Abstract
OBJECTIVES:
We investigated four components of the Wnt signaling pathway in medulloblastomas. Medulloblastoma is the most common type of malignant pediatric brain tumor, and the Wnt signaling pathway has been shown to be activated in this type of tumor.
METHODS:
Sixty-one medulloblastoma cases were analyzed for β-catenin gene (CTNNB1) mutations, β-catenin protein expression via immunostaining and Wnt signaling pathway-related gene expression. All data were correlated with histological subtypes and patient clinical information.
RESULTS:
CTNNB1 sequencing analysis revealed that 11 out of 61 medulloblastomas harbored missense mutations in residues 32, 33, 34 and 37, which are located in exon 3. These mutations alter the glycogen synthase kinase-3β phosphorylation sites, which participate in β-catenin degradation. No significant differences were observed between mutation status and histological medulloblastoma type, patient age and overall or progression-free survival times. Nuclear β-catenin accumulation, which was observed in 27.9% of the cases, was not associated with the histological type, CTNNB1 mutation status or tumor cell dissemination. The relative expression levels of genes that code for proteins involved in the Wnt signaling pathway (CTNNB1, APC, AXIN1 and WNT1) were also analyzed, but no significant correlations were found. In addition, large-cell variant medulloblastomas presented lower relative CTNNB1 expression as compared to the other tumor variants.
CONCLUSIONS:
A small subset of medulloblastomas carry CTNNB1 mutations with consequent nuclear accumulation of β-catenin. The Wnt signaling pathway plays a role in classic, desmoplastic and extensive nodularity medulloblastoma variants but not in large-cell medulloblastomas.
Keywords: β-catenin, Gene Expression, Immunohistochemistry, Medulloblastoma, Wnt Pathway
INTRODUCTION
Medulloblastomas are malignant neuroepithelial tumors of the cerebellum and the most frequent pediatric primary malignant intracranial neoplasm. They exhibit a tendency to metastasize via cerebrospinal fluid (CSF) pathways (1,2). The 2007 World Health Organization (WHO) classification defines five histological subtypes of medulloblastoma: classic, desmoplastic/nodular, extensively nodular, anaplastic and large-cell (3). Medulloblastomas are high-grade embryonal tumors (WHO grade IV), and the deregulation of various signaling pathways, such as the Shh, Notch and Wnt pathways, involved in the normal development of the cerebellum have been described as being involved in their progression (4). Many authors have combined data from clinical, pathologic and molecular analyses to identify four to six distinct medulloblastoma variants (5-8), although the current consensus is that there are only four core molecular subgroups of medulloblastomas (9,10). Furthermore, a subset of medulloblastomas associated with Wnt signaling pathway activation has been associated with classical histology and good prognosis (11). Wnt proteins, a group of secreted proteins, regulate the cytoplasmic levels of β-catenin, a component of the adherens junctions of mammalian epithelial cells that, through α-catenin, link cadherin cell-surface adhesion molecules to the actin cytoskeleton (12). Adenomatous polyposis coli (APC) protein and AXIN1, in a complex with glycogen synthase kinase-3β (GSK3β), prevent β-catenin from entering the nucleus (13). GSK3β phosphorylates β-catenin and thereby targets it for recognition by ubiquitination and consequent degradation (14-16). In contrast, mutations in exon 3 of the β-catenin gene (CTNNB1) in codons that code for phosphorylation sites confer resistance to phosphorylation and guide the translocation of β-catenin to the nucleus, where it acts as a co-activator of the Tcf/Lef family of DNA-binding proteins and leads to the upregulation of target genes (17-21).
A systematic analysis of a series of 61 sporadic medulloblastoma cases was performed to correlate histological subtype and clinical data. β-catenin protein expression analysis and mutational analysis of CTNNB1 was first conducted, and a subsequent gene expression analysis of the Wnt pathway components was performed.
METHODS
Tumor samples, DNA and RNA extraction
The casuistic samples consisted of 61 medulloblastoma patients who received surgeries from the neurosurgery group of the Department of Neurology, Hospital das Clínicas, School of Medicine, University of São Paulo. Forty-one samples were collected during resection and were immediately snap-frozen and stored in liquid nitrogen for DNA and RNA extraction. The tissue samples were classified according to the morphology of the tumoral area of higher degree of malignancy following the WHO classification (3). Patients up to 18 years of age were considered pediatric cases. Patient data concerning tumor cell spread in cerebrospinal fluid (CSF), tumor localization, degree of resection, as well as the overall and progression-free survival time were analyzed. Tumor DNA was extracted from all samples using the standard phenol/chloroform method. Total RNA was extracted from each sample using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The evaluation of RNA quantification and purification was carried out by measuring the absorbance at 260 and 280 nm. A260/280 ratios in the range of 1.8-2.0 were considered satisfactory for purity standards. Denaturing agarose gel electrophoresis was used to assess the quality of the samples. This study received the approval of the local Ethical Committee of the Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo. Informed consent was obtained from all patients or their legal guardians.
