Abstract
Aims: Protein S-bacillithiolations are mixed disulfides between protein thiols and the bacillithiol (BSH) redox buffer that occur in response to NaOCl in Bacillus subtilis. We used BSH-specific immunoblots, shotgun liquid chromatography (LC)–tandem mass spectrometry (MS/MS) analysis and redox proteomics to characterize the S-bacillithiolomes of B. subtilis, B. megaterium, B. pumilus, B. amyloliquefaciens, and Staphylococcus carnosus and also measured the BSH/oxidized bacillithiol disulfide (BSSB) redox ratio after NaOCl stress. Results: In total, 54 proteins with characteristic S-bacillithiolation (SSB) sites were identified, including 29 unique proteins and eight proteins conserved in two or more of these bacteria. The methionine synthase MetE is the most abundant S-bacillithiolated protein in Bacillus species after NaOCl exposure. Further, S-bacillithiolated proteins include the translation elongation factor EF-Tu and aminoacyl-tRNA synthetases (ThrS), the DnaK and GrpE chaperones, the two-Cys peroxiredoxin YkuU, the ferredoxin–NADP+ oxidoreductase YumC, the inorganic pyrophosphatase PpaC, the inosine-5′-monophosphate dehydrogenase GuaB, proteins involved in thiamine biosynthesis (ThiG and ThiM), queuosine biosynthesis (QueF), biosynthesis of aromatic amino acids (AroA and AroE), serine (SerA), branched-chain amino acids (YwaA), and homocysteine (LuxS and MetI). The thioredoxin-like proteins, YphP and YtxJ, are S-bacillithiolated at their active sites, suggesting a function in the de-bacillithiolation process. S-bacillithiolation is accompanied by a two-fold increase in the BSSB level and a decrease in the BSH/BSSB redox ratio in B. subtilis. Innovation: Many essential and conserved proteins, including the dominant MetE, were identified in the S-bacillithiolome of different Bacillus species and S. carnosus using shotgun-LC-MS/MS analyses. Conclusion: S-bacillithiolation is a widespread redox control mechanism among Firmicutes bacteria that protects conserved metabolic enzymes and essential proteins against overoxidation. Antioxid. Redox Signal. 18, 1273–1295.
Introduction
The reduced cytoplasm is maintained by low-molecular-weight (LMW) thiol-redox buffers, such as glutathione (GSH), present in eukaryotes and in Escherichia coli (34). Redox buffers protect bacteria against reactive oxygen species (ROS) generated during respiration or produced by activated macrophages during infections (1). ROS cause reversible intramolecular and intermolecular disulfides of proteins as well as mixed disulfides between protein thiols and LMW thiols (S-thiolations). In eukaryotes, protein S-glutathionylation is the most important post-translational thiol modification caused by ROS, and has been shown to control redox-sensing transcription factors and to protect active-site cysteine (Cys) residues of essential metabolic enzymes against irreversible oxidation to Cys-sulfinic or sulfonic acids (8, 13, 51). As an effective redox control mechanism, S-glutathionylation must meet several criteria: (i) reversibility, (ii) specificity to active site Cys, (iii) change in protein function/activity, and (iv) induction by ROS or reactive nitrogen species (RNS). S-glutathionylation also serves as a form of GSH storage to prevent the export of oxidized glutathione disulfide (GSSG) under oxidative stress (8). Many eukaryotic proteins such as α-ketoglutarate dehydrogenase, glyceraldehyde 3-phosphate dehydrogenase, ornithine δ-aminotransferase, pyruvate kinase, heat-specific chaperones, and regulatory proteins (c-Jun and NF-κB) are reversibly inactivated or activated by S-glutathionylation (8, 21). In E. coli, the oxidative stress-responsive regulator OxyR is activated by S-glutathionylation in vitro (22), while the activities of glyceraldehyde-3-phosphate dehydrogenase, methionine synthase, and 3′-phosphoadenosine-5′-phosphosulfate reductase are inhibited by S-glutathionylation in E. coli (3, 19, 29).
