and E058ΔiucA by RT-PCR. Templates: lanes 1, 3, 5: cDNA derived from total RNA of E058. Lane 7: total RNA from E058 without activation of RT. Lanes 8, 10, 12: cDNA derived from total RNA of E058ΔiucA. Lane 14: total RNA from E058ΔiucA without activation of RT. Lanes 2, 4, 6: pAPEC-O2-ColV-like DNA from E058. Lanes 9, 11, 13: pAPEC-O2-ColV-like DNA from E058ΔiucA. Primers: Lanes 1, 2, 8, 9: GF/GR. Lanes 3, 4, 10, 11: iucA-F/iucA-R. Lanes 5, 6, 7, 12, 13, 14: BF/BR (Table 2). (B). Detection of iucA gene transcription in E058 and E058ΔiucAΔiutA by RT-PCR. Templates: lanes 1, 3, 5: cDNA derived from total RNA of E058. Lane 7: total RNA from E058 without activation of RT. Lanes 8, 10, 12: cDNA derived from total RNA of E058ΔiucAΔiutA. Lane 14: total RNA from E058ΔiucAΔiutA without activation of RT. Lanes 2, 4, 6: pAPEC-O2-ColV-like DNA from E058. Lanes 9, 11, 13: pAPEC-O2-ColV-like DNA from E058ΔiucAΔiutA. Primers: Lanes 1, 2, 8, 9: GF/GR. Lanes 3, 4, 10, 11: iucA-F/iucA-R. Lanes 5, 6, 7, 12, 13, 14: BF/BR (Table 2). (C). Detection of iutA gene transcription in E058 and E058ΔiucAΔiutA by RT-PCR. Templates: lanes 1, 3, 5: cDNA derived from total RNA of E058. Lane 7: total RNA from E058 without activation of RT. Lanes 8, 10, 12: cDNA derived from total RNA of E058ΔiucAΔiutA. Lane 14: total RNA from E058ΔiucAΔiutA without activation of RT. Lanes 2, 4, 6: pAPEC-O2-ColV-like DNA from E058. Lanes 9, 11, 13: pAPEC-O2-ColV-like DNA from E058ΔiucAΔiutA. Primers: Lanes 1, 2, 8, 9: DF/DR. Lanes 3, 4, 10, 11: iutA-F/iutA-R. Lanes 5, 6, 7, 12, 13, 14: O2F/O2R (Table 2). (D). Detection of iucC gene transcription in E058 and E058ΔiucC by RT-PCR. Templates: lanes 1, 3, 5: cDNA derived from total RNA of E058. Lane 7: total RNA from E058 without activation of RT. Lanes 8, 10, 12: cDNA derived from total RNA of E058ΔiucC. Lane 14: total RNA from E058ΔiucC without activation of RT. Lanes 2, 4, 6: pAPEC-O2-ColV-like DNA from E058. Lanes 9, 11, 13: pAPEC-O2-ColV-like DNA from E058ΔiucC. Primers: Lanes 1, 2, 8, 9: BF/BR. Lanes 3, 4, 10, 11: iucC-F/iucC-R. Lanes 5, 6, 7, 12, 13, 14: DF/DR (Table 2). (E). Detection of iucC gene transcription in E058 and E058ΔiucCΔiutA by RT-PCR. Templates: lanes 1, 3, 5: cDNA derived from total RNA of E058. Lane 7: total RNA from E058 without activation of RT. Lanes 8, 10, 12: cDNA derived from total RNA of E058ΔiucCΔiutA. Lane 14: total RNA from E058ΔiucCΔiutA without activation of RT. Lanes 2, 4, 6: pAPEC-O2-ColV-like DNA from E058. Lanes 9, 11, 13: pAPEC-O2-ColV-like DNA from E058ΔiucCΔiutA. Primers: Lanes 1, 2, 8, 9: BF/BR. Lanes 3, 4, 10, 11: iucC-F/iucC-R. Lanes 5, 6, 7, 12, 13, 14: DF/DR (Table 2). (F). Detection of iutA gene transcription in E058 and E058ΔiucCΔiutA by RT-PCR. Templates: lanes 1, 3, 5: cDNA derived from total RNA of E058. Lane 7: total RNA from E058 without activation of RT. Lanes 8, 10, 12: cDNA derived from total RNA of E058ΔiucCΔiutA. Lane 14: total RNA from E058ΔiucCΔiutA without activation of RT. Lanes 2, 4, 6: pAPEC-O2-ColV-like DNA from E058. Lanes 9, 11, 13: pAPEC-O2-ColV-like DNA from E058ΔiucCΔiutA. Primers: Lanes 1, 2, 8, 9: DF/DR. Lanes 3, 4, 10, 11: iutA-F/iutA-R. Lanes 5, 6, 7, 12, 13, 14: O2F/O2R (Table 2). A 200 bp marker (Takara) was used as the molecular size standard (M).
