Abstract
Loss of Prdm16 expression in the mouse leads to a complete cleft of the secondary palate. In the current report, changes in gene expression in the secondary palates of Prdm16−/− fetuses were determined in an attempt to reveal the mechanism(s) leading to failure of palate closure in these mice. Defined, pathway-based, PCR arrays were conducted to analyze the expression of genes associated with the extracellular matrix and the TGF-β and BMP signaling networks, perturbations of which can lead to palatal clefting. Loss of Prdm16 expression in the secondary palate leads to alterations in numerous genes within these groups, many of which have been linked to chondrogenesis and osteogenesis. The expression of several genes linked to bone development was significantly changed in the developing secondary palate. Analysis of gene expression in the mandibles of Prdm16−/− fetuses revealed similar alterations in the same gene set. These data suggest that one function of Prdm16 is the regulation of genes that play a role in differentiation of mesenchymal cells into chondro/osteocytes.
Keywords: craniofacia, mandible, palate, chondrogenesis, osteogenesis
Introduction
In mice, the secondary palate develops from bilateral, medially directed outgrowths of the maxillary processes beginning on gestation day (GD)11.5. These primordia, termed palatal processes, first grow vertically on either side of the tongue until GD13.5 and consist of mesenchymal cells embedded in a loosely organized extracellular matrix (ecm) and enclosed within a stratified epithelium. This early development is characterized by robust cell proliferation and synthesis of extracellular matrix (ECM) proteins. On GD14.0–14.5, the palatal processes undergo a series of morphogenetic movements that result in their reorientation above the tongue bringing the medial edge epithelium of each process into contact forming a midline epithelial seam. The mechanisms driving this reorientation are not known, but it has been suggested that the ECM is important for this process (Morris-Wiman and Brinkley, 1993). The palatal processes then meet at the midline and fuse. The resultant medial edge epithelial seam (MES) is removed through a combination of apoptosis, cell migration, and epithelial to mesenchymal transformation, however, there is controversy over the contribution of each (Jin and Ding, 2006, Xu, et al., 2006). Further development of the palate consists of continued growth to keep pace with overall craniofacial expansion, and regional differentiation into the bony (anterior) and muscular (posterior) soft palate. Disruption of any steps up to and including fusion can lead to a cleft palate.
Recent studies have demonstrated that Prdm16 gene knockout mice have a complete cleft of the secondary palate (Bjork, et al., 2010). PRDM16 is a member of a family of at least 16 structurally related proteins characterized by a PRDI-BF1 and RIZ1 homologous domain). PRDM16 has two putative DNA-binding domains and may function as a transcriptional coactivator/repressor (Kajimura, et al., 2008). PRDM16 was recently demonstrated to be a master regulator of brown adipocyte differentiation from Myf5-positive mesenchymal precursors, leading to inhibition of myogenic differentiation and promotion of brown fat formation (Kajimura, et al., 2008). We have shown that PRDM16 forms a complex with both TGF-β and BMP-specific Smad proteins (Warner, et al., 2007). While the functional role of this interaction is not known, regulation of TGF-β- and BMP-induced gene expression by PRDM16, through modulating the strength and/or extent of the response, is a possible mechanism of action.
To determine the mechanism by which PRDM16 regulates development of the secondary palate and/or orofacial tissue, analysis of alterations in the expression of genes associated with the extracellular matrix, critical for growth and elevation of the palatal processes, and signaling molecules in the TGF-β/BMP signaling pathways was conducted. The TGF-β/BMP pathway was chosen because of an established importance for palate development and because PRDM16 may functionally interact to alter patterns of expression of genes in these pathways (Warner, et al. 2007).
Materials and methods
Animals
Strain B6;129S5-Prdm16Gt683Lex/Mmcd mice were purchased from the Mutant Mouse Regional Resource Center at the University of California at Davis and were housed in our animal facility. In this strain, Prdm16 expression was inactivated by a gene-trap that inserted the gene for β-galactosidase between exon 1 and 2. To obtain embryos/fetuses of the appropriate gestational age, a single heterozygous male was housed overnight with two nulliparous heterozygous females. The presence of a vaginal plug was taken as evidence of mating and designated GD0.5. Pregnant females were euthanized by CO2 asphyxiation followed by cervical dislocation on GD11.5–15.5 and embryos/fetuses removed and immediately chilled on ice. Limb buds were used for genotyping using the EZ Fast Tissue/Tail PCR Genotyping Kit (EZ BioResearch, St. Louis, MO). PCR primers sequences were: wild-type, 5’ primer (5’-ACAGGCGAGGAACTGTATGAAAGG-3’); 3’ primer (5’-CCATCTGAGGTCGTCTGAAACTGG-3’); and mutant (5’-AAATGGCGTTACTTAAGCTAGCTTGC-3’). All procedures for the humane use and handling of mice were approved by the University of Louisville Institutional Animal Care and Use Committee and encompass guidelines as set out in the EC Directive 86/609/EEC for animal experimentation.
