Abstract
Interleukin-10 (IL-10) is a multifunctional cytokine which participates in the development and progression of various malignant tumors. To date, a number of case–control studies were conducted to detect the association between IL-10-592C>A polymorphism and cancer risk in humans. However, the results of these studies on the association remain conflicting. In an effort to solve this controversy, we performed a meta-analysis based on 70 case–control studies from 65 articles, including 16 785 cancer cases and 19 713 controls. We used odds ratios (ORs) with 95% confidence intervals (CIs) to assess the strength of the association. The overall results suggested that the variant homozygote genotype AA of the IL-10-592C>A polymorphism was associated with a moderately decreased risk of all cancer types (OR = 0.90, 95% CI = 0.83–0.98 for homozygote comparison, OR = 0.92, 95% CI = 0.86–0.98 for recessive model). In the stratified analyses, the risk remained for studies of smoking-related cancer, Asian populations and hospital-based studies. These results suggested that the IL-10-592C>A polymorphism might contribute to the cancer susceptibility, especially in smoking-related cancer, Asians and hospital-based studies. Further studies are needed to confirm the relationship.
Introduction
Cancer is a major public health problem in the world. Evidences support an important role for genetics in determining risk for cancer. Association studies are appropriate for searching susceptibility genes involved in cancer [1].
Interleukin-10 (IL-10) is a multifunctional cytokine which participate in the development and progression of various malignant tumors [2]. It has anti-inflammatory and immunosuppressive activities including the ability to downregulate the expression of macrophage costimulatory molecule. The impact of IL-10 on macrophage function appears to play a role in the growth of blood vessel, as studies showed that IL-10 might contribute to the regulation of angiogenesis in many kinds of tumors [3], [4]. As an immunosuppressive molecule allowing tumor to escape from immune surveillance, IL-10 might act as a potential tumor promoter which results in a more aggressive behavior of malignant cells. Conversely, due to its immune-stimulating and anti-angiogenic properties, IL-10 is supposed to prevent or reduce the growth and distant spread of tumor [5]–[7].It has been indicated that IL-10 overexpression as well as deficiency was found under different pathophysiological conditions depending on the cancers analyzed [8].
IL-10 is encoded by a gene located on chromosome 1 (1q31–1q32) [9]. The IL-10 promoter is highly polymorphic and three important single nucleotide polymorphisms (SNPs) in the promoter region which influence the transcription of IL-10 messenger RNA and the expression of IL-10 in vitro are rs1800896(-1082A>G), rs1800871 (-819C>T), and rs1800872(-592C>A) [10], [11]. It has been reported that the -1082A>G and haplotype (-1082_-819_-592) were associated with differential production of the protein in stimulated cells, with the ATA haplotype leading to decreased IL-10 expression and the GCC haplotype increased IL-10 expression [12].
To date, a number of case-control studies were conducted to investigate the association between IL-10 -592 C>A and cancer risk in humans [13]–[77]. However, the results of these studies remain conflicting rather than conclusive. So, we performed the present meta-analysis to evaluate the association between IL-10 -592 C>A and cancer risk.
Materials and Methods
Identification and Eligibility of Relevant Studies
We searched the electronic literature from Pubmed for all relevant reports (the last search update was Oct 16, 2012), using the key words: (“interleukin-10” or “IL-10” or “IL10”) and (“variant” or “variation” or “polymorphism”) and (“cancer” or “tumor” or “carcinoma” or “malignancy”). The search was limited to English language papers. In addition, studies were identified by a manual search of the reference lists of reviews and retrieved studies. Studies were selected if there was available data for the IL-10 -592C>A polymorphism with cancer risk in a case-control design including retrospective or prospective and nested case-control studies. As studies with the same population by different investigators or overlapping data by the same authors were found, the most recent or complete articles with the largest numbers of subjects were included. Studies included in our meta-analysis have to meet the following criteria: (i) evaluation of the IL-10 -592C>A polymorphism and cancer risk, (ii ) use a case-control design (retrospective or prospective and nested case-control) and (iii ) contain available genotype frequency. Major reasons for exclusion of studies were (i ) only case population and (ii ) duplicate of previous publication.
Data Extraction
Two of the authors extracted all data independently complying with the selection criteria and reached a consensus on all items. In the present study, the following characteristics were collected: the first author’s last name, year of publication, country of origin, ethnicity, frequencies of genotyped in cases and controls, source of control groups (population- or hospital-based controls) and cancer type. For studies including subjects of different ethnic groups, data were extracted separately for each ethnic group whenever possible [43]. Different ethnic descents were categorized as European, Asian, or African or mixed (composed of an admixture of different ethnic groups). Meanwhile, studies investigating more than one kind of cancer were counted as individual data sets only in subgroup analyses by cancer type [15], [36], [39], [59]. One study which was duplicate of previous publication were excluded from the analysis [78].
Statistical Analysis
The strength of the association between the IL-10 -592C>A polymorphism and cancer risk was measured by odds ratios (ORs) with 95% confidence intervals (CIs). The statistical significance of the pooled OR was determined using the Z-test. Pooled estimates of the OR were obtained by calculating a weighted average of OR from each study [79]. First, we estimated cancer risks with the CA and AA genotypes, compared with the wild-type CC homozygote, and then evaluated the risks associated with CA/AA versus CC and AA versus CC/CA, assuming the dominant and recessive effects of the variant A allele, respectively. Stratified analyses were also performed by cancer types (if one cancer type contained less than three individual studies, it was combined into ‘other cancers’ group), ethnicity and source of controls. In consideration of the possibility of heterogeneity across the studies, I 2 was applied to assess heterogeneity between studies [80]. Values from single study were combined using models of both fixed effects and random effects [81]. We used a fixed effects model when I 2 was equal or less than 50%, and a random effects model when I 2 was greater than 50%. In the absence of heterogeneity, the two methods provide identical results, because the fixed effects model, using the Mantel–Haenszel’s method, assumes that studies are sampled from populations with the same effect size, making an adjustment to the study weights according to the in-study variance; whereas the random-effects model using the DerSimonian and Laird’s method assumes that studies are taken from populations with varying effect sizes, calculating the study weights both from in-study and between-study variances, considering the extent of variation or heterogeneity. Sensitivity analyses were performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled OR. Funnel plots and Egger’s linear regression test were used to provide diagnosis of the potential publication bias [82]. All analyses were conducted using Stata software (version8.0; StataCorp LP, College Station, TX), and all tests were two sided.
