Abstract
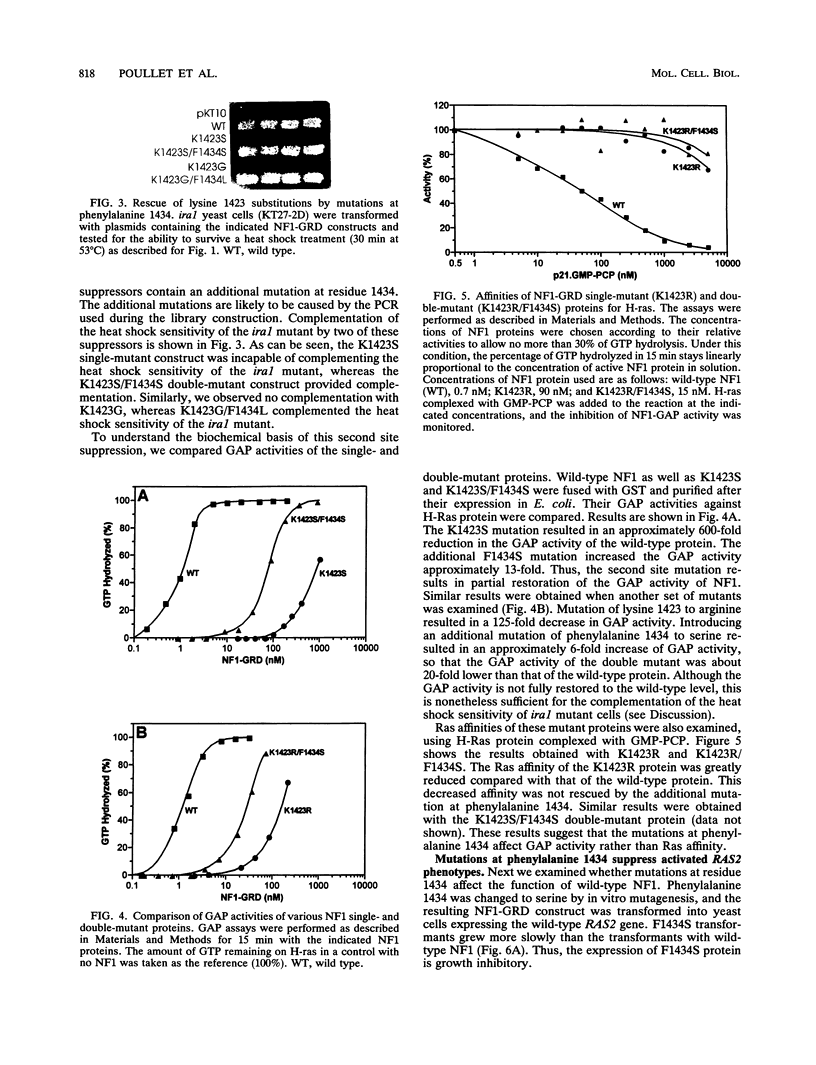
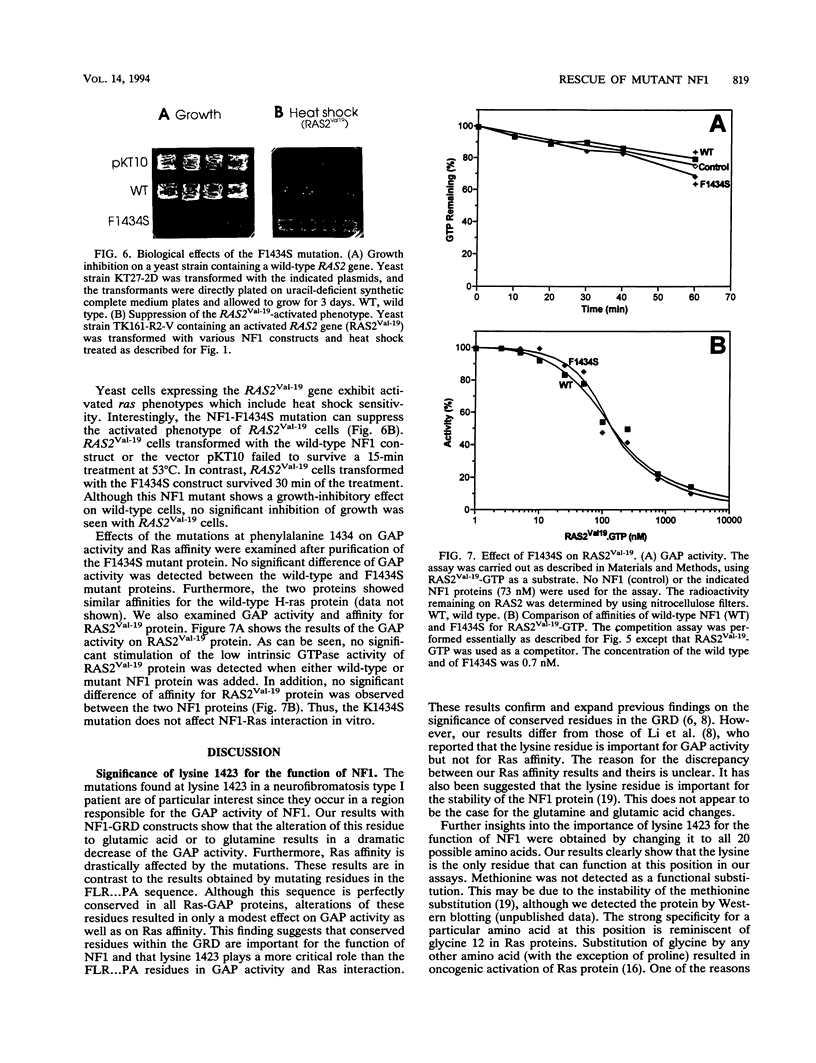
Lysine 1423 of neurofibromin (neurofibromatosis type I gene product [NF1]) plays a crucial role in the function of NF1. Mutations of this lysine were detected in samples from a neurofibromatosis patient as well as from cancer patients. To further understand the significance of this residue, we have mutated it to all possible amino acids. Functional assays using yeast ira complementation have revealed that lysine is the only amino acid that produced functional NF1. Quantitative analyses of different mutant proteins have suggested that their GTPase-activating protein (GAP) activity is drastically reduced as a result of a decrease in their Ras affinity. Such a requirement for a specific residue is not observed in the case of other conserved residues within the GAP-related domain. We also report that another residue, phenylalanine 1434, plays an important role in NF1 function. This was first indicated by the finding that defective NF1s due to an alteration of lysine 1423 to other amino acids can be rescued by a second site intragenic mutation at residue 1434. The mutation partially restored GAP activity in the lysine mutant. When the mutation phenylalanine 1434 to serine was introduced into a wild-type NF1 protein, the resulting protein acquired the ability to suppress activated phenotypes of RAS2Val-19 cells. This suppression, however, does not involve Ras interaction, since the phenylalanine mutant does not stimulate the intrinsic GTPase activity of RAS2Val-19 protein and does not have an increased affinity for Ras proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen L. B., Ballester R., Marchuk D. A., Chang E., Gutmann D. H., Saulino A. M., Camonis J., Wigler M., Collins F. S. A conserved alternative splice in the von Recklinghausen neurofibromatosis (NF1) gene produces two neurofibromin isoforms, both of which have GTPase-activating protein activity. Mol Cell Biol. 1993 Jan;13(1):487–495. doi: 10.1128/mcb.13.1.