Abstract
Artificial transcription activator-like (TAL) effector-based activators (TALE activators) have broad utility but previous studies suggest that these monomeric proteins often possess low activities. Here we demonstrate that TALE activators can robustly function individually or in synergistic combinations to increase expression of endogenous human genes over wide dynamic ranges. These findings will encourage applications of TALE activators for research and therapy and guide design of novel monomeric TAL effector-based fusion proteins.
Rapid advances in Xanthomonas-derived transcription activator-like (TAL) effector technology have enabled any researcher to construct customizable DNA-binding domains with broad potential uses for targeted alteration of gene sequence or expression. Highly conserved 33–35 amino acid TAL effector repeat domains each bind one nucleotide of DNA with specificity dictated by two hypervariable residues.1 This one-to-one code enables the design of proteins with desired DNA-binding specificities by simply joining TAL effector repeats into an array. Recently, considerable effort has been focused on dimeric TAL effector nucleases (TALENs), artificial proteins composed of customized TAL effector repeats fused to a nuclease domain, which enable targeted modification of endogenous genes in a variety of organisms and cell types.2 Engineered TAL effectors have also been fused to heterologous transcriptional activation domains to construct artificial monomeric TAL effector activators (TALE activators) (Supplementary Fig. 1).3–11 However, in contrast to the high efficiencies reported for dimeric TALENs, the activities of monomeric TALE activators reported thus far have often been disappointingly modest at best in their abilities to increase target gene expression. For example, 22 of 26 published TALE activators (for which quantitative information is available)3–6, 8, 9 failed to induce target endogenous gene expression by five-fold or more (Supplementary Table 1). The use of multiple different architectures makes it difficult to ascertain what parameters may or may not influence the activities of the TALE activators tested in these previous studies (Supplementary Table 1).
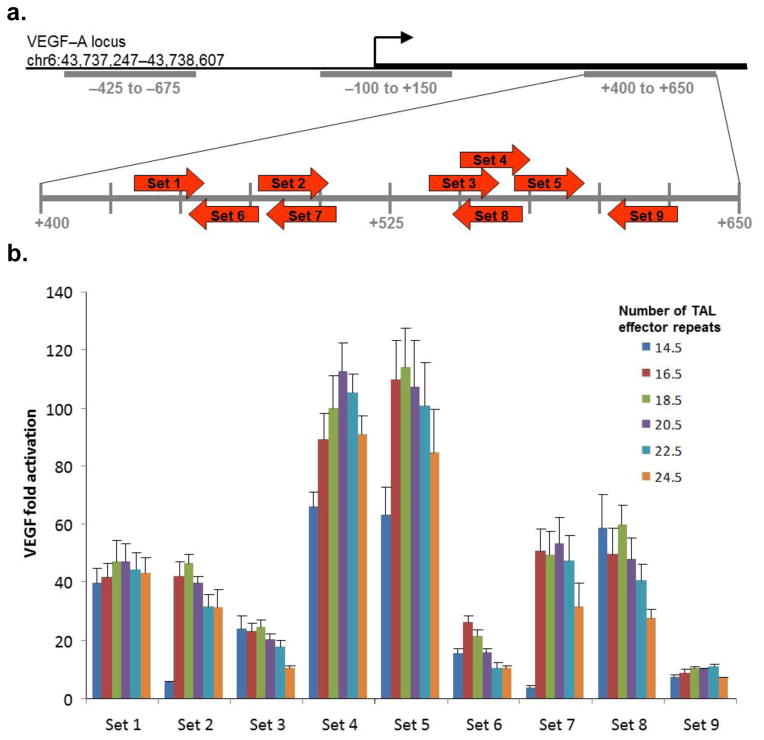
Here we leveraged our recently developed Fast Ligation-based Automatable Solid-phase High-throughput (FLASH) assembly method12 to systematically test the activities of TALE activators. In initial experiments, we constructed a large series of TALE activators composed of variable numbers of TAL effector repeats and tested their abilities to stimulate expression of the endogenous human VEGF-A gene. We targeted nine regions that all lie within a DNase I hyper-sensitive site (HSS) located ~500 bp downstream of the VEGF-A transcription startpoint (Fig. 1a) because previously published work with artificial zinc finger-based transcriptional activators has shown that targeting sequences in HSSs greatly enhances success rates.13 We constructed sets of six variable-length TALE activators (composed of 14.5, 16.5, 18.5, 20.5, 22.5, or 24.5 TAL effector repeats) for each of the nine target regions. All 54 proteins were built on a common architecture similar to one previously described3 but that harbors a VP64 (instead of a VP16) activation domain (Methods and Supplementary Figure 1). Strikingly, we found that 53 of these 54 TALE activators induced significant increases in VEGF-A protein expression ranging from 5.3- to 114-fold (average of 44.3-fold activation) (Fig. 1b) and that these activities do not appear to depend on which strand of DNA is bound. We do not know precisely why we observed variability in the range of fold-activations but possibilities include variable protein expression levels or differences in the DNA-binding activities of the arrays. Regardless of mechanism, our results suggest that TALE activators can function efficiently when they are targeted to a DNase I HSS.