Results
Characteristics of Studies
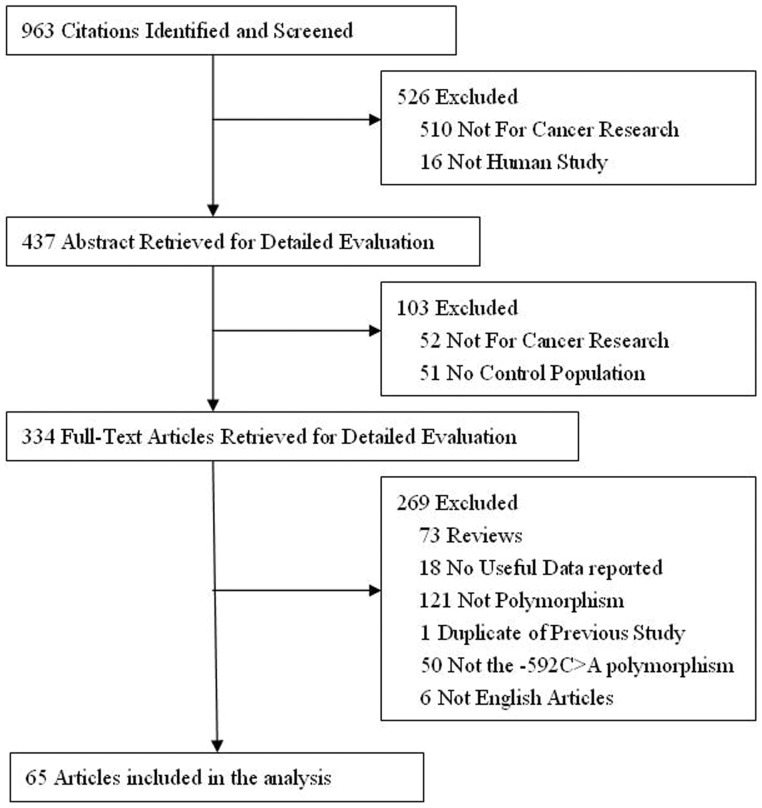
Finally, a total of 70 studies from 65 articles that included a total of 16 785 cancer cases and 19 713 controls met the inclusion criteria (Fig. 1). Study characteristics are summarized in Table 1. For the IL-10-592C>A polymorphism, there were 2 studies of African descendents, 20 studies of Asian descendents and 37 studies of European descendents. In our meta-analysis, most of the cancer types were gastric cancer. Cancers were confirmed histologically or pathologically in most studies. The distribution of genotype in the controls of the studies was in agreement with Hardy-Weinberg equilibrium for all except five studies [14], [20], [21], [27], [72], which were further tested in the sensitivity analyses.
Figure 1. Studies identified with criteria for inclusion and exclusion.
Table 1. Characteristics of studies included in the meta-analysis.
| First author | Year | Cancer type | Country | Ethnicity | Source of control groups | Cases/controls | P of HWE |
| Howell | 2001 | Melanoma | UK | European | HB | 165/158 | 0.69 |
| MartíNez-Escribano | 2002 | Melanoma | Spain | European | HB | 42/48 | 0.58 |
| Roh | 2002 | Cervical Cancer | Korea | Asian | HB | 144/179 | 0.72 |
| El–Omar | 2003 | Esophageal Cancer | USA | Mixed | PB | 161/210 | 0.43 |
| El–Omar | 2003 | Gastric Cancer | USA | Mixed | PB | 314/210 | 0.43 |
| Heneghan | 2003 | Hepatocellular Carcinoma | China | Asian | HB | 98/97 | 0.02 |
| Munro | 2003 | Hodgkin Lymphoma | UK | European | HB | 147/110 | 0.11 |
| Wu | 2003 | Gastric Cancer | Taiwan | Asian | HB | 220/230 | 0.23 |
| Basturk | 2004 | Renal Cell Cancer | Turkey | Mixed | HB | 29/50 | 0.32 |
| Maranda | 2004 | Diffuse Large B-Cell Lymphoma | France | European | HB | 199/112 | 0.54 |
| Savage | 2004 | Esophageal Cancer | China | Asian | PB | 119/386 | 0.38 |
| Savage | 2004 | Gastric Cancer | China | Asian | PB | 84/386 | 0.38 |
| Zhang | 2004 | Basal Cell Carcinoma | China | European | HB | 241/260 | 0.32 |
| Alonso | 2005 | Melanoma | Spain | European | HB | 98/100 | 0.21 |
| Alpízar-Alpízar | 2005 | Gastric Cancer | Costa Rica | Mixed | HB | 45/45 | 0.01 |
| Guzowski | 2005 | Breast Cancer | USA | Mixed | HB | 50/25 | 0.71 |
| Guzowski | 2005 | Leukemia | USA | Mixed | HB | 17/25 | 0.71 |
| Langsenlehner | 2005 | Breast Cancer | Australia | European | PB | 500/496 | 0.82 |
| Lee | 2005 | Gastric Cancer | Korea | Asian | HB | 122/120 | 0.06 |
| Macarthur | 2005 | Colorectal Cancer | Scotland | European | PB | 258/403 | 0.46 |
| Mazur | 2005 | Myeloma | Poland | European | HB | 54/50 | 0.22 |
| Shih | 2005 | Non-small Cell Lung Cancer | Taiwan | Asian | HB | 154/205 | 0.91 |
| Tseng | 2005 | Hepatocellular Carcinoma | Taiwan | Asian | HB | 208/184 | 0.57 |
| Zambon | 2005 | Gastric Cancer | Italy | European | HB | 129/644 | 0.70 |
| Zoodsma | 2005 | Cervical Cancer | Netherlands | European | PB | 654/606 | 0.21 |
| Braicu | 2006 | Ovarian Cancer | Germany | European | HB | 147/129 | 0.90 |
| Crivello | 2006 | Colorectal Cancer | Italy | European | HB | 62/124 | 0.72 |
| Kamangar | 2006 | Gastric Cancer | Filand | European | PB | 112/208 | 0.78 |
| Lan | 2006 | Non-Hodgkin’s Lymphoma | USA | European | PB | 482/563 | 0.03 |
| Pratesi | 2006 | Nasopharyngeal Carcinoma | Italy | European | PB | 89/130 | 0.27 |
| Purdue | 2006 | Non-Hodgkin’s Lymphoma | Australia | European | PB | 540/489 | 0.87 |
| Scola | 2006 | Breast Cancer | Italy | European | HB | 84/106 | 0.07 |
| Sicinschi | 2006 | Gastric Cancer | Mexico | European | HB | 181/369 | 0.38 |
| Sugimoto | 2006 | Gastric Cancer | Japan | Asian | HB | 105/168 | 0.42 |
| Cozar | 2007 | Colon Cancer | Spain | European | HB | 95/175 | 0.39 |
| Cozar | 2007 | Renal Cell Cancer | Spain | European | HB | 127/175 | 0.39 |
| Eder | 2007 | Prostate Cancer | Australia | European | PB | 547/545 | 0.44 |
| Garcia-Gonzalez | 2007 | Gastric Cancer | Spain | European | HB | 404/404 | 0.08 |
| Gonullu | 2007 | Breast Cancer | Turkey | Mixed | HB | 38/24 | 0.59 |
| Ivansson | 2007 | Cervical Cancer | Sweden | European | HB | 1282/288 | 0.33 |
| Purdue | 2007 | Testicular Germ Cell Tumors | USA | European | PB | 505/604 | 0.28 |
| Vogel | 2007 | Basal Cell Carcinoma | Denmark | European | PB | 304/315 | 0.92 |
| Wei | 2007 | Nasopharyngeal Carcinoma | China | Asian | HB | 198/210 | 0.84 |
| Cacev | 2008 | Colon Cancer | Croatia | European | PB | 160/160 | 0.40 |
| Colakogullari | 2008 | Lung Cancer | Turkey | Mixed | HB | 44/59 | 0.74 |
| Crusius | 2008 | Gastric Cancer | Netherlands | European | PB | 237/1122 | 0.05 |
| Erdogan | 2008 | Thyroid Cancer | Turkey | Mixed | HB | 42/113 | 0.81 |
| Faupel-Badger | 2008 | Prostate Cancer | Filand | European | PB | 511/386 | 0.54 |
| Kube | 2008 | Non-Hodgkin’s Lymphoma | Germany | European | HB | 500/236 | 0.35 |
| Vogel | 2008 | Lung Cancer | Denmark | European | PB | 403/744 | 0.34 |
| Yao | 2008 | Oral Cancer | China | Asian | HB | 280/300 | 0.81 |
| Zabaleta | 2008 | Prostate Cancer | USA | African | HB | 67/128 | 0.19 |
| Zabaleta | 2008 | Prostate Cancer | USA | European | HB | 479/401 | 0.44 |
| Ando | 2009 | Gastric Cancer | Japan | Asian | HB | 330/190 | 0.06 |
| Kang | 2009 | Gastric Cancer | Korea | Asian | HB | 333/332 | 0.59 |
| Schoof | 2009 | Melanoma | Germany | European | HB | 164/162 | 1.00 |
| Tsilidis | 2009 | Colorectal Cancer | USA | Mixed | PB | 203/361 | 0.58 |
| Wang | 2009 | Prostate Cancer | USA | Mixed | PB | 255/255 | 0.64 |
| Hart | 2010 | Non-small Cell Lung Cancer | Norway | European | PB | 434/433 | 0.01 |
| Kong | 2010 | Breast Cancer | China | Asian | HB | 315/322 | 0.01 |
| Liu | 2010 | Prostate Cancer | China | Asian | HB | 262/270 | 0.48 |
| Vancleave | 2010 | Prostate Cancer | USA | African | HB | 189/651 | 0.06 |
| Li | 2011 | Hepatocellular Carcinoma | China | Asian | PB | 150/347 | − |
| Liu | 2011 | Gastric Cancer | China | Asian | HB | 234/243 | 0.77 |
| Shekari | 2011 | Cervical Cancer | India | European | HB | 200/200 | 0.05 |
| Zeng | 2011 | Gastric Cancer | China | Asian | HB | 151/153 | 0.15 |
| Andersen | 2012 | Colorectal Cancer | Denmark | European | PB | 378/775 | 0.33 |
| He | 2012 | Gastric Cancer | China | Asian | HB | 196/248 | 0.10 |
| Pooja | 2012 | Breast Cancer | India | European | HB | 200/200 | 0.08 |
| Zhang | 2012 | Non-Hodgkin’s Lymphoma | China | Asian | PB | 514/557 | 0.87 |
HB, hospital based; HWE: Hardy–Weinberg Equilibrium; PB, population based.
