Abstract
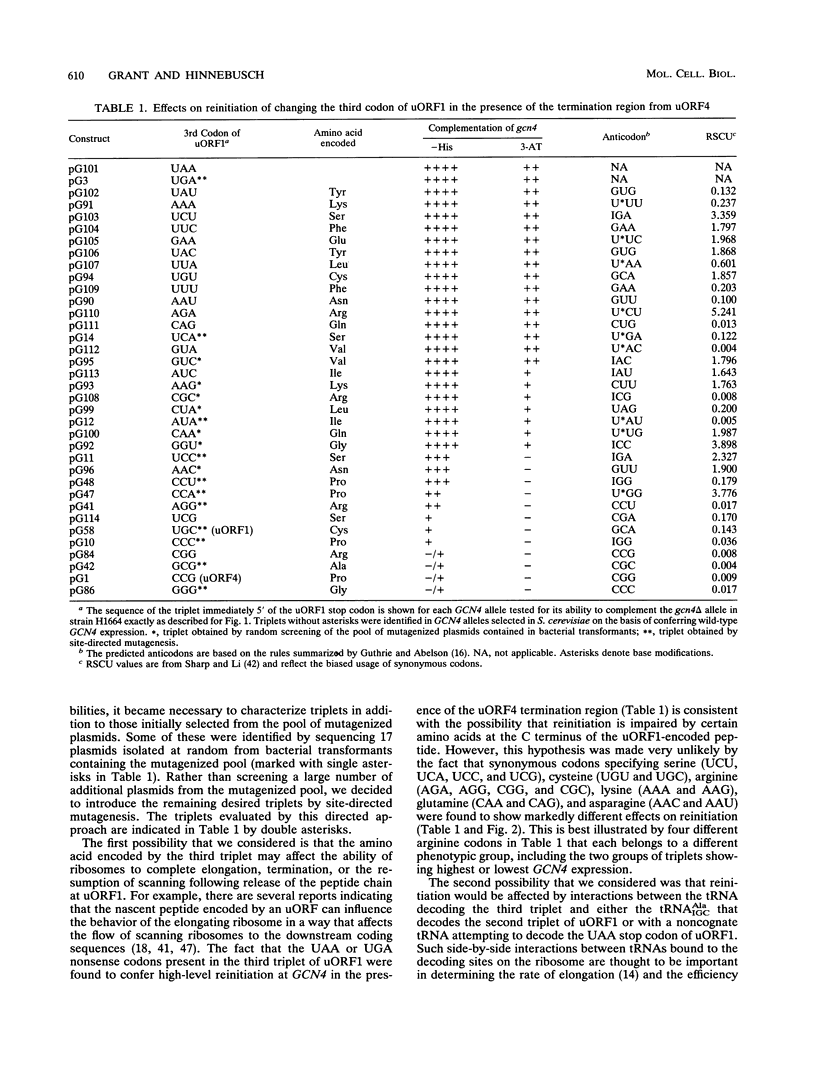
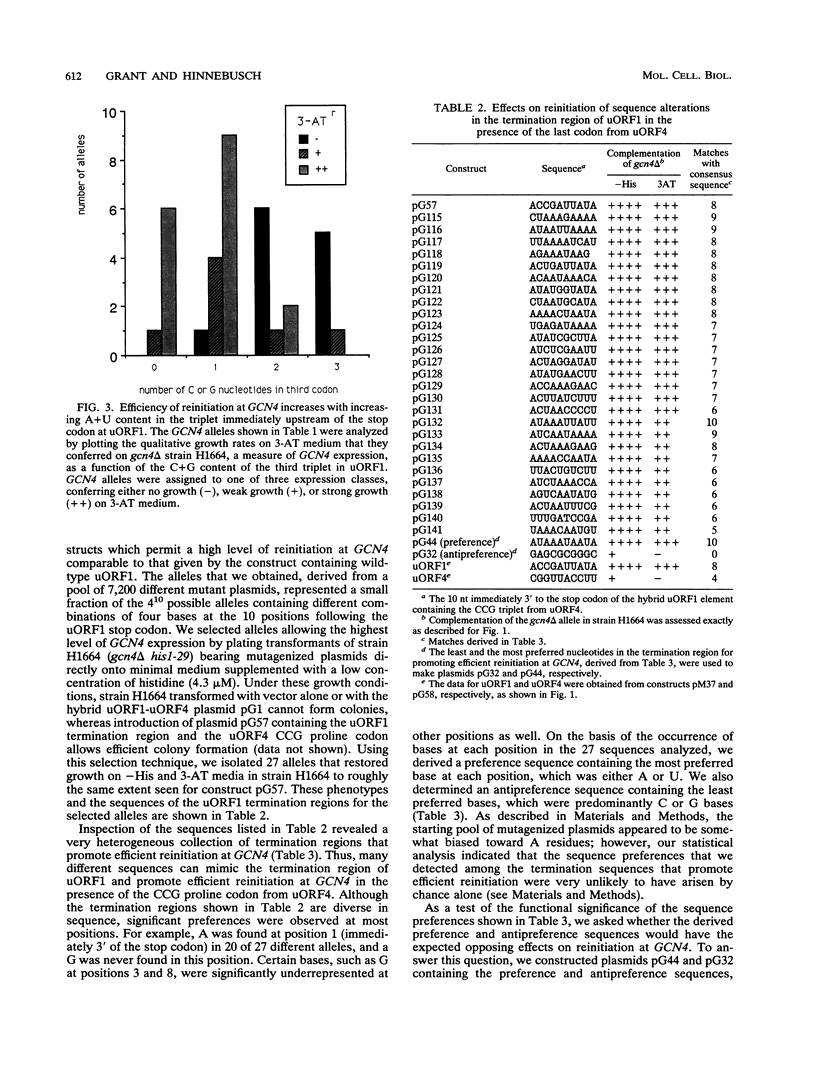
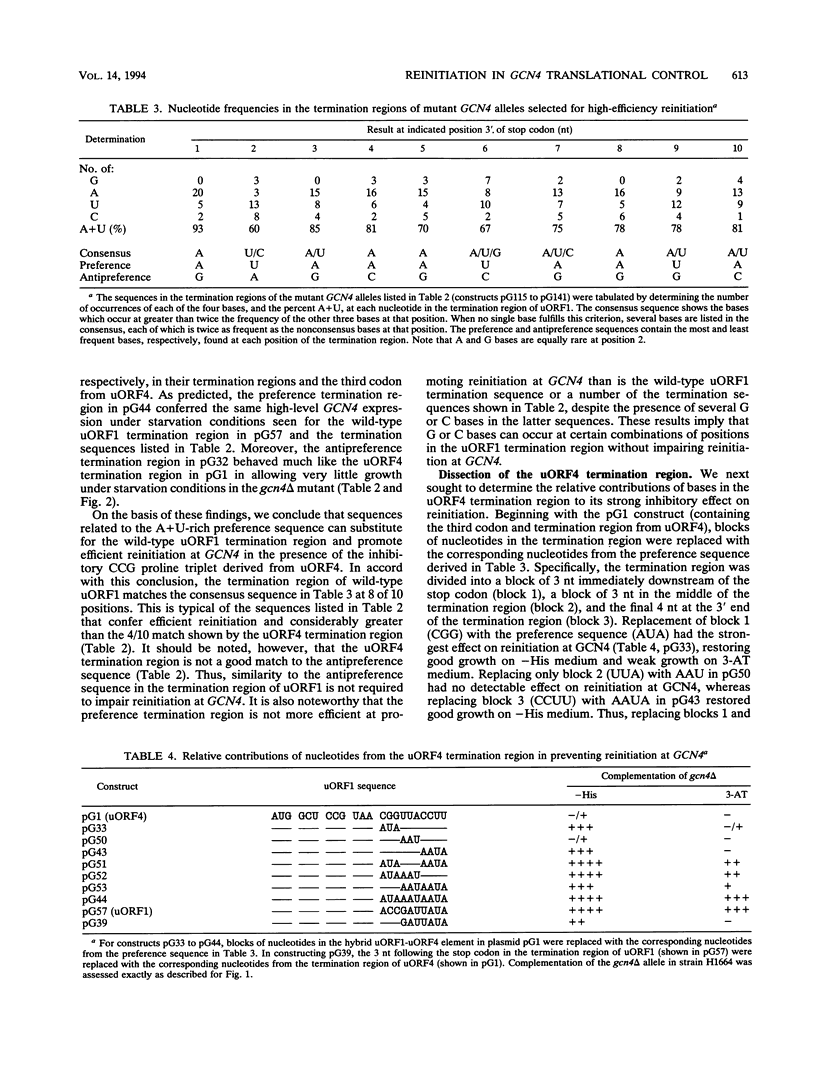
Translational control of the GCN4 gene involves two short open reading frames in the mRNA leader (uORF1 and uORF4) that differ greatly in the ability to allow reinitiation at GCN4 following their own translation. The low efficiency of reinitiation characteristic of uORF4 can be reconstituted in a hybrid element in which the last codon of uORF1 and 10 nucleotides 3' to its stop codon (the termination region) are substituted with the corresponding nucleotides from uORF4. To define the features of these 13 nucleotides that determine their effects on reinitiation, we separately randomized the sequence of the third codon and termination region of the uORF1-uORF4 hybrid and selected mutant alleles with the high-level reinitiation that is characteristic of uORF1. The results indicate that many different A+U-rich triplets present at the third codon of uORF1 can overcome the inhibitory effect of the termination region derived from uORF4 on the efficiency of reinitiation at GCN4. Efficient reinitiation is not associated with codons specifying a particular amino acid or isoacceptor tRNA. Similarly, we found that a diverse collection of A+U-rich sequences present in the termination region of uORF1 could restore efficient reinitiation at GCN4 in the presence of the third codon derived from uORF4. To explain these results, we propose that reinitiation can be impaired by stable base pairing between nucleotides flanking the uORF1 stop codon and either the tRNA which pairs with the third codon, the rRNA, or sequences located elsewhere in GCN4 mRNA. We suggest that these interactions delay the resumption of scanning following peptide chain termination at the uORF and thereby lead to ribosome dissociation from the mRNA.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A. A., Deacon N. J. The 3'-terminal primary structure of five eukaryotic 18S rRNAs determined by the direct chemical method of sequencing. The highly conserved sequences include an invariant region complementary to eukaryotic 5S rRNA. Nucleic Acids Res. 1980 Oct 10;8(19):4365–4376. doi: 10.1093/nar/8.19.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baim S. B., Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988 Apr;8(4):1591–1601. doi: 10.1128/mcb.8.