Abstract
Central to understanding mechanotransduction in the knee meniscus is the characterization of meniscus cell mechanics. In addition to biochemical and geometric differences, the inner and outer regions of the meniscus contain cells that are distinct in morphology and phenotype. This study investigated the regional variation in meniscus cell mechanics in comparison to articular chondrocytes and ligament cells. It was found that the meniscus contains two biomechanically distinct cell populations, with outer meniscus cells being stiffer (1.59 ± 0.19 kPa) than inner meniscus cells (1.07 ± 0.14 kPa). Additionally, it was found that both outer and inner meniscus cell stiffnesses were similar to ligament cells (1.32 ± 0.20 kPa), and articular chondrocytes showed the highest stiffness overall (2.51 ± 0.20 kPa). Comparison of compressibility characteristics of the cells showed similarities between articular chondrocytes and inner meniscus cells, as well as between outer meniscus cells and ligament cells. These results show that cellular biomechanics vary regionally in the knee meniscus, and that meniscus cells are biomechanically similar to ligament cells. The mechanical properties of musculoskeletal cells determined in this study may be useful for the development of mathematical models or the design of experiments studying mechanotransduction in a variety of soft tissues.
Keywords: Cytocompression, mechanics, cartilage, meniscus, musculoskeletal
Introduction
Understanding mechanotransduction in the knee meniscus can help elucidate the mechanisms by which meniscus cells maintain healthy tissue or mount a healing response following injury. Located between the femoral condyles and the tibial plateau in the knee joint, the menisci are semi-lunar, wedge-shaped fibrocartilaginous tissues (McDermott et al. 2004). Each meniscus functions to increase congruence between articulating surfaces, distribute load, and absorb shock during normal movements such as walking or running (Walker and Erkman 1975, Walker and Hajek 1972). Because of its unique shape, meniscus tissue is exposed to a variety of load types including compression, tension, and shear (Fithian et al. 1990, Skaggs et al. 1994, Sweigart and Athanasiou 2005, Sweigart et al. 2004). These forces are also borne by the resident cells in the tissue, which affect their gene expression and synthetic properties (Upton et al. 2008, Upton et al. 2006). As the regional mechanical properties of meniscus cells are unknown, however, it is difficult to predict how forces at the tissue level are translated to the cells and what effect these forces have on tissue homeostasis or remodeling.
As biochemically distinct regions exist in the inner and outer portions of the meniscus, the mechanical properties of meniscus cells may also vary regionally. Different cell types are known to reside in the inner and outer meniscus regions; the inner region contains rounded, articular chondrocyte-like cells that produce collagen type II and aggrecan, whereas outer meniscus cells display many cellular processes and are the main producers of collagen type I (Aspden et al. 1985, Gabrion et al. 2005, McDevitt and Webber 1990, Skaggs et al. 1994, Valiyaveettil et al. 2005). Regional variations in cellular mechanics have been documented previously in different zones of articular cartilage (Darling et al. 2006) and in different regions of the intervertebral disc (Guilak et al. 1999), which also correspond with known differences in cellular morphology and synthetic profiles. If, in addition to phenotypic differences, outer and inner meniscus cells show distinct mechanical properties, these cells may deform differently in response to the same mechanical load, causing varying phenotypic and synthetic changes.
It is also important to understand meniscus cell mechanics in the context of other musculoskeletal cells in the knee joint. As articular cartilage and ligament represent opposite ends of the musculoskeletal soft tissue spectrum, it is likely that cells from the meniscus may have similar properties to cells from these tissues. Identifying key similarities and differences in the mechanical properties of these cells can aid in the development of theoretical models of the meniscus, and inform further studies on the effects of mechanical stimulation on meniscus cells.
Typically, single cells are mechanically tested using one of four techniques: micropipette aspiration (Guilak et al. 2000, Hochmuth 2000, Theret et al. 1988), atomic force microscopy (AFM) (Darling et al. 2010, Darling et al. 2007, Darling et al. 2006, Ikai 2009, You and Yu 1999), cytoindentation,(Koay et al. 2003, Shin and Athanasiou 1999) or unconfined compression (Leipzig and Athanasiou 2008, Leipzig et al. 2006, Ofek et al. 2009, Shieh and Athanasiou 2007). Because of the size of the pipette or probe used in micropipette aspiration and atomic force microscopy, these techniques are most useful for measuring the mechanical properties of subcellular components such as portions of the cellular membrane. Cytoindentation offers some advantages over these techniques as the probe is larger than that used in AFM, and the cylindrical probe geometry allows for simpler modeling of the viscoelastic mechanical behavior of the cell. Unconfined compression, however, uses a probe that is much larger than the cell, giving it the unique ability to measure bulk cellular properties. This is advantageous because bulk cellular deformation likely occurs during physiologic loading experienced by meniscus cells in situ. Bulk cytomechanics have been measured previously using a cytocompression device, which has been used to successfully detect differences in the mechanical properties of a variety of cell types (Feng et al. 2010, Koay et al. 2008, Ofek et al. 2009, Ofek et al. 2009). Thus, application of this technique is able to detect differences in cellular mechanics in a variety of situations, and presents a useful method of measuring the bulk mechanical properties of meniscus cells.
In light of the lack of information on meniscus cell mechanics, the overall objective of this study is to characterize the biomechanical properties of meniscus cells and compare them to those of articular chondrocytes and ligament cells. To determine if regional variations in meniscus cellular mechanics exist, mechanical properties of inner meniscus cells as well as outer meniscus cells are also measured and compared. Unconfined compression stress-relaxation tests are performed using a cytocompression device to measure bulk cell stiffness, Poisson’s ratio, and recovery characteristics of each cell type. It is hypothesized that inner and outer meniscus cells have distinct mechanical properties, and that inner cells are more similar to articular chondrocytes while outer cells have properties more similar to ligament cells.
Materials and Methods
Cell isolation and seeding
Five bovine knee joints from two week old animals were obtained 24 hours after sacrifice and patellar ligaments, articular cartilage from the femoral head, and medial menisci were harvested using aseptic surgical techniques. The outer and inner portions of each medial meniscus were dissected from the tissue and processed separately. Ligament, cartilage, inner meniscus, and outer meniscus tissue was minced to approximately 1 mm2 pieces, and digested in Dulbecco’s Modified Eagle Medium with Glutamax (DMEM) [Invitrogen, Carlsbad, CA], 1% penicillin/streptomycin/fungizone (P/S/F) [Lonza, Basel, Switzerland], 1% non-essential amino acids (NEAA) [Invitrogen], and 0.2% w/v collagenase type 2 [Worthington, Lakewood, NJ]. Digestion was carried out overnight at 37°C with gentle shaking, after which cells were counted and frozen in liquid nitrogen in DMEM containing 20% fetal bovine serum (FBS) [Atlanta Biologicals, Lawrenceville, GA] and 10% dimethyl sulfoxide [Sigma-Aldrich, St. Louis, MO].