Quantitative Synthesis
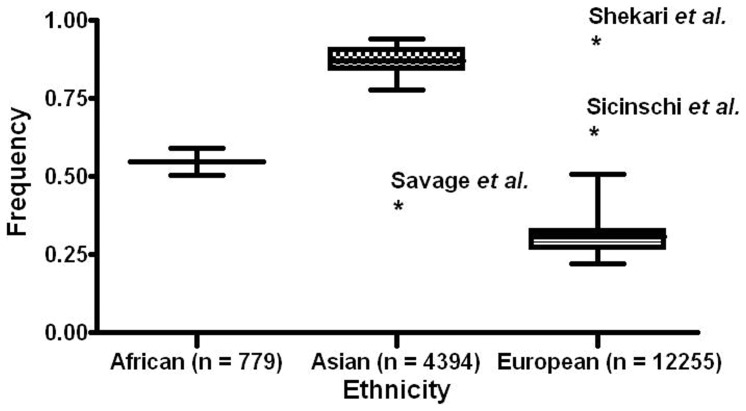
There was a wide variation in the A allele frequency of the polymorphism among the controls across different ethnicities. For Asian populations, the IL-10-592C>A A allele frequency was 0.87 (95% CI = 0.85–0.90), which was significantly higher than that in European populations (0.31, 95% CI = 0.29–0.32, P<0.001) (Fig. 2). Overall, individuals with AA genotype had a 0.90 fold lower cancer risk compared with the CC genotype (OR = 0.90, 95% CI = 0.83–0.98, I 2 = 18.00%). In addition, significant main effects was also observed in a recessive model (OR = 0.92, 95% CI = 0.86–0.98, I 2 = 25.80%). In a stratified analysis by specific cancer type, we also found decreased risk among studies of smoking-related cancer (OR = 0.77, 95% CI = 0.62–0.96 for AA versus CC, I 2 = 15.80% for heterogeneity; OR = 0.87, 95% CI = 0.76–0.99 for CA/AA versus CC, I 2 = 0.00% for heterogeneity). According to ethnicity, significantly decreased risks were also found among the Asian population (OR = 0.79, 95% CI = 0.69–0.91 for AA versus CC, I 2 = 13.30% for heterogeneity; OR = 0.85, 95% CI = 0.75–0.97 for CA/AA versus CC, I 2 = 0.00% for heterogeneity; OR = 0.87, 95% CI = 0.80–0.95 for AA versus CC/CA, I 2 = 34.40% for heterogeneity). In the stratified analysis by source of control groups, we found that the variant genotypes were associated with a significantly decreased risk in hospital-based controls in all genetic model (OR = 0.92, 95% CI = 0.85–0.99 for CA versus CC, I 2 = 0.00% for heterogeneity; OR = 0.86, 95% CI = 0.77–0.96 for AA versus CC, I 2 = 9.30% for heterogeneity; OR = 0.91, 95% CI = 0.85–0.98 for CA/AA versus CC, I 2 = 0.00% for heterogeneity; OR = 0.91, 95% CI = 0.84–0.98 for AA versus CC/CA, I 2 = 26.60% for heterogeneity). However, no significant associations were found for European and African populations. According to source of controls, no significant associations were observed in population-based studies (Table 2).
Figure 2. Frequencies of the variant alleles among controls stratified by ethnicity.
Asterisks represent outliers.
Table 2. Stratified analyses of the IL-10 -592C>A polymorphism on cancer risk.