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990 Nov 11;18(21):6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990 Apr 25;18(8):2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener D. R., Ray S. C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991 Jun 25;19(12):3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992 Jan 2;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Donahue T. F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M. J., Galpine A., Austin S. A., Panniers R., Henshaw E. C., Duncan R., Hershey J. W., Pollard J. W. Regulation of polypeptide chain initiation in Chinese hamster ovary cells with a temperature-sensitive leucyl-tRNA synthetase. Changes in phosphorylation of initiation factor eIF-2 and in the activity of the guanine nucleotide exchange factor GEF. J Biol Chem. 1987 Jan 15;262(2):767–771. [PubMed] [Google Scholar]
- Craigen W. J., Lee C. C., Caskey C. T. Recent advances in peptide chain termination. Mol Microbiol. 1990 Jun;4(6):861–865. doi: 10.1111/j.1365-2958.1990.tb00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F. Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993 Apr 25;21(8):1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992 Feb 7;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Folley L. S., Yarus M. Codon contexts from weakly expressed genes reduce expression in vivo. J Mol Biol. 1989 Oct 5;209(3):359–378. doi: 10.1016/0022-2836(89)90003-x. [DOI] [PubMed] [Google Scholar]
- Gafner J., Robertis E. M., Philippsen P. Delta sequences in the 5' non-coding region of yeast tRNA genes. EMBO J. 1983;2(4):583–591. doi: 10.1002/j.1460-2075.1983.tb01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J. W. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Hill J. R., Morris D. R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem. 1993 Jan 5;268(1):726–731. [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa S., Kaji A. Molecular cloning and expression of ribosome releasing factor. J Biol Chem. 1989 Nov 25;264(33):20054–20059. [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987 Oct;7(10):3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986 May;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P. F., Hinnebusch A. G. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev. 1989 Aug;3(8):1217–1225. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Harashima S., Hinnebusch A. G. A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc Natl Acad Sci U S A. 1987 May;84(9):2863–2867. doi: 10.1073/pnas.84.