Complementation studies and the result of aerobactin production
For complementation studies, the native iucA gene was amplified using primers Re-iucA-F/Re-iucA-R (Table 2) and subcloned into the pGEX-6P-1 vector by digestion with restriction enzymes BamH I and EcoR I and ligation. The recombinant plasmid pGEX-6P-1-iucA was electroporated into E058ΔiucA to complement the deleted iucA gene, and colonies on LB agar containing ampicillin were tested for the presence of iucA using primers Re-iucA-F/Re-iucA-R (Table 2). A positive clone was named Re-E058ΔiucA. Using the same method, Re-E058ΔiucC was generated except that the restriction enzymes were EcoR I and Sal I. In testing for aerobactin production, the wild-type strain E058, the mutant E058ΔiutA and complementation strains Re-E058ΔiucA and Re-E058ΔiucC showed the ability to produce aerobactin, while the E058ΔiucA, E058ΔiucAΔiutA, E058ΔiucC and E058ΔiucCΔiutA mutants lost the ability to produce aerobactin (Data not shown).
Characterization of the mutants
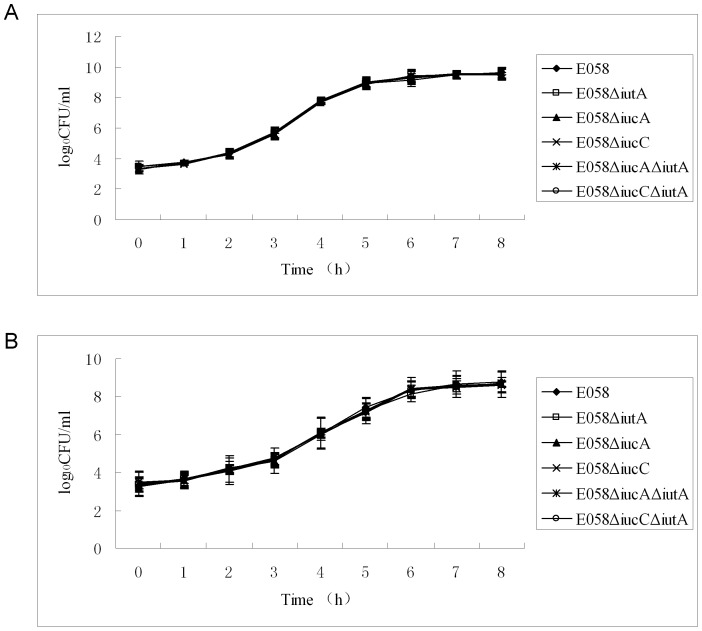
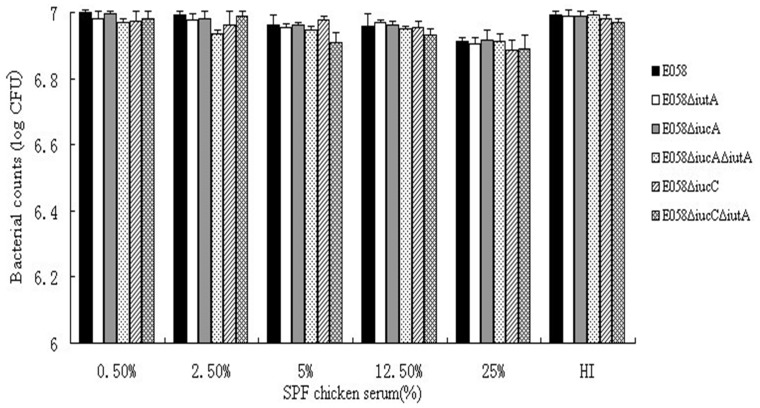
Growth curves, colonies, bactericidal assay and ingestion capability of the E058ΔiucA, E058ΔiucC, E058ΔiutA, E058ΔiucAΔiutA and E058ΔiucCΔiutA mutants were similar to those of the parental strain E058. Compared to the parent strain, no significant differences in the growth during the logarithmic phase were observed for all five mutants grown in LB rich medium and under iron limitation (Figure 2). Results of the bactericidal assay showed that the abilities of the E058ΔiutA, E058ΔiucA, E058ΔiucAΔiutA, E058ΔiucC and E058ΔiucCΔiutA mutants to survive in SPF chicken serum were not affected (Figure 3). The APEC E058 mutants (E058ΔiutA, E058ΔiucA, E058ΔiucAΔiutA, E058ΔiucC and E058ΔiucCΔiutA) had ingestion ratios (0.23%, 0.24%, 0.19%, 0.23% and 0.20%, respectively) which were not apparently different from that of the parental E058 (0.23%). Thus, the deletion of either the single gene of iutA, iucA or iucC, the double genes of iucA/iutA or iucC/iutA had no effect on the ingestion of APEC E058 into HD-11 cells.