| Variables | n a | Cases/Controls | CA versus CC | AA versus CC | CA/AA versus CC(dominant) | AA versus CC/CA(recessive) | ||||
| OR (95% CI) | I 2 (%) | OR (95% CI) | I 2 (%) | OR (95% CI) | I 2 (%) | OR (95% CI) | I 2 (%) | |||
| Total | 70 | 16785/19713 | 0.98 (0.93–1.03) | 0.00 | 0.90 (0.83–0.98) | 18.00 | 0.97 (0.92–1.01) | 0.80 | 0.92 (0.86–0.98) | 25.80 |
| Cancer types | ||||||||||
| Breast cancer | 6 | 1187/1173 | 0.94 (0.78–1.12) | 0.00 | 0.77 (0.59–1.02) | 0.00 | 0.90 (0.76–1.07) | 0.00 | 0.83 (0.66–1.05) | 8.60 |
| Cervical cancer | 4 | 2280/1273 | 1.12 (0.95–1.33) | 39.40 | 1.22 (0.88–1.68) | 0.00 | 1.15 (0.97–1.35) | 19.60 | 1.19 (0.94–1.50) | 0.00 |
| Colorectal cancer | 6 | 1156/1998 | 1.07 (0.92–1.25) | 19.40 | 0.97 (0.56–1.68)b | 50.50 | 1.06 (0.92–1.23) | 27.50 | 0.93 (0.54–1.60)b | 50.30 |
| Gastric cancer | 16 | 3197/5072 | 0.95 (0.84–1.07) | 0.00 | 0.91 (0.77–1.08) | 42.90 | 0.94 (0.83–1.05) | 4.20 | 0.91 (0.76–1.10)b | 56.60 |
| Lung cancer | 4 | 1035/1441 | 0.94 (0.78–1.12) | 44.40 | 0.69 (0.34–1.38)b | 64.60 | 0.91 (0.77–1.09) | 28.20 | 0.74 (0.39–1.39)b | 69.60 |
| Hepatocellularcarcinoma | 3 | 456/628 | 0.88 (0.52–1.50) | 44.50 | 0.76 (0.45–1.27) | 0.00 | 0.85 (0.57–1.24) | 0.00 | 0.83 (0.60–1.15) | 0.00 |
| Melanoma | 4 | 469/468 | 1.09 (0.83–1.43) | 0.00 | 1.18 (0.71–1.98) | 44.70 | 1.11 (0.86–1.43) | 0.00 | 1.14 (0.69–1.88) | 48.60 |
| Non-HodgkinLymphoma | 5 | 2235/1957 | 0.95 (0.82–1.10) | 0.00 | 0.79 (0.61–1.00) | 0.00 | 0.92 (0.80–1.06) | 0.00 | 0.83 (0.69–1.00) | 0.00 |
| Prostate cancer | 7 | 2310/2636 | 1.01 (0.89–1.15) | 36.60 | 1.04 (0.84–1.30) | 0.00 | 1.03 (0.91–1.16) | 43.20 | 1.02 (0.85–1.24) | 0.00 |
| Other cancer | 15 | 2460/3067 | 0.89 (0.79–1.01) | 0.00 | 0.85 (0.69–1.04) | 0.00 | 0.89 (0.80–1.00) | 0.00 | 0.93 (0.78–1.11) | 0.00 |
| Smoking-relatedcancer | 11 | 2038/2902 | 0.88 (0.77–1.00) | 6.70 | 0.77 (0.62–0.96) | 15.80 | 0.87 (0.76–0.99) | 0.00 | 0.85 (0.71–1.00) | 24.20 |
| Ethnicity | ||||||||||
| African | 2 | 256/779 | 0.98 (0.72–1.34) | 0.00 | 0.88 (0.58–1.35) | 0.00 | 0.96 (0.71–1.28) | 0.00 | 0.89 (0.60–1.32) | 0.00 |
| Asian | 20 | 4217/5127 | 0.90 (0.78–1.03) | 0.00 | 0.79 (0.69–0.91) | 13.30 | 0.85 (0.75–0.97) | 0.00 | 0.87 (0.80–0.95) | 34.40 |
| European | 37 | 11114/12430 | 0.99 (0.94–1.05) | 12.30 | 0.96 (0.86–1.08) | 26.50 | 0.99 (0.94–1.05) | 12.20 | 0.98 (0.88–1.09) | 30.00 |
| Mixed | 11 | 1198/1377 | 0.97 (0.82–1.15) | 0.00 | 1.04 (0.75–1.45) | 0.00 | 0.98 (0.84–1.16) | 0.00 | 1.05 (0.76–1.45) | 0.00 |
| Source of controls | ||||||||||
| Hospital based | 46 | 8871/9022 | 0.92 (0.85–0.99) | 0.00 | 0.86 (0.77–0.96) | 9.30 | 0.91 (0.85–0.98) | 0.00 | 0.91 (0.84–0.98) | 26.60 |
| Population based | 24 | 7914/10691 | 1.02 (0.96–1.09) | 19.20 | 0.96 (0.84–1.09) | 31.30 | 1.01 (0.95–1.08) | 27.20 | 0.94 (0.84–1.06) | 27.00 |
Number of comparisons.
Random-effects estimate.
Smoking-related cancers: lung, esophageal, nasopharyngeal, oral and renal cell cancers.
Bold values indicate significant difference.
Test for Heterogeneity
In the sub-analysis of gastric cancer, there was significant heterogeneity for recessive model comparison (AA versus CC/CA: I 2 = 56.60% for heterogeneity). Then, we assessed the source of heterogeneity for recessive model comparison (AA versus CC/CA) by ethnicity and source of controls. As a result, neither ethnicity (χ2 = 5.06, df = 2, P = 0.08) nor source of controls (χ2 = 0.01, df = 1, P = 0.91) was found to contribute to substantial heterogeneity.
Sensitivity Analyses
Sensitivity analyses indicated that two independent studies by Wu et al. [50] was the main origin of heterogeneity in the sub-analysis of gastric cancer. The heterogeneity was effectively decreased or removed by exclusion of the study (AA versus CC/CA: I 2 = 46.90%). Although the genotype distribution in five studies did not follow Hardy-Weinberg equilibrium, the corresponding pooled ORs were not materially altered by including the studies. In addition, no other single study influenced the pooled OR qualitatively, as indicated by sensitivity analyses, suggesting that the results of this meta-analysis are stable.
Publication Bias
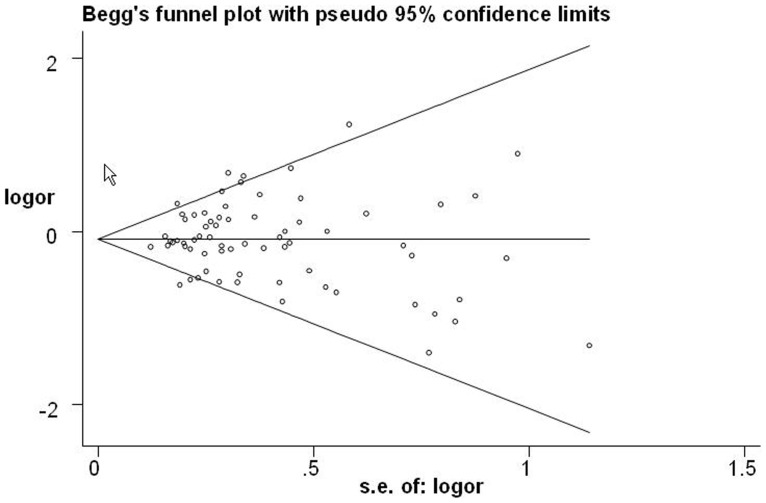
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of literatures. As shown in Fig. 3, the shapes of the funnel plots did not reveal any evidence of obvious asymmetry in all comparison models. Thus, Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not show any evidence of publication bias (t = −1.38, P = 0.172 for CA versus CC; t = −0.86, P = 0.390 for AA versus CC; t = −1.99, P = 0.051 for AA/CA versus CC; t = −0.12, P = 0.903 for AA versus CC/CA).
Figure 3. Begg’s funnel plot for publication bias test (AA vs. CC/CA).
Each point represents a separate study for the indicated association. Log [OR], natural logarithm of odds ratio. Horizontal line, mean effect size.
Discussion
The present meta-analysis investigated the association between the IL-10-592C>A polymorphisms and cancer risk, based on 70 published case–control studies from 65 articles. The results provided evidence that the IL-10-592C>A polymorphism was associated with a significant decrease in overall cancer risk. The variant homozygote genotype AA of the IL-10-592C>A polymorphism, was associated with a modest but significantly decreased risk in homozygote comparison and recessive model. In the stratified analysis, the risk remained for studies of smoking-related cancer, Asian populations and hospital-based studies. Given the important roles of IL-10 in carcinogenesis, it is biological plausible that IL-10 polymorphism might modulate the risk of cancer.
A common [ATA] haplotype is formed by polymorphisms at positions -1082,-819 and -592 in the promoter of IL-10 gene [83], [84]. It has been reported that the -592 A variant, the -1082 A variant as well as the [ATA] haplotype was associated with lower IL-10 expression [51], [85], [86]. Thus, the -592 A variant can be regarded as a low-producer allele of the IL-10 gene. Research has showed that increased serum IL-10 levels could facilitate development of tumors by suppressing the expression of MHC class I and II antigens [87] and preventing tumor antigen presentation to CD8-cytotic T lymphocytes. It has been revealed that the homozygous IL-10-592AA genotype, indicating homozygosity for the [ATA] haplotype, was protective against breast cancer [51]. Similarly, elevated serum levels of IL-10 were found in non-small cell lung cancer patients; moreover, IL-10 serum levels were shown to be higher in patients with metastatic disease when compared to those with undisseminated cancer [88]. Consistently, we also found that individuals with the AA genotype which exhibits low production of IL-10 were associated with a lower cancer risk than participants with the CC genotype in our meta-analysis.