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Jackson B. M., Miller P. F., Hinnebusch A. G. The first and fourth upstream open reading frames in GCN4 mRNA have similar initiation efficiencies but respond differently in translational control to change in length and sequence. Mol Cell Biol. 1988 Dec;8(12):5439–5447. doi: 10.1128/mcb.8.12.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Hijazi K. A., Göringer H. U., Dahlberg A. E. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain V. M. Initiation of protein synthesis in mammalian cells. Biochem J. 1986 May 1;235(3):625–637. doi: 10.1042/bj2350625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985 Mar;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Persson B. C. Modification of tRNA as a regulatory device. Mol Microbiol. 1993 Jun;8(6):1011–1016. doi: 10.1111/j.1365-2958.1993.tb01645.x. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Karpen J. W., Kaji A. Further characterization of ribosome releasing factor and evidence that it prevents ribosomes from reading through a termination codon. J Biol Chem. 1981 Jun 10;256(11):5798–5801. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiss M. R., Degnin C. R., Geballe A. P. Translational control of human cytomegalovirus gp48 expression. J Virol. 1991 Dec;65(12):6782–6789. doi: 10.1128/jvi.65.12.6782-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Valle R. P., Morch M. D. Stop making sense: or Regulation at the level of termination in eukaryotic protein synthesis. FEBS Lett. 1988 Aug 1;235(1-2):1–15. doi: 10.1016/0014-5793(88)81225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G. B. Translational control of protein synthesis by tRNA unrelated to changes in tRNA concentration. J Mol Evol. 1973;2(2-3):199–204. doi: 10.1007/BF01654000. [DOI] [PubMed] [Google Scholar]
- Werner M., Feller A., Messenguy F., Piérard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987 Jun 19;49(6):805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]
- Wolin S. L., Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988 Nov;7(11):3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Boeke J. D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H., Donahue T. F. Control of translation initiation in Saccharomyces cerevisiae. Mol Microbiol. 1992 Jun;6(11):1413–1419. doi: 10.1111/j.1365-2958.1992.tb00861.x. [DOI] [PubMed] [Google Scholar]