Figure 1.
Activities of 54 variable length TALE activators targeted to the endogenous human VEGF-A gene. (a) Schematic depicting the human VEGF-A promoter region. The transcription startpoint is indicated with a black arrow and previously published DNase I hypersensitive regions13 are shown as grey bars. The DNase I hypersensitive region located between positions +400 to +650 relative to the transcription start site has been expanded, with red arrows indicating the locations and orientations of the 26 bp sites bound by the longest length TALE-activator (harboring 24.5 TAL effector repeats) in each set. (b) Activation of VEGF-A protein expression in 293 cells by 54 variable-length TALE activators. Cells were transfected in triplicate with plasmids encoding each TALE-activator and then assayed as described in Methods. Mean fold-activation values are shown with error bars representing standard errors of the mean. All activators tested (except the 14.5-repeat activator from set 7) induced fold-activation of VEGF-A expression to a value significantly greater than 1, as determined by a one-sided, paired t-test (P < 0.05).
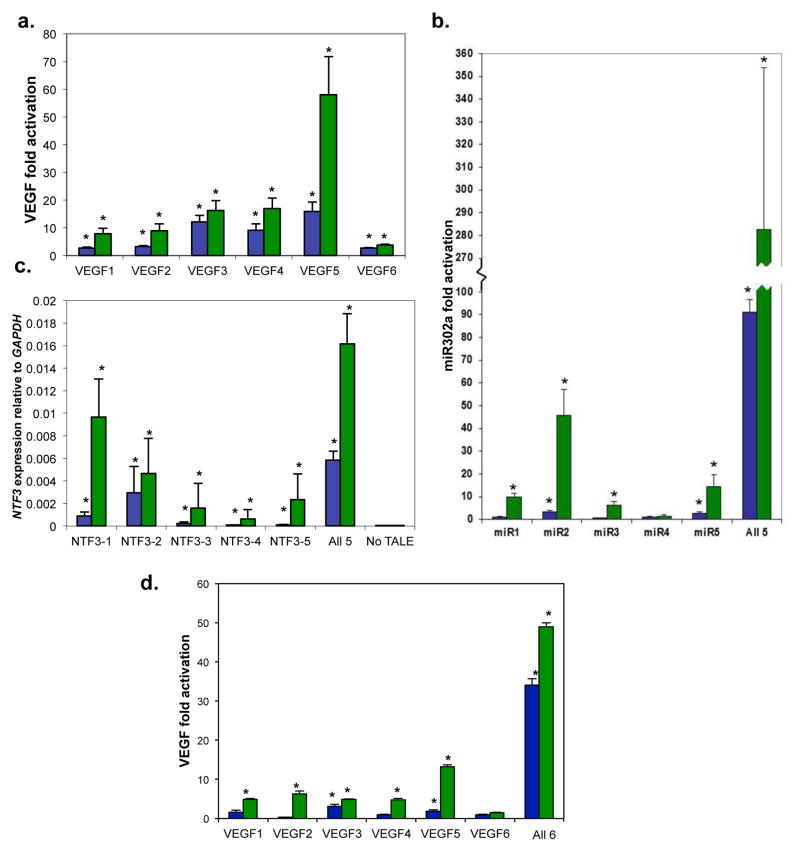
To further test the robustness of TALE activators, we built proteins targeted to six additional sites in the human VEGF-A promoter, five sites in the human NTF3 promoter, and five sites in the microRNA miR-302/367 cluster promoter. All of these TALE activators were composed of 16.5 or 17.5 TAL effector repeats and were targeted to sites within DNase I HSSs (Supplementary Fig. 2 and Methods). Notably, all six TALE activators targeted to VEGF-A and four of five activators targeted to the miR-302/367 cluster induced significant increases of target gene expression in human HEK293 and primary BJ fibroblasts, respectively (Fig. 2a and b). Because NTF3 mRNA is expressed at an essentially undetectable level in the HEK293 cells used for our experiments, we were unable to quantify fold-activation values for proteins targeted to this gene, but all five TALE activators significantly increased expression of NTF3 relative to a GAPDH control (Fig. 2c). Overall, 15 of these 16 TALE activators (~94%) induced significant increases in expression of their endogenous gene targets (Fig. 2a–c).