Prior to each cytocompression experiment, cells were thawed, counted, and resuspended in DMEM containing 1% P/S/F, 1% NEAA, 10% FBS, and 50 µg/mL ascorbate-2-phosphate. Cells were then seeded onto cut-glass slides at a concentration of 200,000 cells/mL, as described previously (Koay et al. 2008, Ofek et al. 2009). After incubation at 37°C for 1.5 hours, slides were gently washed with media leaving only cells attached to the glass surface. Individual cells with rounded morphology were then subjected to compression, and videos of the compression events were recorded.
Unconfined compression of single cells
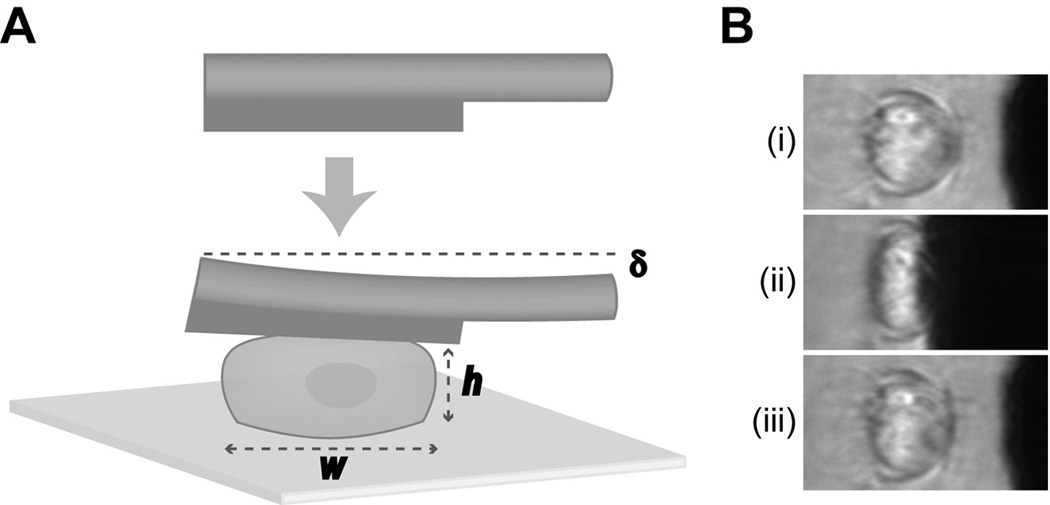
As previously described, a cytocompression device was used to subject single chondrocytes, inner meniscus cells, outer meniscus cells, or ligament cells to unconfined compression stress-relaxation tests at 10–50% strain levels (Koay et al. 2008, Ofek et al. 2009, Shieh et al. 2006). A sample size of n = 30 was used for each group in this study, based on power analysis performed for previous studies using this cytocompression method (Ofek et al. 2009). A schematic representation of this procedure is shown in Figure 1. Briefly, glass slides seeded with cells were placed on an inverted microscope such that the objective viewed side profiles of the cells. A tungsten probe, 50.8 µm in diameter and driven by a piezoelectric motor, was brought approximately 5–10 µm from the top of each cell, and was used to compress the cell at a rate of 4 µm/second (Figure 1A). Based on the probe’s distance from the cell, the cell’s stiffness, and the prescribed distance the probe was to travel toward the cell (15 µm), each cell was exposed to a different amount of strain. After moving the prescribed distance, the probe was held in place for 30 seconds to allow the cell to equilibrate, and then retracted at a rate of 4 µm/second (Figure 1B). Each compression event was recorded using a CCD camera, and recovery behavior following probe retraction was recorded for 45 seconds.
Fig. 1. Schematic of cytocompression experiments.
(A) Cells were seeded onto a glass slide and compressed at a rate of 4 µm/second by a tungsten probe. The height (h) and width (w) of the cell, as well as the probe deflection (δ) was measured at various stages of each compression event and used to calculate cellular biomechanical properties. (B) Representative frames of an inner meniscus cell seeded onto a glass slide initially (i), at equilibrium compression (ii), and during the recovery stage (iii), of a cytocompression event
Video capture and image analysis
Recordings of cells compressed and recovering from compression were analyzed as previously described (Ofek et al. 2009). Briefly, individual compression events were recorded using MetaMorph image analysis software [Olympus, Center Valley, CA], and cellular dimensions were measured throughout each event. Cell height and width was measured initially, at equilibrium compression, and during cellular recovery following release from compression. Additionally, the distance the probe traveled during each compression event was measured. A pixel to micron ratio was determined for each session by recording the unimpeded probe as it moved through a prescribed distance of 15 µm.
Biomechanical measurements
Cantilever beam theory was employed to determine the stress applied to each cell based on probe’s deflection, stiffness, and moment of inertia, as previously described (Koay et al. 2003, Ofek et al. 2009). The reaction force, F, on the cell was described as:
where the constants E (Young’s modulus), I (moment of inertia), and L (length of the probe) were 394.5 GPa, 3.27×10−19 m4, and 31.5 cm, respectively. The deflection of the cantilever (δ) was determined by comparing the prescribed and actual displacement of the probe for each compression event. The applied stress (σa) was determined by dividing F by the approximate contact area of the cell with the probe, as in our previous studies (Koay et al. 2008, Leipzig and Athanasiou 2005). To determine cell stiffness, applied stress was plotted against the applied axial strain at equilibrium for each compression event, and a linear regression was performed (see Figure 3). The slope of each regression line in Figure 3 indicates the stiffness of each cell type. Cellular height and width dimensions initially, at equilibrium compression, and at equilibrium recovery were also used to determine lateral (εl) and axial (εa) strains. Residual axial strain (εr) was calculated as (hi-hf)/hi, where hi is the initial cell height and hf is the cell height following recovery from the compression event. From these strain measures, the apparent Poisson’s ratio (ν) was calculated as ν = −(εl / εa). To determine compressibility, cells were approximated as ellipsoids with identical width and depth dimensions, and cellular volume was approximated as V = πhd2/6. Cell volume was calculated initially (Vinit), at equilibrium compression (Veq), and following recovery (Vrec). As previously described (Ofek et al. 2009), the recovered volume fraction was then defined as Vrec/Vinit, and the apparent compressibility as (βa) as (Vinit – Veq)/(σaVinit). The characteristic recovery time (τ) for each cell was also determined by modeling axial strain of the cell over time as an exponential decay function:
where A is the recovery coefficient, and t is the time in seconds.
