Abstract
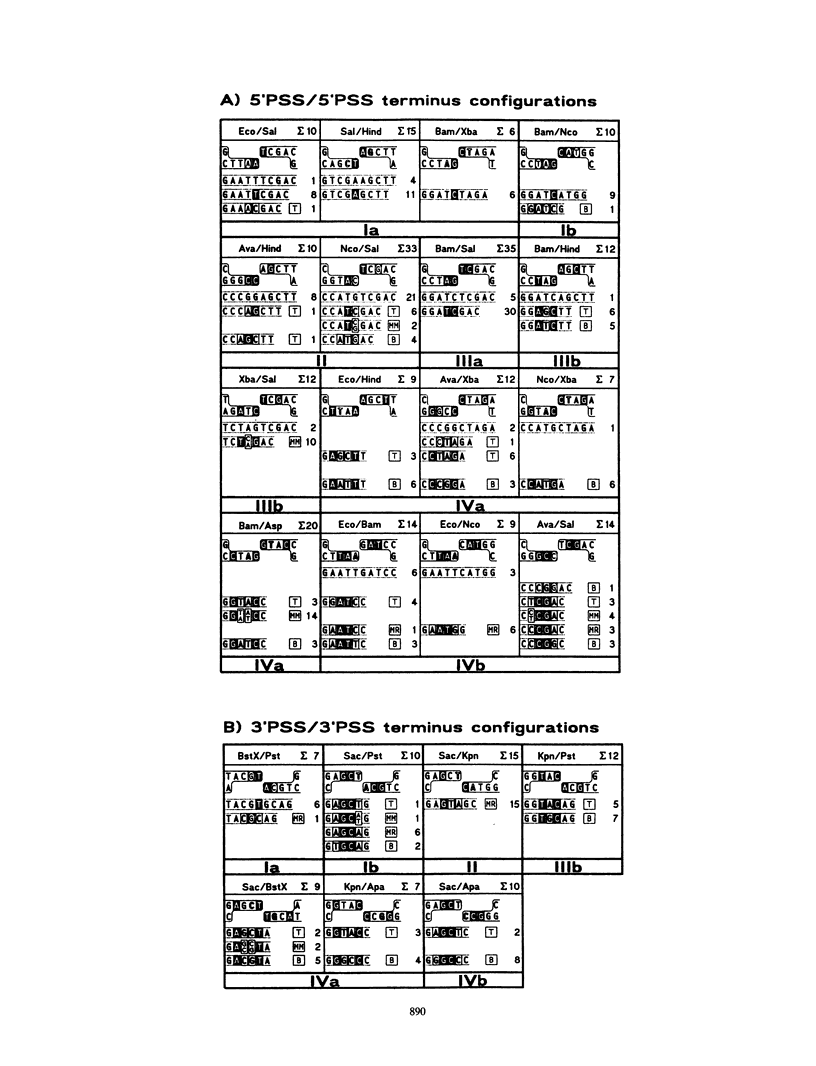
Rejoining of nonhomologous DNA termini plays a central role in processes of illegitimate recombination. In Xenopus egg extracts, DNA ends with noncomplementary 4-nucleotide antiparallel single-strand protrusions are assumed to be joined by formation of short mismatched overlap intermediates. The extents of these overlaps may be set by single fortuitously matching base pairs and determine the patterns of subsequent gap filling and nick ligation. Under conditions of alternative overlap settings, rules for the most probable joining pathway and the effects of mismatches on junction formation were analyzed. We show that in certain cases, fill-in and ligation converting overlap intermediates into covalently closed junctions may proceed in the presence of unrepaired mismatches, whereas in other cases, completion of junction formation is preceded by removal of mismatches. Results are discussed in relation with "alignment" proteins postulated to structurally support overlap heteroduplexes during junction formation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gahlmann R., Doerfler W. Integration of viral DNA into the genome of the adenovirus type 2-transformed hamster cell line HE5 without loss or alteration of cellular nucleotides. Nucleic Acids Res. 1983 Nov 11;11(21):7347–7361. doi: 10.1093/nar/11.21.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination gets a break. Trends Genet. 1992 Dec;8(12):408–412. doi: 10.1016/0168-9525(92)90322-u. [DOI] [PubMed] [Google Scholar]
- Grilley M., Holmes J., Yashar B., Modrich P. Mechanisms of DNA-mismatch correction. Mutat Res. 1990 Sep-Nov;236(2-3):253–267. doi: 10.1016/0921-8777(90)90009-t. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Exonucleolytic proofreading. Cell. 1988 Jun 17;53(6):837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- Lieber M. R. The mechanism of V(D)J recombination: a balance of diversity, specificity, and stability. Cell. 1992 Sep 18;70(6):873–876. doi: 10.1016/0092-8674(92)90237-7. [DOI] [PubMed] [Google Scholar]
- Linn S. How many pols does it take to replicate nuclear DNA? Cell. 1991 Jul 26;66(2):185–187. doi: 10.1016/0092-8674(91)90608-2. [DOI] [PubMed] [Google Scholar]
- Mosbaugh D. W., Linn S. Excision repair and DNA synthesis with a combination of HeLa DNA polymerase beta and DNase V. J Biol Chem. 1983 Jan 10;258(1):108–118. [PubMed] [Google Scholar]
- Munz P. L., Young C. S. End-joining of DNA fragments in adenovirus transfection of human cells. Virology. 1991 Jul;183(1):160–169. doi: 10.1016/0042-6822(91)90129-y. [DOI] [PubMed] [Google Scholar]
- North P., Ganesh A., Thacker J. The rejoining of double-strand breaks in DNA by human cell extracts. Nucleic Acids Res. 1990 Nov 11;18(21):6205–6210. doi: 10.1093/nar/18.21.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Vielmetter W. Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res. 1988 Feb 11;16(3):907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randahl H., Elliott G. C., Linn S. DNA-repair reactions by purified HeLa DNA polymerases and exonucleases. J Biol Chem. 1988 Sep 5;263(25):12228–12234. [PubMed] [Google Scholar]
- Roth D. B., Chang X. B., Wilson J. H. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989 Jul;9(7):3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Porter T. N., Wilson J. H. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985 Oct;5(10):2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986 Dec;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal H., Pfeiffer P., Klein M., Gehrmann P., Scheid A. Use of DNA end joining activity of a Xenopus laevis egg extract for construction of deletions and expression vectors for HIV-1 Tat and Rev proteins. Gene. 1993 Feb 28;124(2):275–280. doi: 10.1016/0378-1119(93)90405-r. [DOI] [PubMed] [Google Scholar]
- Thacker J., Chalk J., Ganesh A., North P. A mechanism for deletion formation in DNA by human cell extracts: the involvement of short sequence repeats. Nucleic Acids Res. 1992 Dec 11;20(23):6183–6188. doi: 10.1093/nar/20.23.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thode S., Schäfer A., Pfeiffer P., Vielmetter W. A novel pathway of DNA end-to-end joining. Cell. 1990 Mar 23;60(6):921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winegar R. A., Lutze L. H., Rufer J. T., Morgan W. F. Spectrum of mutations produced by specific types of restriction enzyme-induced double-strand breaks. Mutagenesis. 1992 Nov;7(6):439–445. doi: 10.1093/mutage/7.6.439. [DOI] [PubMed] [Google Scholar]