Tobacco smoking is a well-established risk factor for cancers of many organs, including lung, esophagus, oral cavity, pharyngeal and kidney [89]–[92]. Tobacco use has been proven to affect the immune system and influence the production of IL-10 [93]. Also, studies showed that smokers have impaired T lymphocyte suppressor cell function and decreased natural killer cell activity compared with non-smokers. Furthermore, IL10 might protect tumors by inhibiting cytotoxic T lymphocyte (CTL)-mediated tumor-specific cell lysis [87], [94]. Further studies are needed to investigate the relationship.
Our results indicated that the AA variant genotype was associated with decreased risk in smoking-related cancer but not for breast cancer, cervical cancer, colorectal cancer, esophageal cancer, gastric cancer, melanoma, lung cancer, hepatocellular carcinoma, Non-Hodgkin lymphoma or prostate cancer. One possible explanation for the discrepancy is that carcinogenic mechanism underlying the etiology may differ by different tumor sites and that the IL-10 genetic variants may play a different role in different cancers. Even at the same tumor site, considering the relatively small sample size in some studies and the possible small effect size of this genetic polymorphism to cancer, the discrepancy will become apparent since some of these studies may have insufficient statistical power to detect a small but real association. For example, there were only three studies included in the analysis with limited sample size for esophageal cancer, with 456 cases and 628 controls, so the results may be capricious and should be interpreted with caution.
In the subgroup analysis by ethnicity, we found an evidence for the association between the IL-10-592C>A polymorphism and cancer risk among Asians but not among Europeans or Africans, suggesting a possible role of ethnic differences in genetic backgrounds and the environment they lived in (Table 2). Several concerns may account for it. First, nine of the 20 Asian studies investigated gastric cancer (weighted 36.68% and 42.24% in comparison of AA versus CC and AA versus CC/CA), whereas only five out of the 37 studies focused on gastric cancer in the European population (weighted 18.02% and 18.48% in comparison of AA versus CC and AA versus CC/CA). Second, the prevalence of variant allele of IL-10-592 C>A polymorphism among the controls varies markedly with ethnicity. For Asian populations, the IL-10-592C>A A allele frequency was 0.87 (95% CI = 0.85–0.90), which was significantly higher than that in European populations (0.31, 95% CI = 0.29–0.32, P<0.001). Other factors such as different matching criteria, selection bias and adjustment in the statistical analyses, misclassification on disease status and genotyping method might also play a role. In addition, there are only two reported studies and limited number of patients was available for African, which limited us to detect stable effects in this population. Additional studies are warranted to further validate ethnic difference in the effect of the IL-10-592C>A polymorphism on cancer risk, especially in Africans.
When stratifying the source of controls, a moderate strength was observed in hospital-based controls, but not the population-based controls. The discrepancy may result from a differential effect of selection criteria in different cancers, as well as the weight of each study, which was dictated by sample size in the meta-analysis. Another reason may be that the hospital-based studies have some inherent selection biases as such controls may just represent a sample of ill-defined reference population and may not be very representative of the study population or the general population, particularly when the genotypes under investigation were associated with the disease conditions that the hospital-based controls may have.
One of the most important goals of the meta-analysis is to identify the source of heterogeneity. In the sub-analysis of gastric cancer, there was significant heterogeneity for recessive model comparison. Thus, we stratified the studies on gastric cancer according to ethnicity and source of controls. Through analysis, it was found that neither ethnicity nor source of controls contribute to substantial heterogeneity. It is possible that other unmeasured characteristics in different study populations and/or inherited limitations of the recruited studies may partially contribute to the observed heterogeneity.
The meta-analysis has some strengths and limitations. This article is potentially limited in several ways. First, our result was based on unadjusted estimates, while a more precise analysis should be conducted adjusted by other factors such as age, sex, smoking and drinking status. Lack of information for data analysis might cause serious confounding bias. In addition, lacking the original data of the included studies limited our further evaluation of the potential interactions because gene–gene, gene–environment interactions and even different polymorphic loci of the same gene might modulate cancer risk. Second, some of the studies had a very small sample size and did not have adequate power to detect the possible risk for IL-10-592C>A polymorphism and the observed significant ORs in some studies of small sample size may be false association. Third, misclassifications on genotypes and disease status may influence the results because the quality control of genotyping was not well documented in some studies and cases in several studies were also not confirmed by pathology or other gold standard methods. Nonetheless, advantages in our meta-analysis should also be acknowledged. First, the statistical power of the analysis was greatly increased, because a substantial number of cases and controls were pooled from different studies. Second, the quality of case–control studies included in this meta-analysis was satisfactory according to our selection criteria. Third, we did not detect any publication bias indicating that the whole pooled result should be unbiased.
In conclusion, our meta-analysis suggests that the IL-10-592C>A polymorphism was associated with a significant decrease in overall cancer risk, especially in smoking-related cancer, Asians and hospital-based studies. However, large studies using standardized unbiased genotyping methods, enrolling precisely defined cancer patients and well-matched controls, especially in African populations, with more detailed individual and environmental data are warranted to validate the results of our meta-analysis.
Funding Statement
These authors have no support or funding to report.
