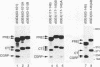Abstract
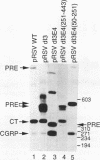
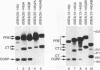
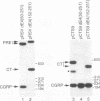
The calcitonin (CT)/calcitonin gene-related peptide I (CGRP-I) gene (CALC-I gene) is subject to alternative tissue-specific processing of its primary transcript. CT mRNA is the predominant mRNA produced in thyroid C cells, whereas CT gene-related peptide I mRNA is the main product in neurons of the central and peripheral nervous systems. The CT-specific exon 4 is surrounded by weak processing sites. In this study we have investigated whether exon 4 sequences are involved in the tissue-specific selection of the exon 4 splice acceptor site. The results indicate that two separate elements, termed A and B, in the 5' part of exon 4 are required for production of CT-specific RNA. These sequences are located between nucleotides 67 and 88 (A) and nucleotides 117 and 146 (B) relative to the 5' end of exon 4. Variation of the distance between these sequence elements and the 3' splice site of exon 4 does not change the processing choice. These sequence elements are functionally equivalent. CT-specific splicing requires the presence of both sequence A and B or duplicates of either sequence element in exon 4. The effect of these sequences on the RNA processing choice is overruled by mutation of the CT-specific uridine branch acceptor nucleotide into a commonly preferred adenosine residue.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adema G. J., Baas P. D. A novel calcitonin-encoding mRNA is produced by alternative processing of calcitonin/calcitonin gene-related peptide-I pre-mRNA. J Biol Chem. 1992 Apr 15;267(11):7943–7948. [PubMed] [Google Scholar]
- Adema G. J., Baas P. D. Deregulation of alternative processing of Calcitonin/CGRP-I pre-mRNA by a single point mutation. Biochem Biophys Res Commun. 1991 Aug 15;178(3):985–992. doi: 10.1016/0006-291x(91)90989-k. [DOI] [PubMed] [Google Scholar]
- Adema G. J., Bovenberg R. A., Jansz H. S., Baas P. D. Unusual branch point selection involved in splicing of the alternatively processed Calcitonin/CGRP-I pre-mRNA. Nucleic Acids Res. 1988 Oct 25;16(20):9513–9526. doi: 10.1093/nar/16.20.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adema G. J., van Hulst K. L., Baas P. D. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 1990 Sep 25;18(18):5365–5373. doi: 10.1093/nar/18.18.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara S. G., Evans R. M., Rosenfeld M. G. Calcitonin/calcitonin gene-related peptide transcription unit: tissue-specific expression involves selective use of alternative polyadenylation sites. Mol Cell Biol. 1984 Oct;4(10):2151–2160. doi: 10.1128/mcb.4.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M., Piñol-Roma S., Staknis D., Dreyfuss G., Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992 Jul;12(7):3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovenberg R. A., Adema G. J., Jansz H. S., Baas P. D. Model for tissue specific Calcitonin/CGRP-I RNA processing from in vitro experiments. Nucleic Acids Res. 1988 Aug 25;16(16):7867–7883. doi: 10.1093/nar/16.16.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovenberg R. A., Moen T. C., Jansz H. S., Baas P. D. In vitro splicing analysis of mini-gene constructs of the alternatively processed human calcitonin/CGRP-I pre-mRNA. Biochim Biophys Acta. 1989 Jul 7;1008(2):223–233. doi: 10.1016/0167-4781(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Bovenberg R. A., van de Meerendonk W. P., Baas P. D., Steenbergh P. H., Lips C. J., Jansz H. S. Model for alternative RNA processing in human calcitonin gene expression. Nucleic Acids Res. 1986 Nov 25;14(22):8785–8803. doi: 10.1093/nar/14.22.8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis K. C., Baker B. S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989 Mar 24;56(6):997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Chain A. C., Zollman S., Tseng J. C., Laski F. A. Identification of a cis-acting sequence required for germ line-specific splicing of the P element ORF2-ORF3 intron. Mol Cell Biol. 1991 Mar;11(3):1538–1546. doi: 10.1128/mcb.11.3.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986 Mar 28;231(4745):1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Cooper T. A. In vitro splicing of cardiac troponin T precursors. Exon mutations disrupt splicing of the upstream intron. J Biol Chem. 1992 Mar 15;267(8):5330–5338. [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote G. J., Nguyen I. N., Berget S. M., Gagel R. F. Calcitonin exon sequences influence alternative RNA processing. Mol Endocrinol. 1990 Nov;4(11):1744–1749. doi: 10.1210/mend-4-11-1744. [DOI] [PubMed] [Google Scholar]
