Summary
Purine nucleoside phosphorylase (PNP) is a target for leukemia, gout and autoimmune disorders. Dynamic motion of catalytic site loops has been implicated in catalysis, but experimental evidence was lacking. We replaced catalytic site groups His257 or His64 with 6-fluoro-tryptophan (6FW) as site-specific NMR probes. Conformational adjustments in the 6FW-His257-helical and His64-6FW-loop regions were characterized in PNP phosphate bound enzyme and in complexes with catalytic site ligands, including transition state analogues. Chemical shift and line-shape changes associated with these complexes revealed dynamic coexistence of several conformational states in these regions in phosphate bound enzyme and altered or single conformations in other complexes. These conformations were also characterized by X-ray crystallography. Specific 19F-Trp labels and X-ray crystallography provide multidimensional characterization of conformational states for free, catalytic and inhibited complexes of human PNP.
INTRODUCTION
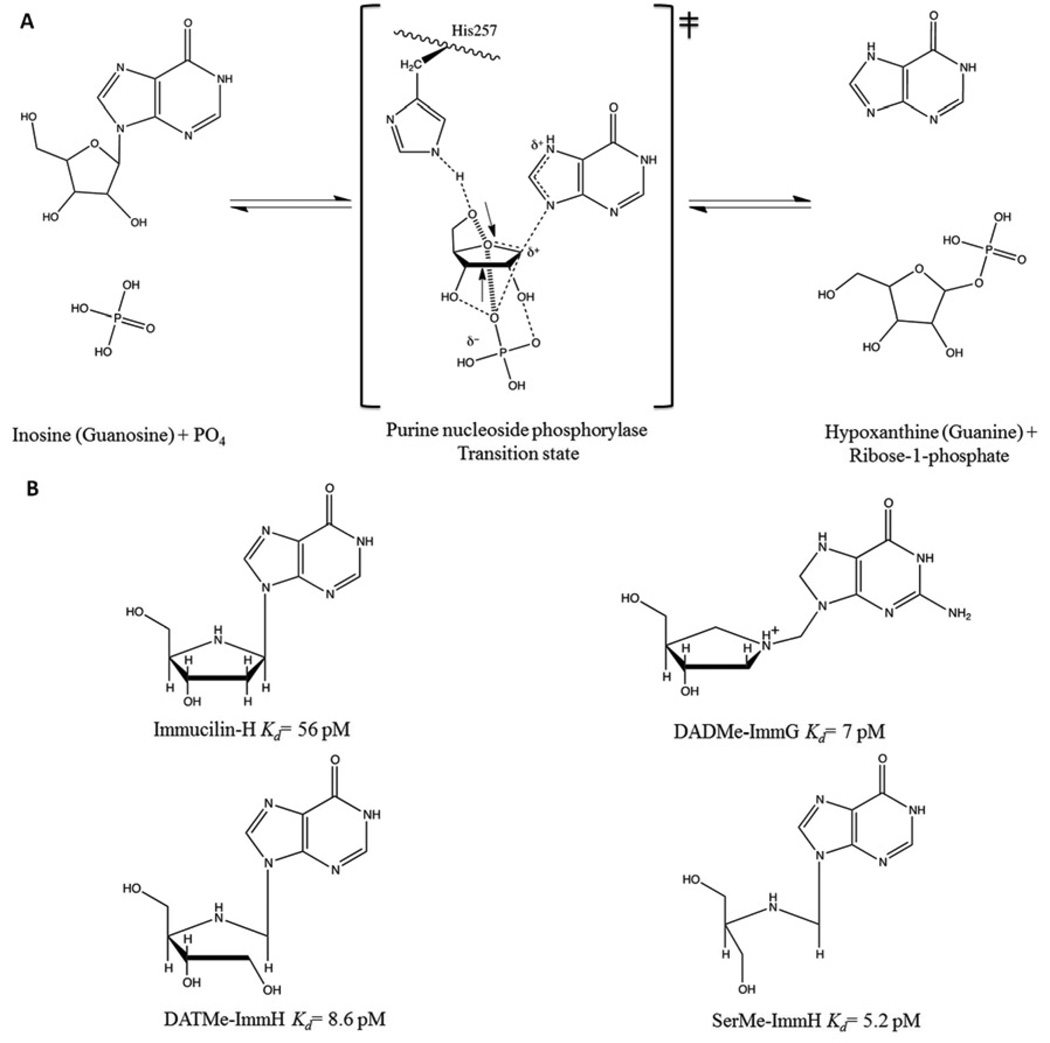
Human purine nucleoside phosphorylase (PNP) is essential for the purine recycling pathway involving the degradation and synthesis of DNA and RNA. It catalyzes the reversible phosphorolysis of purine (deoxy)-ribonucleosides to the corresponding purine base and (2’-deoxy) ribose 1-phosphate (Fig. 1A) (Schramm, 2005; Stoeckler et al., 1978; Stoeckler et al., 1980). The genetic deficiency of human PNP activity induces apoptosis in activated T-cells while other cells and tissues remain unaffected (Giblett et al., 1975). The transition state structures of bovine and human PNPs have been solved by a combination of experimentally determined kinetic isotope effects coupled with theoretical calculations (Kline and Schramm, 1993; Lewandowicz and Schramm, 2004). Stable chemical mimics of these transition states have produced four generations of picomolar transition state analogue inhibitors (Ho et al., 2010) (Figure 1B). Two PNP inhibitors are in clinical trials for leukemia and gout and a third shows antimalarial effects in Aotus primates (Balakrishnan et al., 2010; Holister et al., 2011; Cassera et al., 2011).
Figure 1. Phosphorolysis reaction catalyzed by human PNP and chemical structure of four generations of PNP inhibitors.
(A) The reaction occurs via ribosyl C1-migration from base to phosphate nucleophile to effect nucleophilic displacement by electrophile migration. The mechanism is SN1-like through fully formed ribocation transition state. Ribose 1-phosphate and the purine base are products. (B) Four generation of transition state analog inhibitors, ImmH, DADMeImmG, DATMeImmH, and SerMeImmH.
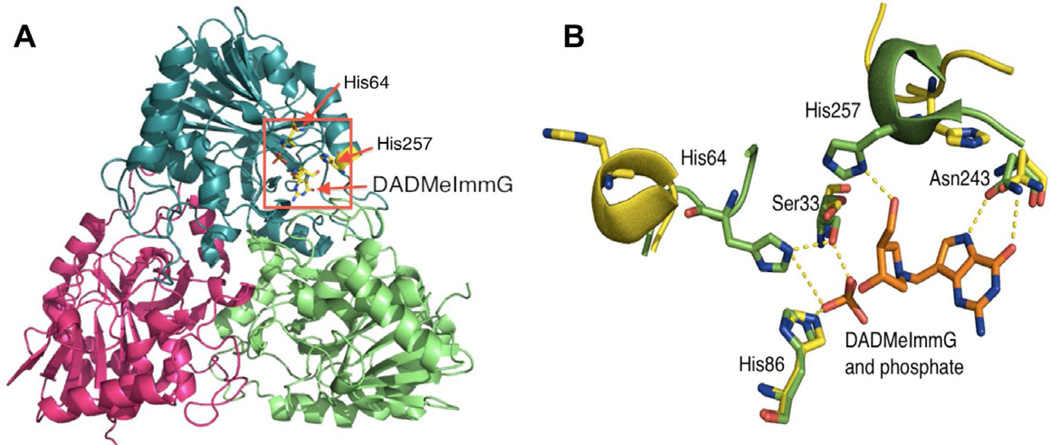
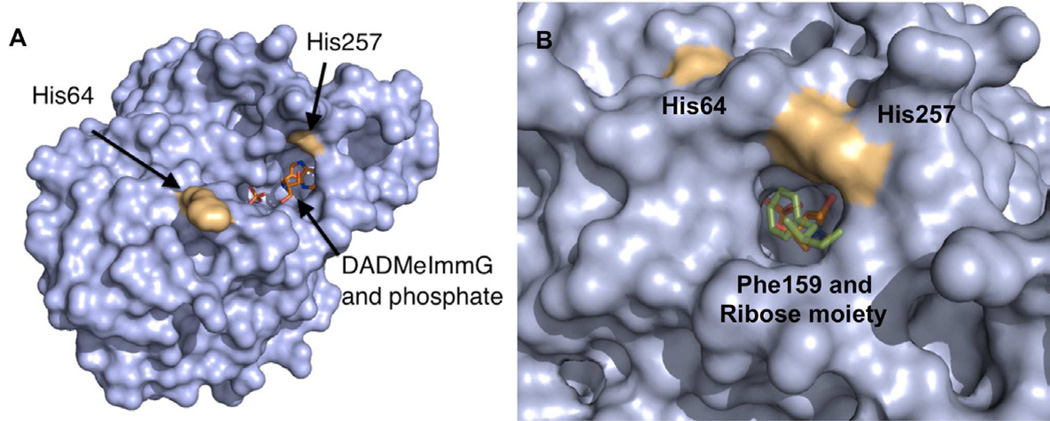
Human PNP is a homotrimer (~32 KD per monomer) with catalytic sites located near inter subunit interfaces (Fig. 2A). Crystal structures of human PNP in complex with substrates and transition state (TS) analogues (Figure 1B) have defined the active site residues in contact with the purine nucleoside and the phosphate nucleophile, confirming that the reaction occurs in a three-centered mechanism (purine-ribocation-phosphate) (de Azevedo et al., 2003; Koellner et al., 1997; Rinaldo-Matthis et al., 2008; Shi et al., 2004). The PNP active site residues interacting closely with substrate (inosine) or transition state analogue (DADMeImmG) are Asn243, Glu201, His257, Phe200, Tyr88, Met219, and Phe159 (from the adjacent subunit). Residues Ser33, His 64, Arg84, His86, Ala116, Tyr192 and Ser220 interact with the phosphate nucleophile (Fig. 2B shows only residues Asn243, His257, Ser33, His64, and His86 for clarity).