References
- 1. Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- 2. Mocellin S, Marincola F, Rossi CR, Nitti D, Lise M (2004) The multifaceted relationship between IL-10 and adaptive immunity: putting together the pieces of a puzzle. Cytokine Growth Factor Rev 15: 61–76. [DOI] [PubMed] [Google Scholar]
- 3. Huang S, Ullrich SE, Bar-Eli M (1999) Regulation of tumor growth and metastasis by interleukin-10: the melanoma experience. J Interferon Cytokine Res 19: 697–703. [DOI] [PubMed] [Google Scholar]
- 4. Fortis C, Foppoli M, Gianotti L, Galli L, Citterio G, et al. (1996) Increased interleukin-10 serum levels in patients with solid tumours. Cancer Lett 104: 1–5. [DOI] [PubMed] [Google Scholar]
- 5. Mocellin S, Panelli MC, Wang E, Nagorsen D, Marincola FM (2003) The dual role of IL-10. Trends Immunol 24: 36–43. [DOI] [PubMed] [Google Scholar]
- 6. Sharma N, Luo J, Kirschmann DA, O’Malley Y, Robbins ME, et al. (1999) A novel immunological model for the study of prostate cancer. Cancer Res 59: 2271–2276. [PubMed] [Google Scholar]
- 7. Stearns ME, Wang M, Hu Y, Garcia FU, Rhim J (2003) Interleukin 10 blocks matrix metalloproteinase-2 and membrane type 1-matrix metalloproteinase synthesis in primary human prostate tumor lines. Clin Cancer Res 9: 1191–1199. [PubMed] [Google Scholar]
- 8. Asadullah K, Sterry W, Volk HD (2003) Interleukin-10 therapy–review of a new approach. Pharmacol Rev 55: 241–269. [DOI] [PubMed] [Google Scholar]
- 9. Spits H, de Waal Malefyt R (1992) Functional characterization of human IL-10. Int Arch Allergy Immunol 99: 8–15. [DOI] [PubMed] [Google Scholar]
- 10. Kingo K, Ratsep R, Koks S, Karelson M, Silm H, et al. (2005) Influence of genetic polymorphisms on interleukin-10 mRNA expression and psoriasis susceptibility. J Dermatol Sci 37: 111–113. [DOI] [PubMed] [Google Scholar]
- 11. Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, et al. (1997) An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 24: 1–8. [DOI] [PubMed] [Google Scholar]
- 12. Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, et al. (1998) Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci U S A 95: 9465–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Purdue MP, Sakoda LC, Graubard BI, Welch R, Chanock SJ, et al. (2007) A case-control investigation of immune function gene polymorphisms and risk of testicular germ cell tumors. Cancer Epidemiol Biomarkers Prev 16: 77–83. [DOI] [PubMed] [Google Scholar]
- 14. Hart K, Landvik NE, Lind H, Skaug V, Haugen A, et al. (2011) A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer 71: 123–129. [DOI] [PubMed] [Google Scholar]
- 15. Guzowski D, Chandrasekaran A, Gawel C, Palma J, Koenig J, et al. (2005) Analysis of single nucleotide polymorphisms in the promoter region of interleukin-10 by denaturing high-performance liquid chromatography. J Biomol Tech 16: 154–166. [PMC free article] [PubMed] [Google Scholar]
- 16. Gonullu G, Basturk B, Evrensel T, Oral B, Gozkaman A, et al. (2007) Association of breast cancer and cytokine gene polymorphism in Turkish women. Saudi Med J 28: 1728–1733. [PubMed] [Google Scholar]
- 17. Tsilidis KK, Helzlsouer KJ, Smith MW, Grinberg V, Hoffman-Bolton J, et al. (2009) Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes Control 20: 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J, Song B, Bai X, Liu W, Li Z, et al. (2010) Association of genetic polymorphisms in the interleukin-10 promoter with risk of prostate cancer in Chinese. BMC Cancer 10: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faupel-Badger JM, Kidd LC, Albanes D, Virtamo J, Woodson K, et al. (2008) Association of IL-10 polymorphisms with prostate cancer risk and grade of disease. Cancer Causes Control 19: 119–124. [DOI] [PubMed] [Google Scholar]
- 20. Alpizar-Alpizar W, Perez-Perez GI, Une C, Cuenca P, Sierra R (2005) Association of interleukin-1B and interleukin-1RN polymorphisms with gastric cancer in a high-risk population of Costa Rica. Clin Exp Med 5: 169–176. [DOI] [PubMed] [Google Scholar]
- 21. Kong F, Liu J, Liu Y, Song B, Wang H, et al. (2010) Association of interleukin-10 gene polymorphisms with breast cancer in a Chinese population. J Exp Clin Cancer Res 29: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee JY, Kim HY, Kim KH, Kim SM, Jang MK, et al. (2005) Association of polymorphism of IL-10 and TNF-A genes with gastric cancer in Korea. Cancer Lett 225: 207–214. [DOI] [PubMed] [Google Scholar]
- 23. Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, et al. (2009) Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate 69: 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tseng LH, Lin MT, Shau WY, Lin WC, Chang FY, et al. (2006) Correlation of interleukin-10 gene haplotype with hepatocellular carcinoma in Taiwan. Tissue Antigens 67: 127–133. [DOI] [PubMed] [Google Scholar]
- 25. Scola L, Vaglica M, Crivello A, Palmeri L, Forte GI, et al. (2006) Cytokine gene polymorphisms and breast cancer susceptibility. Ann N Y Acad Sci 1089: 104–109. [DOI] [PubMed] [Google Scholar]
- 26. Crusius JB, Canzian F, Capella G, Pena AS, Pera G, et al. (2008) Cytokine gene polymorphisms and the risk of adenocarcinoma of the stomach in the European prospective investigation into cancer and nutrition (EPIC-EURGAST). Ann Oncol 19: 1894–1902. [DOI] [PubMed] [Google Scholar]
- 27. Lan Q, Zheng T, Rothman N, Zhang Y, Wang SS, et al. (2006) Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood 107: 4101–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schoof N, von Bonin F, Konig IR, Mossner R, Kruger U, et al. (2009) Distal and proximal interleukin (IL)-10 promoter polymorphisms associated with risk of cutaneous melanoma development: a case–control study. Genes Immun 10: 586–590. [DOI] [PubMed] [Google Scholar]
- 29.Zeng X, Li Y, Liu T, Zhang J (2011) Diverse H. pylori strains, IL-10 promoter polymorphisms with high morbidity of gastric cancer in Hexi area of Gansu Province, China. Mol Cell Biochem. [DOI] [PubMed]
- 30.Shekari M, Kordi-Tamandani DM, Malekzadeh K, Sobti RC, Karimi S, et al.. (2011) Effect of Anti-inflammatory (IL-4, IL-10) Cytokine Genes in Relation to Risk of Cervical Carcinoma. Am J Clin Oncol. [DOI] [PubMed]
- 31. Kube D, Hua TD, von Bonin F, Schoof N, Zeynalova S, et al. (2008) Effect of interleukin-10 gene polymorphisms on clinical outcome of patients with aggressive non-Hodgkin’s lymphoma: an exploratory study. Clin Cancer Res 14: 3777–3784. [DOI] [PubMed] [Google Scholar]
- 32. Sugimoto M, Furuta T, Shirai N, Nakamura A, Kajimura M, et al. (2007) Effects of interleukin-10 gene polymorphism on the development of gastric cancer and peptic ulcer in Japanese subjects. J Gastroenterol Hepatol 22: 1443–1449. [DOI] [PubMed] [Google Scholar]
- 33. Sicinschi LA, Lopez-Carrillo L, Camargo MC, Correa P, Sierra RA, et al. (2006) Gastric cancer risk in a Mexican population: role of Helicobacter pylori CagA positive infection and polymorphisms in interleukin-1 and -10 genes. Int J Cancer 118: 649–657. [DOI] [PubMed] [Google Scholar]