PCR Arrays
The secondary palates from GD13.5 mice were microdissected and RNA purified using the RNeasy RNA purification kit (Qiagen, Valencia, CA). cDNAs were synthesized and semi-quantitative real-time PCR arrays performed according to the manufacturer’s instructions (SABiosciences, Frederick, MD). The Sequence Detection System 7000 system from Applied Biosystems (Gaithersburg, MD) was used to perform \real-time PCR. The data was analyzed using the RT2 Profiler PCR Array data analysis web-based software (SABiosciences, Frederick, MD). Three independent RNA preparations from 3–6 fetuses each were used and averaged to obtain the final results.
RT-PCR
RNA was purified from GD12.5–14.5 palatal tissue or GD11.5–15.5 mandibles from wild-type and Prdm16−/− fetuses with the Qiagen RNeasy system (Qiagen, Valencia, CA) and cDNAs prepared using the Superscript II reverse transcription system (Invitrogen, Carlsbad, CA). Probe-primer pairs to specific genes were purchased from Applied Biosystems (Gaithersburg, MD) and data normalized to the signal from the 18S ribosomal RNA gene.
Skeleton staining with alcian blue and alizarin red
Mouse fetuses were isolated on GD14.5 and processed for cartilage and bone staining with alcian blue and alizarin red, respectively. Fetuses were skinned and fixed in 95% ethanol for 24 hours. Specimens were stained with 0.015% alcian blue 8GS and 0.005% alizarin red S (both from Sigma Chemical Co., St. Louis, MO) in 0.9N acetic acid and 60% ethanol for 24 hours at 37 °C, and then cleared with 1% KOH for 8 hours followed by 20% glycerol/1% KOH for several days. Specimens were then processed through graded glycerol solutions before photographing. Six wild-type and 3 Prdm16−/− fetuses were used for this analysis.
Western blotting
Secondary palate and mandible tissue was isolated from GD14.5 wild-type and Prdm16−/− fetuses and sonicated in cell extraction buffer (Invitrogen) supplemented with protease/phosphatase inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Forty µg of total protein was separated on a 4–12% acrylamide Bis-Tris gel (Invitrogen) and electroblotted onto PVDF nylon membranes (Immobilon P, Millipore, Billerica, MA). Membranes were probed with antibodies to osteopontin. This monoclonal antibody (MPIIIB10) was developed by M. Solursh and A. Franzen and was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA). As a loading control, blots were reprobed with an HRP-conjugated Rabbit monoclonal antibody to β-tubulin (Cell Signaling Technology, Danvers, MA). The protein concentration of each sample was determined by the micro-BCA assay (Thermo Scientific Pierce, Rockford, IL). Blots were developed using the ECL Plus Western blotting detection system (GE Healthcare Lifesciences, Pittsburgh, PA). Images were collected with the ImageQuant LAS4000 biomolecular imager equipped with a 6.3 megapixel CCD camera and the data quantitated using ImageQuant TL 7.0 analysis software (GE Healthcare Lifesciences).
Statistical analysis
InStat3 (Graphpad Software, Inc., LaJolla, CA) was used to perform statistical tests.
Results and Discussion
Differential expression of ECM genes
Mice with a genetic deletion of the Prdm16 gene exhibit a complete cleft of the secondary palate (Bjork, et al., 2010). During normal development of the secondary palate, the palatal processes remodel to a position above the tongue on ~GD14.0. In Prdm16−/− mice, the palatal processes fail to undergo this remodeling and remain vertical lateral to the tongue. The cellular or molecular mechanism(s) underlying this phenotype are not known. Because the ECM has been shown to be important for normal palate development (de Oliveira Demarchi, et al., 2010) and possibly for elevation of the palatal processes (Morris-Wiman and Brinkley, 1993), the expression of a panel of 84 genes associated with the ECM was determined using a PCR-based array from SABiosciences. The complete list of genes is presented in Table S1. Among the panel of ECM associated genes analyzed, only four were differentially expressed in Prdm16−/− palatal tissue when compared to wild-type palates (Table 1). These four genes, each of which was expressed at a higher level in Prdm16−/− palatal tissue, were Col2a1, Hapln1, Mmp-11, and Sgce, however, only the expression of Hapln1 reached statistical significance (p<0.05, one-way ANOVA).
Table 1.
Increased expression of ECM genes in Prdm16−/− palate tissue
| Gene | Fold-Increase |
|---|---|
| Col2a1 | 1.9 ± 0.4 |
| Hapln1 | 2.0 ± 0.6* |
| Mmp11 | 1.8 ± 0.2 |
| Sgce | 1.6 ± 0.1 |
Total RNA was purified from secondary palates isolated from wild-type and Prdm16−/− littermates and cDNA synthesized and then analyzed by PCR array for the expression of 84 genes linked to ECM metabolism. The data are expressed as fold-increase ± S.D. (n = 3) in expression in palate tissue from E13.5 Prdm16−/− embryos compared to wild-type embryos.
p<0.05, one-way ANOVA.