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov 16;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Bernards A., Haase V. H., Murthy A. E., Menon A., Hannigan G. E., Gusella J. F. Complete human NF1 cDNA sequence: two alternatively spliced mRNAs and absence of expression in a neuroblastoma line. DNA Cell Biol. 1992 Dec;11(10):727–734. doi: 10.1089/dna.1992.11.727. [DOI] [PubMed] [Google Scholar]
- Buchberg A. M., Cleveland L. S., Jenkins N. A., Copeland N. G. Sequence homology shared by neurofibromatosis type-1 gene and IRA-1 and IRA-2 negative regulators of the RAS cyclic AMP pathway. Nature. 1990 Sep 20;347(6290):291–294. doi: 10.1038/347291a0. [DOI] [PubMed] [Google Scholar]
- Gaul U., Mardon G., Rubin G. M. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992 Mar 20;68(6):1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- Gutmann D. H., Boguski M., Marchuk D., Wigler M., Collins F. S., Ballester R. Analysis of the neurofibromatosis type 1 (NF1) GAP-related domain by site-directed mutagenesis. Oncogene. 1993 Mar;8(3):761–769. [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr 15;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Li Y., Bollag G., Clark R., Stevens J., Conroy L., Fults D., Ward K., Friedman E., Samowitz W., Robertson M. Somatic mutations in the neurofibromatosis 1 gene in human tumors. Cell. 1992 Apr 17;69(2):275–281. doi: 10.1016/0092-8674(92)90408-5. [DOI] [PubMed] [Google Scholar]
- Marchuk D. A., Saulino A. M., Tavakkol R., Swaroop M., Wallace M. R., Andersen L. B., Mitchell A. L., Gutmann D. H., Boguski M., Collins F. S. cDNA cloning of the type 1 neurofibromatosis gene: complete sequence of the NF1 gene product. Genomics. 1991 Dec;11(4):931–940. doi: 10.1016/0888-7543(91)90017-9. [DOI] [PubMed] [Google Scholar]
- Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O'Connell P., Cawthon R. M. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990 Nov 16;63(4):843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Mitts M. R., Bradshaw-Rouse J., Heideman W. Interactions between adenylate cyclase and the yeast GTPase-activating protein IRA1. Mol Cell Biol. 1991 Sep;11(9):4591–4598. doi: 10.1128/mcb.11.9.