cDNA synthesis
A conventional reverse transcription reaction was performed to yield single-stranded cDNA from 1 μg of total RNA that was previously treated with one unit of DNase I (FPLC-pure, GE Healthcare, Piscataway, NJ). The reaction used random and oligo(dT) primers (Invitrogen, Carlsbad, CA) for extension, RNase inhibitor (RNase OUT, Invitrogen) and SuperScript III reverse transcriptase, according to the manufacturer's recommended protocol (Invitrogen). The resulting cDNA was then treated with one unit of RNase H (GE Healthcare) and diluted with TE buffer. A pooled normal cerebellum total RNA sample was obtained from a cerebellum pool of 24 males and females (age 16-70 years) (Clontech, Mountain View, CA).
β-catenin immunohistochemical staining
For the immunohistochemistry (IHC) analysis, 5-μm sections were deparaffinized and subjected to pressure cooking epitope retrieval by steaming for 4 minutes, as measured from the initiation of boiling in a citrate buffer (10 mM, pH 6.0) (22). Anti-β-catenin mouse monoclonal antibody (BD Transduction Laboratories, San Jose, CA) was diluted (1:400) in buffer consisting of 1% albumin, 0.1% NaN3 and PBS. The primary antibody was incubated overnight at 4 °C and then detected following incubation with a biotinylated anti-mouse secondary antibody (Dako, Carpinteria, CA) followed by a biotin-streptavidin peroxidase complex (StreptABC Complex/HRP Kit, Dakocytomation, Glostrup, Denmark) (23). Colon cancer tissue was used as a positive control. The sections were then counterstained with Harris hematoxylin, and a semi-quantitative analysis was performed by two independent observers (RS and SKNM). The localization of β-catenin was considered nuclear when, in addition to a cytoplasmic/membrane reaction, more than 10% of tumor cell nuclei were positively stained.
Gene expression analysis
The relative expression levels of the Wnt pathway genes were determined using quantitative real-time PCR (QT-PCR). The following primers and fluorescence-labeled probes were purchased from Applied Biosystems (Foster City, CA): CTNNB1 (Hs00991820_g1), AXIN1 (Hs00394718_m1), APC (Hs01568269_m1) and WNT1 (Hs01011249_g1). These desired primers and probe sequences were obtained from references in the NCBI dbSNP database. GUSB and HPRT1 were used as endogenous controls. All samples and controls were tested in duplicate. The PCR mixtures (10 μl) contained 3 μl of cDNA, 1 μl of primer and probe and 6 μl of TaqMan Universal Master Mix (Applied Biosystems). PCR reactions were carried out in an ABI Prism 7500 real-time thermal cycler with “FAM no quencher” selected as the detector with the following program: 50 °C for 5 min, followed by polymerase activation at 95 °C for 10 min and a second step of 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Values were normalized relative to the internal housekeeping controls. The equation 2-ΔΔCt was applied to calculate the relative expression of CTNNB1, AXIN1, APC and WNT1 in tumor samples versus the non-neoplastic cerebellum pool, where ΔCt = target gene Ct – the geometric mean of GUSB and HPRT1 Ct and ΔΔCt = ΔCt tumor - ΔCt cerebellum (24). The values of gene expression for the normal cerebellum pool were set to 1. Gene expression values above the median were considered overexpression for the purposes of this analysis.
Statistical analysis
The statistical analyses of the relative gene expression levels according to histological classification were performed using the Kruskal-Wallis test. The Dunn multiple comparison post-test was applied to compare the differences between medulloblastoma groups. A bivariate analysis that measured the strengths of association between two genes was performed using Spearman's Rho correlation method. Associations among the categorical variables were tested using the χ2 test. In cases where at least one of the expected frequencies was below 5, Fisher's exact test was applied. Overall survival was calculated as the interval between the date of initial diagnosis and the date of death or the last follow-up, and progression-free survival was measured from the date of initial diagnosis to the date of first recurrence (the return of disease or the signs and symptoms of disease after a period of improvement). The Log rank test was used for univariate analysis to estimate the differences in survival time for variables, according to the Kaplan-Meier method. Calculations were performed using SPSS software, version 15.0 (Chicago, IL), and STATA, version 7 (STATA Corp., College Station, TX). Values of p<0.05 were considered statistically significant for all analyses (including the Dunn multiple comparison post-test).