Figure 2. Structural elements of the human PNP homotrimer.
(A) The ternary complex with one of the subunits bound to DADMeImmG and phosphate. The red rectangle shows the location of the active site near the subunit interface. (B) Catalytic site residues of human PNP with an overlap of two crystal structures. The positions of active site residues of human PNP in the sulfate bound form (yellow) are overlapped with the ternary complex with bound DADMeImmG and phosphate (green).
A comparison of the sulfate-bound hPNP (PDB ID: 1M73) and the phosphate + DADMeImmG-bound hPNP (PDB ID: 3PHB) crystal structures reveal two major conformational alterations upon binding of substrates or transition state analogues (Fig. 2B). The His257-helix and His64-loop adopt an open conformation and are directed away from the active site in the sulfate-bound structure, while the inhibitor + phosphate tertiary complex shows a closed conformation, with these residues closer to the active site and within H-bonding distance of ligands (Fig. 2B). His257 of PNP is in hydrogen bond distance of the 5’-OH of the ribosyl group and is a key residue in positioning the “oxygen stack” (O5’-O4’-Op) that contributes to catalysis through a vibrational promoting mode of the three oxygen atoms. This “oxygen stack” has been proposed to provide electron density changes to destabilize the ribosyl group to form the carbocation transition state (TS) and thereby enhance departure of the purine based leaving group (Figs. 1A and 2B) (Murkin et al., 2007; Saen-Oon et al., 2008). His64 is located in an unstructured and flexible loop region that changes between open, intermediate and closed conformations depending on the ligand bound to the active site. The range of motion of this loop is from ~17Å away from the active site in guanine-bound PNP (open loop conformation), to ~12.5Å away from the active site in a phosphate bound form (intermediate loop conformation) and finally to a phosphate + inhibitor-bound form where His64 is in hydrogen bond distance of the phosphate nucleophile (closed loop conformation) (Figs. 2B, 3C and 3D). These differences observed in the crystal structures suggest flexibility for these regions during the catalytic cycle (Murkin et al., 2007; Murkin et al., 2008). Molecular dynamic calculations (QM/MM) of human PNP also predicts these regions to be flexible (Hirschi et al., 2010).
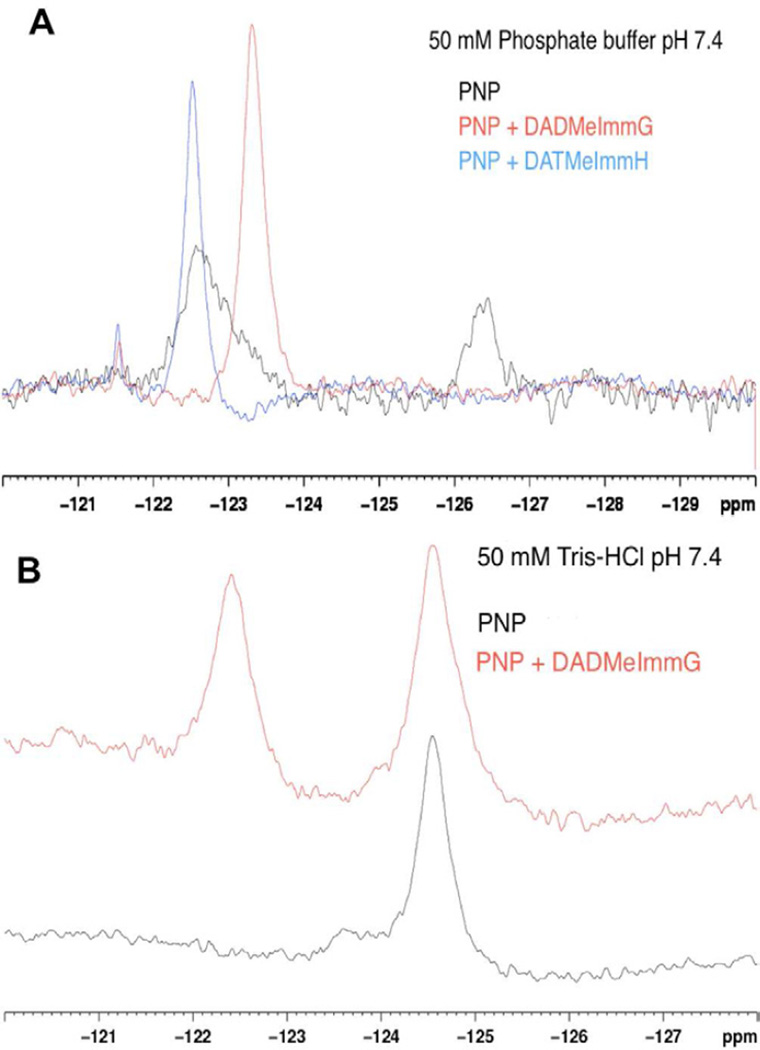
Figure 3. 19F-NMR spectra of H257-6FW-leuko PNP.
(A) 19F-NMR spectrum of H257-6FW-leuko PNP bound to phosphate (black line), 19F-NMR spectrum of H257-6FW-leuko PNP bound to DADMeImmG and phosphate (red line) and 19F-NMR spectrum of H257-6FW-leuko PNP bound to DATMeImmH and phosphate (blue line). (B) 19F-NMR spectrum of H257-6FW-leuko PNP bound to guanine/hypoxanthine in Tris-HCl buffer pH 7.4 (black line) and 19F-NMR spectrum of H257-6FW-leuko PNP bound to guanine/hypoxanthine and DADMeImmG in Tris-HCl buffer pH 7.4 (red line).
Laser-induced temperature-jump spectroscopy previously characterized the conformational dynamics of the catalytic site Phe159 loop located near the ribosyl region of bound reactants (Ghanem et al., 2009). A Phe159Trp mutation in Leuko-PNP (Trp-free PNP, where the three native tryptophan residues were replaced by tyrosines (Ghanem et al., 2008)) was used as a reporter group to probe the motions of the catalytic site loop. Here, we introduce a single 6-fluoro-tryptophan (6FW) into the H257W-Leuko-PNP and H64W-Leuko-PNP mutants at the His257 and His64 positions respectively. These fluorinated PNP mutants (H257-6FW-Leuko-PNP and H64-6FW-Leuko-PNP) were employed as active site reporters for 19F NMR. A powerful application of 19F NMR is to probe proteins of known structure in solution where the sensitivity of the 19F resonance to its molecular environment can reveal structural and kinetic features of protein conformation and dynamics (Kitevski-LeBlanc and Prosser, 2012). Structural changes in the His257-helical and His64-loop regions were monitored by incorporation of 6FW side chains. NMR spectral changes in ligand-bound PNP complexes revealed the coexistence of distinct conformational states for both regions (His257-helix and His64-loop). X-ray crystal structures of the same ligand bound protein complexes provide structural benchmarks to complement the 19F-NMR dynamic observations.
RESULTS
Steady state kinetics and transition state analogue inhibition
The effects of mutations and incorporation of 6FW on the steady state kinetics of inosine phosphorolysis and DADMeImmG inhibition were compared to native and Leuko PNP (Table 1). The H257W-Leuko-PNP mutant has a kcat reduced by ~16 fold and a Km increased by ~2 fold compared to native PNP and a kcat reduced by ~12 fold and a Km increased by ~3 fold when compared to Leuko-PNP (Table 1). Similarly, the H64W-Leuko-PNP has a kcat reduced by ~6 fold and a Km increased by ~4 fold when compared to native PNP and a kcat reduced by ~5 fold and a Km increased by ~5 fold when compared to Leuko-PNP (Table 1). Similar effects on the kinetic parameters for inosine phosphorolysis were observed for the fluorinated mutants ([H257-6FW-leuko-PNP and H64-6FW-Leuko-PNP), indicating that the incorporation of 6FW has only modest effects on the catalytic activity.
Table 1.