Differential expression of TGF-β/BMP signaling pathway genes
Several of the major signaling pathways important for embryonic development have been directly shown to play a role in normal palate development in mice. Of these, the pathway activated by the gene superfamily that includes TGF-β and BMP has been studied the most thoroughly to date and each has been shown to regulate ECM metabolism (Roberts, et al., 1986, Shum, et al., 2003). Additionally, it has been shown that PRDM16 binds to both TGF-β and BMP-linked Smads (Warner, et al., 2007), thereby potentially regulating transcription of downstream genes. Therefore, the expression of a panel of 84 genes linked to these two signaling pathways was determined by PCR array analysis. The complete list of genes is presented in Table S2. The expression of 5 genes was increased greater than 1.5-fold in palatal tissue from Prdm16−/− mice compared to wild-type littermates (Table 2). These genes include Bmp1, Gdf7 (Bmp12), Gdf6 (Bmp13), Gsc, and Il-6. All increases, with the exception of Bmp1, were statistically significant when compared to wild-type levels (Gdf6 and Il-6, p<0.05; Gdf7and Gsc, p<0.01, one-way ANOVA).
Table 2.
Increased expression of TGF-β/BMP genes in Prdm16−/− palate tissue
Total RNA was purified from secondary palates isolated from wild-type and Prdm16−/− littermates and cDNA synthesized and then analyzed by PCR array for the expression of 84 genes in the TGF-β and BMP signaling pathways. The data are expressed as fold-increase ± S.E.M. in expression in palate tissue from E13.5 Prdm16−/− embryos compared to wild-type embryos. Three independent litters were used.
p<0.05 or
p<0.01, one-way ANOVA
Differential expression of genes linked to chondrogenesis and osteogenesis in the secondary palate
Because the genes that exhibited changes in expression in Prdm16−/− palatal tissue had in common a role in chondro- and/or osteogenesis, the expression of additional genes important for these two processes was also determined. Genes selected for analysis were categorized as early (Sox9, Col2a1), mid (Acan, Has2, Runx2, Shox2) and late (Osx, Opn, Akp) stages of chondro- and osteogenic differentiation (Goldring, et al., 2006). The data are presented as gene expression level in wild-type or Prdm16−/− palatal tissue on either GD13.5 or GD14.5 relative to the expression level on GD12.5 (Table 3). The largest change in expression was found for Col2a1, Osx, Shox2, and Opn with the remainder of the genes tested displaying little differences between wild-type and Prdm16−/− samples. In wild-type samples, the expression of Col2a1 was increased by 4.9 ± 0.6-fold on GD13.5 (compared to GD12.5) and by 2.8 ± 0.5-fold on GD14.5. In Prdm16−/− samples, Col2a1 expression was significantly higher than wild-type levels on both GD13.5 and GD14.5 (>40-fold and >25-fold, respectively). Conversely, while Osx expression in Prdm16−/− fetuses was elevated when compared to GD12, expression was significantly less on GD13.5 and GD14.5 in samples from Prdm16−/− fetuses when compared to wild-type. The expression of Shox2 in Prdm16−/− samples was 2-fold higher on GD13.5 and almost 3-fold higher on GD14.5. Finally, the expression of Opn was significantly less on GD14.5 in Prdm16 mutant palates with no significant difference observed on GD13.5. Therefore, the expression of two genes important for early and mid stages of chondrogenesis (Col2a1 and Shox2) were both significantly increased, while two gene markers for later stages (Osx and Opn) were decreased. PRDM16 may indirectly regulate the expression of these genes because a recent study to identify gene targets in palate mesenchymal cells by ChIP-Chip failed to identify any of these genes (Warner, et al., 2012). However, in that study, cultured palate mesenchymal cells were used and may not have identified all genes regulated by PRDM16 in the intact secondary palate. Further experiments will be needed to determine if any of these genes are directly regulated. In addition, the concomitant rise in expression of both Col2a1 and Shox2 are likely not linked because in Shox2 knockout mice, there was no change in expression or distribution of Col2a1 (Cobb, et al., 2006). In this same report, Shox2 was shown to be upstream of Runx2. However, we saw no change in the expression of the latter in palate tissue (Table 3). The simultaneous decrease in Osx and Opn expression may be linked, however. Nakashima et al. found that Opn expression in the periosteum bone layer was decreased in Osx-null mouse mutants (Nakashima, et al., 2002).
TABLE 3.