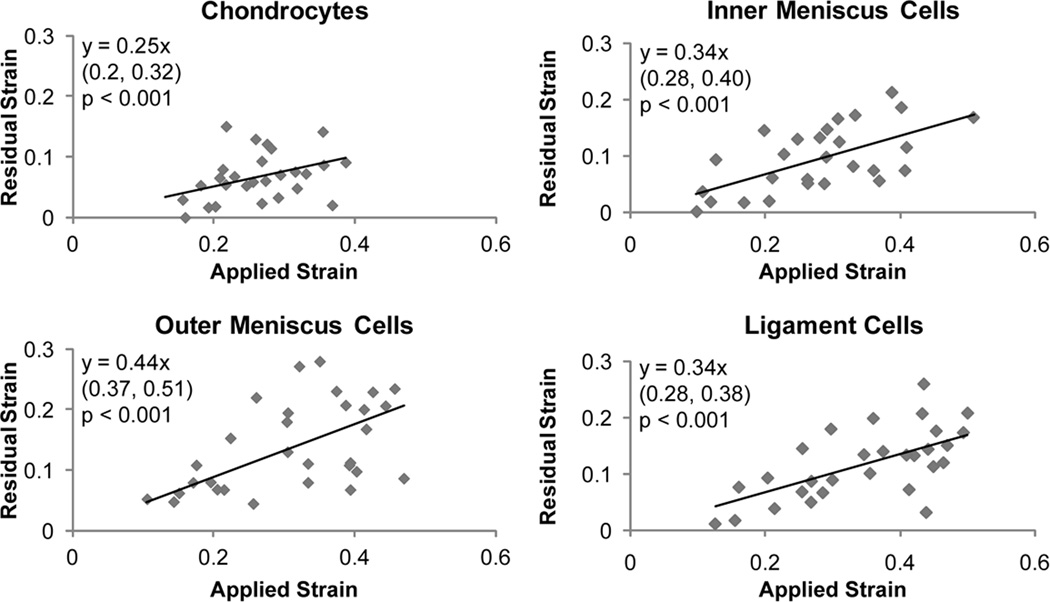
Fig. 3. Correlation of residual vs. applied strain of compressed cells.
Linear regression analysis was performed on residual strain versus applied strain data for chondrocytes, inner and outer meniscus cells, and ligament cells. Significant correlations were observed for all cell types (p < 0.001). The equation obtained from linear regression is shown in each graph with 95% confidence bounds for the slope in parentheses below. Positive correlations were observed for all cell types, but chondrocytes displayed a statistically lower slope than outer meniscus cells
Cell stiffness and Poisson's ratio calculated from measurements of the cell and probe during each compression event are listed in Table 1, and stress-strain plots for each cell type are shown in Figure 2. In Table 1, biomechanical properties not connected by the same letter are statistically significant from each other. All linear regression models applied to the stress-strain curves for each cell type had significance values less than 0.001.
Table 1.
Mechanical characteristics of musculoskeletal cells*
| Mechanical Property |
Articular Chondrocytes |
Inner Meniscus Cells |
Outer Meniscus Cells |
Ligament Cells |
|---|---|---|---|---|
| Cell Stiffness (kPa) | 2.51 ± 0.2 a | 1.07 ± 0.14 c | 1.59 ± 0.19 b | 1.32 ± 0.2 bc |
| Poisson's Ratio | 0.5 ± 0.27 | 0.48 ± 0.21 | 0.54 ± 0.26 | 0.46 ±0.17 |
| Recovered Volume (%) | 98.2 ± 6.6 a | 94 ± 6.8 ab | 92.7 ± 6.8 b | 96.2 ± 7 ab |
| Recovery Time (sec) | 7.15 ± 4.96 | 8.21 ± 3.74 | 7.34 ± 3.22 | 10.73 ± 6.09 |
All values are presented as mean ± standard deviation, except cell stiffness which is presented as slope ± 95% confidence bounds (see Figure 3)
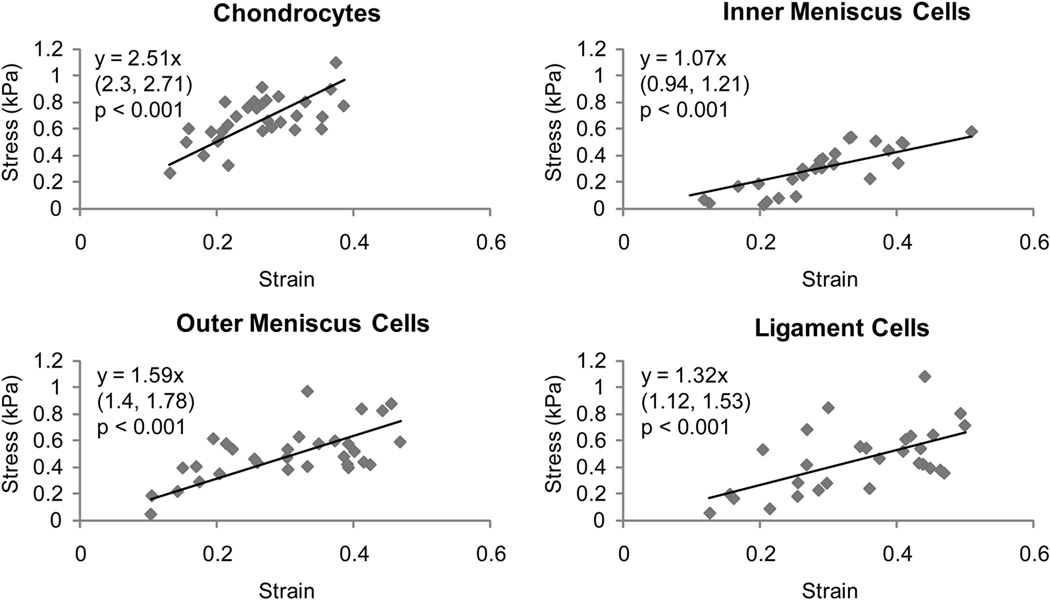
Fig. 2. Stress versus strain correlations for different cell types.