Comparison of the steady state kinetic constants and inhibition constants for native, leuko, H257W-leuko, H64W-leuko, H257-6FW-leuko and H64-6FW-leuko PNPs with inosine as a substrate and DADMelmmG as an inhibitor.
| WTPNP | Leuko PNP |
H257W Leuko |
H257-6FW Leuko |
H64W Leuko |
H64-6FW Leuko |
|
|---|---|---|---|---|---|---|
| Kcat(s−1) | 47 ± 0.12 | 36 ± 1 | 3 ± 0.1 | 3 ± 0.1 | 8 ± 0.16 | 7.9 ± 0.22 |
| Km (uM) | 68 ± 8 | 50 ± 3 | 136 ± 12 | 121 ± 14 | 259 ± 39 | 240 ± 19 |
| Kcat/Km | 6.9 × 105 | 7.2 × 105 | 2.2 × 104 | 2.5 × 104 | 3.1 × 104 | 3.3 × 104 |
| DADMelmmG inhibition | ||||||
| Ki(pM) | 100 ± 10 | 376 ± 63 | 151 ± 16 | 119 ± 6 | 109 ± 17 | 80 ± 4 |
| Ki*(pM) | 14 ± 2 | 9 ± 1 | 110 ± 10 | NA | 27 ± 6 | NA |
DADMeImmG inhibition constants (Ki and Ki*) for the H257W-Leuko-PNP mutant were increased by ~0.6 and ~7 fold when compared to native PNP and reduced by ~3 fold and increased by ~12 fold respectively when compared to the Leuko-PNP enzyme (Table 1). In the case of the H64W-Leuko-PNP mutant, the Ki for DADMeImmG was unaltered when compared to native PNP and reduced by ~3 fold when compared to Leuko-PNP while the Ki for DADMeImmG was increased by 2 fold when compared to native PNP and increased by ~3 fold when compared to Leuko-PNP (Table 1). Remarkably, slow onset inhibition (Ki*) was not observed in either of the fluorinated mutants (H257-6FW-Leuko-PNP and H64-6FW-Leuko-PNP) while the Ki was essentially unaltered in both fluorinated enzymes (relative to the unfluorinated parent molecules). A similar loss of slow onset inhibition with Immucillin-H has also been observed in H257 mutants (Murkin et al., 2007). Despite the four amino acid substitutions and the incorporation of a fluorinated amino acid (the percentage of 6FW incorporation was ~75%, as determined by mass spectrometry) on an active site location, the H257W-Leuko-PNP and H64W-Leuko-PNP mutants are active and provide useful active site probes to study the dynamics and environment of the 257-helix and the 64-loop regions.
19F-NMR of H257-6FW-Leuko-PNP
The 19F-NMR spectrum of phosphate bound H257-6FW-Leuko-PNP in 50 mM potassium phosphate buffer at pH 7.4 shows the presence of two distinct and clearly resolved signals at −122.7 ppm and −126.4 ppm (Fig. 3A, black line). As there is a single fluorine label in this mutant, the 6FW in the 257-helix region adopts two different conformations with distinct environments. Preliminary paramagnetic relaxation effect (PRE) experiments using Omniscan showed a larger effect on intensity on the signal at −122.7 suggesting that this conformation is more solvent exposed (data not shown). The integrated area of these two distinct NMR signals is ~ 3:1 indicating that the −122.7 ppm conformation is favored over the −126.4 ppm conformation.
When a nuclei experiences different chemical environments on the NMR timescale it is in chemical exchange. The rate chemical exchange is affected by temperature. At low temperature, interconversion slows beyond the NMR range and the 19F spectrum consists of distinct peaks. Increased temperature broadens, then merge the peaks into an average position. No evidence of chemical exchange between these conformations was observed after collecting 19F-NMR spectra of H257-6FW-Leuko-PNP at 288 K, 298 K, and 310 K. This supports slow conformational exchange, conformers that are not in exchange, or different conformations in each monomer of the PNP trimer. Negative cooperativity between PNP subunits is known to occur upon binding of inhibitors (Ghanem et al., 2009; Miles et al., 1998) showing that conformational alterations in one subunit can be transmitted to neighboring subunits. Thus, the two signals observed in 50 mM phosphate, pH 7.4 could arise from equilibrium between two major conformations located in separate subunits.
Enzymes achieve their reaction rate enhancement by lowering the energetic barrier to the transition state through dynamic interactions (Schramm, 2005). Transition state analogue inhibitors are designed to mimic the geometry and electrostatics of the transition state and convert the dynamic interactions into thermodynamic stable states. Structural rearrangements associated with transition state analogue inhibitor binding are expected to mimic the geometry of the enzyme during catalysis. The effect of ligand binding on the conformation of the His257-region was examined in an equimolar complex of the transition state analogue inhibitor DADMeImmG with the H257-6FW-Leuko-PNP mutant in phosphate buffer. In this complex, the two signals observed in the PNP-phosphate complex disappear and a single signal at −123.3 ppm appears (Figure 3A, red line). A similar behavior is observed in a complex of PNP-DATMeImmH-phosphate, but here, the single signal observed upon inhibitor binding is at −122.5 ppm (Figure 3A, blue line). These small differences in chemical shift between the DADMeImmG and DATMeImmH complexes represent small differences in the chemical environments of the fluorine probe in each complex. Upon binding of the transition state analogue, the fluorine probes located in the three monomers experience identical environments in the reorganized catalytic sites.
In PNP-phosphate, the H257-6FW-region is flexible and able to exchange between two conformations or exist in different stable conformations to produce two distinct signals in the 19F-NMR spectrum (Figure 3A, black line). In PNP-inhibitor-phosphate complexes, the H257-6FW-region is locked into a single conformation by interactions with the tightly bound transition state analogue inhibitor and phosphate. Similar 19F-NMR effects occur upon binding of substrates and inhibitors in other systems (Hoeltzli and Frieden, 1994; Liu et al., 2012b; Salopek-Sondi and Luck, 2002).
Phosphate alone is known to induce structural changes in PNP (Ghanem et al., 2008; Ghanem et al., 2009) and we tested the behavior of the fluorine NMR spectrum of the H257-6FW-Leuko-PNP mutant in the absence of phosphate (50 mM Tris-HCl buffer, pH = 7.4) to compare it with PNP-phosphate. The spectrum of H257-6FW-Leuko-PNP without phosphate or inhibitor shows a major sharp signal at −124.6 ppm and a minor broad signal at −123.6 ppm (Fig. 3B). These almost overlapping signals suggest two conformational states with relatively similar environments. Upon addition of an equimolar amount of DADMeImmG, the minor, broad signal at −123.6 ppm disappears and is replaced by a sharper signal that appears at−122.4 ppm while the −124.6 ppm signal remains unaffected (Figure 3B, red line). The lack of a full conversion to a single signal upon binding of inhibitor suggests a reduced affinity for DADMeImmG in the absence of phosphate.
Native and Leuko-PNPs co-purify with tightly bound hypoxanthine to give a stoichiometry of ~ 0.7 to 1.0 per enzyme monomer. Phosphate binding facilitates the release of hypoxanthine (Edwards et al., 2009; Ghanem et al., 2008; Ghanem et al., 2009). Likewise, H257-6FW-Leuko-PNP co-purifies with bound guanine and hypoxanthine with a stoichiometry of 0.8 to 1.0 per enzyme monomer (data not shown). The guanine content suggests a greater affinity for this base relative to hypoxanthine for this mutant. Crystal structures of H257W-Leuko-PNP without phosphate (see below) also show guanine or hypoxanthine bound to the phosphate-free protein but not to the phosphate-bound protein. Altogether, these observations suggest that the sharper signal at −124.6 ppm arises from a 257-region with a restricted movement due to a tightly bound guanine and the broad signal at −123.6 ppm arises from a flexible 257-region in the apo protein (or subunits). This idea is supported by the −123.6 ppm peak being readily affected by the binding of inhibitor (Fig. 3B) and that phosphate and charcoal treatment (known to partially strip PNP from hypoxantine or guanine) of H257-6FW-Leuko-PNP followed by dialysis against 50 mM Tris-HCl buffer pH = 7.4, reduced the intensity of the −124.6 ppm signal and the guanine content of this protein (data not shown).
X-ray crystallography of [6-F-Trp]-H257W-Leuko PNP
Crystal structures were determined for H257W-Leuko-PNP in Tris-HCl buffer pH = 7.4 (PDB ID 4ECE), H257WLeuko-PNP bound to phosphate and DADMeImmG (PDB ID 4EB8), H257-6FW-Leuko-PNP bound to phosphate and DADMeImmG (PDB ID 4EAR) and H257W-Leuko-PNP bound to phosphate (PDB ID 4GKA) to aid in the interpretation of the NMR spectra (Table 2) (Figs. 4A, 4B and S1). An overlap of the crystal structures of the H257W-Leuko-PNP bound to DADMeImmG and phosphate, H257W-Leuko-PNP in Tris-HCl buffer pH = 7.4 (guanine bound), and the H257W-Leuko-PNP in phosphate buffer pH = 7.4 (phosphate bound) is shown in Fig. 4A (green, yellow and blue respectively). Also, an overlap of the WT PNP and H257W-Leuko-PNP both bound to DADMeImmG and phosphate is shown in the supplemental section (Figure S1). The overall structures of these proteins overlap well except for the active site residues Ser33, His64 and Trp257, which show significant conformational rearrangements between the different ligand-bound forms. These changes are essentially the same as the ones observed in native PNPs (Figure 2B). Ser33 is located at the edge of a β-sheet region that “slides” ~ 4 Å towards the active site upon binding of phosphate and inhibitor to form a catalytic triad with phosphate, His86 and His64 (Figure 4A and 4B). His64 is located in an unstructured and flexible loop region that is positioned ~17 Å away from the active site in the guanine-bound PNP. This residue moves ~5 Å towards the active site upon binding of phosphate, and becomes organized at the active site in the PNP-inhibitor-phosphate tertiary complex where it interacts with Ser33 and His86 to form the Ser33-His64-His86 catalytic triad that is proposed to stabilize the phosphate in the active site (Fig. 4A and 4B).