Figure 2.
Activities of 16 TALE activators targeted to the endogenous human VEGF-A, miR-302/367 cluster, and NTF3 genes. For all three gene targets, experiments were performed in triplicate with TALE activators harboring either the VP64 (green bars) or NF-KB p65 (blue bars) activation domain. Error bars represent standard errors of the mean. (a) VEGF-A-targeted TALE activators. Fold-activation values of VEGF-A protein were determined as described in Methods. Asterisks indicate activators that induced fold-activation of VEGF-A significantly greater than 1, as determined by a one-sided, paired t-test (P < 0.05). (b) miR-302/367-targeted TALE activators. Fold-activation values of miR-302a transcript were determined as described in Methods. Asterisks indicate activators that induced fold-activation of miR-302a transcript levels to a level significantly greater than 1 as determined by a one-sided, paired t-test (P < 0.05). (c) NTF3-targeted TALE activators. Expression levels of NTF3 mRNA relative to GAPDH mRNA are shown. Asterisks indicate activators that induced significant elevation of NTF3 transcript levels relative to a control as determined by a one-sided, paired t-test (P < 0.05). (d) Synergistic activation of VEGF-A. Experiments were performed as in (a) except that the amount of DNA used for individual TALE activators was six-fold lower. Asterisks indicate activators that induced fold-activation of VEGF-A significantly greater than 1, as determined by a one-sided, paired t-test (P < 0.05).
We next explored whether replacing the VP64 domain in our TALE activators with an NF-KB p65 domain would lead to consistently higher or lower levels of target gene expression. For the 15 target sites for which we obtained active TALE activators, we found that the mean fold-activation induced was lower with the NF-KB p65 TALE activators than with their matched VP64 counterparts (Fig. 2a–c). We conclude that substitution with an NF-KB p65 domain can provide a general approach for reducing the fold-activation induced by TALE activators bearing a VP64 domain.
Because we were able to generate more than one active TALE activator for each target gene, we also sought to test whether multiple proteins working simultaneously on a single promoter could induce even greater levels of fold-activation. Naturally occurring transcriptional activators can exhibit synergy – that is, the fold-activation observed in the presence of multiple proteins is higher than the additive effects of the individual proteins.14 Activator synergy enables both combinatorial and graded control of transcription in eukaryotes. We tested combinations of five VP64 or p65 TALE activators for their abilities to activate the miR-302/267 cluster or the NTF3 gene. Despite the fact that each TALE-activator plasmid was present at one-fifth the level used when tested individually, we found that multiple proteins induced substantially elevated target gene expression (Fig. 2bc). Synergistic activation was observed with VP64 and p65 activators on the miR-302/367 cluster (Fig. 2b) and with p65 activators on the NTF3 gene (Fig. 2c). We also tested combinations of six VP64 or p65 TALE activators for their abilities to activate the VEGF-A gene but did not observe synergistic increases in expression (data not shown). We hypothesized that the lack of observed synergy might be due to the different amounts of TALE activator-encoding plasmids used in the individual and combination experiments. Consistent with this, we found that both the VP64 and p65 TALE activators could mediate synergistic increases in VEGF-A expression under conditions where the same amounts of activator plasmids were used when tested individually or in combinations (Fig. 2d). We conclude that both VP64 and p65 TALE activators can function synergistically to induce even higher levels of endogenous gene expression than can be achieved using individual activators.
Our ability to robustly construct highly active TALE activators contrasts with the collective results of previous studies that described proteins with often much lower activities on endogenous gene targets. We found that 62 of 65 (~95%) VP64 TALE activators (for which we could calculate fold-activation values) increased expression of their target gene by five-fold or more. Potential explanations for our higher success rate include our targeting of DNase I HSSs or the sequence architecture of the TALE DNA-binding domains used (Supplementary Discussion). Our findings also expand the types of genes and the range of DNA sequences that can be targeted by TALE activators. To our knowledge, our results provide the first demonstration that it is possible to activate a non-coding gene, thereby broadening the range of potential targets for TALE activators. In addition, analysis of our data suggests that there are no significant limitations in the range of sequences we can successfully target (Supplementary Discussion and Supplementary Table 2). Taken together, our results provide strong experimental support for the idea that TALE activators can be used to control the expression of essentially any gene.
