Abstract
The interaction of autotaxin with its substrates leads to the production of lysophosphatidic acids (LPA), bioactive lipids with an emerging prominent role in inflammation and cancer. Two papers in this issue tell the previously unknown story of autotaxin, from substrate discrimination to highly efficient local delivery of LPA to target receptors.
LPA has evoked gradually increasing interest since the early 1990s, when accumulating evidence suggested its role as a major serum growth factor–like molecule that signals through G protein–coupled receptors (GPCR)1. This unleashed efforts to characterize the source of LPA, which pointed to autotaxin (ATX), a protein first identified as an ‘autocrine motility factor’ secreted by melanoma cells1,2. The effects of ATX can recapitulate many of the hallmarks of cancer, including cell proliferation, survival, invasion, migration and neovascularization, and it has been implicated in numerous cancer types including breast, ovarian and melanoma3. ATX, also known as ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2), is a secreted lysophospholipase D (lysoPLD) that hydrolyzes lysophosphatidylcholine (LPC) to form LPA. The significant role that ATX appears to play in cancer and inflammation prompted a series of projects aimed at the structural and functional characterization of the ATX-LPA axis.
A longstanding challenge has been understanding how LPA is produced and its level controlled in the cellular microenvironment. In addition, the determinants of the substrate specificity of ATX remained unknown. Two papers in this issue4,5 use a range of structural and biochemical techniques to derive overlapping and complementary information on the structure and function of ATX, from substrate recognition to product delivery to target receptors.
Hausmann et al.4 resolved the crystal structure of rat ATX (which shares 93% sequence identity with human ATX). The structure reveals that ATX is made up of several domains: two consecutive somatomedin B–like domains (SMB1 and SMB2), followed by a catalytic (phosphodiesterase) domain and a nuclease-like domain. The protein has a compact, robust architecture, with the central catalytic domain interacting extensively with the two SMB domains on one side, and with the nuclease-like domain on the other (Fig. 1).
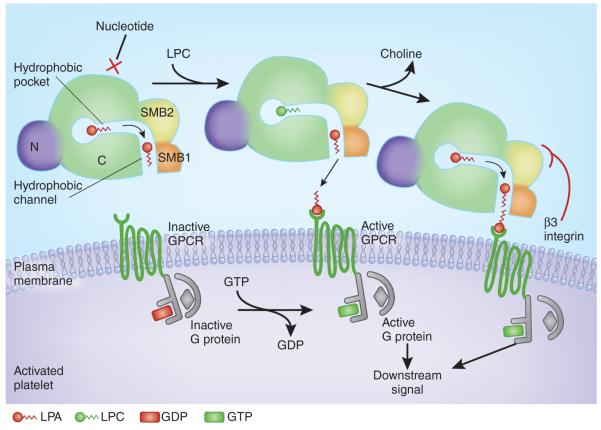
Figure 1.
Model for substrate recognition by ATX, LPA production and local receptor supersaturation. The surface of an activated platelet is shown, with the cartoon of a cross-section through the ATX protein and with domains labeled (C, catalytic domain; N, nuclease-like domain). Nucleotides cannot bind the active site when phospholipid substrates are engaged. A hydrophobic pocket in ATX accommodates LPC acyl chains and allows substrate discrimination. A channel between the catalytic site and SMB1 likely binds and shuttles LPA products before local delivery to the target receptor. Interaction of the SMB2 domain from ATX with a β3 integrin on the surface of the platelet may induce a conformational change in ATX that facilitates local release of LPA product at the cognate GPCR. This would achieve a high local specific concentration of product at the receptor site, effectively supersaturating the receptor relative to the rest of the platelet microenvironment and eliciting intracellular downstream signals. A flat surface at the hydrophobic channel entrance facilitates local interfacing with the target cell membrane.
ATX has the ability to hydrolyze both nucleotide and phospholipid substrates in vitro; its catalytic domain is most similar to that of the nucleotide pyrophosphatase of bacterium Xanthomonas axonopodis (XaNPP), whereas the nuclease-like domain lacks enzymatic activity. Structural comparison to XaNPP revealed that ATX lacks a stretch of 18 amino acids—an insertion loop that forms a pocket to accommodate nucleotides in XaNPP and the other ENPP family members. The absence of this loop in ATX opens a gate to a hydrophobic pocket that accommodates the acyl chains of lysophospholipids (Fig. 1). Mutants with mutations in the hydrophobic pocket showed reduced lysoPLD activity and altered selectivity for acyl chain length when compared to wild-type ATX. These observations indicate that both nucleotides and lipid substrates share the same binding site but the acyl chain of lipids forms additional interactions with the hydrophobic pocket of ATX, thus explaining the higher affinity of ATX for lysophospholipids than for nucleotide substrates.
The mechanism of ATX interaction with target cells has been elusive. The SMB domains of ATX are homologous to the SMB domain of vitronectin, whose Arg-Gly-Asp (RGD) motif binds integrins to form a complex with the plasminogen activator inhibitor and the urokinase-type plasminogen activator; this complex is thought to regulate tumor cell adhesion and migration6. ATX binds activated platelets, suggesting that it could be recruited by integrins and thus that the SMB domains of ATX may be functional7. However, mutagenesis analysis identified only one specific mutation, away from the RGD site in the SMB2 domain, that significantly affected binding to activated platelets4, suggesting that the binding of ATX to integrins also involves sites distinct from RGD within the SMB2 domain.