Innovation.
Recently, we identified six S-bacillithiolated proteins in Bacillus subtilis in response to NaOCl stress (7). The methionine synthase MetE was inactivated by S-bacillithiolation causing methionine auxotrophy. Here, we have studied the conserved S-bacillithiolome among four different Bacillus species and Staphylococcus carnosus using bacillithiol (BSH)-specific immunoblots, shotgun liquid chromatography–tandem mass spectrometry analysis, and redox proteomics. We identified 54 S-bacillithiolated proteins, including eight conserved proteins (MetE, AroA, GuaB, TufA, PpaC, SerA, YphP, and YumC) and 29 unique proteins involved in amino acid and cofactor biosynthesis, nucleotide metabolism, translation, protein quality control, redox, and antioxidant functions. S-bacillithiolation is accompanied by an increased oxidized bacillithiol disulfide (BSSB) level and a decreased BSH/BSSB ratio. Together, our data support a major role of the BSH redox buffer in redox control and protection of conserved and essential proteins against irreversible oxidation by S-bacillithiolations in Firmicutes bacteria.
Gram-positive bacteria do not produce GSH. Among the actinomycetes, mycothiol is the predominant LMW thiol that serves analogous roles to GSH (20, 39). In low-GC Gram-positive bacteria, such as Bacillus and Staphylococcus species, bacillithiol (BSH) serves as a redox buffer that was structurally characterized as Cys-GlcN-malate (12, 16, 40). Mutants with defects in BSH biosynthesis displayed strong sensitivities to the epoxide antibiotic fosfomycin and to NaOCl stress (7, 12). In Bacillus subtilis, the redox-sensing MarR-type repressor OhrR forms mixed disulfides with BSH (S-bacillithiolations) in response to organic hydroperoxides and NaOCl stress (7, 26). S-bacillithiolation of OhrR alleviates repression of transcription of the ohrA peroxiredoxin gene as a protection mechanism against NaOCl toxicity (7). Besides OhrR, we have recently identified by liquid chromatography (LC)–tandem mass spectrometry (MS/MS) analysis five cytoplasmic proteins with S-bacillithiolations after NaOCl treatment (7). Hypochloric acid shows very fast reaction rates with Cys residues (k2=3×107 M−1 s−1) and rapidly leads to irreversible oxidation products, such as sulfinic and sulfonic acids (9, 15, 42, 50). Thus, S-bacillithiolation serves to protect active-site Cys residues of key metabolic enzymes against overoxidation. S-bacillithiolation of the methionine synthase MetE at the active site Cys730 caused a methionine starvation phenotype in NaOCl-treated cells, presumably to slow down translation while the OhrA peroxiredoxin removes the toxic oxidant (7).
In this work, we have identified proteome-wide conserved S-bacillithiolations in various industrially important Bacillus and Staphylococcus species. We used BSH-specific immunoblot analyses, redox proteomic approaches, and shotgun-LC-MS/MS analyses to identify essential and conserved proteins in the S-bacillithiolome that have been previously identified in the S-glutathionylome in eukaryotes. The protein bacillithiolations were accompanied by an increased level of oxidized bacillithiol disulfide (BSSB) and a decreased BSH/BSSB redox ratio in B. subtilis.