Figure 2. Growth curves of E058 wild-type strain and its mutants.
(A) The E058(□), E058ΔiutA(▪), E058ΔiucA(▴), E058ΔiucC(×), E058ΔiucAΔiutA (*) and E058ΔiucCΔiutA (•) strains were grown in LB broth at 37°C, respectively, and their growth curves were determined by measuring viable counts (CFU ml−1). (B) Growth curves of the E058 and its mutants in LB containing 200 µM 2,2′-dipyridyl (DIP). The data represent averages of three independent assays.
Figure 3. Bactericidal activity of SPF chicken serum against the wild type strain and mutants.
Strains E058 (black bar), E058ΔiutA (white bar), E058ΔiucA (gray bar), E058ΔiucAΔiutA (gray dots), E058ΔiucC (gray oblique lines), E058ΔiucCΔiutA (gray grids). HI represents the group of heat-inactivated 25% SPF chicken serum used as controls for each strain tested. Data represent averages of three independent assays.
Deletions of iutA, iucA, iucC, iucA/iutA and iucC/iutA attenuate APEC E058 virulence in vivo
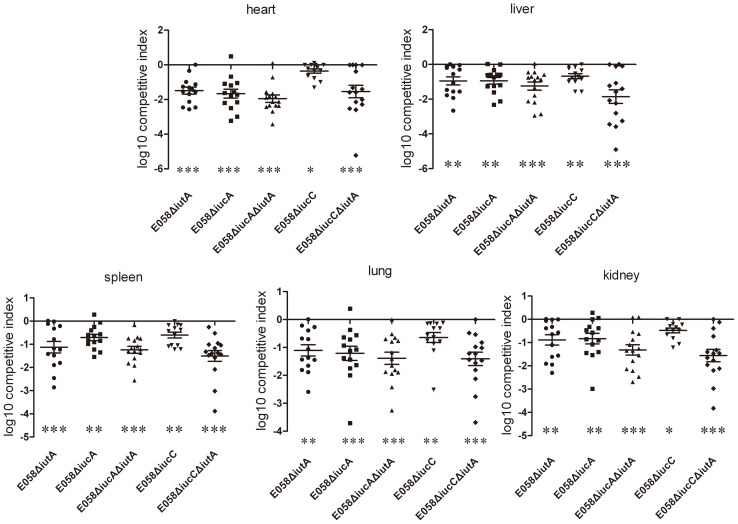
To determine the effects of iutA, iucA, iucC, iucA/iutA and iucC/iutA on the virulence of APEC E058 in vivo, competitive co-infections were carried out. As shown in Figure 4, at 24 h post-challenge, the E058ΔiutA mutant was significantly attenuated in the liver, lung and kidney (P<0.01) and was attenuated to an even greater degree in the heart and spleen (P<0.001) (Figure 4). The E058ΔiucA mutant was significantly attenuated in the liver, spleen and kidney (P<0.01), and especially in the heart and lung (P<0.001) (Figure 4). The E058ΔiucC mutant was also significantly attenuated in the heart and kidney (P<0.05) and exhibited more significant levels of attenuation in the liver, spleen and lung (P<0.01). Both double mutants (E058ΔiucAΔiutA and E058ΔiucCΔiutA) were significantly attenuated in all five tested tissues (P<0.001) (Figure 4).