Linear regression analysis was performed on stress-strain data for chondrocytes, inner and outer meniscus cells, and ligament cells. Significant correlations were observed for all cell types (p < 0.001). For each graph, the equation obtained from linear regression is shown with 95% confidence bounds for the slope in parentheses below. Chondrocytes showed the highest cell stiffness (slope) than all other cell types, while ligament cell stiffness was not statistically different from either meniscus cell type. Inner meniscus cells, however, were statistically less stiff than outer meniscus cells
Immunocytochemistry
Cells from each group were fluorescently stained for cell nuclei, actin, focal adhesion kinase (FAK), and microtubules. Each cell type was seeded onto a glass slide at a concentration of 0.25×106 cells/mL, and incubated at 37°C for 1.5 hours. Following incubation, seeded cells were washed with warm PBS and fixed with 3.7% formaldehyde for 15 minutes. After fixation, cells were washed with PBS and made permeable via incubation with 0.1% Triton X-100 for 15 minutes. Cells were then incubated with Image-iT™ FX Enhancer [Invitrogen] for 30 minutes, and then stained for actin using CF594 Phalloidin [Biotium, Hayward, CA] for 20 minutes. For microtubule staining, cells were incubated with an anti-α-tubulin antibody [cat. # 322500, Invitrogen] for 1 hour, and for FAK staining, cells were incubated with pp125FAK anti-FAK antibody [Sigma] for 1 hour. Secondary antibodies specific to each primary, and conjugated to a 488nm fluorophore, were incubated with the cells for 1 hour. Cell nuclei were then stained with 10 µM Hoechst 33342 for 7 minutes. Each slide was mounted using Prolong Gold Antifade medium [Invitrogen]. Images of cells were taken using a fluorescence microscope, and exposure times for each fluorophore were optimized for articular chondrocytes and kept constant for capturing images from all groups.
Data analysis
A one-way ANOVA with p < 0.05 was used to compare cell height and width across cell types, and a Tukey’s post-hoc test was used to determine significant differences between groups. Linear regression analysis was performed with the data analysis package in Microsoft® Office Excel® 2007, one-way ANOVAs were performed in JMP® 7.0.1, and exponential fits were carried out in MATLAB® 7.11. Linear regression analysis was performed to determine if significant correlations of stress, residual strain, and apparent compressibility existed as a function of applied axial strain and to test if these correlations varied with cell type. A correlation was determined significant if p < 0.05. For significant correlations, differences between slopes were detected by comparing 95% confidence intervals as done previously (Koay et al. 2008, Ofek et al. 2009).
Results
Compressive behavior
Comparisons of the 95% confidence intervals from the linear regression analysis of each cell type (Table 1 and Figure 2) showed that the articular chondrocytes were the stiffest of the four cell types, followed by the outer meniscus cells and inner meniscus cells, respectively. The stiffness of the ligament cells was not statistically different from either the inner or the outer meniscus cells, and was significantly lower than the articular chondrocytes. Average apparent Poisson’s ratios of the four cell types were not statistically different from each other (Table 1).
Recovery behavior
Cell morphological changes following compression were recorded, and recovered volume, characteristic recovery time, residual strain, and apparent compressibility were calculated for each cell. The average percent recovered volume was highest for the articular chondrocytes, and lowest for the outer meniscus cells (Table 1). Percent recovered volume for inner meniscus cells and ligament cells was intermediate but was not statistically different from articular chondrocytes or outer meniscus cells. The average characteristic time to recovery was not statistically different between cell types, and was on the order of 7 to 10 seconds for all cell types.
Linear regression analysis of residual strain versus applied strain and apparent compressibility versus applied strain revealed significant linear correlations for all cell types (Figures 3 and 4, respectively). Regression analysis of residual strain versus applied strain showed positive linear correlations for all cell types (Figure 4). The slope of the fitted line for articular chondrocyte residual versus applied strain was found to be significantly lower than the slope for outer meniscus cells, as determined by comparing 95% confidence intervals. Slopes for inner meniscus cells and ligament cells were similar to each other and not statistically different from either the chondrocytes or outer meniscus cells (Figure 3).
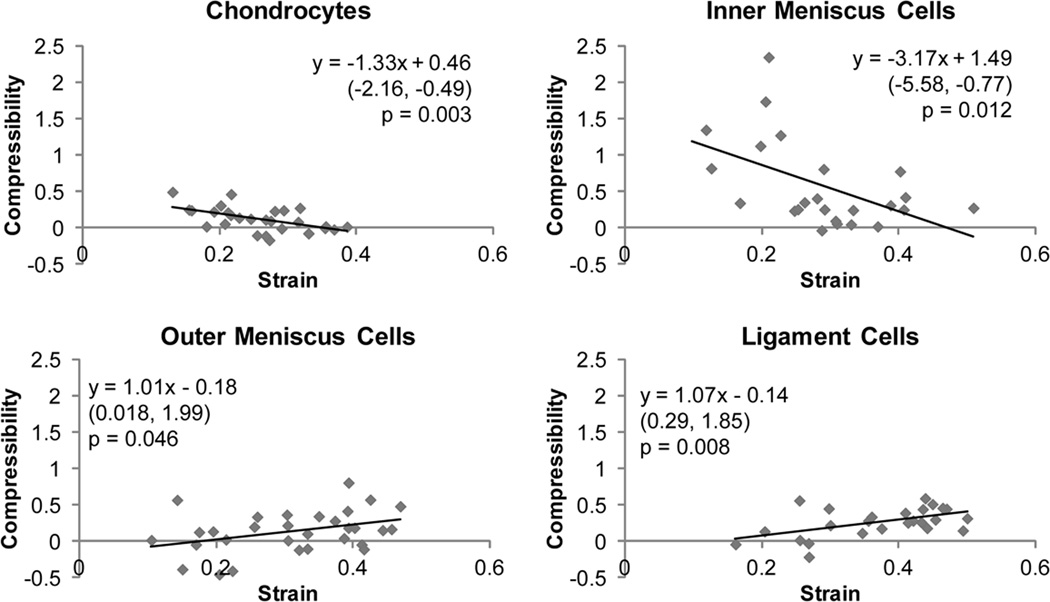
Fig. 4. Apparent compressibility versus applied strain correlations.
Linear regression analysis was performed on compressibility versus applied strain data for chondrocytes, inner and outer meniscus cells, and ligament cells. Significant correlations were observed for all cell types (p < 0.05). The equation obtained from linear regression is shown in each graph with 95% confidence bounds for the slope in parentheses below. Positive correlations were observed for outer meniscus cells and ligament cells, while negative correlations were observed for chondrocytes and inner meniscus cells
Regression analysis of apparent compressibility versus applied strain showed positive linear correlations for articular chondrocytes and inner meniscus cells, and negative correlations for outer meniscus cells and ligament cells (Figure 4). It should be noted that some of the data points for cellular compressibility were negative, indicating the cells increased in volume with applied strain. These negative values are likely indicative of errors due to the assumption that the cells have equal width and depth dimensions. Comparison of the 95% confidence intervals for each regression line showed that chondrocyte and inner meniscus cell slopes were not statistically different from each other but were statistically different from outer meniscus cell and ligament cell slopes. Outer meniscus cell and ligament cell slopes were also not statistically different from each other.