RESULTS
A total of 61 cases of medulloblastomas were available for analysis, consisting of 38 pediatric cases (mean age 7.6 years) and 23 adult cases (mean age 30.8 years). There were 25 females and 36 males. Tumors with midline localization were more frequent in patients under 18 years of age, with 89.5% of pediatric cases exhibiting midline localization versus 56.5% of adult cases (p = 0.005). Forty-four cases were of the classic type (72.1%), 10 were large-cell (16.4%), four were desmoplastic (6.6%) and three demonstrated extensive nodularity (4.9%), according to the WHO histological classification. Medulloblastomas with extensive nodularity and desmoplastic cases were grouped for statistical analyses. Table 1 presents information concerning the mean age at diagnosis, tumor cell dissemination to the CSF and median overall and progression-free survival time for the three groups according to histological classification. Large-cell type medulloblastomas prevailed among younger patients, who presented lower overall and progression-free survival times as compared to patients with other medulloblastoma variants (p = 0.002 and p = 0.004, respectively).
Table 1.
Data from all analyzed patients, including age at diagnosis, tumor cell spread, overall and progression-free survival and β-catenin immunohistochemistry results.
| Histological classification | ||||
| Classic | Desmoplastic/extensive nodularity | Large-cell | p-value | |
| Age at diagnosis (years) | 17.5±12.9 | 18.4±14.0 | 9.7±7.7 | 0.192a |
| CSF spread | ||||
| Yes | 38 (86.4%) | 7 (100%) | 10 (100%) | 0.496b |
| No | 6 (13.6%) | 0 | 0 | |
| Overall survival (mo) | 85.5±9.9 | 123.4±27.0 | 27.7±11.7 | 0.002c |
| Progression-free | ||||
| survival (mo) | 56.6±7.6 | 79.5±18.0 | 15.1±8.6 | 0.004c |
| Nuclear β-catenin | ||||
| negative | 31 (70.5%) | 6 (85.7%) | 7 (70.0%) | 0.810b |
| positive | 13 (29.5%) | 1 (14.3%) | 3 (30.0%) | |
| CTNNB1 mutation | ||||
| negative | 36 (81.8%) | 6 (85.7%) | 8 (80.0%) | 0.496b |
| positive | 8 (18.2%) | 1 (14.3%) | 2 (20.0%) | |
| Total | 44 (72.1%) | 7 (11.5%) | 10 (16.4%) | |
For age, overall survival and progression-free survival, mean ± SD; mo, months. aKruskal Wallis; bFisher's exact test; cLog rank test.
Nuclear staining of β-catenin was identified in 17 out of 61 (27.9%) medulloblastomas (13 classic, one desmoplastic and three large-cell cases) (Figure 1). There was no association between β-catenin nuclear positivity and histological type (Table 1), age or gender. Cytoplasmic β-catenin staining was detected in all cases.
Figure 1.
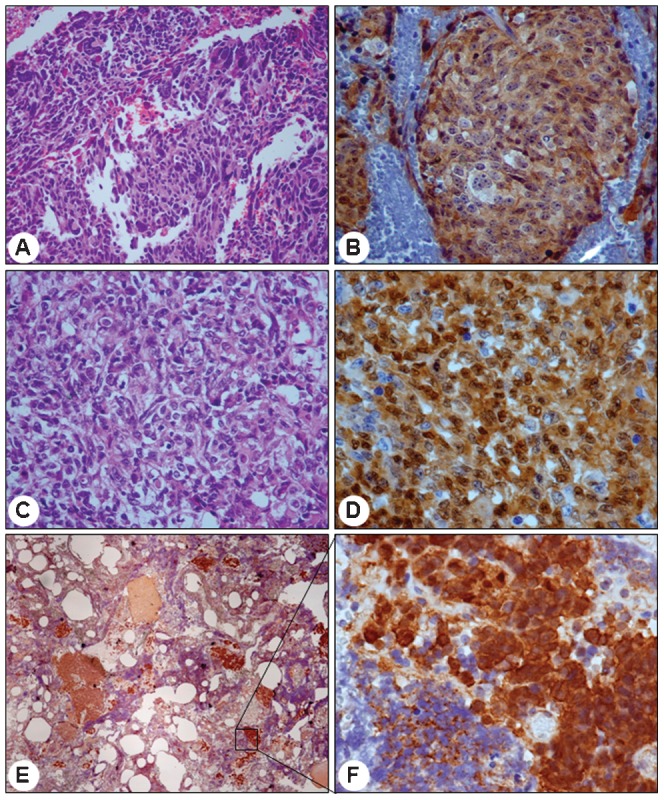
catenin IHC findings in three representative medulloblastoma cases. Large-cell variant presenting anaplastic cells in HE (A, 200x), with the majority of cells presenting positive cytoplasmic β-catenin reactions but few cell showing positive nuclear reactions (B, 400x). Large-cell variant with abundant clear cells in HE (C, 400x) and several positive nuclei intermingled with a small number of cells presenting a negative cytoplasmic reaction (D, 600x). Extensive nodularity variant presenting an islet of positive cells (E, 40x) with strong nuclear reactivity but lower cytoplasmic reactivity (F, 600x).