Figure 4. Competition assays between wild-type E058 and mutants E058ΔiutA, E058ΔiucA, E058ΔiucC, E058ΔiucAΔiutA or E058ΔiucCΔiutA inoculated simultaneously.
Negative competitive index (CI) values indicate a decreased capacity of the mutants to compete for growth with the wild-type strain. Horizontal bars indicate the mean log10CI values. Each data point represents a sample from an individual chicken. Statistically significant differences in CI values between E058 and its mutants are indicated with asterisks (* P<0.05, ** P<0.01, *** P<0.001).
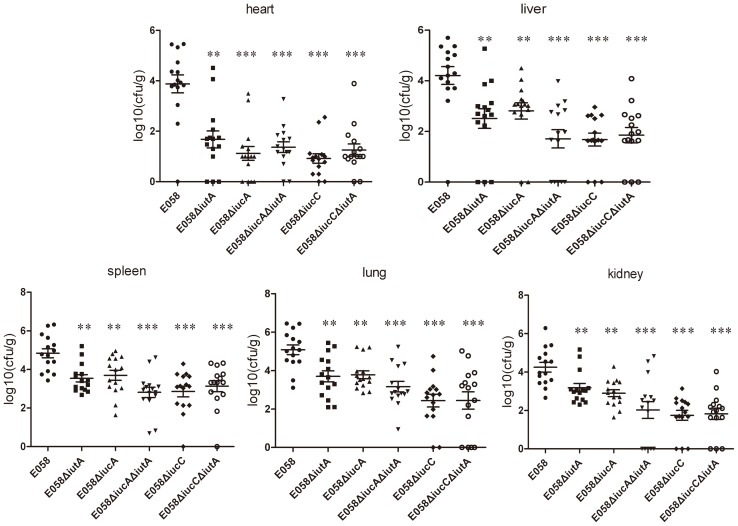
When birds were infected with the E058ΔiutA, E058ΔiucA, E058ΔiucAΔiutA, E058ΔiucC and E058ΔiucCΔiutA mutants, distinct reductions in the number of bacteria recovered from all tested tissues were observed compared to those of birds infected with E058. At 24 h after infection with the E058 parent strain, maximum colonization was observed in the lung (1.1×105 CFU.g−1), and minimal colonies were obtained in the heart (8.7×103 CFU.g−1), while colony numbers in the spleen (6.3×104 CFU.g−1), liver (2.1×104 CFU.g−1) and kidney (3.2×104 CFU.g−1) were intermediate (Figure 5). Compared to the E058 wild-type strain, the mutant E058ΔiutA colony numbers were significantly decreased in the lung (6.5×103 CFU.g−1) (P<0.01), heart (70 CFU.g−1) (p<0.01), kidney (2.6×103 CFU.g−1) (p<0.01), liver (5.88×102 CFU.g−1) (p<0.01) and spleen (5.9×103 CFU.g−1) (P<0.01) and colonization with the E058ΔiucA mutant was significantly lower in the lung (7.8×103 CFU.g−1) (P<0.01), spleen (6.8×103 CFU.g−1) (P<0.01), heart (12 CFU.g−1) (P<0.001), liver (8.1×102 CFU.g−1) (P<0.01) and kidney (9.0×102 CFU.g−1) (P<0.01) (Figure 5). Levels of the E058ΔiucAΔiutA mutant colonized in the lung (3.6×103 CFU.g−1), spleen (1.02×103 CFU.g−1), heart (14 CFU.g−1), liver (82 CFU.g−1) and kidney (96 CFU.g−1) were sharply decreased compared to those of the E058 parent strain (P<0.001) (Figure 5). Interestingly, numbers of the E058ΔiucC mutant in the lung (4.3×102 CFU.g−1), spleen (8.6×102 CFU.g−1), heart (9 CFU.g−1), liver (67 CFU.g−1) and kidney (64 CFU.g−1) also dropped dramatically compared to those of E058 (P<0.001) (Figure 5). Not surprisingly, the E058ΔiucCΔiutA mutant colonized in the lung (4.2×102 CFU.g−1), spleen (1.3×103 CFU.g−1), heart (17 CFU.g−1), liver (90 CFU.g−1) and kidney (93 CFU.g−1) also decreased remarkably compared to E058 (P<0.001) (Figure 5). On average the mutant strains were reisolated at levels of 10–1000 times lower than the wild-type strain from the internal organs tested at 24 h post-challenge. These results indicated that iutA and both iucA and iucC genes are involved in the process of systemic infection of APEC E058.