Immunocytochemistry
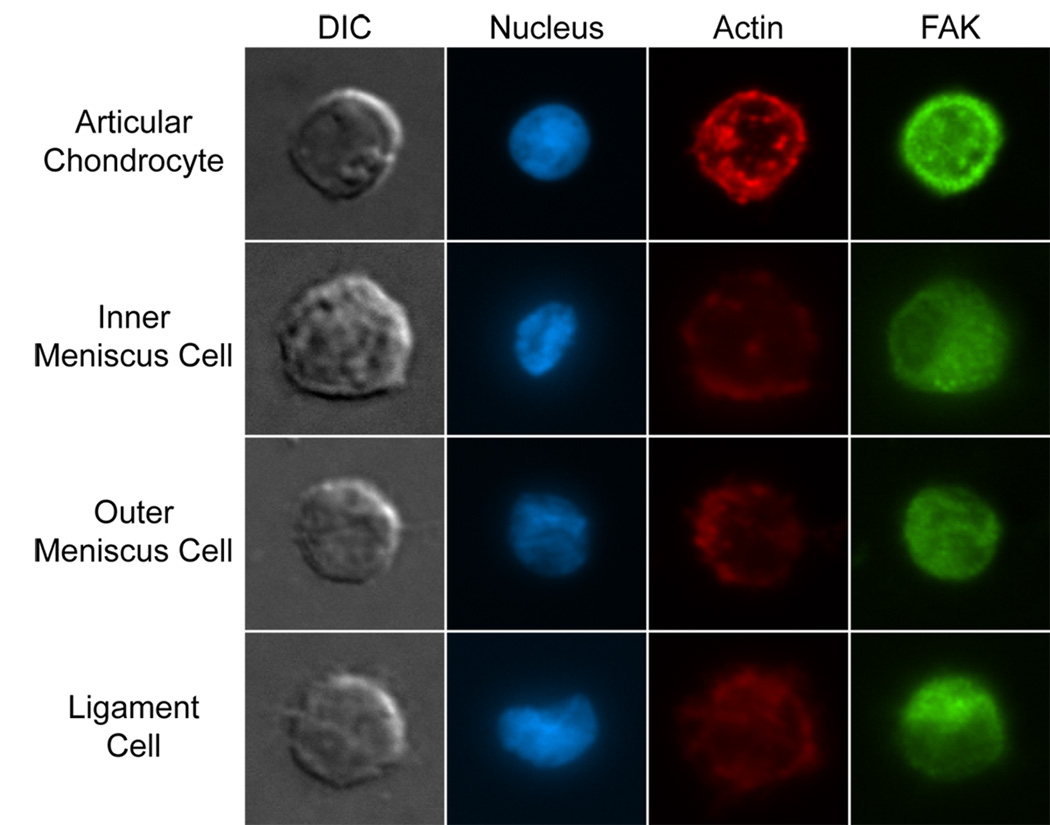
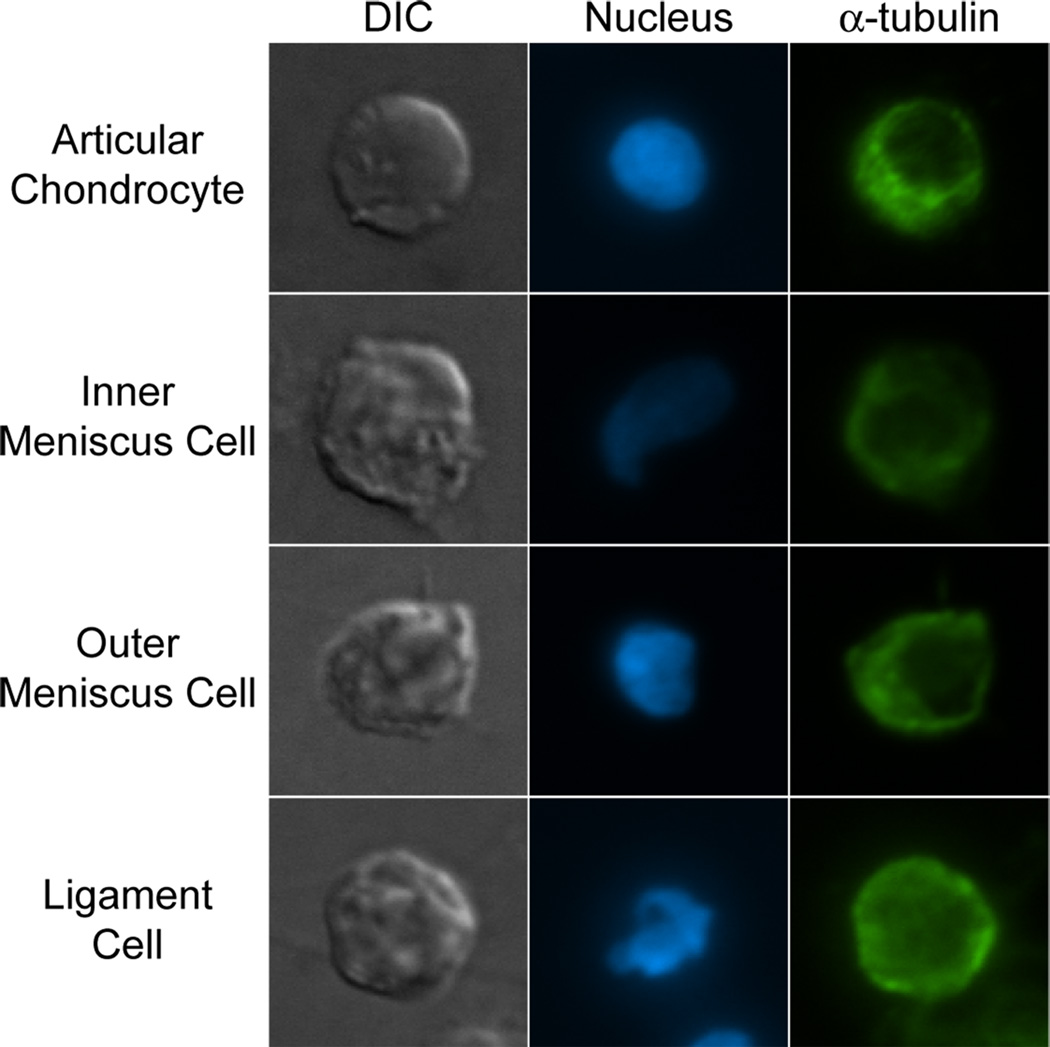
Representative cells stained for actin and FAK or microtubules are shown in figures 5 and 6, respectively. All cells stained positively for actin, FAK, and microtubules. Articular chondrocytes showed distinct, cortical actin staining, while inner and outer meniscus cells and ligament cells showed more diffuse staining for actin (Figure 5). FAK staining (Figure 5) and microtubule staining (Figure 6) appeared to be similar for all cell groups.