Furthermore, the SMB2 domain of ATX abuts the hydrophobic binding pocket and a nearby narrow tunnel. Bound LPA inhibits other substrates such as nucleotides from binding, but not LPC, thus playing a role in substrate discrimination8. LPA-mediated inhibition of nucleotide hydrolysis was effective in wild-type ATX but not in mutants lacking both SMB domains, suggesting that the SMB2 domain is involved in recognition of LPC and possibly in product release. Interaction of SMB2 with β3 integrins, thus recruiting ATX to cell surfaces, has the potential to induce a conformational change that would localize LPA release from ATX in the proximity of its GPCR (Fig. 1).
In a second, complementary article, Nishimasu et al.5 report the crystal structures of mouse ATX and its complex with LPA. Again, extensive interdomain interactions were demonstrated between the catalytic domain, the two SMB domains and the nuclease-like domain; residues involved in these interface interactions were conserved among different species, and their disruption leads to failure of catalytic function9. These authors also observed the absence of the same insertion loop in ATX compared to XaNPP, providing free access in ATX to a hydrophobic pocket that can accommodate lysophospholipids. The authors found that this hydrophobic pocket is blocked by a conserved Trp-Pro-Gly (WPG) motif in the loop of XaNPP and almost all known ENPP family members (with the exception of ENPP2), indicating that they all lack lysoPLD activity and selectively hydrolyze nucleotides instead.
Nishimasu et al. subsequently solved the crystal structure of ATX in complex with LPAs of different acyl chain lengths and saturation. For 14:0-LPA, the lipid tail is accommodated in the hydrophobic pocket, whose depth is optimal for this acyl length and could explain the LPC chain length preference of ATX (14:0 > 16:0 > 18:0). Longer LPC chains can also fit in the hydrophobic pocket of ATX, provided that unsaturated bonds introduce bends in the acyl chain. The authors replaced individual residues forming the hydrophobic pocket, and found that the resulting mutants retained activity toward nucleotides. In contrast, the activity toward LPC was remarkably reduced, and differences in the fatty acids preferences were observed, demonstrating the importance of the hydrophobic pocket for lipid tail recognition. In cellular assays, ATX mutants that completely lacked lysoLPD activity failed to stimulate cell motility, whereas mutants with residual lysoPLD activity stimulated cell motility as effectively as wild-type ATX, suggesting that the amount of LPA produced by wild-type ATX is in excess for this function, possibly due to a mechanism of highly efficient local delivery of the product to the target receptor.
An elongated hydrophobic channel was observed in the interface between SMB1 and the catalytic domain. Crystallographic and mass spectrometric analyses and modeling revealed that the ‘free-form’ ATX probably contains LPA molecules in this channel, which would effectively provide a second binding site for LPA. The presence of the insertion loop in XaNPP precludes the formation of the hydrophobic channel, so the authors inserted the additional 19 amino acids from XaNPP into mouse ATX. The resulting mutants showed reduced but still considerable lysoPLD activity, suggesting that they retain the hydrophobic pocket needed to bind substrate and generate LPA. Though the insertion mutants had lysoPLD activity equivalent to that of the hydrophobic pocket mutants, they showed significantly impaired cell motility–stimulating activity, arguing that an intact hydrophobic channel could play a functional role, perhaps by means of a shuttle mechanism that would allow efficient LPA delivery directly to its target receptor. They also note that ATX has a flat molecular surface on the side of the channel entrance, which may be suitable for a close interfacing with the plasma membrane (Fig. 1); thus, the capture of ATX mediated by the β3 integrin receptor and the release of the LPA products through the hydrophobic channel to cell-surface LPA receptors may jointly regulate LPA signaling at the cell surface.
The disruption of the newly described process by which ATX locally supersaturates target receptors with LPA product is a rich avenue for therapeutic intervention that could benefit cancer patients; for example, small molecules could be designed to disrupt the binding of ATX to cell-surface receptors such as β3 integrin and thus move LPA generation away from the receptors. Compounds that can insert into and block the hydrophobic channel represent yet another avenue that could be explored for inhibiting ATX activity.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Contributor Information
Adel Tabchy, Department of Systems Biology, Kleberg Center for Molecular Markers, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA..
Gabor Tigyi, Department of Physiology, University of Tennessee Health Science Center, Memphis, Tennessee, USA..
Gordon B Mills, Department of Systems Biology, Kleberg Center for Molecular Markers, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA..
References
- 1.Mills GB, et al. Nat. Rev. Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 2.Stracke ML, et al. J. Biol. Chem. 1992;267:2524–2529. [PubMed] [Google Scholar]
- 3.Liu S, et al. Cancer Cell. 2009;15:539–550. doi: 10.1016/j.ccr.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haussman J, et al. Nat. Struct. Mol. Biol. 2011;18:198–204. doi: 10.1038/nsmb.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimasu H, et al. Nat. Struct. Mol. Biol. 2011;18:205–212. doi: 10.1038/nsmb.1998. [DOI] [PubMed] [Google Scholar]
- 6.Deng G, et al. J. Biol. Chem. 1996;271:12716–12723. doi: 10.1074/jbc.271.22.12716. [DOI] [PubMed] [Google Scholar]
- 7.Pamuklar Z, et al. J. Biol. Chem. 2009;284:7385–7394. doi: 10.1074/jbc.M807820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Meeteren LA, et al. J. Biol. Chem. 2005;280:21155–21161. doi: 10.1074/jbc.M413183200. [DOI] [PubMed] [Google Scholar]
- 9.Jansen S, et al. J. Biol. Chem. 2009;284:14296–14302. doi: 10.1074/jbc.M900790200. [DOI] [PMC free article] [PubMed] [Google Scholar]