Figure 5. Colonization and persistence of wild-type strain E058 and its mutants E058ΔiutA, E058ΔiucA, E058ΔiucC, E058ΔiucAΔiutA and E058ΔiucCΔiutA.
Data are presented as log10 CFU.g−1 of bacteria from tissues, and horizontal bars represent mean values. Each data point represents a sample from an individual chicken. Statistically significant differences are indicated with asterisks (** P<0.01, *** P<0.001), as determined by the Mann Whitney U test.
Discussion
Many types of bacteria have several iron uptake systems to survive and grow in different host environments [26]. In the virulence assay, the mutants were significantly attenuated in the observed tissues. Sequestration of iron results in an extremely low free iron concentration that would limit bacterial growth in this complicated environment in vivo [25]. The ability to obtain iron from the host determines the pathogenicity of APEC strains and may be increased by the aerobactin and iron system [27].
Iron has many vital functions in bacteria and is a cofactor for a large number of enzymes [28]. The enterochelin system is one of the iron uptake systems that is chromosomally encoded and appears less able to compete with transferrin in vivo, although it is very efficient in vitro [29]. By contrast, the aerobactin system is considered to be efficient in vivo and is involved in the invasive properties of human enteroinvasive E. coli and Shigella species [30], [31]. The aerobactin operon encoded by either plasmids or chromosomes in several species of enteric organisms [6], [15], [20], [31], [32], [33], [34], [35] contains genes responsible for the synthesis of the hydroxamate siderophore aerobactin (iuc) and for ferric aerobactin uptake (iut) [13], [14], [35]. Both components are strongly induced under iron starvation. Because aerobactin genes are present in virulent strains and absent from avirulent strains [3], they are regarded to play independent roles in the pathogenicity of APEC, especially in deep tissue damage, and its persistent infection [11]. The five genes encoding aerobactin are localized to an operon, which is present in the ColV plasmid [17]. The iucC gene is related to the aerobactin synthetase reaction, whereas the deletion of iucA will result in a severe polar effect [36]. A previous study from our laboratory [23] and independent reports from others [21], [37] have provided a general overview of the aerobactin operon and several iron uptake system [38], [39], [40], [41]. To date, many studies have focused on the characterization of the iuc genes [29], [35], [36], [42], but the relationship between the iucC gene and the pathogenicity of APEC has not been clarified.
In this study, the E058ΔiucA, E058ΔiucC, E058ΔiucAΔiutA and E058ΔiucCΔiutA mutants with deletions of the iucA, iucC, iucA/iutA and iucC/iutA genes, respectively, lost the capacity to produce aerobactin, while the complementation of E058ΔiucA and E058ΔiucC restored their ability to produce aerobactin similar to the E058 parent strain. Meanwhile, the mutant E058ΔiutA also showed the capacity to produce aerobactin. In the acquisition of iron, APEC O2 is known to contain at least two other chromosomal operons (yersinabactin and enterobactin) [17]. Although we deleted the aerobactin receptor gene iutA, the other two chromosomal operons (yersinabactin and enterobactin) may have helped to compensate for the loss of iutA. In addition, iucA or iucC is responsible for the synthesis of the hydroxa siderophore aerobactin. While the deletion of one or both of these two genes may have resulted in the failure to produce aerobactin (hydroxamate type siderophore), it may not have influenced the other two chromosomal operons (yersinabactin and enterobactin) in the acquisition of iron.
In LB broth and under conditions with limited iron, there was no obvious difference in the growth of the mutants and wild-type parent. During the assays of bacterial ingestion into HD-11 cells, no obvious difference was observed between the wild-type E058 and its mutants. These findings correspond to results of a previous study on E058ΔiucB and E058ΔiucBΔiutA [23].