Fig. 5. Actin and FAK immunocytochemistry.
Articular chondrocytes, inner and outer meniscus cells, and ligament cells were seeded onto glass slides and fluorescently stained for cell nuclei, actin, and focal adhesion kinase (FAK). Cells from all groups stained positively for actin and FAK, and FAK staining was similar across cell types. Actin staining was most defined in articular chondrocytes, and more diffuse in meniscus and ligament cells
Fig. 6. α-tubulin immunocytochemistry.
Single articular chondrocytes, inner and outer meniscus cells, and ligament cells were fluorescently stained for microtubules using an anti-α-tubulin antibody. All cells stained positively for microtubules, with no gross differences noted
Discussion
This study compared the biomechanical properties of cells from different meniscus regions to articular chondrocytes and patellar ligament cells. Overall, it was found that meniscus cells were similar in biomechanical properties and cytoskeletal staining to ligament cells, although outer meniscus cells proved to be stiffer than inner meniscus cells. Additionally, articular chondrocytes were found to be significantly stiffer and show more distinct actin staining than all other cell types studied, suggesting that biomechanical properties of cells may correlate to their tissues of origin. To our knowledge, this study is the first to identify two mechanically distinct subpopulations of cells in the meniscus which correspond to known regional biochemical variations. These results may aid in further characterization of musculoskeletal cells, providing key information for mathematical models or mechanical stimulation experiments that seek to understand tissue mechanics and mechanotransduction.
Unconfined compression of inner and outer meniscus cells showed that mechanical properties of meniscus cells are unique to their region of origin. Specifically, outer meniscus cells were found to be stiffer than inner meniscus cells, but also showed more compressibility with applied strain compared with inner cells. Immunocytochemistry performed on these cells showed similar staining of actin and microtubules for inner and outer meniscus cells, indicating that they have similar cytoskeletal makeup and positive FAK staining confirmed that the cells were actively attached to the glass surface. As the actin cytoskeleton has been implicated as the largest contributor to cell stiffness (Ofek et al. 2009), and outer and inner meniscus cells showed similar actin staining, the increased stiffness of these cells may be a result of a more organized actin cytoskeleton in the outer cells acting to resist axial compression. Outer meniscus cells were also found to have similar compressibility characteristics to ligament cells, while inner meniscus cells were more similar in this respect to chondrocytes. These compressibility similarities follow phenotypic and primary loading pattern similarities, with outer meniscus cells and ligament cells being more fibroblastic and primarily experiencing tensile loads, and inner meniscus cells and articular chondrocytes showing chondrocytic characteristics while experiencing mainly compressive loads (Upton et al. 2006). As cells are primarily filled with water, the increased compressibility with strain of outer meniscus and ligament cells may indicate a more permeable cell membrane in these cells, allowing more water to escape under higher strains. Further investigation is needed to confirm these theories, but these observations provide an interesting groundwork for correlating compressibility characteristics with cell type. Taken together, the differences observed in inner and outer meniscus cell mechanics further illustrate previously identified morphological and phenotypic differences of meniscus cells, and show that the apparent compressibility of cells may be linked to primary loading patterns in tissues.
Overall, it appeared that meniscus cells as a whole shared more biomechanical similarities to ligament cells than to articular chondrocytes. As hypothesized, outer meniscus cells and ligament cells showed similarities in nearly all biomechanical measures. Additionally, and in contrast to the proposed hypothesis, inner meniscus cells were also similar in stiffness and residual strain characteristics to ligament cells. These similarities were confirmed by fluorescent staining of the actin cytoskeleton, a main contributor to cell stiffness as described above, showing similar diffuse staining for meniscus cells and ligament cells, as compared to the disctinct cortical actin staining of articular chondrocytes. Further, meniscus tissue is similar to ligament tissue in terms of collagen organization, with collagen primary oriented in the circumferential direction in all regions of the meniscus and along the direction of loading in ligaments. The loads experienced by these two tissues are also similar, as tension is generated in both tissues along the axis of collagen orientation. In contrast, primary loading in articular cartilage is compressive. Although inner meniscus tissue bears some resemblance to articular cartilage, containing collagen type II and more sulfated glycosaminoglycans than the outer meniscus, the similarities in inner meniscus cell mechanics to ligament cells may indicate that tensile loading experienced by inner cells is important to their overall phenotype. Thus, differences in stiffness between articular chondrocytes and meniscus or ligament cells seem to be correlated with the actin cytoskeleton and the loading patterns experienced by the cells in their native environment.
Despite differences in cell stiffness, all cell types showed similar characteristic recovery times (Table 1). Additionally, microtubule staining of all cell types showed similar intensity and distribution (Figure 6). Previous research investigating the mechanical role of cytoskeletal elements has shown that microtubules are essential for cellular recovery following axial compression, as disruption of these elements leads to a 100% increase in characteristic recovery time (Ofek et al. 2009). Given the importance of microtubules in cellular recovery behavior, the similarities observed in microtubule staining between cell types supports their similar characteristic recovery times.
The differences in cellular mechanics demonstrated in this study may also indicate that tissues with similar properties may contain cells with similar biomechanical properties. This is in agreement with previous work using AFM to compare the mechanics of different cell types, which reported a correlation between cell stiffness and tissue properties (Darling et al. 2008). The higher stiffness of articular chondrocytes compared with meniscus and ligament cells also closely matches the regional mechanical differences observed in intervertebral disc (IVD) cells (Guilak et al. 1999) which reside in different biochemical environments. In the IVD, nucleus pulposus cells have been found to be approximately 3-fold stiffer than annulus fibrosus cells. In the present study, articular chondrocytes were found to be approximately 2-fold stiffer than the other cell types tested. As the nucleus pulposus contains similar biochemical components to articular cartilage, namely high sulfated GAG and collagen type II content, and the annulus fibrosus is made up of fibrous tissue like the meniscus or patellar ligament, the present results agree with the correlation of cell stiffness and tissue properties. These correlations may help predict the biomechanical properties of cells from other musculoskeletal tissues such as tendon, allowing for more accurate biomechanical models to be constructed.