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakafuku M., Nagamine M., Ohtoshi A., Tanaka K., Toh-e A., Kaziro Y. Suppression of oncogenic Ras by mutant neurofibromatosis type 1 genes with single amino acid substitutions. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6706–6710. doi: 10.1073/pnas.90.14.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi T., Lee P. S., Oka K., Levin V. A., Tanase S., Morino Y., Saya H. Differential expression of two types of the neurofibromatosis type 1 (NF1) gene transcripts related to neuronal differentiation. Oncogene. 1991 Sep;6(9):1555–1559. [PubMed] [Google Scholar]
- Schaber M. D., Garsky V. M., Boylan D., Hill W. S., Scolnick E. M., Marshall M. S., Sigal I. S., Gibbs J. B. Ras interaction with the GTPase-activating protein (GAP). Proteins. 1989;6(3):306–315. doi: 10.1002/prot.340060313. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Colby W. W., Capon D. J., Goeddel D. V., Levinson A. D. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984 Nov 1;312(5989):71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Lin B. K., Wood D. R., Tamanoi F. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):468–472. doi: 10.1073/pnas.88.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Boguski M., Riggs M., Rodgers L., Wigler M. sar1, a gene from Schizosaccharomyces pombe encoding a protein that regulates ras1. Cell Regul. 1991 Jun;2(6):453–465. doi: 10.1091/mbc.2.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmüller L., Wittinghofer A. Expression of the GTPase activating domain of the neurofibromatosis type 1 (NF1) gene in Escherichia coli and role of the conserved lysine residue. J Biol Chem. 1992 May 25;267(15):10207–10210. [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Xu G. F., O'Connell P., Viskochil D., Cawthon R., Robertson M., Culver M., Dunn D., Stevens J., Gesteland R., White R. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990 Aug 10;62(3):599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]