In the single-strain infection model, compared to E058, colonization levels of the E058ΔiucA and E058ΔiutA mutants were reduced in all five tested tissues (P<0.01 or P<0.001), and that of the E058ΔiucC mutant was remarkably reduced in the above-mentioned tissues (P<0.001). The same results were observed for both the E058ΔiucAΔiutA and E058ΔiucCΔiutA double mutants.
The capacity of the E058ΔiutA, E058ΔiucA, E058ΔiucAΔiutA, E058ΔiucC and E058ΔiucCΔiutA mutants to compete for growth in 3-week-old SPF chicken tissues with the E058 wild-type strain was also evaluated in a co-infection model. Compared to its parental strain, the E058ΔiutA mutant showed a significant reduction of bacterial counts in the heart, liver, kidney, spleen and lung (P<0.05 or P<0.01). The E058ΔiucA mutant showed a remarkable reduction in all the tissues mentioned above (P<0.01 or P<0.001). The E058ΔiucC mutant was also significantly attenuated in all the tissues mentioned above (P<0.05 or P<0.01). Furthermore, highly significant reductions of both the E058ΔiucAΔiutA and E058ΔiucCΔiutA double mutants were observed in all five tested tissues (P<0.001). The co-infection assay demonstrated that the deletion of either the iutA, iucA or iucC gene reduced the virulence of the APEC strain E058. However, it seemed a greater degree of colonization defects in an independent challenge versus a co-challenge experiment. As the individual infection results showed a 2-log decrease in colonization for most of the mutants in most tissues (10–1000-fold). Whereas, the co-challenge experiment results showed a 1-log fitness defect for most mutants in all tissues. These results implied that the exogenous siderophores synthesized by the wild-type strain may be to a certain extent complement the effect of the siderophore synthesis mutations in a co-infection model. Similarly, a ΔiucBΔentD double mutant, defective in synthesis of both siderophores, was rescued by co-infection with a wild-type strain in the mouse UTI model [43].
The pathogenicity of mutants with deletion of either the whole iucABCDiutA operon or the single gene iutA in APEC χ7122 is attenuated [11]. Meanwhile, the effect of the single gene iucA or iucC, and the double genes iucAiutA or iucCiutA on the virulence of APEC remains to be determined. Tivendale et al. demonstrated that the iucA gene could be amplified only from the three most virulent strains, E3, E30 and E956, but not from the less virulent strains, E133, E1043 and E1292. As discussed by these authors, the identification of potential virulence genes in avian colibacillosis can be attempted by using different approaches, such as to generate isogenic mutants of these genes and determine their virulence in birds [10]. In this study, we used this approach to explore the effects of iucA and iucC genes on the pathogenicity of the APEC strain E058. In a previous study, we demonstrated that either the iucB gene or iutA gene is likely involved directly or indirectly in iron uptake, with no obvious synergistic effect between these two genes, as it relates to the pathogenicity of APEC E058 [23]. To our knowledge, the present study is the first to report the contribution of iucC to the pathogenesis of APEC and the effects of double mutations in iucA/iutA or iucC/iutA on APEC virulence compared to that of each simple mutation. Taken together, the above results demonstrate that the iucA and iucC genes, which participate in aerobactin synthesis, play an important role in the pathogenicity of the APEC strain E058.
Acknowledgments
We thank P. H. Williams for providing the E. coli LG1522, and Lisa K. Nolan for providing the E. coli LG1522, E. coli 23559 and E. coli 23561. We also thank Prof. Xin'an Jiao (Jiangsu Key Laboratory of Zoonosis of Yangzhou University) for kindly providing the cell line HD-11.