Expression of chondrogenesis and osteogenesis genes in Wild-type and Prdm16−/− palate tissue
| EARLY | ||||
|---|---|---|---|---|
| GD13.5 | GD14.5 | |||
| +/+ | −/− | +/+ | −/− | |
| Col2a1 | 4.9±0.6 | 42.8±0.6 | 2.8±0.5 | 25.6±2.2 |
| Sox9 | −1.5±0.6 | nc | nc | −1.7±0.1 |
| MID | ||||
| Runx2 | 1.9±0.1 | 1.5±0.3 | 1.7±0.4 | nc |
| Acan | 2.1±0.7 | 3.0±0.3 | nc | nc |
| Has-2 | nc | nc | −2.3±1.1 | nc |
| Shox2 | 29.0±1.6 | 58.0±0.2 | 26.0±1.1 | 76.0±4.0 |
| LATE | ||||
| Osx/Sp7 | 23.7±3.6 | 8.8±0.8 | 20.8±0.4 | 14.4±1.1 |
| Opn | 3.5±0.1 | 4.6±0.3 | 12.2±0.2 | 2.5±0.3 |
| Akp | 8.5±0.4 | 4.1±2.3 | 11.4±0.6 | 8.3±0.2 |
Total RNA was extracted from secondary palates isolated from wild-type and Prdm16−/− fetuses on GD12.5–14.5 and cDNAs synthesized. The expression of the indicated genes was determined by semi-quantitative real-time PCR (Taqman) with pre-designed probe/primer sets from Applied Biosystems and represent genes from early (proliferation/condensation), mid (differentiation), and late (ossification) events during chondro/osteogenesis. The data were normalized to the expression of the 18S rRNA gene and are expressed as fold-change compared to expression on GD12.5 ± S.D. (n = 3).
Indicates P < 0.01 (+/+ vs. −/−; one-way ANOVA). nc, no change.
Prdm16 knockout leads to altered mandibular chondrogenesis and osteogenesis
In addition to cleft palate, Prdm16−/− fetuses also have a smaller mandible (Bjork, et al., 2010). Because proper development of the mandible is also important for repositioning of the tongue to allow the palatal processes to remodel, make contact, and fuse, we performed an analysis, similar to that described above for palatal tissue, of the expression of genes important for all stages of chondrogenesis and osteogenesis in GD11.5–15.5 mandibles from wild-type and Prdm16−/− embryos/fetuses. These data are reported in Table 4. Genes important for early (Col1a1, Col2a1, Sox9), mid (Runx2, Acan, Lp-1, Has-2, Shox2) and late (Osx, Satb2, Opn, Akp) stages of chondro- and osteogenic differentiation were analyzed. The expression Col1a1, an early marker of chondrogenesis, was increased in mandible tissue of Prdm16−/− fetuses on GD13.5 and GD15.5, compared to levels in wild-type tissue. With the exception of Has-2 and Shox2, there was an increase in expression of markers for processes involved in mid phases of chondrogenic determination in Prdm16−/− mandibular tissue. Has-2 expression levels were not altered, however, Shox2 levels were reduced on GD12.5, GD14.5, and GD15.5 in Prdm16−/− mandibular tissue. Runx2, Acan, and LP-1 levels were significantly elevated in Prdm16−/− mandibular tissue, particularly on GD15.5. Finally, the late markers, Osx and Opn were significantly reduced in Prdm16−/− mandibular tissue, relative to wild-type levels, however, the expression of Akp was more complex: increasing on GD12.5 decreasing on GD13.5 and GD15.5, with no significant change on GD14.5. The premature elevation of many of these markers suggests that the normal program of chondrogenesis was perturbed leading to stunted forward expansion of Meckel’s cartilage.
TABLE 4.