Direct sequence analysis of the PCR amplification products revealed heterozygous missense mutations of CTNNB1 exon 3 in 11 out of 61 (18.0%) medulloblastomas. Three were detected at codon 32 (GAC>AAC (two cases) and GAC>TAC (one case), as well as seven at codon 33 (TCT>TAT (five cases) and TCT>TGT (two cases) and one at codon 34 (GGA>AGA (one case). The detected CTNNB1 mutations had no statistically significant relationship to any specific histological type (eight classic, two large-cell and one desmoplastic case) (Table 1) or age range (eight pediatric and three adult cases). Among the 11 tumors harboring CTNNB1 mutations, eight cases presented nuclear β-catenin staining, five of which were the classic type, one was desmoplastic and two were of the large-cell type. Furthermore, there was no correlation between CTNNB1 mutation status and CSF tumor cell dissemination, as only one out of four patients with dissemination presented mutation.
We also analyzed CTNNB1, APC, AXIN1 and WNT1 mRNA expression levels by QT-PCR in 41 medulloblastoma cases. WNT1 expression could not be detected in most of the cases, and the data are not presented here. However, the expression levels of CTNNB1, APC and AXIN1 were analyzed and compared to a non-tumorous cerebellum pool. The medulloblastoma samples presented a higher CTNNB1 expression in most cases as compared to normal cerebellum tissue (range 0.02-6.76, median value 1.55). Furthermore, eighteen cases (43.9%) presented overexpression of CTNNB1. In contrast, APC (range 1.22-40.57, median value 5.40) and AXIN1 (range 0.18-3.04, median value 0.89) expression levels presented a wide range of variation. A total of 14 cases (34.1%) presented APC overexpression, while AXIN1 overexpression was detected in 16 cases (39.0%).
We next carried out a Spearman's Rho analysis of the relative expression values for CTNNB1, AXIN1 and APC, although no significant correlation was found. Moreover, the overexpression status of each gene did not correlate with the age at diagnosis, gender, CTNNB1 mutation status, β-catenin nuclear staining, localization of the tumor, dissemination or overall and progression-free survival time.
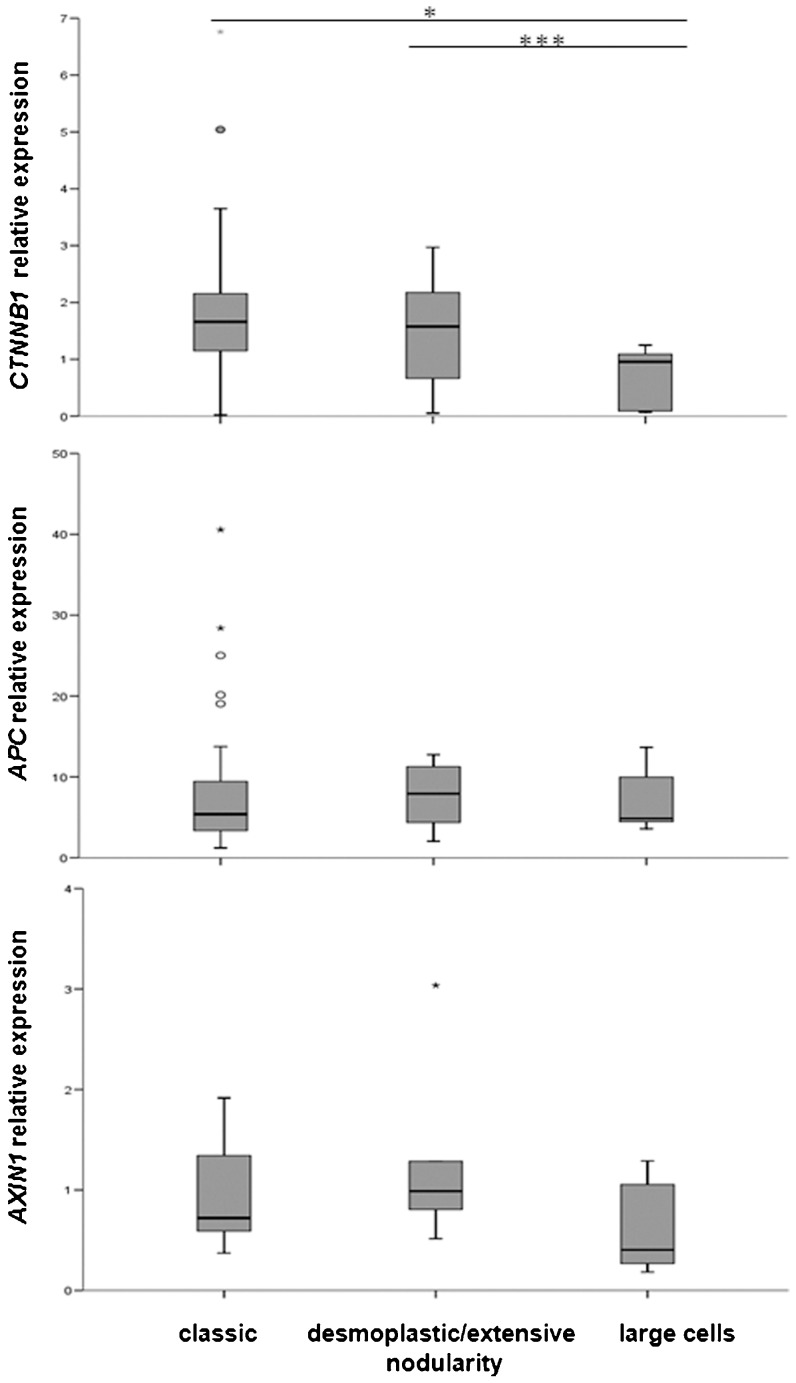
We also analyzed the relative expression of these three genes and their relation to the histological classification of the medulloblastomas. CTNNB1 presented differences in relative expression between the large-cell and classic variants (p<0.05) and also between the large-cell and desmoplastic/extensive nodularity variants (p<0.0005), whereas the APC and AXIN1 relative expression levels did not present differences among the three medulloblastoma groups analyzed (Figure 2).
Figure 2.