Table 2.
Data collection and structural refinement statistics.
| H257W-leukoPNP- DADMe-ImmG-PO4 (PDB ID 4EB8) |
H257W(f)-leukoPNP- DADMe-ImmG-PO4 (PDB ID 4EAR) |
H257W-leukoPNP- PO4 (PDB ID 4GKA) |
H257W-leukoPNP- guanine (PDB ID 4ECE) |
||
|---|---|---|---|---|---|
| Data collection statistics | |||||
| Space group | P212121 | P212121 | P21 | P21 | |
| Unit cell parameters | a (Å) | 55.75 | 55.92 | 53.88 | 77.55 |
| b (Å) | 131.04 | 131.10 | 233.67 | 129.68 | |
| c (Å) | 137.50 | 137.68 | 70.08 | 104.35 | |
| β (°) | - | - | 97.33 | 105.29 | |
| Temperature (K) | 100 | 100 | 100 | 100 | |
| Wavelength (Å) | 1.075 | 1.075 | 1.075 | 1.075 | |
| Resolution (Å) | 50–2.30 (2.38–2.30) | 50–1.70 (1.73–1.70) | 50–2.20 (2.28–2.20) | 50–2.60 (2.69–2.60) | |
| Rmerge (%) | 9.7 (82.8) | 5.8 (91.4) | 10.3 (87.6) | 7.0 (87.2) | |
| Completeness (%) | 99.9 (100) | 98.3 (97.4) | 100 (100) | 100 (100) | |
| I/σI | 20.8 (2.2) | 47.3 (2.6) | 19.4 (2.1) | 26.2 (2.0) | |
| Unique reflections | 45728 (4476) | 110270 (5375) | 86349 (8627) | 61417 (6104) | |
| Redundancy | 6.0 (5.9) | 8.0 (8.0) | 4.2 (4.2) | 4.2 (4.2) | |
| Mosaicity (°) | 0.9 | 0.4 | 0.5 | 0.3 | |
| B-factor from Wilson plot (A2) | 43 | 28 | 33 | 69 | |
| Refinement statistics | |||||
| Resolution (Å) | 47.48–2.30 | 41.24–1.70 | 45.29–2.20 | 49.04–2.60 | |
| Total number of reflections | 45558 | 110107 | 86296 | 61129 | |
| Working set: number of reflections | 43257 | 104515 | 81976 | 58023 | |
| Rfactor (%) | 18.11 | 17.47 | 18.80 | 22.15 | |
| Test set: number of reflections | 2301 | 5592 | 4320 | 3106 | |
| Rfree (%) | 22.76 | 20.29 | 23.20 | 26.10 | |
| Protein atoms | 6722 (A,B,C)1 | 6753 (A,B,C)1 | 12645 (A,B,C,D,E,F)1 | 13261 (A,B,C,D,E,F)1 | |
| Water atoms | 324 | 579 | 404 | 306 | |
| DADMe-ImmG atoms | 60 | 60 | - | - | |
| Phosphate atoms | 15 | 15 | 30 | - | |
| Guanine atoms | - | - | - | 66 | |
| Ethylene glycol atoms | 8 | - | - | - | |
| Glycerol atoms | 12 | ||||
| Geometry statistics | |||||
| Rmsd (bond distance) (Å) | 0.01 | 0.01 | 0.01 | 0.01 | |
| Rmsd (bond angle) (°) | 1.51 | 1.49 | 1.40 | 1.35 | |
| RmsdB | |||||
| Main chain bonded atoms (Å2) | 1.99 | 1.38 | 1.90 | 1.87 | |
| Side chain bonded atoms (Å2) | 2.74 | 2.25 | 2.48 | 2.26 | |
| Average B | |||||
| Main chain atoms (Å2) | 45.70 | 36.18 | 45.82 | 59.27(A,B,C)/85.49(D,E,F) | |
| Side chain atoms (Å2) | 49.76 | 41.36 | 50.38 | 62.45(A,B,C)/88.54(D,E,F) | |
| Water atoms (Å2) | 45.51 | 45.63 | 42.48 | 55.14 | |
| DAD-Me-ImmG atoms (Å2) | 32.68 | 26.06 | - | - | |
| Phosphate atoms (Å2) | 32.05 | 26.29 | 40.34 | - | |
| Guanine atoms (Å2) | - | - | - | 48.63 | |
| Ethylene glycol atoms (Å2) | 47.98 | - | - | - | |
| Glycerol atoms (Å2) | - | - | 47.83 | - | |
| Ramachandran plot | |||||
| Most favored region (%) | 93.1 | 93.5 | 93.5 | 93.3 | |
| Additionally allowed regions (%) | 6.3 | 5.9 | 6.1 | 5.9 | |
| Generously allowed regions (%) | 0.1 | 0.1 | 0 | 0.3 | |
| Disallowed regions (%)2 | 0.4 | 0.4 | 0.4 | 0.4 | |
ABC chains form one timer and DEF belong 10 another trimer
Thr221 of each monomer is close to allowed regions in the Ramachandran plot. In the crystal structures, Thr221 locates in one of the active site loops
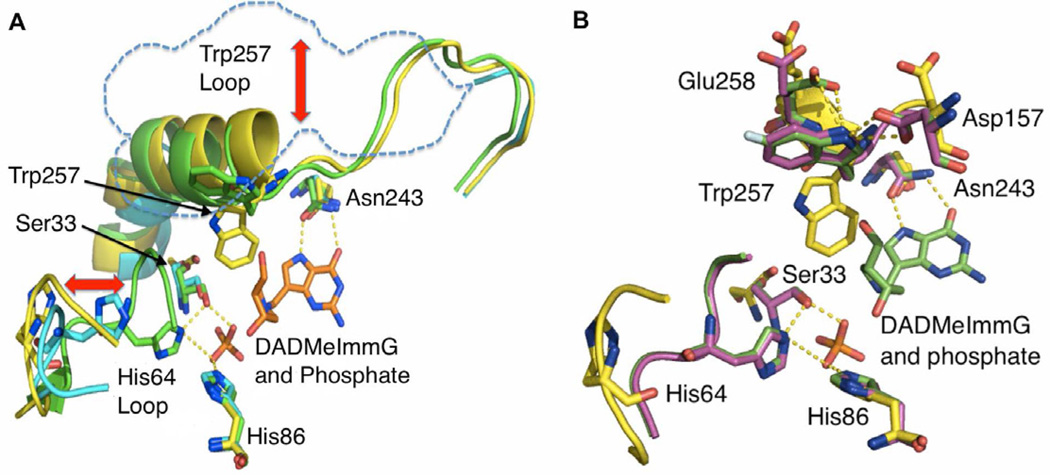
Figure 4. Human PNP complexes; structural overlays of the catalytic site.
(A). Overlap of the active site residues of H257W-Leuko PNP bound to hypoxanthine (yellow), H257W-Leuko PNP bound to phosphate (light blue) and H257W-Leuko PNP bound to DADMeImmG and phosphate (green). Double-headed red arrows denote movements in the His257-region and His64 loop. (B) Overlap of the positions of active site residues of H257W-Leuko PNP bound to hypoxanthine (yellow), H257W-Leuko PNP bound to DADMeImmG and phosphate (green) and H257-6FW-leuko PNP bound to DADMeImmG and phosphate (purple). See also Figure S1 and Movie S1.
Similarly, Trp257 adopts different conformations depending on which ligand is bound to the active site. Interestingly, no electron density is observed for the 257-region in the crystal structure of H257W-Leuko-PNP bound to phosphate, indicating a highly mobile or disordered region (Fig. 4A, dashed blue line). The structures agree with the 19F-NMR data showing two signals in the phosphate bound sample. In Tris-HCl buffer (no phosphate) the PNP active sites are partially occupied (~0.8/monomer) with guanine and hypoxanthine and in this case the Trp257 side chain faces the active site (Fig. 4A and 4B). In the absence of a ribose moiety and phosphate the bulky Trp side chain fits nicely into the active site cavity. Shielding the hydrophobic Trp side chain from the polar solvent environment should stabilize this conformation. In the tertiary complex with phosphate and inhibitor, the fully occupied active site excludes the tryptophan side chain from the active site towards the polar solvent (Fig. 4A and 4B). This solvent-exposed conformation is stabilized by hydrogen bond interactions with Glu258 and Asp157 from the neighboring subunit. These residues face away from the Trp side chain in the guanine-bound form of PNP but flip towards the Trp side chain upon inhibitor and phosphate binding (Fig. 4B).
The crystal structure of H257-6FW-Leuko-PNP bound to phosphate and inhibitor almost perfectly overlap with its unfluorinated homologue except for the Glu258 residue. The electronegative fluorine atom apparently decreases the hydrogen bonding character of the indole nitrogen in the fluorinated tryptophan thus reducing the strength of its interaction with Glu258. Despite this small difference, the overall similarity of these structures (Fig. 4B) demonstrates that the fluorine atom does not significantly perturb the structure of H257W-Leuko-PNP.
PNP crystal structures with phosphate and DADMe-ImmG (4EB8, 4EAR, Table 2), show the His64-loop in the closed conformation with W257 pointing away from the active site. Without phosphate and DADMe-ImmG but with guanine (4ECE, Table 2), the His64-loop has an open conformation and W257 is pointing towards the active site. With the PNP-phosphate structure (4GKA, Table 2) the W257-region did not show up in the electron-density map.