We have shown that TALE activators can be used to regulate target genes across a wide dynamic range of expression, an important capability that will enable a broader range of applications for this technology. Our studies suggest multiple potential approaches that might be used to fine-tune the level of gene expression induced by TALE activators (Supplementary Discussion). The finding that TALE activators can synergistically activate transcription further broadens the range of gene expression changes that can be achieved with this platform and raises the exciting possibility that target genes might be made responsive to multiple inputs, as has recently been shown with engineered zinc finger transcription factors.15 The greater targeting range of engineered TALE activators relative to artificial zinc finger activators provides a substantial advantage for enabling synthetic biology applications in which artificial circuits are designed to interface with endogenous genes.
In this report, we have demonstrated that TALE activators should function efficiently to regulate essentially any protein-coding or non-coding gene in human cells. This capability provides a useful complement to cDNA overexpression or RNAi-based regulation strategies for studying gene function and to previously described synthetic biology strategies for regulating endogenous gene expression16–20. An important area for future investigation will be the potential off-target effects of TALE activators in human cells (Supplementary Discussion). Our successes with TALE activators in human cells should encourage use of these proteins in other cell types and organisms. More importantly, our findings should inspire the generation of other monomeric TAL effector-based fusion proteins that might be used to rationally alter expression and/or the epigenetic status of genes. Thus, our findings should stimulate efforts to expand the repertoire of engineered TAL effector-based tools available for research, synthetic biology, and therapeutic applications.
Methods
Selection of TALE-activator binding sites
For the human VEGF-A gene, target sites were chosen that fall within DNase I hypersensitive sites previously described for 293 cells.13 For the NTF3 and miR-302/367 cluster genes, target sites were chosen within digital DNase I hypersensitive regions identified from University of Washington ENCODE data using the UCSC genome browser (http://genome.ucsc.edu/);21 we targeted these regions because they have been identified as DNase I hypersensitive sites in multiple different cell types and we therefore reasoned that these areas had a high probability of being in open chromatin.
Construction of TALE Activators
DNA fragments encoding TAL effector repeat arrays were generated using the Fast Ligation-based Automatable High-throughput Assembly (FLASH) method as previously described.12 These fragments were cloned using overhangs generated by digestion with BsmBI restriction enzyme into expression vectors containing an EF1α promoter and the Δ152 N-terminal and +95 C-terminal TALE-derived domains from the previously described TALE activator NT-L+95.3 NF-KB p65 and VP64 activation domains were fused directly to the C-terminal end of the +95 domain and all fusion proteins harbor a nuclear localization signal.
Cell Culture and Transfection
Human Flp-In T-REx 293 cells and primary human BJ fibroblasts were maintained in Advanced DMEM supplemented with 10% FBS, 1% penicillin-streptomycin and 1% Glutamax (Life Technologies). Cells were transfected using either Lipofectamine LTX (Life Technologies) or Nucleofection (Lonza) according to manufacturer’s instructions. Briefly, for experiments targeting VEGF-A and NTF3 expression, 160,000 Flp-In T-REx 293 cells were seeded in 24-well plates and transfected the following day with 300 ng of plasmid encoding the TALE activator (except in the reduced-concentration VEGF-A experiments (Figure 2d) in which 50 ng of plasmid encoding TALE activator and 250 ng of a control plasmid expressing only the activation domain were transfected), 30 ng of pmaxGFP plasmid (Lonza), 0.5 μl Plus Reagent and 1.65 μl Lipofectamine LTX. For the synergy experiments with VEGF-A and NTF3, we transfected cells with 300 ng of TALE activator-encoding plasmids (50 ng of each of six TALE activator-encoding plasmids for VEGF-A and 60 ng of each of five TALE-activator-encoding plasmids for NTF3). For experiments targeting miR-302/367 cluster expression, 5×105 BJ fibroblasts were Nucleofected with 10 μg of plasmid encoding TALE activator and 500 ng of pmaxGFP plasmid using the NHDF kit (Lonza) and program U-023 on the Nucleofector 2b device. For the synergy experiment with miR-302/367, we transfected cells with 10 μg of TALE activator-encoding plasmids (2 μg of each of five TALE activator-encoding plasmids).
ELISA Assays
Flp-In TREx 293 cells were transfected with plasmids encoding TALE activators targeted to the human VEGF-A gene. All transfections were performed in triplicate. Cell media was harvested 40 hours after transfection and secreted VEGF-A protein levels in the media were assayed using a Human VEGF-A ELISA kit (R&D Systems). All samples were measured according to the manufacturer’s instructions. Fold-activation values were calculated by dividing mean VEGF-A levels from media harvested from cells transfected with plasmids expressing TALE activators by mean VEGF-A levels from cells transfected with plasmid expressing only the VP64 or p65 activation domain.