Relative expression levels of the CTNNB1, AXIN1 and APC genes in medulloblastoma cases according to histological classification. * p<0.05; *** p<0.0005.
DISCUSSION
The association between the canonical Wnt pathway and medulloblastoma has been explored ever since the identification of mutations in tumor suppressor genes (25-27). Subsequently, other proteins related to this pathway have also been studied to evaluate their participation in sporadic medulloblastomas. In the present work, we evaluated protein expression (by immunohistochemistry) and genes (mutation status and expression levels) involved in the Wnt signaling pathway in a series of 61 medulloblastoma cases. When we analyzed the survival time according to the histological type of the medulloblastoma, patients with large-cell variant tumors had a significantly poorer outcome than did patients with other variants, corroborating previously described findings (4,9,28).
Interestingly, when the expression level of genes involved in the Wnt signaling pathway was examined, no expression or very low WNT1 transcript levels were detected. These results are in accordance to the very low frequency (<1%) of WNT1 expression described by Yokota et al. (29), and these results also corroborate the role of other members of the Wnt protein family in medulloblastomas. In contrast, the downstream targets CTNNB1, APC and AXIN1 were overexpressed in 43.9%, 34.1% and 39.0%, respectively, of medulloblastoma cases in the present series. Of note, the large-cell medulloblastoma variant presented significantly lower expression levels of CTNNB1 than the other variants. These findings suggest that the Wnt medulloblastoma subgroup rarely contains the large-cell variant, and these results are similar to a recent consensus concerning medulloblastoma molecular subclassification (9,10). Cytoplasmic β-catenin immunoreactivity was demonstrated in all cases, which is in accordance with previous studies (25,30,31). However, these results were independent of CTNNB1 expression level or mutational status, which suggests that β-catenin expression might be regulated by other pathway proteins, such as ERBB receptors or SUFU protein. ERBB1/2 receptors interact directly with β-catenin, leading to its phosphorylation and consequent cytoplasmic level increase (32), whereas SUFU acts as a negative regulator of the Wnt pathway by reducing β-catenin in the nucleus via exporting it into the cytoplasm for degradation (26,33). Furthermore, β-catenin binds to FAM, a deubiquitinating enzyme also known as USP9X (ubiquitin specific peptidase 9, X-linked), which inhibits the degradation of β-catenin, leading to its accumulation in the cytoplasm (34,35). Additionally, APC and AXIN1 can shuttle β-catenin from the nucleus to the cytoplasm (36,37).