Mathematical models of the mechanical deformation of single cells have already been developed for articular cartilage (Kim et al. 2010) and the intervertebral disc (Cao et al. 2011), and the knee meniscus under axial compression, (Upton et al. 2006) giving new insights into the translation of tissue strains to cell strains and indicating potential stimulation parameters for tissue engineering. With respect to the knee meniscus, previous research has used information from modeling to demonstrate that mechanotransduction exists in meniscus cells, showing that inner and outer meniscus cells respond to cyclic tensile loading with increased nitric oxide production and total protein synthesis (Upton et al. 2006). Meniscus explants have also been shown to respond to 2% cyclic compressive strain with increased aggrecan gene expression (Aufderheide and Athanasiou 2006), but other research has shown decreased collagen and increased proteinase gene expression levels following a 0.08–0.16 MPa dynamic compression regimen (Upton et al. 2003). These data indicate that there may be optimal levels of strain to apply to meniscus cells to achieve protein synthesis, beyond which catabolic processes are initiated. Thus, as meniscus cells have demonstrated sensitivity to mechanical stimulation, understanding their mechanical properties can help to further tailor the stimulation of engineered constructs to initiate particular mechanotransductive events for achieving functional tissue properties.
To test the bulk mechanical properties of cells in this study, several types of cellular manipulation were employed, such as enzymatic isolation and short-term monolayer culture. However, the effects of these processes on the mechanical properties of the cells are not well understood, and therefore present limitations that must be taken into account when interpreting the present results. One of the main limitations in this study is that cells must be removed from their native microenvironment to be tested. This neglects the natural load transmitting function of the cell’s territorial matrix, which likely plays an important role in cellular mechanotransduction in situ. Additionally, to test bulk cellular properties, cells were seeded for a short period of time to maintain a rounded morphology. While native chondrocytes and inner meniscus cells are normally spherical, this is not the native morphology of outer meniscus cells and ligament cells. However, while not wholly representative of native morphology, the measured mechanical properties of meniscus cells and ligament cells may be useful for determining appropriate mechanical stimulation regimens for tissue engineered constructs which often use large concentrations of isolated cells. It should also be noted that cells were harvested from a skeletally immature knee joint and cryopreserved prior to testing. Though the mechanical properties of chondrocytes observed here were on the same scale as those obtained from skeletally mature chondrocytes that had not been cryopreserved (Leipzig and Athanasiou 2005, Ofek et al. 2009), future studies should investigate whether these biomechanical distinctions remain constant with age and are affected by cryopreservation. The results from this study should therefore be considered along with these caveats, but provide a strong basis for further investigation into the mechanical correlations between cells and their native environments. Characterization of the mechanical properties of single meniscus cells is an important step toward understanding mechanotransduction in the meniscus. This study demonstrated that meniscus cell mechanics vary regionally in the tissue, and that meniscus cells are most biomechanically similar to ligament cells. The mechanical properties measured in this study will be useful parameters for constructing accurate biomechanical models of the knee meniscus and for designing experiments to deliver precise mechanical stimulation to tissue engineered constructs.
Acknowledgments
We would like to thank the National Science Foundation Rice-Houston Alliance for Graduate Education and the Professoriate (NSF-AGEP) for their generous support of this work. We would also like to acknowledge funding from the National Institutes of Health R01AR047839.
Abbreviations
- MC
meniscus cell
- FAK
focal adhesion kinase
- AFM
atomic force microscopy
- GAG
glycosaminoglycan
References
- Aspden RM, Yarker YE, Hukins DW. Collagen orientations in the meniscus of the knee joint. J Anat. 1985;140(Pt 3):371–380. [PMC free article] [PubMed] [Google Scholar]
- Aufderheide AC, Athanasiou KA. A direct compression stimulator for articular cartilage and meniscal explants. Ann Biomed Eng. 2006;34:1463–1474. doi: 10.1007/s10439-006-9157-x. [DOI] [PubMed] [Google Scholar]
- Cao L, Guilak F, Setton LA. Three-dimensional finite element modeling of pericellular matrix and cell mechanics in the nucleus pulposus of the intervertebral disk based on in situ morphology. Biomech Model Mechanobiol. 2011;10:1–10. doi: 10.1007/s10237-010-0214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling EM, Topel M, Zauscher S, Vail TP, Guilak F. Viscoelastic properties of human mesenchymally-derived stem cells and primary osteoblasts, chondrocytes, and adipocytes. J Biomech. 2008;41:454–464. doi: 10.1016/j.jbiomech.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling EM, Wilusz RE, Bolognesi MP, Zauscher S, Guilak F. Spatial mapping of the biomechanical properties of the pericellular matrix of articular cartilage measured in situ via atomic force microscopy. Biophys J. 2010;98:2848–2856. doi: 10.1016/j.bpj.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling EM, Zauscher S, Block JA, Guilak F. A thin-layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: do cell properties reflect metastatic potential? Biophys J. 2007;92:1784–1791. doi: 10.1529/biophysj.106.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling EM, Zauscher S, Guilak F. Viscoelastic properties of zonal articular chondrocytes measured by atomic force microscopy. Osteoarthritis Cartilage. 2006;14:571–579. doi: 10.1016/j.joca.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Feng Y, Ofek G, Choi DS, Wen J, Hu J, Zhao H, Zu Y, Athanasiou KA, Chang CC. Unique biomechanical interactions between myeloma cells and bone marrow stroma cells. Prog Biophys Mol Biol. 2010;103:148–156. doi: 10.1016/j.pbiomolbio.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Fithian DC, Kelly MA, Mow VC. Material properties and structure-function relationships in the menisci. Clin Orthop Relat Res. 1990:19–31. [PubMed] [Google Scholar]