Expression of chondrogenesis and osteogenesis genes in Wild-type and Prdm16−/− mandibles
| EARLY | ||||||||
|---|---|---|---|---|---|---|---|---|
| GD12.5 | GD13.5 | GD14.5 | GD15.5 | |||||
| +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | −/− | |
| Col1a1 | 2.2±0.1 | 3.5±0.3 | 8.7±0.2 | 12.0±0.5 | 9.3±0.1 | 8.6±0.4 | 10.6±0.2 | 17.4±0.6 |
| Col2a1 | 1.3±0.1 | 1.5±0.9 | 3.2±0.2 | 5.0±0.1 | 2.6±0.2 | 1.3±1.0 | 2.3±1.3 | 4.3±0.1 |
| Sox9 | 1.5±0.3 | 1.1±1.1 | 1.5±0.4 | 1.1±1.1 | 1.1±0.3 | 1.1±1.1 | 1.2±0.8 | 1.3±0.8 |
| MID | ||||||||
| Runx2 | 1.2±0 | 1.6±0.5 | 1.6±0.2 | 3.7±0.1 | 1.1±0.6 | 1.9±0.4 | 1.2±1.4 | 3.0±0.4 |
| Acan | 5.1±1.7 | 5.6±1.0 | 4.8±1.6 | 16.3±1.8 | 8.7±2.1 | 14.6±1.7 | 19.3±3.4 | 30.3±2.8 |
| LP-1 | 2.9±0.1 | 3.1±0.3 | 1.4±0.4 | 1.5±0.5 | 2.80±.3 | 6.2±0.4 | 4.0±0.2 | 9.8±0.5 |
| Has-2 | 2.8±0.7 | 1.6±0.4 | 1.8±0.3 | 1.1±1.1 | 2.70±.2 | 1.6±0.4 | 1.8±0.3 | 1.4±0.4 |
| 1404 | 6.4±0.1 | 2.7±0.2 | 2.2±0.2 | 1.1±1.1 | 3.0±0.2 | 1.6±0.6 | 3.3±0.2 | 1.4±0.7 |
| LATE | ||||||||
| Osx/Sp7 | 5.2±0.1 | 4.2±0 | 7.6±0.3 | 8.1±0 | 8.6±0.4 | 8.4±0 | 13.5±0.2 | 8.9±0.2 |
| Satb2 | 1.7±0.2 | 1.2±0.7 | 1.0±1.0 | 1.4±0.5 | 1.5±0.3 | 1.0±0 | 1.8±0.4 | 1.1±1.1 |
| Opn | 4.1±1.4 | 1.5±0.5 | 3.5±0.8 | 1.3±1.1 | 37.3±2.6 | 29.4±1.1 | 92.4±5.7 | 49.9±1.6 |
| Akp | 4.9±1.4 | 7.7±0.2 | 10.2±0.4 | 1.5±0.3 | 10.8±0.5 | 12.0±0.3 | 17.9±0.6 | 10.3±0.4 |
| MUSCLE-SPECIFIC | ||||||||
| Myogenin | 4.3±0.3 | 2.3±0.5 | 3.2±0.2 | 1.1±1.1 | 5.9±0.3 | 4.0±0.6 | 1.6±0.7 | 4.8±0.6 |
Mandibles were dissected on GD11.5-GD15.5 from wild-type and Prdm16−/− embryos/fetuses, RNA prepared and cDNAs synthesized and analyzed by real-time PCR for the expression of early (proliferation/condensation), mid (differentiation), and late (ossification) processes of chondro/osteogenesis. The data are expressed as the relative level (mean ± S.D.) on days 12.5–15.5 of gestation compared to the level on day 11.5.
P < 0.01, one-way ANOVA. The data are representative of 2 independent sets of samples assayed in triplicate.
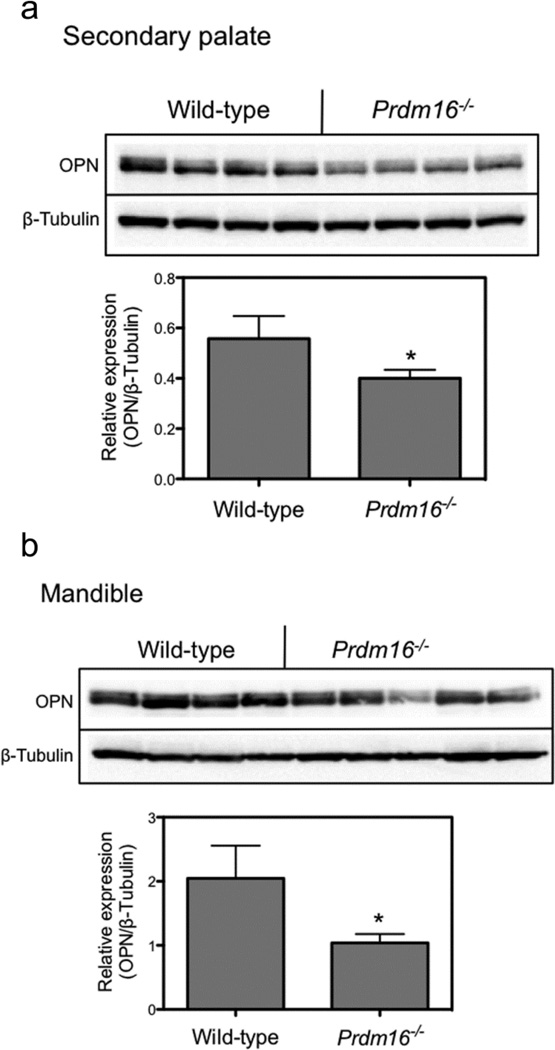
Western blot analysis of the expression of osteopontin in GD14.5 palate and mandible tissue was performed to determine if protein levels reflected the decrease in mRNA expression. As expected from the RT-PCR experiments, the expression of OPN was reduced in Prdm16−/− palate tissue compared to wild-type levels (Fig. 1A). In order to quantitate the extent of decrease, the blots were probed with antibodies to β-tubulin and the signal from OPN was normalized to that from β-tubulin and is presented in the bar graph. By RT-PCR, the expression of OPN was decreased ~45% in Prdm16−/− palate tissue (Table 3) and by ~30% by Western blot. In mandible tissue, although there was some variation in expression of OPN in Prdm16−/− fetuses, on average there was a 50% decrease in signal (Fig. 1B) compared to ~20% by RT-PCR (Table 4). Therefore, a late marker for chondrogenesis was found to be decreased at the mRNA and protein level. Furthermore, we recently demonstrated that the expression of Opn was also reduced in Prdm16−/− palates as assayed by RNA in situ hybridization (Warner, et al., 2012).