Funding Statement
This work was supported by the National Natural Science Foundation of China Grant 30972196, 30771604 and 30471281, and National Program for High Technology Research and Development in China Grant 2003 AA222141 to Song Gao. This study was also supported by the program for Changjiang Scholars and Innovative Research Team in University (PCSIRT0978) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dho M, Lafont JP (1984) Adhesive properties and iron uptake ability in Escherichia coli lethal and nonlethal for chicks. Avian Dis 28: 1016–1025. [PubMed] [Google Scholar]
- 2. Knobl T, Baccaro MR, Moreno AM, Gomes TA, Vieira MA, et al. (2001) Virulence properties of Escherichia coli isolated from ostriches with respiratory disease. Vet Microbiol 83: 71–80. [DOI] [PubMed] [Google Scholar]
- 3. Lafont JP, Dho M, D'Hauteville HM, Bree A, Sansonetti PJ (1987) Presence and expression of aerobactin genes in virulent avian strains of Escherichia coli . Infect Immun 55: 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ngeleka M, Brereton L, Brown G, Fairbrother JM (2002) Pathotypes of avian Escherichia coli as related to tsh-, pap-, pil-, and iuc-DNA sequences, and antibiotic sensitivity of isolates from internal tissues and the cloacae of broilers. Avian Dis 46: 143–152. [DOI] [PubMed] [Google Scholar]
- 5. Vidotto MC, Muller EE, de Freitas JC, Alfieri AA, Guimaraes IG, et al. (1990) Virulence factors of avian Escherichia coli . Avian Dis 34: 531–538. [PubMed] [Google Scholar]
- 6. Colonna B, Nicoletti M, Visca P, Casalino M, Valenti P, et al. (1985) Composite IS1 elements encoding hydroxamate-mediated iron uptake in FIme plasmids from epidemic Salmonella spp . J Bacteriol 162: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bullen JJ (1981) The significance of iron in infection. Rev Infect Dis 3: 1127–1138. [DOI] [PubMed] [Google Scholar]
- 8. Finkelstein RA, Sciortino CV, Mcintosh MA (1983) Role of iron in microbe-host interactions. Rev Infect Dis 5 Suppl 4 S759–777. [DOI] [PubMed] [Google Scholar]
- 9. Weinberg ED (1984) Iron withholding: a defense against infection and neoplasia. Physiol Rev 64: 65–102. [DOI] [PubMed] [Google Scholar]
- 10. Tivendale KA, Allen JL, Ginns CA, Crabb BS, Browning GF (2004) Association of iss and iucA, but not tsh, with plasmid-mediated virulence of avian pathogenic Escherichia coli . Infect Immun 72: 6554–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dozois CM, Daigle F, Curtiss R 3rd (2003) Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA 100: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, et al. (2006) Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun 74: 6287–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carbonetti NH, Williams PH (1984) A cluster of five genes specifying the aerobactin iron uptake system of plasmid ColV-K30. Infect Immun 46: 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gross R, Engelbrecht F, Braun V (1984) Genetic and biochemical characterization of the aerobactin synthesis operon on pColV. Mol Gen Genet 196: 74–80. [DOI] [PubMed] [Google Scholar]
- 15. Bindereif A, Neilands JB (1985) Aerobactin genes in clinical isolates of Escherichia coli . J Bacteriol 161: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sabri M, Leveille S, Dozois CM (2006) A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152: 745–758. [DOI] [PubMed] [Google Scholar]
- 17. Johnson TJ, Siek KE, Johnson SJ, Nolan LK (2006) DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J Bacteriol 188: 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kariyawasam S, Johnson TJ, Nolan LK (2006) The pap operon of avian pathogenic Escherichia coli strain O1: K1 is located on a novel pathogenicity island. Infect Immun 74: 744–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gibson F, Magrath DI (1969) The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim Biophys Acta 192: 175–184. [DOI] [PubMed] [Google Scholar]
- 20. Williams PH (1979) Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli . Infect Immun 26: 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skyberg JA, Johnson TJ, Nolan LK (2008) Mutational and transcriptional analyses of an avian pathogenic Escherichia coli ColV plasmid. BMC Microbiol 8: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao L, Gao S, Huan H, Xu X, Zhu X, et al. (2009) Comparison of virulence factors and expression of specific genes between uropathogenic Escherichia coli and avian pathogenic E. coli in a murine urinary tract infection model and a chicken challenge model. Microbiology 155: 1634–1644. [DOI] [PubMed] [Google Scholar]