- Gabrion A, Aimedieu P, Laya Z, Havet E, Mertl P, Grebe R, Laude M. Relationship between ultrastructure and biomechanical properties of the knee meniscus. Surg Radiol Anat. 2005;27:507–510. doi: 10.1007/s00276-005-0031-6. [DOI] [PubMed] [Google Scholar]
- Guilak F, Tedrow JR, Burgkart R. Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun. 2000;269:781–786. doi: 10.1006/bbrc.2000.2360. [DOI] [PubMed] [Google Scholar]
- Guilak F, Ting-Beall HP, Baer AE, Trickey WR, Erickson GR, Setton LA. Viscoelastic properties of intervertebral disc cells. Identification of two biomechanically distinct cell populations. Spine (Phila Pa 1976) 1999;24:2475–2483. doi: 10.1097/00007632-199912010-00009. [DOI] [PubMed] [Google Scholar]
- Hochmuth RM. Micropipette aspiration of living cells. J Biomech. 2000;33:15–22. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- Ikai A. A Review on: Atomic Force Microscopy Applied to Nano-mechanics of the Cell. Adv Biochem Eng Biotechnol. 2009 doi: 10.1007/10_2008_41. [DOI] [PubMed] [Google Scholar]
- Kim E, Guilak F, Haider MA. An axisymmetric boundary element model for determination of articular cartilage pericellular matrix properties in situ via inverse analysis of chondron deformation. J Biomech Eng. 2010;132:031011. doi: 10.1115/1.4000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay EJ, Ofek G, Athanasiou KA. Effects of TGF-beta1 and IGF-I on the compressibility, biomechanics, and strain-dependent recovery behavior of single chondrocytes. J Biomech. 2008;41:1044–1052. doi: 10.1016/j.jbiomech.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Koay EJ, Shieh AC, Athanasiou KA. Creep indentation of single cells. J Biomech Eng. 2003;125:334–341. doi: 10.1115/1.1572517. [DOI] [PubMed] [Google Scholar]
- Leipzig ND, Athanasiou KA. Static compression of single chondrocytes catabolically modifies single-cell gene expression. Biophys J. 2008;94:2412–2422. doi: 10.1529/biophysj.107.114207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipzig ND, Athanasiou KA. Unconfined creep compression of chondrocytes. J Biomech. 2005;38:77–85. doi: 10.1016/j.jbiomech.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Leipzig ND, Eleswarapu SV, Athanasiou KA. The effects of TGF-beta1 and IGF-I on the biomechanics and cytoskeleton of single chondrocytes. Osteoarthritis Cartilage. 2006;14:1227–1236. doi: 10.1016/j.joca.2006.05.013. [DOI] [PubMed] [Google Scholar]
- McDermott ID, Sharifi F, Bull AM, Gupte CM, Thomas RW, Amis AA. An anatomical study of meniscal allograft sizing. Knee Surg Sports Traumatol Arthrosc. 2004;12:130–135. doi: 10.1007/s00167-003-0366-7. [DOI] [PubMed] [Google Scholar]
- McDevitt CA, Webber RJ. The ultrastructure and biochemistry of meniscal cartilage. Clin Orthop Relat Res. 1990:8–18. [PubMed] [Google Scholar]
- Ofek G, Willard VP, Koay EJ, Hu JC, Lin P, Athanasiou KA. Mechanical characterization of differentiated human embryonic stem cells. J Biomech Eng. 2009;131:061011. doi: 10.1115/1.3127262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Wiltz DC, Athanasiou KA. Contribution of the cytoskeleton to the compressive properties and recovery behavior of single cells. Biophys J. 2009;97:1873–1882. doi: 10.1016/j.bpj.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh AC, Athanasiou KA. Dynamic compression of single cells. Osteoarthritis Cartilage. 2007;15:328–334. doi: 10.1016/j.joca.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Shieh AC, Koay EJ, Athanasiou KA. Strain-dependent recovery behavior of single chondrocytes. Biomech Model Mechanobiol. 2006;5:172–179. doi: 10.1007/s10237-006-0028-z. [DOI] [PubMed] [Google Scholar]
- Shin D, Athanasiou K. Cytoindentation for obtaining cell biomechanical properties. J Orthop Res. 1999;17:880–890. doi: 10.1002/jor.1100170613. [DOI] [PubMed] [Google Scholar]
- Skaggs DL, Warden WH, Mow VC. Radial tie fibers influence the tensile properties of the bovine medial meniscus. J Orthop Res. 1994;12:176–185. doi: 10.1002/jor.1100120205. [DOI] [PubMed] [Google Scholar]
- Sweigart MA, Athanasiou KA. Tensile and compressive properties of the medial rabbit meniscus. Proc Inst Mech Eng H. 2005;219:337–347. doi: 10.1243/095441105X34329. [DOI] [PubMed] [Google Scholar]
- Sweigart MA, Zhu CF, Burt DM, DeHoll PD, Agrawal CM, Clanton TO, Athanasiou KA. Intraspecies and interspecies comparison of the compressive properties of the medial meniscus. Ann Biomed Eng. 2004;32:1569–1579. doi: 10.1114/b:abme.0000049040.70767.5c. [DOI] [PubMed] [Google Scholar]
- Theret DP, Levesque MJ, Sato M, Nerem RM, Wheeler LT. The application of a homogeneous half-space model in the analysis of endothelial cell micropipette measurements. J Biomech Eng. 1988;110:190–199. doi: 10.1115/1.3108430. [DOI] [PubMed] [Google Scholar]
- Upton ML, Chen J, Guilak F, Setton LA. Differential effects of static and dynamic compression on meniscal cell gene expression. J Orthop Res. 2003;21:963–969. doi: 10.1016/S0736-0266(03)00063-9. [DOI] [PubMed] [Google Scholar]
- Upton ML, Gilchrist CL, Guilak F, Setton LA. Transfer of macroscale tissue strain to microscale cell regions in the deformed meniscus. Biophys J. 2008;95:2116–2124. doi: 10.1529/biophysj.107.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton ML, Guilak F, Laursen TA, Setton LA. Finite element modeling predictions of region-specific cell-matrix mechanics in the meniscus. Biomech Model Mechanobiol. 2006;5:140–149. doi: 10.1007/s10237-006-0031-4. [DOI] [PubMed] [Google Scholar]
- Upton ML, Hennerbichler A, Fermor B, Guilak F, Weinberg JB, Setton LA. Biaxial strain effects on cells from the inner and outer regions of the meniscus. Connect Tissue Res. 2006;47:207–214. doi: 10.1080/03008200600846663. [DOI] [PubMed] [Google Scholar]
- Valiyaveettil M, Mort JS, McDevitt CA. The concentration, gene expression, and spatial distribution of aggrecan in canine articular cartilage, meniscus, and anterior and posterior cruciate ligaments: a new molecular distinction between hyaline cartilage and fibrocartilage in the knee joint. Connect Tissue Res. 2005;46:83–91. doi: 10.1080/03008200590954113. [DOI] [PubMed] [Google Scholar]
- Walker PS, Erkman MJ. The role of the menisci in force transmission across the knee. Clin Orthop Relat Res. 1975:184–192. doi: 10.1097/00003086-197506000-00027. [DOI] [PubMed] [Google Scholar]
- Walker PS, Hajek JV. The load-bearing area in the knee joint. J Biomech. 1972;5:581–589. doi: 10.1016/0021-9290(72)90030-9. [DOI] [PubMed] [Google Scholar]
- You HX, Yu L. Atomic force microscopy imaging of living cells: progress, problems and prospects. Methods Cell Sci. 1999;21:1–17. doi: 10.1023/a:1009876320336. [DOI] [PubMed] [Google Scholar]