DADMeImmG titration of H257-6FW-Leuko-PNP
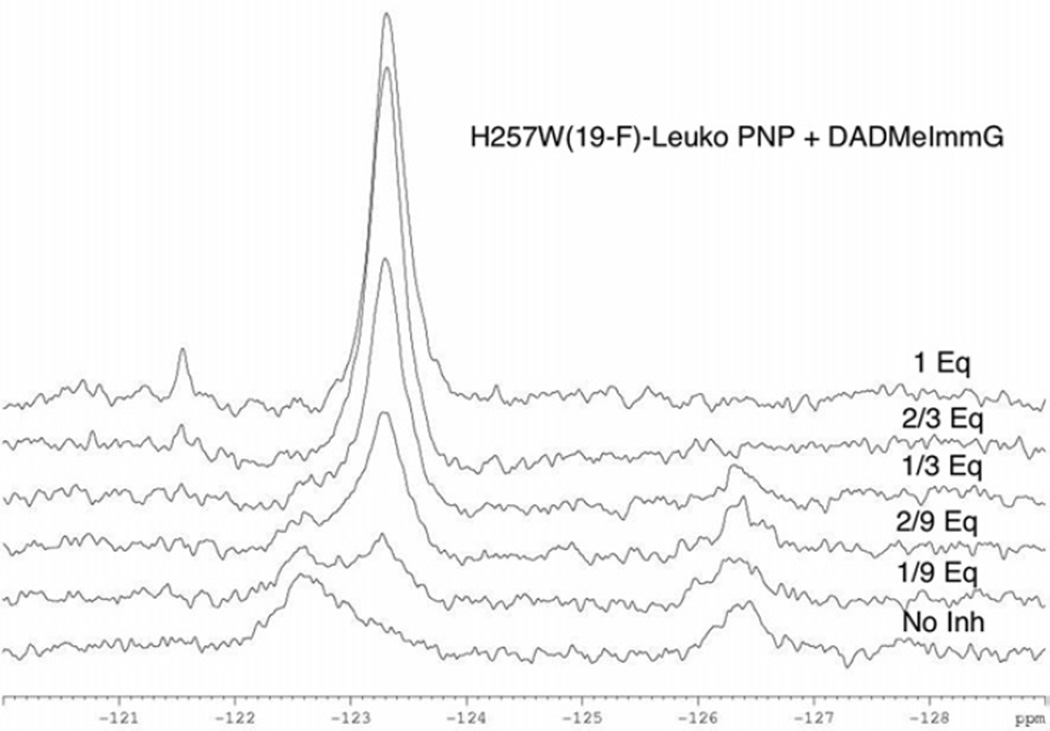
The 19F-NMR spectrum of H257-6FW-Leuko-PNP in 50 mM phosphate buffer, pH 7.4 shows two distinct signals at −122.7 ppm and −126.4 ppm (Fig. 3A, black line). These two signals disappear and a new signal at −123.3 ppm appears upon binding of the transition state inhibitor DADMeImmG (Fig. 3A, red line). A sub-stoichiometric titration of the transition state analogue inhibitor DADMeImmG into H257-6FW-Leuko-PNP showed the intensity of the signal at −122.7 ppm to decrease and the new signal at −123.3 ppm to appear while the signal at −126.4 ppm remained unchanged at 1/9 catalytic site stoichiometry of inhibitor (Fig. 5). As more equivalents of inhibitor are added this pattern continues until one monomer (1/3 equivalents of DADMeImmG) of the trimeric enzyme is saturated. With only one of the three catalytic sites filled, the signal at −122.7 ppm is reduced to a small shoulder while the signal at −123.3 ppm increases in intensity, and the signal at −126.4 ppm is slightly reduced in intensity. When two of the three sites are filled (2/3 equivalents of DADMeImmG), the initial signals at −122.7 ppm and −126.4 ppm disappear leaving behind only the new sharp signal at −123.3 ppm. Thus the structural change for 6FW is completed at the third site by cooperative changes induced from the neighboring subunits. Addition of the third equvalent of DADMeImmG causes no additional changes in linewidth or intensity.
Figure 5. DADMeImmG titration of H257-6FW-leuko PNP.
19F-NMR spectral changes of H257-6FW-leuko PNP upon the addition of sub-stoichiometric amounts of DADMeImmG in the presence of 50 mM phosphate pH 7.4.
The two initial signals at −122.7 ppm and −126.4 ppm are different conformations of the 257-region or different positions of the 6FW-257 side chain. As the −122.7 ppm signal is the first to be titrated, the spectra suggest that this conformation more readily responds to inhibitor saturation while the conformation represented by the −126.4 ppm signal is more directly linked to inhibitor binding (Saturation equals binding to all (3) active sites. Binding refers to a single binding event to one of the active sites.). The sub-stoichiometric saturation of the enzyme demonstrates that the conformational status of the flexible 257-region is transmitted between subunits.
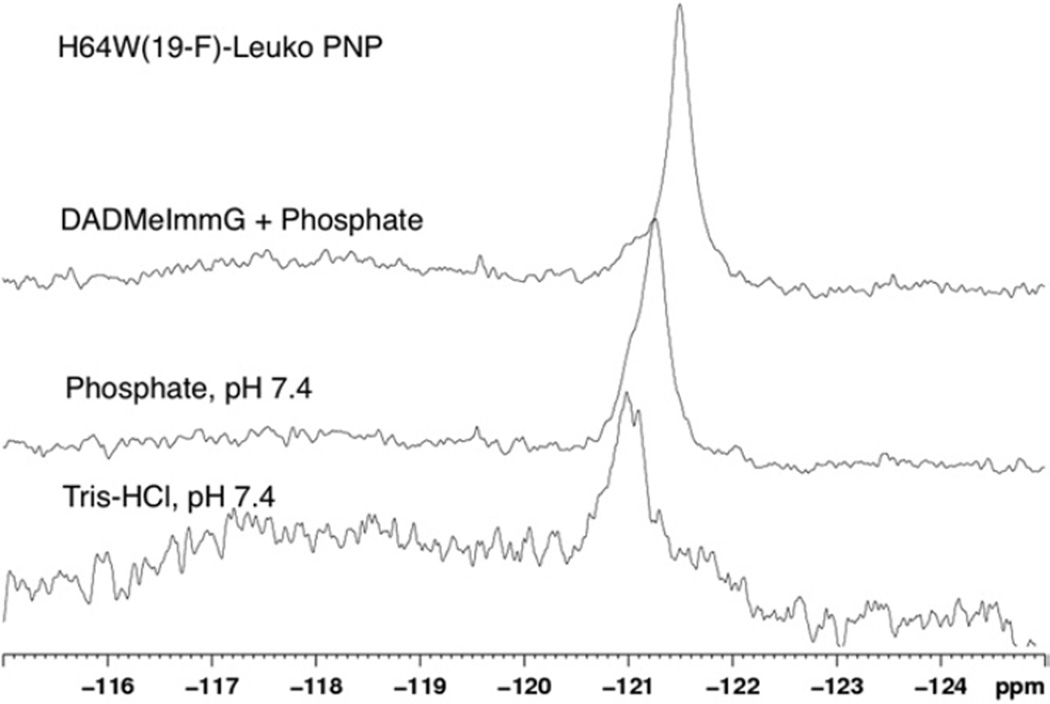
19F-NMR of H64-6FW-Leuko-PNP
His64 is located in an unstructured and flexible random-coil region that changes its position depending on the ligand bound at the active site (Fig. 4A and 4B). The range of motion of this loop includes a fully open conformation in the guanine bound form to a fully closed loop conformation in the phosphate and inhibitor bound complex, where His64 hydrogen bonds with the phosphate. Upon phosphate binding, this loop adopts an intermediate conformation between the open and closed forms (Fig. 4A and 4B). The dynamic characteristics of the conformations were examined by 19F-NMR on the H64-6FW-Leuko-PNP mutant with bound guanine, with phosphate, and with phosphate and inhibitor bound (Fig. 6). One signal was observed at −121 ppm for the guanine bound complex and a signal at −121.3 ppm with a shoulder at −121 ppm was observed for the phosphate bound complex. No evidence of chemical exchange was observed after collecting 19F-NMR spectra at 288 K, 298 K, and 310 K. The single signal for both of these complexes and the lack of a temperature effect suggests this loop to be in fast chemical exchange. The broad signal reflects the average conformation adopted by this flexible loop. Previous QM/MD calculations predicted this loop to be the most mobile region of PNP (Hirschi et al., 2010), and this is supported by the NMR results. Human PNP structures from the PDB reveals the His64 loop region to show consistently high B-factors. The mobility of this region is supported by the 19F-NMR spectrum of H64-6FW-Leuko-PNP bound to one equivalent of inhibitor and phosphate showing a single sharp signal at −121.5 ppm with a shoulder at −121 ppm reminiscent of the signal observed in the guanine-PNP. The shoulder suggests that even under saturating inhibitor conditions (Table 1) this loop is still able to move away from the inhibitor-bound conformation.
Figure 6. 19F-NMR spectra of H257-6FW-leuko PNP.
19F-NMR spectrum of H257-6FW-leuko PNP bound to guanine/hypoxanthine (bottom), 19F-NMR spectrum of H257-6FW-leuko PNP bound to phosphate (middle), 19F-NMR spectrum of H257-6FW-leuko PNP bound to DADMeImmG and phosphate (top).