- 34. Garcia-Gonzalez MA, Lanas A, Quintero E, Nicolas D, Parra-Blanco A, et al. (2007) Gastric cancer susceptibility is not linked to pro-and anti-inflammatory cytokine gene polymorphisms in whites: a Nationwide Multicenter Study in Spain. Am J Gastroenterol 102: 1878–1892. [DOI] [PubMed] [Google Scholar]
- 35. Yao JG, Gao LB, Liu YG, Li J, Pang GF (2008) Genetic variation in interleukin-10 gene and risk of oral cancer. Clin Chim Acta 388: 84–88. [DOI] [PubMed] [Google Scholar]
- 36. Cozar JM, Romero JM, Aptsiauri N, Vazquez F, Vilchez JR, et al. (2007) High incidence of CTLA-4 AA (CT60) polymorphism in renal cell cancer. Hum Immunol 68: 698–704. [DOI] [PubMed] [Google Scholar]
- 37. Mazur G, Bogunia-Kubik K, Wrobel T, Karabon L, Polak M, et al. (2005) IL-6 and IL-10 promoter gene polymorphisms do not associate with the susceptibility for multiple myeloma. Immunol Lett 96: 241–246. [DOI] [PubMed] [Google Scholar]
- 38. Howell WM, Turner SJ, Bateman AC, Theaker JM (2001) IL-10 promoter polymorphisms influence tumour development in cutaneous malignant melanoma. Genes Immun 2: 25–31. [DOI] [PubMed] [Google Scholar]
- 39. El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, et al. (2003) Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124: 1193–1201. [DOI] [PubMed] [Google Scholar]
- 40. Cacev T, Radosevic S, Krizanac S, Kapitanovic S (2008) Influence of interleukin-8 and interleukin-10 on sporadic colon cancer development and progression. Carcinogenesis 29: 1572–1580. [DOI] [PubMed] [Google Scholar]
- 41. Alonso R, Suarez A, Castro P, Lacave AJ, Gutierrez C (2005) Influence of interleukin-10 genetic polymorphism on survival rates in melanoma patients with advanced disease. Melanoma Res 15: 53–60. [DOI] [PubMed] [Google Scholar]
- 42. VanCleave TT, Moore JH, Benford ML, Brock GN, Kalbfleisch T, et al. (2010) Interaction among variant vascular endothelial growth factor (VEGF) and its receptor in relation to prostate cancer risk. Prostate 70: 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zabaleta J, Lin HY, Sierra RA, Hall MC, Clark PE, et al. (2008) Interactions of cytokine gene polymorphisms in prostate cancer risk. Carcinogenesis 29: 573–578. [DOI] [PubMed] [Google Scholar]
- 44. Eder T, Mayer R, Langsenlehner U, Renner W, Krippl P, et al. (2007) Interleukin-10 [ATA] promoter haplotype and prostate cancer risk: a population-based study. Eur J Cancer 43: 472–475. [DOI] [PubMed] [Google Scholar]
- 45. Zoodsma M, Nolte IM, Schipper M, Oosterom E, van der Steege G, et al. (2005) Interleukin-10 and Fas polymorphisms and susceptibility for (pre)neoplastic cervical disease. Int J Gynecol Cancer 15 Suppl 3282–290. [DOI] [PubMed] [Google Scholar]
- 46. Pratesi C, Bortolin MT, Bidoli E, Tedeschi R, Vaccher E, et al. (2006) Interleukin-10 and interleukin-18 promoter polymorphisms in an Italian cohort of patients with undifferentiated carcinoma of nasopharyngeal type. Cancer Immunol Immunother 55: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Erdogan M, Karadeniz M, Ozbek M, Ozgen AG, Berdeli A (2008) Interleukin-10 gene polymorphism in patients with papillary thyroid cancer in Turkish population. J Endocrinol Invest 31: 750–754. [DOI] [PubMed] [Google Scholar]
- 48. Wei YS, Kuang XH, Zhu YH, Liang WB, Yang ZH, et al. (2007) Interleukin-10 gene promoter polymorphisms and the risk of nasopharyngeal carcinoma. Tissue Antigens 70: 12–17. [DOI] [PubMed] [Google Scholar]
- 49. Lech-Maranda E, Baseggio L, Bienvenu J, Charlot C, Berger F, et al. (2004) Interleukin-10 gene promoter polymorphisms influence the clinical outcome of diffuse large B-cell lymphoma. Blood 103: 3529–3534. [DOI] [PubMed] [Google Scholar]
- 50. Wu MS, Wu CY, Chen CJ, Lin MT, Shun CT, et al. (2003) Interleukin-10 genotypes associate with the risk of gastric carcinoma in Taiwanese Chinese. Int J Cancer 104: 617–623. [DOI] [PubMed] [Google Scholar]
- 51. Langsenlehner U, Krippl P, Renner W, Yazdani-Biuki B, Eder T, et al. (2005) Interleukin-10 promoter polymorphism is associated with decreased breast cancer risk. Breast Cancer Res Treat 90: 113–115. [DOI] [PubMed] [Google Scholar]
- 52. Roh JW, Kim MH, Seo SS, Kim SH, Kim JW, et al. (2002) Interleukin-10 promoter polymorphisms and cervical cancer risk in Korean women. Cancer Lett 184: 57–63. [DOI] [PubMed] [Google Scholar]
- 53. Martinez-Escribano JA, Moya-Quiles MR, Muro M, Montes-Ares O, Hernandez-Caselles T, et al. (2002) Interleukin-10, interleukin-6 and interferon-gamma gene polymorphisms in melanoma patients. Melanoma Res 12: 465–469. [DOI] [PubMed] [Google Scholar]
- 54. Munro LR, Johnston PW, Marshall NA, Canning SJ, Hewitt SG, et al. (2003) Polymorphisms in the interleukin-10 and interferon gamma genes in Hodgkin lymphoma. Leuk Lymphoma 44: 2083–2088. [DOI] [PubMed] [Google Scholar]
- 55. Ioana Braicu E, Mustea A, Toliat MR, Pirvulescu C, Konsgen D, et al. (2007) Polymorphism of IL-1alpha, IL-1beta and IL-10 in patients with advanced ovarian cancer: results of a prospective study with 147 patients. Gynecol Oncol 104: 680–685. [DOI] [PubMed] [Google Scholar]
- 56. Vogel U, Christensen J, Wallin H, Friis S, Nexo BA, et al. (2008) Polymorphisms in genes involved in the inflammatory response and interaction with NSAID use or smoking in relation to lung cancer risk in a prospective study. Mutat Res 639: 89–100. [DOI] [PubMed] [Google Scholar]
- 57. Purdue MP, Lan Q, Kricker A, Grulich AE, Vajdic CM, et al. (2007) Polymorphisms in immune function genes and risk of non-Hodgkin lymphoma: findings from the New South Wales non-Hodgkin Lymphoma Study. Carcinogenesis 28: 704–712. [DOI] [PubMed] [Google Scholar]
- 58. Kamangar F, Abnet CC, Hutchinson AA, Newschaffer CJ, Helzlsouer K, et al. (2006) Polymorphisms in inflammation-related genes and risk of gastric cancer (Finland). Cancer Causes Control 17: 117–125. [DOI] [PubMed] [Google Scholar]
- 59. Savage SA, Abnet CC, Haque K, Mark SD, Qiao YL, et al. (2004) Polymorphisms in interleukin -2, -6, and -10 are not associated with gastric cardia or esophageal cancer in a high-risk chinese population. Cancer Epidemiol Biomarkers Prev 13: 1547–1549. [PubMed] [Google Scholar]
- 60. Liu J, Song B, Wang JL, Li ZJ, Li WH, et al. (2011) Polymorphisms of interleukin-10 promoter are not associated with prognosis of advanced gastric cancer. World J Gastroenterol 17: 1362–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zambon CF, Basso D, Navaglia F, Belluco C, Falda A, et al. (2005) Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine 29: 141–152. [DOI] [PubMed] [Google Scholar]
- 62. Crivello A, Giacalone A, Vaglica M, Scola L, Forte GI, et al. (2006) Regulatory cytokine gene polymorphisms and risk of colorectal carcinoma. Ann N Y Acad Sci 1089: 98–103. [DOI] [PubMed] [Google Scholar]
- 63. He B, Pan Y, Xu Y, Nie Z, Chen L, et al. (2012) Increased risk for gastric cancer in carriers of the lymphotoxin-alpha+252G variant infected by Helicobacter pylori. Genet Test Mol Biomarkers 16: 9–14. [DOI] [PubMed] [Google Scholar]