Figure 1. Western blot analysis of the expression of OPN in mouse fetal palate and mandible tissue.
Prdm16−/− and wild-type palate and mandible tissue was dissected from GD14.5 fetuses and processed for Western blot as detailed in Materials and Methods section. Panel A. Expression of OPN and β-tubulin (loading control) in secondary palate tissue lysates. Four separate fetuses were analyzed for each genotype. The bar graph represents the quantitation of the expression of OPN relative to that for β-tubulin. In palate tissue isolated from Prdm16−/− fetuses, the expression of OPN was reduced by ~30% compared to wild-type tissue. Panel B. Expression of OPN in GD14.5 mandible lysates. Four wild-type and 5 Prdm16−/− fetuses were collected and analyzed. Although the expression levels of OPN were variable in Prdm16−/− fetuses, there was a significant decrease in expression. The bar graph demonstrates that the normalized expression of OPN was reduced by ~ 50% in Prdm16−/− mandible tissue. For each graph, the data are presented as the mean ± S.D. of 4–5 fetuses. (*, p < 0.05, one-way ANOVA).
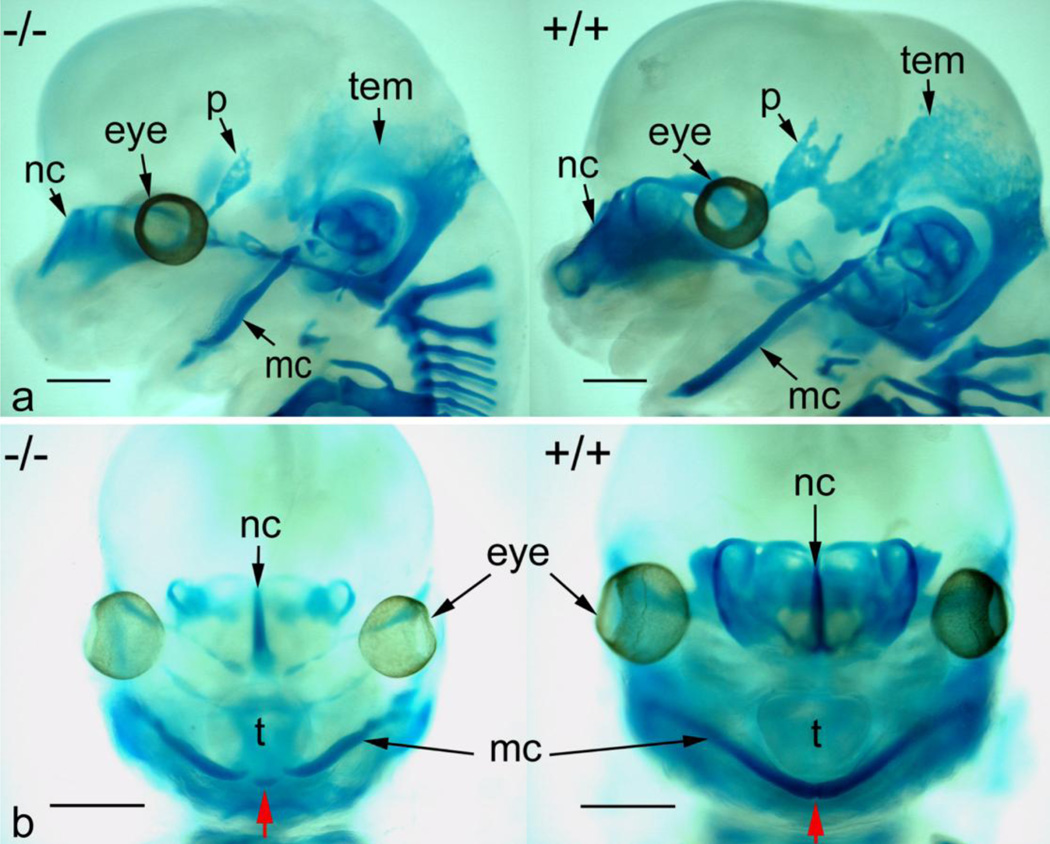
We also performed skeletal staining of GD14.5 fetuses with alcian blue (cartilage) and alizarin red (bone) (Fig. 2). Because there is little osteogenesis on GD14.5, there were no alizarin red-positive ossification centers observed. In addition to generalized reduction in alcian blue staining in forming cranial bones (parietal and temporal, for example), there was a significant reduction in both chondrogenesis and anterior growth of Meckel’s cartilage in Prdm16−/− fetuses (red arrow, panel B).