In the crystal structure of H257W-Leuko-PNP with phosphate, two monomers of the trimer have an open conformation for loop-64, and in the third monomer, the 64-loop has a closed conformation similar to that seen in the crystal structures with phosphate and inhibitor (DADMe-ImmG). In the “closed conformation monomer” there was a symmetry-related monomer next to the 64-loop; with the other two monomers having an open-conformation 64-loop. There are no other monomers or symmetry related monomers in close proximity. This type of crystal packing forces the loop-64 to stay closed or the closed 64-loop of one monomer allowed the type of crystal packing seen with this crystal structure. Therefore, there is crystallographic ambiguity for phosphate in relation to the positioning of the 64-loop.
Discussion
19F-NMR as a structural probe
Structural changes upon ligand binding by 19F-NMR complemented with X-ray crystallography have been used in several protein systems including D-galactose chemosensory receptor (Luck and Falke, 1991), dihydrofolate reductase (Hoeltzli and Frieden, 1994), leucine-specific binding protein (Salopek-Sondi and Luck, 2002) calmodulin (Kitevski-LeBlanc et al., 2009) and G-protein coupled receptors (Liu et al., 2012a). These examples demonstrated the utility of 19F-NMR as a tool for defining structure-activity relationships. Insertion of specific 6FWs as catalytic site probes to understand transition state analogue effects on catalytic site functional loops and dynamics, coupled with kinetic and crystallographic analyses, represents a departure from previous uses of protein 19F-NMR.
The 19F-NMR and crystallographic data establish the ligand-dependent structural changes and implicate these conformational movements in the catalytic cycle. The His257 and His64 regions of the catalytic sites adopt “open” or “closed” conformations depending on the ligands bound to the active site. The His64-loop undergoes the more dramatic conformational alteration (Fig. 4A and 4B). These observations support the molecular dynamic simulation predictions that the His64-loop is the most mobile part of hPNP followed by the His257-region (Hirschi et al., 2010). The calculations also predicted different degrees of mobility for the His257-region and His64-loop depending on the bound ligand. High mobility was predicted for the substrates inosine and phosphate and for the first-generation transition state analogue immucilin-H. Second and third-generation transition state analogues like DADMeImmH and DATMeImmH appear to cause more restricted conformations (Hirschi et al., 2010). Examination of the His257-region and His64-loop in human PNP structures in the PDB shows that the His257-region faces away from the active site in the sulfate bound complex but orients towards the active site when substrate or transition state analogues are bound. The His64-loop is positioned distal to the active site with substrates or immucillin-H bound, but adopts a closed conformation with the transition state analogues DADMeImmG and DATMeImmH that more closely resemble the transition state of human PNP.
Relevant catalytic site motions
Our F-tryptophan structural data for human PNP suggest a sequential order for the dynamic motions of the His257-region and His64-loop during catalysis. Previous fluorometric titrations, pre-steady-state-kinetics, equilibrium ultrafiltration binding and laser T-jump fluorimetry support a random path for formation of the catalytically relevant tertiary complex; PNP-nucleoside-phosphate (Ghanem et al., 2008). Upon binding of phosphate and nucleoside (inosine or guanosine), the His257-region moves towards the active site cavity to facilitate the 5’-OH nucleoside anchoring and formation of the oxygen stack structure; H257:Nδ-O5’-O4’-Op. These interactions of the His257-region in turn drive the His64-loop into the closed conformation and formation of the Ser33-His64-His86 catalytic triad (Fig. 4A and 4B). Interactions of PNP with the phosphate are known to strongly polarize the phosphate to create a more highly reactive oxygen nucleophile (Deng et al., 2004). These conformational states provide a favorable arrangement for loss of the N-ribosidic bond via the His257 interactions and capture of the ribocation via the His64-loop activation of the phosphate nucleophile. A movie showing the relevant catalytic site motions of human PNP upon binding of phosphate and transition state inhibitor was generated using the morph server (Krebs & Gerstein., 2000) to show the active site conformational rearrangements (link to movie here).
Transition state formation
Nucleoside phosphorolysis by human PNP involves the formation of a fully developed ribocation transition state (Lewandowicz & Schramm, 2004). Past the transition state barrier, C1 of the ribocation migrates to the activated, immobilized phosphate anion to complete the reaction coordinate in a motion termed ‘nucleophilic displacement by electrophile migration’ (Fedorov et al., 2001). Proper positioning and organization of the His64 and His257 regions of the catalytic site are required for both steps. Closure of catalytic site loops and flaps are often thought to protect reactive intermediates from solvent participation. Ribocations are sensitive to water attack, which would cause D-ribose formation instead of the desired ribose 1-phosphate product in PNP. Although PNP can form ribose from nucleosides when phosphate is not present, the transition state lifetime of PNP when phosphate is present is around 10 fsec (Saen-Oon et al., 2008), too short to permit water diffusion and requiring the exquisitely accurate placement of phosphate, ribocation and purine base in the catalytic site. Disruption of the His257 loop by His257Gly does not lead to ribose formation but causes N3-isoinosine formation, a chemical consequence of catalytic site misalignment (Ghanem et al., 2009a). Open and closed representations of the active site of human PNP with docked DADMeImmG and phosphate show the closed conformation of the active site region of human PNP to effectively enclose the substrates during catalysis, with a small opening near the ribose moiety that is capped by Phe200 contributed from the neighboring subunit (Fig. 7). These structural changes generate the appropriate geometry for catalysis.
Figure 7. Surface representation of the active site region of human PNP.
(A) Surface representation of the open conformation of the active site of human PNP with DADMeImmG and Phosphate. (B) Surface representation of the closed conformation of the active site of human PNP with DADMeImmG and phosphate and also showing Phe159 from the neighboring subunit (green stick model). See also Movie S1.
Product release
Following catalysis, product release requires both the His257-region and the His64-loop to open for the release of α-D-ribose 1-phosphate. This step is efficient and fast relative to purine base release, the rate-limiting step in human PNP (Ghanem et al., 2009b). The motions defined here are therefore linked to the overall rate of catalysis by human PNP. There are no equivalent dynamic motions related to release of the purine ring, making its release the slowest step in the overall catalytic cycle. The relatively tight binding of purine bases is also evident in the tightly bound purine bases found in purified PNP protein. The catalytic site titration experiments monitored here by 19F-NMR also give insights into the negative cooperativity known for binding of transition state analogues to PNPs (Miles et al., 1998). Structural changes at adjacent sites result in conformational changes that occur in a negative cooperative manner to partially occlude access of next-ligand binding, and give rise to the observed binding profile.
Significance
Dynamic motions in enzyme catalysis occur on time scales ranging from msec for protein conformational rearrangements to fsec for single bond vibrations. How each time domain is linked to enzyme function is in debate (Hammes-Schiffer and Benkovic, 2006). The catalytic turnover rate of human PNP is on the msec timescale but the transition state lifetime is only 10 fsec, thus slower conformational changes limit the catalytic cycle (Saen-Oon et al., 2008b). Crystal structures and computational dynamics suggested that two regions of the protein, near His257 and His64 are the most flexible (Hirschi et al., 2010). The specific 6FW NMR probes analyzed by NMR and crystal structures show at least two distinct conformations for these parts of the PNP protein. One is open to allow the substrate to bind and the other is closed to permit close interactions with the reactants bound in the catalytic site to permit formation of the transition state. As far as our knowledge goes, our 19F-NMR experiments presented here are the first direct evidence of the presence of multiple active site conformations in human PNP in solution These flexible portions of the protein open and close fast relative to the overall msec catalytic turnover rate. The rate limiting step in the human PNP catalysis is release of the purine ring. There is no highly flexible protein element to help clear the catalytic site of the purine ring. This slow purine release is a design characteristic of human PNP, readily evident in the co-purification of PNP and tightly bound guanine or hypoxanthine at the catalytic sites.
Experimental Procedures
Site directed mutagenesis
Mutagenesis was performed using the QuickChange Lghtning site-directed mutagenesis kit from Stratagene (La Jolla, CA). Pairs of complementary primers (synthesized and purified by Invitrogen) were designed to introduce the required mutations in the Leuko-PNP gene cloned into the pCRT7/NT-TOPO (Invitrogen) plasmid (Ghanem et al., 2008). The oligonucleotide pairs (mutated codons are in boldface) were as follows: H257W-fw 5’ GAAAGCCTGGAGAAGGCCAACTGGGAAGAAGTCTTAGCAGCTGGC 3’, H257W-rv 5’ GCCAGCTGCTAAGACTTCTTCCCAGTTGGCCTTCTCCAGGCTTTC 3’ and H64W-fw 5’ GAAGTACAGTGCCAGGTTGGGCTGGCCGACTGGTG 3’, H64W-rv 5’ CACCAGTCGGCCAGCCCAACCTGGCACTGTACTTC 3’. Mutagenesis was performed according to the manufacturer’s protocol, and the reaction products were transformed into the E. coli BL21(DE3)pLysS (invitrogen) strain for selection and expression purposes. The presence of the mutated codons in the hPNP gene was confirmed by DNA sequencing.