- 64. Ando T, Ishikawa T, Kato H, Yoshida N, Naito Y, et al. (2009) Synergistic effect of HLA class II loci and cytokine gene polymorphisms on the risk of gastric cancer in Japanese patients with Helicobacter pylori infection. Int J Cancer 125: 2595–2602. [DOI] [PubMed] [Google Scholar]
- 65. Kang JM, Kim N, Lee DH, Park JH, Lee MK, et al. (2009) The effects of genetic polymorphisms of IL-6, IL-8, and IL-10 on Helicobacter pylori-induced gastroduodenal diseases in Korea. J Clin Gastroenterol 43: 420–428. [DOI] [PubMed] [Google Scholar]
- 66. Shih CM, Lee YL, Chiou HL, Hsu WF, Chen WE, et al. (2005) The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer 50: 291–297. [DOI] [PubMed] [Google Scholar]
- 67. Colakogullari M, Ulukaya E, Yilmaztepe Oral A, Aymak F, Basturk B, et al. (2008) The involvement of IL-10, IL-6, IFN-gamma, TNF-alpha and TGF-beta gene polymorphisms among Turkish lung cancer patients. Cell Biochem Funct 26: 283–290. [DOI] [PubMed] [Google Scholar]
- 68. Macarthur M, Sharp L, Hold GL, Little J, El-Omar EM (2005) The role of cytokine gene polymorphisms in colorectal cancer and their interaction with aspirin use in the northeast of Scotland. Cancer Epidemiol Biomarkers Prev 14: 1613–1618. [DOI] [PubMed] [Google Scholar]
- 69. Zhang Z, Liu W, Jia X, Gao Y, Hemminki K, et al. (2004) Use of pyrosequencing to detect clinically relevant polymorphisms of genes in basal cell carcinoma. Clin Chim Acta 342: 137–143. [DOI] [PubMed] [Google Scholar]
- 70. Ivansson EL, Gustavsson IM, Magnusson JJ, Steiner LL, Magnusson PK, et al. (2007) Variants of chemokine receptor 2 and interleukin 4 receptor, but not interleukin 10 or Fas ligand, increase risk of cervical cancer. Int J Cancer 121: 2451–2457. [DOI] [PubMed] [Google Scholar]
- 71. Basturk B, Yavascaoglu I, Vuruskan H, Goral G, Oktay B, et al. (2005) Cytokine gene polymorphisms as potential risk and protective factors in renal cell carcinoma. Cytokine 30: 41–45. [DOI] [PubMed] [Google Scholar]
- 72. Heneghan MA, Johnson PJ, Clare M, Ho S, Harrison PM, et al. (2003) Frequency and nature of cytokine gene polymorphisms in hepatocellular carcinoma in Hong Kong Chinese. Int J Gastrointest Cancer 34: 19–26. [DOI] [PubMed] [Google Scholar]
- 73. Li Y, Chang SC, Goldstein BY, Scheider WL, Cai L, et al. (2011) Green tea consumption, inflammation and the risk of primary hepatocellular carcinoma in a Chinese population. Cancer Epidemiol 35: 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Andersen V, Egeberg R, Tjonneland A, Vogel U (2012) Interaction between interleukin-10 (IL-10) polymorphisms and dietary fibre in relation to risk of colorectal cancer in a Danish case-cohort study. BMC Cancer 12: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pooja S, Chaudhary P, Nayak LV, Rajender S, Saini KS, et al. (2012) Polymorphic variations in IL-1beta, IL-6 and IL-10 genes, their circulating serum levels and breast cancer risk in Indian women. Cytokine 60: 122–128. [DOI] [PubMed] [Google Scholar]
- 76. Vogel U, Christensen J, Wallin H, Friis S, Nexo BA, et al. (2007) Polymorphisms in COX-2, NSAID use and risk of basal cell carcinoma in a prospective study of Danes. Mutat Res 617: 138–146. [DOI] [PubMed] [Google Scholar]
- 77. Zhang Y, Wang MY, He J, Wang JC, Yang YJ, et al. (2012) Tumor necrosis factor-alpha induced protein 8 polymorphism and risk of non-Hodgkin’s lymphoma in a Chinese population: a case-control study. PLoS One 7: e37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lech-Maranda E, Baseggio L, Charlot C, Rigal D, Berger F, et al. (2007) Genetic polymorphisms in the proximal IL-10 promoter and susceptibility to non-Hodgkin lymphoma. Leuk Lymphoma 48: 2235–2238. [DOI] [PubMed] [Google Scholar]
- 79.Breslow NE, Day NE (1987) Statistical methods in cancer research. Volume II–The design and analysis of cohort studies. IARC Sci Publ: 1–406. [PubMed]
- 80. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. Bmj 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Midgette AS, Wong JB, Beshansky JR, Porath A, Fleming C, et al. (1994) Cost-effectiveness of streptokinase for acute myocardial infarction: A combined meta-analysis and decision analysis of the effects of infarct location and of likelihood of infarction. Med Decis Making 14: 108–117. [DOI] [PubMed] [Google Scholar]
- 82. Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J (2005) A method for meta-analysis of molecular association studies. Stat Med 24: 1291–1306. [DOI] [PubMed] [Google Scholar]
- 83. Lim S, Crawley E, Woo P, Barnes PJ (1998) Haplotype associated with low interleukin-10 production in patients with severe asthma. Lancet 352: 113. [DOI] [PubMed] [Google Scholar]
- 84. Edwards-Smith CJ, Jonsson JR, Purdie DM, Bansal A, Shorthouse C, et al. (1999) Interleukin-10 promoter polymorphism predicts initial response of chronic hepatitis C to interferon alfa. Hepatology 30: 526–530. [DOI] [PubMed] [Google Scholar]
- 85. Lowe PR, Galley HF, Abdel-Fattah A, Webster NR (2003) Influence of interleukin-10 polymorphisms on interleukin-10 expression and survival in critically ill patients. Crit Care Med 31: 34–38. [DOI] [PubMed] [Google Scholar]
- 86. Hutchinson IV, Pravica V, Hajeer A, Sinnott PJ (1999) Identification of high and low responders to allografts. Rev Immunogenet 1: 323–333. [PubMed] [Google Scholar]
- 87. Matsuda M, Salazar F, Petersson M, Masucci G, Hansson J, et al. (1994) Interleukin 10 pretreatment protects target cells from tumor- and allo-specific cytotoxic T cells and downregulates HLA class I expression. J Exp Med 180: 2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. De Vita F, Orditura M, Galizia G, Romano C, Roscigno A, et al. (2000) Serum interleukin-10 levels as a prognostic factor in advanced non-small cell lung cancer patients. Chest 117: 365–373. [DOI] [PubMed] [Google Scholar]
- 89. Sun T, Miao X, Zhang X, Tan W, Xiong P, et al. (2004) Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96: 1030–1036. [DOI] [PubMed] [Google Scholar]
- 90. Wei Q, Cheng L, Amos CI, Wang LE, Guo Z, et al. (2000) Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J Natl Cancer Inst 92: 1764–1772. [DOI] [PubMed] [Google Scholar]
- 91. Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, et al. (1988) Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res 48: 3282–3287. [PubMed] [Google Scholar]
- 92. Setiawan VW, Stram DO, Nomura AM, Kolonel LN, Henderson BE (2007) Risk factors for renal cell cancer: the multiethnic cohort. Am J Epidemiol 166: 932–940. [DOI] [PubMed] [Google Scholar]
- 93. Vassallo R, Tamada K, Lau JS, Kroening PR, Chen L (2005) Cigarette smoke extract suppresses human dendritic cell function leading to preferential induction of Th-2 priming. J Immunol 175: 2684–2691. [DOI] [PubMed] [Google Scholar]
- 94. Salazar-Onfray F, Petersson M, Franksson L, Matsuda M, Blankenstein T, et al. (1995) IL-10 converts mouse lymphoma cells to a CTL-resistant, NK-sensitive phenotype with low but peptide-inducible MHC class I expression. J Immunol 154: 6291–6298. [PubMed] [Google Scholar]