- Cote G. J., Nguyen I. N., Lips C. J., Berget S. M., Gagel R. F. Validation of an in vitro RNA processing system for CT/CGRP precursor mRNA. Nucleic Acids Res. 1991 Jul 11;19(13):3601–3606. doi: 10.1093/nar/19.13.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote G. J., Stolow D. T., Peleg S., Berget S. M., Gagel R. F. Identification of exon sequences and an exon binding protein involved in alternative RNA splicing of calcitonin/CGRP. Nucleic Acids Res. 1992 May 11;20(9):2361–2366. doi: 10.1093/nar/20.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler J. O., Rossi J. J. Unexpected point mutations activate cryptic 3' splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991 Jul;5(7):1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- Domenjoud L., Gallinaro H., Kister L., Meyer S., Jacob M. Identification of a specific exon sequence that is a major determinant in the selection between a natural and a cryptic 5' splice site. Mol Cell Biol. 1991 Sep;11(9):4581–4590. doi: 10.1128/mcb.11.9.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbrooke M. R., Parker D., McVey J. H., Riley J. H., Sorenson G. D., Pettengill O. S., Craig R. K. Expression of the human calcitonin/CGRP gene in lung and thyroid carcinoma. EMBO J. 1985 Mar;4(3):715–724. doi: 10.1002/j.1460-2075.1985.tb03688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeson R. B., Hedjran F., Yeakley J. M., Guise J. W., Rosenfeld M. G. Alternative production of calcitonin and CGRP mRNA is regulated at the calcitonin-specific splice acceptor. Nature. 1989 Sep 7;341(6237):76–80. doi: 10.1038/341076a0. [DOI] [PubMed] [Google Scholar]
- Eng F. J., Warner J. R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991 May 31;65(5):797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Estes P. A., Cooke N. E., Liebhaber S. A. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J Biol Chem. 1992 Jul 25;267(21):14902–14908. [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Maniatis T. The role of branchpoint and 3'-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991 Feb;5(2):211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- García-Blanco M. A., Jamison S. F., Sharp P. A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989 Dec;3(12A):1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- Gil A., Sharp P. A., Jamison S. F., Garcia-Blanco M. A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991 Jul;5(7):1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guo W., Mulligan G. J., Wormsley S., Helfman D. M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991 Nov;5(11):2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley M. L., Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991 May 17;65(4):579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991 May 10;252(5007):833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hoshijima K., Higuchi I., Sakamoto H., Shimura Y. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8092–8096. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992 Jan 24;68(2):365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- McKeown M. Alternative mRNA splicing. Annu Rev Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- McKeown M., Belote J. M., Boggs R. T. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell. 1988 Jun 17;53(6):887–895. doi: 10.1016/s0092-8674(88)90369-8. [DOI] [PubMed] [Google Scholar]
- McKeown M. Regulation of alternative splicing. Genet Eng (N Y) 1990;12:139–181. doi: 10.1007/978-1-4613-0641-2_9. [DOI] [PubMed] [Google Scholar]
- Nagoshi R. N., Baker B. S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990 Jan;4(1):89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Nagoshi R. N., McKeown M., Burtis K. C., Belote J. M., Baker B. S. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988 Apr 22;53(2):229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Patton J. G., Mayer S. A., Tempst P., Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991 Jul;5(7):1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Roesser J. R., Liittschwager K., Leff S. E. Regulation of tissue-specific splicing of the calcitonin/calcitonin gene-related peptide gene by RNA-binding proteins. J Biol Chem. 1993 Apr 15;268(11):8366–8375. [PubMed] [Google Scholar]
- Rosenfeld M. G., Mermod J. J., Amara S. G., Swanson L. W., Sawchenko P. E., Rivier J., Vale W. W., Evans R. M. Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature. 1983 Jul 14;304(5922):129–135. doi: 10.1038/304129a0. [DOI] [PubMed] [Google Scholar]
- Ryner L. C., Baker B. S. Regulation of doublesex pre-mRNA processing occurs by 3'-splice site activation. Genes Dev. 1991 Nov;5(11):2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Fresco L. D., Rio D. C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5' splice site control U1 snRNP binding. Genes Dev. 1992 Aug;6(8):1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3' end of introns. EMBO J. 1988 Nov;7(11):3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992 Apr 10;256(5054):237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- Tsai A. Y., Streuli M., Saito H. Integrity of the exon 6 sequence is essential for tissue-specific alternative splicing of human leukocyte common antigen pre-mRNA. Mol Cell Biol. 1989 Oct;9(10):4550–4555. doi: 10.1128/mcb.9.10.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu N., Kobayashi H., Miyatake T., Tsuji S. A novel exon mutation in the human beta-hexosaminidase beta subunit gene affects 3' splice site selection. J Biol Chem. 1992 Feb 5;267(4):2406–2413. [PubMed] [Google Scholar]
- Watakabe A., Inoue K., Sakamoto H., Shimura Y. A secondary structure at the 3' splice site affects the in vitro splicing reaction of mouse immunoglobulin mu chain pre-mRNAs. Nucleic Acids Res. 1989 Oct 25;17(20):8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Sakamoto H., Shimura Y. Repositioning of an alternative exon sequence of mouse IgM pre-mRNA activates splicing of the preceding intron. Gene Expr. 1991;1(3):175–184. [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Tanaka K., Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993 Mar;7(3):407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Xu R., Teng J., Cooper T. A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993 Jun;13(6):3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]