In contrast to the cytoplasmic reactivity observed for β-catenin in all cases, nuclear staining of the protein was observed in 17 out of 61 cases (27.9%). One potential explanation for β-catenin nuclear accumulation relates to the form of the protein that is generated by the mutation, which predominates in the nucleus (25). Additionally, CTNNB1 was found to be overexpressed in 43.9% of the medulloblastoma cases analyzed. However, neither CTNNB1 mutation status nor gene expression level could be correlated to protein expression or the nuclear location of β-catenin.
The upregulation of β-catenin observed in our study via gene and protein expression analyses may also have been influenced by the oncoprotein MUC1, which interacts directly with β-catenin to block GSK3β-mediated phosphorylation and consequent β-catenin degradation (38).
The overall mutation rate identified for CTNNB1 (18.0%) in the present series was similar to previously reported results from other studies (39-41). In the present study, all the studied cases presented missense heterozygotic CTNNB1 mutations in highly conserved serine residues that are phosphorylation sites for GSK3β and required for the degradation of β-catenin or at codons that code for amino acids flanking those serine residues. Alteration in one allele is sufficient to induce β-catenin nuclear translocation, which triggers its oncogenic role; therefore, all mutations can be considered dominant activating mutations (17-18). However, the present results confirm that CTNNB1 mutational status might not be the only factor determining the trafficking of β-catenin within the cell. As described previously, accumulation of this protein has also been observed without CTNNB1 mutation (29) and in the presence of mutations in other genes downstream in the Wnt pathway, such as APC and AXIN1. In fact, APC and AXIN1 are important for the export of β-catenin from the nucleus to the cytoplasm (36,37), and nuclear accumulation of β-catenin has been observed in colorectal cancers with APC mutations (37,42,43). A similar mechanism for β-catenin nuclear accumulation might also be expected in medulloblastomas because APC mutations have been described in 4.3% of cases of this type of tumor (44). AXIN1 mutations have also been reported in 5% of medulloblastoma cases (45), and such mutations might also lead to β-catenin nuclear accumulation. Furthermore, the post-APC knockdown accumulation of β-catenin in the nucleus observed in a medulloblastoma cell line provides additional in vitro evidence of the mechanisms underlying alterations in β-catenin trafficking (46).
Additionally, epigenetic silencing of genes coding for the secreted frizzled-related protein (SFRP1, SFRP2 and SFRP3) family members has been described as an additional mechanism that may activate the Wnt signaling pathway in medulloblastomas (47). Furthermore, the SFRP family members code for proteins that can limit Wnt signaling, as these soluble proteins bind to Wnt ligands and sequester them from the frizzled receptors, which are the serpentine receptors responsible for binding to Wnt proteins at the plasma membrane (48).
Few previous studies have focused on the expression level of β-catenin transcripts in medulloblastomas or on the expression of genes coding for the proteins involved in the Wnt signaling pathway. Thus, this study is the first to demonstrate the relative mRNA expression of genes involved in the Wnt pathway in medulloblastomas using QT-PCR. When we analyzed the expression levels of CTNNB1, AXIN1 and APC according to the medulloblastoma histological type, CTNNB1 presented lower relative gene expression levels in large-cell variant tumors than in the classic or desmoplastic/extensive nodularity type tumors. However, Wnt signaling activation has been described most frequently in classic medulloblastomas (4,6), which suggests that these data should be further evaluated with a larger number of samples because desmoplastic, extensive nodularity and large-cell medulloblastomas are not as common as classic medulloblastoma variant tumors. The expression levels of CTNNB1, AXIN1 and APC did not correlate with the β-catenin mutation status, which corroborates previous gene expression microarray analyses (49). Furthermore, the analyzed gene profiles were not significantly associated with tumor cell dissemination or the progression-free and overall survival times.
The results of our study suggest that molecular pathways other than the Wnt pathway are involved in medulloblastoma formation, such as the Sonic hedgehog signaling pathway (SHH subgroup) and the Notch pathway, as well as additional cytogenetic aberrations (5-8). In summary, we demonstrated that only a small subset of medulloblastomas carry the CTNNB1 gene mutation that leads to nuclear accumulation of β-catenin. Thus, components of the Wnt signaling pathway as well as other signaling pathways play important roles in the genetic abnormalities underlying medulloblastomas. The present findings confirm the importance of the Wnt pathway in classic, desmoplastic and extensive nodularity tumor variants but not in the large-cell medulloblastoma variant.
ACKNOWLEDGMENTS
We would like to thank FAPESP (grant 04/12133-6), the Ludwig Institute for Cancer Research, Fundação Faculdade de Medicina and CNPq for financial grants. We thank Valeria Muoio and the other neurosurgeons for their help with collecting samples and the clinical follow up. We also thank the Psychiatry Institute for logistical help concerning the surgical therapies.