Expression purification of unlabeled enzymes
Native, H257W-leuko and H64W-Leuko enzymes were expressed and purified to homogeneity as judged by SDS-PAGE by following the same procedure previously described for native PNP (Lewandowicz and Schramm, 2004).
Expression and purification of 19F-labeled enzymes
19F-labeled enzymes (H257W-leuko and H64W-Leuko) were expressed in E. coli BL21(DE3)pLys (invitrogen) cells containing the hPNP mutant constructs cloned into the pCRT7/NT-TOPO (Invitrogen) plasmid. Cells were grown in M63 minimum medium supplemented with glucose and 100 ug mL−1 ampicillin, at 37°C. 6-Fluoro-DL-tryptophan (Sigma) was added 30 minutes before IPTG induction (OD: 0.8–1) to a final concentration of 0.2 mg mL−1. Cells were allowed to grow for an additional 8 hours and harvested by centrifugation. All 19F-labelled proteins were purified to homogeneity following the same procedure previously described for native PNP (Lewandowicz and Schramm, 2004).
Activity and inhibition assays
Activity assays for native and mutant PNP enzymes with inosine as a substrate were performed as previously described by monitoring the conversion of hypoxanthine to uric acid (ε293 = 12.9 mM−1 cm−1) (Kim et al., 1968) in a coupled assay containing 60 munit of xanthine oxidase and variable concentrations of inosine, in 50 mM phosphate buffer pH 7.4 at 25 °C (Lewandowicz et al., 2003). The slow onset inhibition was measured following the addition of enzyme to complete assay mixtures containing 1 mM inosine and various inhibitor concentrations (Kicska et al., 2002; Rinaldo-Matthis et al., 2007). Enzyme was added to assay mixtures followed by monitoring rates of product formation for 1-2 h to determine both the initial reaction rates and to determine if slow onset inhibition occurred. Protein and inhibitor concentrations were determined by their corresponding extinction coefficients. 23380 M−1 cm−1 for the single tryptophan mutants, 8.92 M−1 cm−1 for DADMeImmG and 9.54 M−1 cm−1 for DATMeImmH.
Enzymatic Digestion of proteins and MALDI-TOF Mass Spectrometry analysis
In order to determine the percentage of incorporation of 6-fluoro tryptophan, wild-type PNP and 6-F-Trp into PNP were digested by trypsin following the standard digestion protocol. The digested peptides were then purified by C18 ZipTip (Millipore) for Mass Spec analysis. MALDI-TOF mass spectrometry analysis of the digested peptides was performed on a Voyager Biospectrometry DE-STR Workstation (Applied Biosystems, Foster City, CA) equipped with a nitrogen laser (330 nm). The instrument was operated in positive ionization mode and measurements were performed in linear mode (mass range m/z 800 to 6000) using α-cyano-4-hydroxycinnamic acid as matrix. A saturated matrix solution was prepared in 80% acetonitrile, 19.9% water and 0.1% TFA (v/v/v). Samples were prepared using the dried droplet method by spotting 1 µl of matrix solution and 1 µl of sample solution onto the target. Spectra from 100 to 300 laser shots were accumulated to produce one spectrum. The instrument was calibrated using the monoisotopic masses of standard peptides: KPQQFFDLM (MH+1(mi) 1095.56), CQDSETRTFY (MH+1(mi) 1249.52), and YGGFMTSEKSQTPLVT (MH+1(mi) 1745.84).
19F NMR Spectroscopy
19F NMR spectra were acquired on a Bruker DRX 600 MHz (Bruker Biospin, Billerica, MA) spectrometer equipped with a 5 mm BBFO probe. Unless otherwise indicated, all 1D spectra were recorded at 25 °C with 23,542 scans (9 hour acquisition). The time-domain NMR data were multiplied with an exponential function with a line-broadening factor of 25 Hz prior to Fourier transformation. The protein concentration for the NMR measurements was in the range of 0.4 – 0.8 mM.
Protein crystallization and data collection
The H257W-leukoPNP was dialyzed against 50 mM Tris buffer at pH 8.5 and 1 mM DTT. The concentration of H257W-hPNP for crystallization was 14 mg/ml in the presence of 1 mM DADMe-ImmG and 5 mM phosphate. Crystals were obtained by sitting-drop vapor diffusion with 20% PEG 5000 monomethyl ether, 100 mM Bis-Tris pH 6.5. Crystals of H257W-leukoPNP-DADMeImmG were transferred to a fresh drop of the crystallization solution containing 20% ethylene glycol and flash-cooled. In the case of H257-6FW-leuko PNP having a fluorinated tryptophan, the protein was dialyzed in the 50 mM Tris, pH 7.5, concentrated to 8.0 mg/ml and DADMe-ImmG and phosphate added to final concentrations of 0.5 mM and 5 mM, respectively. The H257-6FW-leuko PNP crystals grew from 20% PEG 1000 in 100 mM Tris, pH 7.0. Prior to flash-cooling at 100 K, the H257-6FW-leuko PNP-DADMe-ImmG crystal was transferred into the corresponding crystallization solution with 26 % (w/v). When setting up the apo H257W-leukoPNP and H257W-leuko PNP with phosphate crystallization trials, H257W-leukoPNP was treated the same as the H257-6FW-leuko PNP variant. The phosphate bound H257W-leukoPNP crystals grew from 16% (w/v) PEG 8000, 20 % (v/v) glycerol, 160 mM magnesium acetate, 80 mM sodium cacodylate, pH 6.5 solution. These crystals were directly flash-cooled from the crystallization drop. The crystals of H257W-leuko PNP with 5 mM phosphate grew from 10 % (v/v) 2-propanol, 0.2 M zinc acetate, 0.1 M sodium cacodylate, pH 6.5 solution, and were flash cooled in the crystallization solution containing 27 % (v/v) glycerol. All X-ray diffraction data were collected at Beamline X29A at Brookhaven National Laboratory. All data were processed with HKL3000 (Minor et al., 2006) program suite; data processing statistics are provided in Table 2.
Structure determination and refinement
The crystal structures of H257W-leukoPNP-DADMe-ImmG, H257-6FW-leuko PNP-DADMe-ImmG and H257W-leukoPNP with only phosphate were determined by molecular replacement in MOLREP (Vagin and Teplyakov, 2010) in the CCP4 program suite (1994; Potterton et al., 2003) using the published structure of hPNP (PDB:1RR6) as the search model. The phosphate bound H257W-leukoPNP crystal structure was determined by using PHASER (McCoy et al., 2007) and using the same model. All the models were first rebuilt in COOT (Emsley and Cowtan, 2004) and refined in REFMAC5 (Murshudov et al., 1997). The DADMeImmG, phosphate and/or guanine were added at the last stages of refinement using Fo − Fc map. In the case of phosphate bound H257W-leuko PNP, only guanine (and not hypoxanthine) was added because it was the most abundant molecule that copurified with this mutant. The quality of all structures were checked by PROCHECK (Laskowski et al., 1993) and MolProbity (Chen et al., 2010; Davis et al., 2007). The refinement and geometry statistics are summarized in Table 2.
In the case of the H257W-leuko-PNP crystal structure without phosphate and inhibitor (4ECE), there are two trimers in the asymmetric unit. One trimer, consisting of monomers A, B and C (Table 2) has lower average temperature factors (B-factors) for the main-chain atoms than the other trimer (D,E,F). Due to the high B-factors, the electron-density in some regions of a D,E,F-trimer are not well defined. Accordingly, the trimer composed of monomers A, B and C is used for all structural comparisons in this study. In all the comparisons, the B-monomers of various crystal structures were utilized.
Supplementary Material
Highlights.