Figure 2. Alcian blue/alizarin red staining of GD14.5 wild-type and Prdm16−/− fetuses.
Panel A, lateral view of Prdm16−/− (−/−) and wild-type (+/+) fetal heads stained with alcian blue and alizarin red and cleared. Reduced chondrogenesis is evident in the nasal cartilage (nc), parietal bone (p), temporal bone (tem), and Meckel’s cartilage (mc) of Prdm16−/− fetuses. Significant reduction in the length of Meckel’s cartilage is also apparent. Panel B, frontal view of the same specimens in Panel A illustrating the lack of fusion of Meckel’s cartilage in Prdm16−/− fetuses at the anterior-most aspect (red arrow) and an overall reduced intensity of alcian blue staining. Scale bars, 1 mm.
Prdm16−/− mice have a cleft palate because of failure of the palatal processes to reorient. The underlying mechanism for this phenotype is not, however, known, but has been suggested to involve alterations in TGF-β signaling (Bjork, et al., 2010). On GD12.5 and GD13.5, the palatal processes from wild-type and Prdm16−/− mice are morphologically similar (i.e. are of comparable size and shape) with stark differences only apparent beginning on GD14.5 (data not shown). ECM synthesis is critical for elevation of the palatal processes (Morris-Wiman, et al., 2000) and we found that the expression of several genes linked to the ECM was increased. Col2a1 is known to be involved in chondrogenesis and knockout mice have a cleft palate (Barbieri, et al., 2003). That the expression of Col2a1 was significantly increased in Prdm16−/− mice suggests that PRDM16, directly or indirectly, represses its synthesis. Because the expression of Prdm16 decreases from GD12.5–14.5, one function may be to prevent premature synthesis of Col2a1 and/or to limit the extent of synthesis. The significance of Mmp11upregulation in Prdm16−/− palate tissue is not known as no direct link has been established with chondro- or osteogenesis. Interestingly, Sgce has been found in both white adipocytes and in skeletal muscle (Groh, et al., 2009) and, as with Mmp11, no direct role in palate development has been identified.
The TGF-β/BMP family is essential for secondary palate development as revealed by several mouse knockout models (Baek, et al., 2011, Proetzel, et al., 1995, Xu, et al., 2006). Because we have previously shown that PRDM16 interacts with TGF-β/BMP-regulated Smads, it may mediate the signaling activity of these cytokines. Indeed, in Prdm16−/− mice, the expression of several TGF-β/BMP-regulated genes was dysregulated, including Gdf6 and Gsc both of which have been linked to skeletal development, with precise control of expression of the latter critical because either knockout (Rivera-Perez, et al., 1995) or overexpression (Boucher, et al., 2000) leads to craniofacial defects, including cleft palate. Additionally, Il-6 (increased in Prdm16−/− mice) has been shown to inhibit the expression of Col2a1 and Acan expression in ATDC5 chondrogenic precursor cells (Nakajima, et al., 2009).
It is not clear why the expression of some genes essential for chondrogenesis are increased (Col2a1, Shox2) while others are decreased (Osx, Opn) and what impact this has on the developing palate. Based on work published by Yu et al., in a conditional knockout of Shox2 expression in the secondary palate, the expression of Runx2 and Osxwere decreased, suggesting that they are downstream targets (Yu, et al., 2005). If this were true, one would expect that expression should increase when Shox2 is overexpressed (as observed in Prdm16−/− palate tissue). However, there was no effect on the expression of Runx2 and a decreased expression of Osx. It is possible that the combined effect of loss of Prdm16 expression with elevated Shox2 (and other genes) could account for the unexpected results. Alternatively, these genes may be directly regulated by PRDM16.
Based upon these results we propose the hypothesis that PRDM16 regulates the expression of key genes important for chondrogenic differentiation of palate mesenchymal cells, and potentially inhibition of myogenic differentiation in the anterior palate. The expression of myogenin was increased in Prdm16−/− palate tissue (Table 4). Loss of Prdm16 expression may lead to disturbances in the constitution of the ECM to such an extent that normal palatal morphogenetic movements do not occur properly and/or that the underlying signaling pathways are altered (e.g. TGF-β/BMP). In addition to affecting normal development of the secondary palate, loss of Prdm16 expression also disrupts mandibular development by altering the expression of a similar panel of genes as in the palate. Recently, Bjork and colleagues suggested that during development of the mandible in Prdm16 knockout mice there was increased ossification (Bjork, et al., 2010). In our analysis, however, we found no evidence of an increase, rather, we observed a generalized decrease in bone maturation (Fig. 2). The defects in mandibular development may contribute to the cleft palate phenotype in Prdm16−/− fetuses by not allowing proper repositioning of the tongue so that the shelves elevate properly, however, additional experiments are required to test this hypothesis. We are currently working to dissect the contributions from altered mandibular development and defects in signaling mechanisms intrinsic to the secondary palate resulting from loss of Prdm16 expression.