Footnotes
No potential conflict of interest was reported.
REFERENCES
- 1. Giangaspero F, Eberhart CG, Haapasala H, Pietsch T, Wiestler OD, Ellison DW. Louis D N, Ohgaki H, Wiestler O D, Cavenee W K, edMedulloblastoma 2007France: IARC; 132–40.In WHO classification of tumors of the central nervous system [Google Scholar]
- 2.Giordana MT, Schiffer P, Schiffer D. Prognostic factors in medulloblastoma. Child′s Nerv Syst. 1998;14(6):256–62. doi: 10.1007/s003810050221. [DOI] [PubMed] [Google Scholar]
- 3.Louis ND, Ohgaki H, Wiestler DO, Cavenee KW, Burger CP, Jouvet A, et al. The 2007 WHO Classification of tumors of the Central Nervous System. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gulino A, Arcella A, Giangaspero F. Pathological and molecular heterogeneity of medulloblastoma. Curr Opin Oncol. 2008;20(6):668–75. doi: 10.1097/CCO.0b013e32831369f4. [DOI] [PubMed] [Google Scholar]
- 5.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–31. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 6.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3(8):e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–14. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29(11):1424–30. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–84. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clifford SC, Lusher ME, Lindsey JC, Langdon JA, Gilbertson RJ, Straughton D, et al. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5(22):2666–70. doi: 10.4161/cc.5.22.3446. [DOI] [PubMed] [Google Scholar]
- 12.Wodar A, Nusse R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi A. Tumor formation by genetic mutations in the components of the Wnt signaling pathway. Cancer Sci. 2003;94(3):225–9. doi: 10.1111/j.1349-7006.2003.tb01424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morin PJ. Beta-catenin signaling and cancer. Bioessays. 1999;21(12):1021–30. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 15.Wong CM, Fan ST, Ng IO. Beta-catenin mutation and over expression in hepatocellular carcinoma. Cancer. 2001;92(1):136–45. doi: 10.1002/1097-0142(20010701)92:1<136::aid-cncr1301>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 16.Cui J, Zhou X, Liu Y, Tang Z, Romeih M. Wnt signaling in hepatocellular carcinoma: analysis of mutation and expression of beta-catenin, T-cell factor-4 and glycogen synthase kinase 3-beta genes. J Gastroenterol Hepatol. 2003;18(3):280–7. doi: 10.1046/j.1440-1746.2003.02973.x. [DOI] [PubMed] [Google Scholar]
- 17.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 18.Peifer M. Beta-catenin as oncogene: the smoking gun. Science. 1997;275(5307):1752–3. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- 19.Zhurinsky J, Sctutman M, Bem-Ze'ev A. Plakoglobin and β-catenin: protein interactions, regulation and biological roles. J Cell Sci. 2000;113(Pt 18):3127–39. doi: 10.1242/jcs.113.18.3127. [DOI] [PubMed] [Google Scholar]
- 20.Howell BW, Hertz J. The LDL receptor gene family: signalling functions during development. Curr Opin Neurobiol. 2001;11(1):74–81. doi: 10.1016/s0959-4388(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 21.Polaskis P. Wnt signaling and cancer. Genes Dev. 2007;14:1837–51. [PubMed] [Google Scholar]
- 22.Miller K, Auld J, Jessup E, Rhodes A, Ashton-Key M. Antigen unmasking in formalin-fixed routinely processed paraffin was-embedded section by pressure cooking: A comparison with microwave oven heating and traditional methods. Advances Anat Pathol. 1995;2:60–4. [Google Scholar]
- 23.Hsu SM, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: A comparison between ABC and unlabed antibody (PAP) procedure. J Histochem Cytochem. 1981;29(4):577–80. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Eberhart CG, Tihan T, Burger PC. Nuclear localization and mutation of beta-catenin in medulloblastomas. J Neuropathol Exp Neurol. 2000;59(4):333–37. doi: 10.1093/jnen/59.4.333. [DOI] [PubMed] [Google Scholar]
- 26.Marino S. Medulloblastoma: developmental mechanisms out of control. Trends Mol Med. 2005;11(1):17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Reya T, Clevers H. Wnt signaling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 28.Min HS, Lee YJ, Park K, Cho BK, Park SH. Medulloblastoma: histopathologic and molecular markers of anaplasia and biologic behavior. Acta Neuropathol. 2006;112(1):13–20. doi: 10.1007/s00401-006-0073-9. [DOI] [PubMed] [Google Scholar]