19F-tryptophan insertions are NMR reporters for the catalytic site of human PNP
Open and closed catalytic site conformations are faster than catalysis
Catalytic site elements have multiple conformations until the site is filled
Combined NMR and crystallography reveal dynamics and structure for PNP
Acknowledgments
This work was supported by the NIH research grants GM068036 and GM041916 (V.L.S), by a fellowship from the Sigrid Jusélius Foundation (A.M.H.) and by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number K12GM102779. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Data for this study were measured at the beamline X29A at the Case Center for Synchrotron Biosciences (CCSB) located at the National Synchrotron Light Source at Brookhaven National Laboratories, New York. The collection of this data was made possible by the Center for Synchrotron Biosciences grant, P30-EB-009998, from the National Institute of Biomedical Imaging and Bioengineering (NIBIB). The authors thank Drs. Peter C. Tyler and Gary B. Evans of Industrial Research Ltd. for the generous gift of DADMeImmG and DADMeImmH and also Dr. Patskovsky for collecting the data sets for PDB IDs 4EAR and 4GKA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Balakrishnan K, Verma D, O'Brien S, Kilpatrick JM, Chen Y, Tyler BF, Bickel S, Bantia S, Keating MJ, Kantarjian H, et al. Phase 2 and pharmacodynamic study of oral forodesine in patients with advanced, fludarabine-treated chronic lymphocytic leukemia. Blood. 2010;116:886–892. doi: 10.1182/blood-2010-02-272039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassera MB, Hazleton KZ, Merino EF, Obaldia N, 3rd, Ho MC, Murkin AS, DePinto R, Gutierrez JA, Almo SC, Evans GB, et al. Plasmodium falciparum parasites are killed by a transition state analogue of purine nucleoside phosphorylase in a primate animal model. PLoS One. 2011;6:e26916. doi: 10.1371/journal.pone.0026916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computatinoal Project, Number 4. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo WF, Jr, Canduri F, dos Santos DM, Silva RG, de Oliveira JS, de Carvalho LP, Basso LA, Mendes MA, Palma MS, Santos DS. Crystal structure of human purine nucleoside phosphorylase at 2.3A resolution. Biochem. Biophys. Res. Commun. 2003;308:545–552. doi: 10.1016/s0006-291x(03)01431-1. [DOI] [PubMed] [Google Scholar]
- Deng H, Lewandowicz A, Schramm VL, Callender R. Activating the phosphate nucleophile at the catalytic site of purine nucleoside phosphorylase: a vibrational spectroscopic study. J. Am. Chem. Soc. 2004;126:9516–9517. doi: 10.1021/ja049296p. [DOI] [PubMed] [Google Scholar]
- Edwards AA, Mason JM, Clinch K, Tyler PC, Evans GB, Schramm VL. Altered enthalpy-entropy compensation in picomolar transition state analogues of human purine nucleoside phosphorylase. Biochemistry. 2009;48:5226–5238. doi: 10.1021/bi9005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, et al. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;30(4):853–60. doi: 10.1021/bi002499f. 40. [DOI] [PubMed] [Google Scholar]
- Ghanem M, Murkin AS, Schramm VL. Ribocation transition state capture and rebound in human purine nucleoside phosphorylase. Chem. Biol. 2009a;16:971–979. doi: 10.1016/j.chembiol.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem M, Saen-oon S, Zhadin N, Wing C, Cahill SM, Schwartz SD, Callender R, Schramm VL. Tryptophan-free human PNP reveals catalytic site interactions. Biochemistry. 2008;47:3202–3215. doi: 10.1021/bi702491d. [DOI] [PubMed] [Google Scholar]
- Ghanem M, Zhadin N, Callender R, Schramm VL. Loop-tryptophan human purine nucleoside phosphorylase reveals submillisecond protein dynamics. Biochemistry. 2009b;48:3658–3668. doi: 10.1021/bi802339c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giblett ER, Ammann AJ, Wara DW, Sandman R, Diamond LK. Nucleoside-phosphorylase deficiency in a child with severely defective T-cell immunity and normal B-cell immunity. Lancet. 1975;1:1010–1013. doi: 10.1016/s0140-6736(75)91950-9. [DOI] [PubMed] [Google Scholar]
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu. Rev. Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- Hirschi JS, Arora K, Brooks CL, 3rd, Schramm VL. Conformational dynamics in human purine nucleoside phosphorylase with reactants and transition-state analogues. J. Phys. Chem. B. 2010;114:16263–16272. doi: 10.1021/jp108056s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MC, Shi W, Rinaldo-Matthis A, Tyler PC, Evans GB, Clinch K, Almo SC, Schramm VL. Four generations of transition-state analogues for human purine nucleoside phosphorylase. Proc. Natl. Acad. Sci. USA. 2010;107:4805–4812. doi: 10.1073/pnas.0913439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeltzli SD, Frieden C. 19F NMR spectroscopy of [6-19F]tryptophan-labeled Escherichia coli dihydrofolate reductase: equilibrium folding and ligand binding studies. Biochemistry. 1994;33:5502–5509. doi: 10.1021/bi00184a019. [DOI] [PubMed] [Google Scholar]
- Hollister AS, Michael A, Becker MA, Terkeltaub R, Waugh A, Lyman A, Amy Flynt A, FitzPatrick D. BCX4208 synergistically lowers serum uric acid (sUA) levels when combined with allopurinol in patients with gout: results of a phase 2 dose-ranging trial. ACR/ARHP Poster Session B, Abs. 2011:1018. [Google Scholar]
- Kicska GA, Tyler PC, Evans GB, Furneaux RH, Kim K, Schramm VL. Transition state analogue inhibitors of purine nucleoside phosphorylase from Plasmodium falciparum. J. Biol. Chem. 2002;277:3219–3225. doi: 10.1074/jbc.M105905200. [DOI] [PubMed] [Google Scholar]
- Kim BK, Cha S, Parks RE., Jr Purine nucleoside phosphorylase from human erythroyctes. II. Kinetic analysis and substrate-binding studies. J. Biol. Chem. 1968;243:1771–1776. [PubMed] [Google Scholar]
- vski-LeBlanc JL, Prosser RS. Current applications of 19F NMR to studies of protein structure and dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2012;62:1–33. doi: 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Kitevski-LeBlanc JL, Evanics F, Prosser RS. Approaches for the measurement of solvent exposure in proteins by 19F NMR. J. Biomol. NMR. 2009;45:255–264. doi: 10.1007/s10858-009-9359-2. [DOI] [PubMed] [Google Scholar]
- Kline PC, Schramm VL. Purine nucleoside phosphorylase. Catalytic mechanism and transition-state analysis of the arsenolysis reaction. Biochemistry. 1993;32:13212–13219. doi: 10.1021/bi00211a033. [DOI] [PubMed] [Google Scholar]
- Koellner G, Luic M, Shugar D, Saenger W, Bzowska A. Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 A resolution. J. Mol. Biol. 1997;265:202–216. doi: 10.1006/jmbi.1996.0730. [DOI] [PubMed] [Google Scholar]
- Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, McArthur MW, Moss DS, Thorton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lewandowicz A, Schramm VL. Transition state analysis for human and Plasmodium falciparum purine nucleoside phosphorylases. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- Lewandowicz A, Shi W, Evans GB, Tyler PC, Furneaux RH, Basso LA, Santos DS, Almo SC, Schramm VL. Over-the-barrier transition state analogues and crystal structure with Mycobacterium tuberculosis purine nucleoside phosphorylase. Biochemistry. 2003;42:6057–6066. doi: 10.1021/bi0343830. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012b;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. 19F NMR studies of the D-galactose chemosensory receptor. 2. Ca(II) binding yields a local structural change. Biochemistry. 1991;30:4257–4261. doi: 10.1021/bi00231a022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles RW, Tyler PC, Furneaux RH, Bagdassarian CK, Schramm VL. One-third-the-sites transition-state inhibitors for purine nucleoside phosphorylase. Biochemistry. 1998;37:8615–8621. doi: 10.1021/bi980658d. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Murkin AS, Birck MR, Rinaldo-Matthis A, Shi W, Taylor EA, Almo SC, Schramm VL. Neighboring group participation in the transition state of human purine nucleoside phosphorylase. Biochemistry. 2007;46:5038–5049. doi: 10.1021/bi700147b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murkin AS, Tyler PC, Schramm VL. Transition-state interactions revealed in purine nucleoside phosphorylase by binding isotope effects. J. Am. Chem. Soc. 2008;130:2166–2167. doi: 10.1021/ja7104398. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Rinaldo-Matthis A, Murkin AS, Ramagopal UA, Clinch K, Mee SP, Evans GB, Tyler PC, Furneaux RH, Almo SC, Schramm VL. L-Enantiomers of transition state analogue inhibitors bound to human purine nucleoside phosphorylase. J. Amer. Chem. Soc. 2008;130:842–844. doi: 10.1021/ja710733g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo-Matthis A, Wing C, Ghanem M, Deng H, Wu P, Gupta A, Tyler PC, Evans GB, Furneaux RH, Almo SC, et al. Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues. Biochemistry. 2007;46:659–668. doi: 10.1021/bi061515r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saen-Oon S, Ghanem M, Schramm VL, Schwartz SD. Remote mutations and active site dynamics correlate with catalytic properties of purine nucleoside phosphorylase. Biophys. J. 2008a;94:4078–4088. doi: 10.1529/biophysj.107.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saen-Oon S, Quaytman-Machleder S, Schramm VL, Schwartz SD. Atomic detail of chemical transformation at the transition state of an enzymatic reaction. Proc. Natl. Acad. Sci. U S A. 2008b;105:16543–16548. doi: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salopek-Sondi B, Luck LA. 19F NMR study of the leucine-specific binding protein of Escherichia coli: mutagenesis and assignment of the 5-fluorotryptophan-labeled residues. Protein Eng. 2002;15:855–859. doi: 10.1093/protein/15.11.855. [DOI] [PubMed] [Google Scholar]
- Schramm VL. Enzymatic transition states: thermodynamics, dynamics and analogue design. Arch. Biochem. and Biophys. 2005;433:13–26. doi: 10.1016/j.abb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Shi W, Ting LM, Kicska GA, Lewandowicz A, Tyler PC, Evans GB, Furneaux RH, Kim K, Almo SC, Schramm VL. Plasmodium falciparum purine nucleoside phosphorylase: crystal structures, immucillin inhibitors, and dual catalytic function. J. Biol. Chem. 2004;279:18103–18106. doi: 10.1074/jbc.C400068200. [DOI] [PubMed] [Google Scholar]
- Stoeckler JD, Agarwal RP, Agarwal KC, Parks RE., Jr Purine nucleoside phosphorylase from human erythrocytes. Methods Enzymol. 1978;51:530–538. doi: 10.1016/s0076-6879(78)51074-4. [DOI] [PubMed] [Google Scholar]
- Stoeckler JD, Cambor C, Parks RE., Jr Human erythrocytic purine nucleoside phosphorylase: reaction with sugar-modified nucleoside substrates. Biochemistry. 1980;19:102–107. doi: 10.1021/bi00542a016. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.