Quantitative RT-PCR assays
To measure NTF3 mRNA levels, cells were harvested 2 days post-transfection and total RNA was isolated using the TRIzol Plus RNA purification system (Ambion). RNA was reverse transcribed using SuperScript III First-Strand Synthesis SuperMix and oligo-dT primer (Life Technologies). qPCR was then performed using the following Taqman primer/probe sets, as previously described3 except with the modification that the GAPDH probe was labeled with HEX to allow for multiplexing - NTF3 forward primer: 5′-GATAAACACTGGAACTCTCAGTGCAA-3′; NTF3 reverse primer: 5′-GCCAGCCCACGAGTTTATTGT-3′; NTF3 taqman probe: 5′-/56-FAM/CAAACCTAC/ZEN/GTCCGAGCACTGACTTCAGA/3IABkFQ/-3′; GAPDH forward primer: 5′-CCATGTTCGTCATGGGTGTGA-3′; GAPDH reverse primer: 5′-CATGGACTGTGGTCATGAGT-3′; GAPDH taqman probe: 5′-/5HEX/TCCTGCACC /ZEN/ACCAACTGCTTAGCA/3IABkFQ/-3′. All TALE activator-encoding plasmids and control plasmids were introduced into cells by Nucleofection in triplicate and qRT-PCR was performed in triplicate on each sample.
To measure miR-302a transcript levels, cells were harvested 3 days post-transfection and GFP-positive cells were isolated by flow cytometry. Total miRNA was isolated using the mirVana miRNA Isolation Kit (Ambion). Reverse transcription and qPCR were performed according to manufacturer’s instructions using Applied Biosystems Taqman microRNA Assays (cat. #000529 for has-miR-302a and cat. #001006 for RNU48 control). Fold-activation of miR-302a RNA transcripts was calculated by comparing transcript levels from BJ fibroblasts transfected with plasmids encoding TALE activators to transcript levels from BJ fibroblasts transfected with control plasmids expressing only the VP64 or p65 activation domains and using the comparative CT (ΔΔCT) method. All TALE activators and controls were introduced into cells by Nucleofection in triplicate and qRT-PCR for miR302a transcript and small RNA control RNU48 were performed in triplicate on each sample.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health Director’s Pioneer Award DP1 OD006862 (J.K.J.), National Institutes of Health P50 HG005550 (J.K.J.), the Jim and Ann Orr MGH Research Scholar Award (J.K.J.), a National Science Foundation Graduate Research Fellowship (M.L.M.), and National Institutes of Health T32 CA009216 (J.D.S.). We thank J. Foley for technical assistance with construction of TALE-activator plasmids, R. Mylvaganam for technical assistance with flow cytometry, and the Massachusetts General Hospital (MGH) Nucleic Acid Quantitation Core (supported by NIH P30 NS45776) for assistance with performing real-time RT-PCR assays.
Footnotes
Author Contributions
M.L.M., S.J.L., D.R., J.F.A., Y.F., J.D.S., and J.K.J. designed the experiments. M.L.M., S.J.L., D.R., J.F.A., and Y.F. performed experiments. M.L.M. and J.K.J. wrote the manuscript.
Conflict of interest statement
J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- 1.Joung JK, Sander JD. Nat Rev Mol Cell Biol. 2012;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mussolino C, Cathomen T. Curr Opin Biotechnol. 2012;23:644–650. doi: 10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Miller JC, et al. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F, et al. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geissler R, et al. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tremblay JP, Chapdelaine P, Coulombe Z, Rousseau J. Hum Gene Ther. 2012;23:883–890. doi: 10.1089/hum.2012.034. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, et al. Angew Chem Int Ed Engl. 2012;51:8505–8508. doi: 10.1002/anie.201203597. [DOI] [PubMed] [Google Scholar]
- 9.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bultmann S, et al. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cermak T, et al. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyon D, et al. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu PQ, et al. J Biol Chem. 2001;276:11323–11334. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- 14.Carey M, Lin YS, Green MR, Ptashne M. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 15.Khalil AS, et al. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deans TL, Cantor CR, Collins JJ. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 17.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 18.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 19.Culler SJ, Hoff KG, Smolke CD. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 21.Rosenbloom KR, et al. Nucleic Acids Res. 2012;40:D912–917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.