Supplementary Material
Acknowledgements
This work was supported in part by NIH grants DE018215, HD053509, and P20 RR017702 from the COBRE program of the National Center for Research Resources and the NIGMS. Additional support was provided by the Kentucky Science and Engineering Foundation.
Footnotes
Conflict of interest
All authors declare no conflicts of interest, commercial or financial.
References
- Baek JA, Lan Y, Liu H, Maltby KM, Mishina Y, Jiang R. Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev Biol. 2011;350:520–531. doi: 10.1016/j.ydbio.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri O, Astigiano S, Morini M, Tavella S, Schito A, Corsi A, Di Martino D, Bianco P, Cancedda R, Garofalo S. Depletion of cartilage collagen fibrils in mice carrying a dominant negative Col2a1 transgene affects chondrocyte differentiation. Am J Physiol Cell Physiol. 2003;285:C1504–C1512. doi: 10.1152/ajpcell.00579.2002. [DOI] [PubMed] [Google Scholar]
- Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher DM, Schaffer M, Deissler K, Moore CA, Gold JD, Burdsal CA, Meneses JJ, Pedersen RA, Blum M. goosecoid expression represses Brachyury in embryonic stem cells and affects craniofacial development in chimeric mice. Int J Dev Biol. 2000;44:279–288. [PubMed] [Google Scholar]
- Cobb J, Dierich A, Huss-Garcia Y, Duboule D. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc Natl Acad Sci U S A. 2006;103:4511–4515. doi: 10.1073/pnas.0510544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Demarchi AC, Zambuzzi WF, Paiva KB, da Silva-Valenzuela MG, Nunes FD, de Cassia Savio Figueira R, Sasahara RM, Demasi MA, Winnischofer SM, Sogayar MC, Granjeiro JM. Development of secondary palate requires strict regulation of ECM remodeling: sequential distribution of RECK, MMP-2, MMP-3, and MMP-9. Cell Tissue Res. 2010;340:61–69. doi: 10.1007/s00441-010-0931-6. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- Groh S, Zong H, Goddeeris MM, Lebakken CS, Venzke D, Pessin JE, Campbell KP. Sarcoglycan complex: implications for metabolic defects in muscular dystrophies. J Biol Chem. 2009;284:19178–19182. doi: 10.1074/jbc.C109.010728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JZ, Ding J. Analysis of cell migration, transdifferentiation and apoptosis during mouse secondary palate fusion. Development. 2006;133:3341–3347. doi: 10.1242/dev.02520. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes and Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Wiman J, Brinkley L. Rapid changes in the extracellular matrix accompany in vitro palatal shelf remodelling. Anat Embryol (Berl) 1993;188:75–85. doi: 10.1007/BF00191453. [DOI] [PubMed] [Google Scholar]
- Morris-Wiman J, Burch H, Basco E. Temporospatial distribution of matrix metalloproteinase and tissue inhibitors of matrix metalloproteinases during murine secondary palate morphogenesis. Anat Embryol (Berl) 2000;202:129–141. doi: 10.1007/s004290000098. [DOI] [PubMed] [Google Scholar]
- Nakajima S, Naruto T, Miyamae T, Imagawa T, Mori M, Nishimaki S, Yokota S. Interleukin-6 inhibits early differentiation of ATDC5 chondrogenic progenitor cells. Cytokine. 2009;47:91–97. doi: 10.1016/j.cyto.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. Transforming growth factor-β 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development. 1995;121:3005–3012. doi: 10.1242/dev.121.9.3005. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, et al. Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci U S A. 1986;83:4167–4171. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum L, Wang X, Kane AA, Nuckolls GH. BMP4 promotes chondrocyte proliferation and hypertrophy in the endochondral cranial base. Int J Dev Biol. 2003;47:423–431. [PubMed] [Google Scholar]
- Warner DR, Horn KH, Mudd L, Webb CL, Greene RM, Pisano MM. PRDM16/MEL1: A novel Smad binding protein expressed in murine embryonic orofacial tissue. Biochim Biophys Acta. 2007;1773:814–820. doi: 10.1016/j.bbamcr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Warner DR, Mukhopadhyay P, Webb CL, Greene RM, Pisano MM. Chromatin immunoprecipitation-promoter microarray identification of genes regulated by PRDM16 in murine embryonic palate mesenchymal cells. Exp Biol Med (Maywood) 2012;237:387–394. doi: 10.1258/ebm.2012.011258. [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr, Urata MM, Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 2006;297:238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen Y. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.