- 23. Xiong L, Ling J, Gao Q, Zhou Y, Li T, et al. (2012) Construction of iucB and iucBiutA mutants of avian pathogenic Escherichia coli and evaluation of their pathogenicity. Vet Microbiol 159: 420–431. [DOI] [PubMed] [Google Scholar]
- 24. Zaleski A, Scheffler NK, Densen P, Lee FK, Campagnari AA, et al. (2000) Lipooligosaccharide P(k) (Galalpha1-4Galbeta1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect Immun 68: 5261–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li G, Laturnus C, Ewers C, Wieler LH (2005) Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect Immun 73: 2818–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li G, Ewers C, Laturnus C, Diehl I, Alt K, et al. (2008) Characterization of a yjjQ mutant of avian pathogenic Escherichia coli (APEC). Microbiology 154: 1082–1093. [DOI] [PubMed] [Google Scholar]
- 27. Der Vartanian M (1988) Differences in excretion and efficiency of the aerobactin and enterochelin siderophores in a bovine pathogenic strain of Escherichia coli . Infect Immun 56: 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andrews SC, Robinson AK, Rodriguez-Quinones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237. [DOI] [PubMed] [Google Scholar]
- 29. Konopka K, Bindereif A, Neilands JB (1982) Aerobactin-mediated utilization of transferrin iron. Biochemistry 21: 6503–6508. [DOI] [PubMed] [Google Scholar]
- 30. Griffiths E, Stevenson P, Hale TL, Formal SB (1985) Synthesis of aerobactin and a 76,000-dalton iron-regulated outer membrane protein by Escherichia coli K-12-Shigella flexneri hybrids and by enteroinvasive strains of Escherichia coli . Infect Immun 49: 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawlor KM, Payne SM (1984) Aerobactin genes in Shigella spp . J Bacteriol 160: 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDougall S, Neilands JB (1984) Plasmid- and chromosome-coded aerobactin synthesis in enteric bacteria: insertion sequences flank operon in plasmid-mediated systems. J Bacteriol 159: 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valvano MA, Crosa JH (1984) Aerobactin iron transport genes commonly encoded by certain ColV plasmids occur in the chromosome of a human invasive strain of Escherichia coli K1. Infect Immun 46: 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Warner PJ, Williams PH, Bindereif A, Neilands JB (1981) ColV plasmid-specific aerobactin synthesis by invasive strains of Escherichia coli . Infect Immun 33: 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bindereif A, Neilands JB (1983) Cloning of the aerobactin-mediated iron assimilation system of plasmid ColV. J Bacteriol 153: 1111–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Lorenzo V, Bindereif A, Paw BH, Neilands JB (1986) Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol 165: 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forman S, Nagiec MJ, Abney J, Perry RD, Fetherston JD (2007) Analysis of the aerobactin and ferric hydroxamate uptake systems of Yersinia pestis . Microbiology 153: 2332–2341. [DOI] [PubMed] [Google Scholar]
- 38. Watson RJ, Millichap P, Joyce SA, Reynolds S, Clarke DJ (2010) The role of iron uptake in pathogenicity and symbiosis in Photorhabdus luminescens TT01. BMC Microbiol 10: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sabri M, Caza M, Proulx J, Lymberopoulos MH, Bree A, et al. (2008) Contribution of the SitABCD, MntH, and FeoB metal transporters to the virulence of avian pathogenic Escherichia coli O78 strain chi7122. Infect Immun 76: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valdebenito M, Crumbliss AL, Winkelmann G, Hantke K (2006) Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int J Med Microbiol 296: 513–520. [DOI] [PubMed] [Google Scholar]
- 41. Escolar L, Perez-Martin J, de Lorenzo V (2000) Evidence of an unusually long operator for the fur repressor in the aerobactin promoter of Escherichia coli . J Biol Chem 275: 24709–24714. [DOI] [PubMed] [Google Scholar]
- 42. de Lorenzo V, Neilands JB (1986) Characterization of iucA and iucC genes of the aerobactin system of plasmid ColV-K30 in Escherichia coli . J Bacteriol 167: 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Torres AG, Redford P, Welch RA, Payne SM (2001) TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69: 6179–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao S, Liu XF, Zhang RK, Jiao XA, Wen QY, et al. (1999) The isolation and identification of pathogenic Escherichia coli isolates of chicken origin from some regions in China. Acta Vet. Et. Zootech Sinica. 30: 164–171. [Google Scholar]
- 45. Van LP (1996) Transcriptional activation of the chicken lysozyme gene by NF-kappa Bp65 (RelA) and c-Rel, but not by NF-kappa Bp50. J. Biochem. 313: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]