- 29.Yokota N, Nishizawa S, Ohta S, Date H, Sugimura H, Namba H, et al. Role of Wnt pathway in medulloblastoma oncogenesis. Int J Cancer. 2002;101(2):198–201. doi: 10.1002/ijc.10559. [DOI] [PubMed] [Google Scholar]
- 30.Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, et al. β-catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children's Cancer study group brain tumour Committee. J Clin Oncol. 2005;23(31):7951–7. doi: 10.1200/JCO.2005.01.5479. [DOI] [PubMed] [Google Scholar]
- 31.Ellison DW, Kocak M, Dalton J, Megahed H, Lusher ME, Ryan SL, et al. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clinic Oncol. 2011;29(11):1400–7. doi: 10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder JA, Adriance MC, McConnell EJ, Thompson MC, Pockaj B, Gendler SJ. ErbB-beta-catenin complexes are associated with human infiltrating ductal breast and murine mammary tumor virus (MMTV)-Wnt-1 and MMTV-c-Neu transgenic carcinomas. J Biol Chem. 2002;277(25):22692–8. doi: 10.1074/jbc.M201975200. [DOI] [PubMed] [Google Scholar]
- 33.Meng X, Poon R, Zhang X, Cheah A, Ding Q, Hui CC, Alman B. Suppressor of fused negatively regulates beta-catenin signaling. J Biol Chem. 2001;276(43):40113–9. doi: 10.1074/jbc.M105317200. [DOI] [PubMed] [Google Scholar]
- 34.Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999;4(12):757–67. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- 35.Pantaleon M, Kanai-Azuma M, Mattick JS, Kaibuchi K, Kaye PL, Wood SA. FAM deubiquitylating enzyme is essential for preimplantation mouse embryo development. Mech Dev. 2001;109(2):151–60. doi: 10.1016/s0925-4773(01)00551-2. [DOI] [PubMed] [Google Scholar]
- 36.Cong F, Varmus H. Nuclear-cytoplasmic shuttling of axin regulates subcellular localization of beta-catenin. Proc Natl Acad Sci USA. 2004;101(9):2882–7. doi: 10.1073/pnas.0307344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson BR. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat Cell Biol. 2000;2(9):653–60. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- 38.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. Muc1 oncoprotein blocks glycogen synthase kinase 3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65(22):10413–22. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 39.Rogers HA, Miller S, Lowe J, Brundler MA, Coyle B, Grundy RG. An investigation of WNT pathway activation and association with survival in central nervous system primitive neuroectodermal tumours (CNS PNET) Br J Cancer. 2009;100(8):1292–302. doi: 10.1038/sj.bjc.6604979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizem J, Popovic M, Cör A. Expression of Gli1 and PARP1 in medulloblastoma: an immunohistochemical study of 65 cases. J Neurooncol. 2010;103(3):459–67. doi: 10.1007/s11060-010-0431-2. [DOI] [PubMed] [Google Scholar]
- 41.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosin-Arbesfeld R, Townsley F, Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406(6799):1009–12. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- 43.Wechsler-Reya RJ, Scott MP. Expansion of CNS precursor pools: a new role for Sonic Hedgehog. Neuron. 1999;22(1):2–3. doi: 10.1016/s0896-6273(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 44.Huang H, Mahler-Araujo BM, Sankila A, Chimelli L, Yonekawa Y, Kleihues P, Ohgaki HV. APC mutations in sporadic medulloblastomas. Am J Pathol. 2000;156(2):433–7. doi: 10.1016/S0002-9440(10)64747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baeza N, Masuoka J, Kleihues P, Ohgaki H. AXIN1 mutations but not deletions in cerebellar medulloblastomas. Oncogene. 2003;22(4):632–6. doi: 10.1038/sj.onc.1206156. [DOI] [PubMed] [Google Scholar]
- 46.Salaroli R, Russo A, Ceccarelli C, Mina GD, Arcella A, Martinelli GN, et al. Intracellular distribution of beta-catenin in human medulloblastoma cell lines with different degree of neuronal differentiation. Ultrastruct Pathol. 2007;31(1):33–44. doi: 10.1080/01913120601169501. [DOI] [PubMed] [Google Scholar]
- 47.Kongkham PN, Northcott PA, Croul SE, Smith CA, Taylor MD, Rutka JT. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene. 2010;29(20):3017–24. doi: 10.1038/onc.2010.32. [DOI] [PubMed] [Google Scholar]
- 48.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121(Pt 6):737–46. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 49.Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S, Manie E, et al. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical Characteristics. J Pathol. 2009;218(1):86–94. doi: 10.1002/path.2514. [DOI] [PubMed] [Google Scholar]