Results
Defining the conditions of NaOCl stress to induce S-bacillithiolations in the proteome of B. amyloliquefaciens, B. pumilus, B. megaterium, and S. carnosus
To describe more comprehensively the S-bacillithiolome, we focused on four industrially important Bacillus species: B. subtilis 168, B. pumilus SBUG1799, B. amyloliquefaciens FZB42 (5), B. megaterium SBUG1152 (36), and S. carnosus TM300 (43). The B. subtilis, B. pumilus, and B. megaterium strains were able to grow in the Belitsky minimal medium (BMM), and low NaOCl concentrations could be applied to reduce the growth rate: 75 μM NaOCl for B. subtilis (7), 125 μM for B. pumilus, and 80 μM for B. megaterium (Supplementary Fig. S1B, C; Supplementary Data are available online at www.liebertpub.com/ars). For the growth of B. amyloliquefaciens FZB42, we used the BMM with 0.1% casamino acids, and 275–300 μM NaOCl showed a growth-inhibitory effect (Supplementary Fig. S1A). For S. carnosus, growth was only optimal in a complete medium, and 3.25–3.5 mM NaOCl reduced the growth rate in a Luria Broth (LB) medium (Supplementary Fig. S1D). Since amino acids and peptides present in LB also react with NaOCl, much higher NaOCl concentrations were required to inhibit the growth of S. carnosus in the LB medium.
MetE is the major target for S-bacillithiolation in Bacillus species as revealed by nonreducing BSH-specific immunoblot analysis
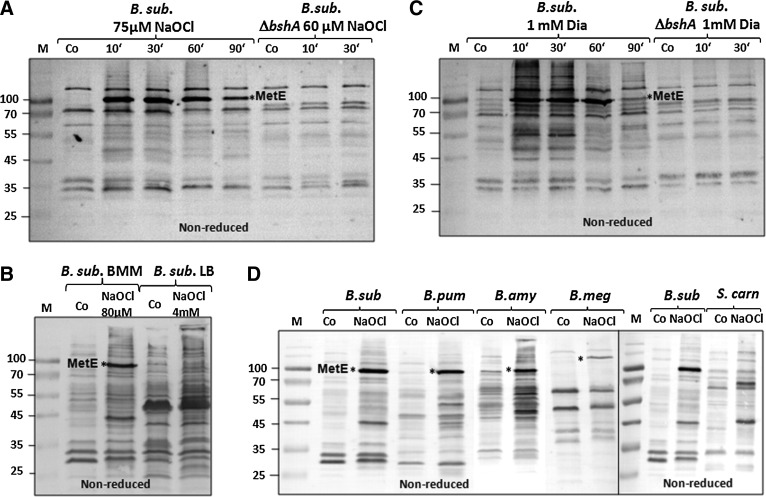
To monitor the extent of S-bacillithiolation after NaOCl stress, BSH-specific antibodies were used to test protein extracts from NaOCl-treated B. subtilis wild-type and bshA-mutant cells in nonreducing immunoblot analyses. The BSH immunoblots showed a very strong 90-kDa band after NaOCl stress, corresponding to MetE-SSB (Fig. 1A). The identity of MetE was confirmed by LC-MS/MS analysis of the specific band from nonreducing sodium dodecyl sulfate (SDS) gels (Supplementary Table S2). S-bacillithiolated MetE, modified at Cys719 and Cys730, was previously identified as the most strongly oxidized protein in the redox proteome of B. subtilis (7). We further confirmed S-bacillithiolation of MetE by comparison of extracts from NaOCl-treated B. subtilis cells grown in the BMM and LB media. In a rich LB medium, MetE is repressed, and thus the MetE-SSB modification is not visible in the BSH immunoblots (Fig. 1B). The MetE-SSB band was also not visible in NaOCl-exposed bshA-mutant cells or in reducing BSH immunoblots (Supplementary Fig. S2). Besides MetE, there were a few weaker bands in the 45–55-kDa range, which represent other protein BSH-mixed disulfides (protein-SSB), since these are absent in the bshA mutant cells. These BSH immunoblot results show that MetE is the most abundant S-bacillithiolated protein in NaOCl-treated B. subtilis cells.
FIG. 1.
NaOCl stress causes widespread S-bacillithiolations in four Bacillus species and Staphylococcus carnosus as shown by nonreducing BSH-specific immunoblot analysis. (A, C) IAM-alkylated protein extracts of B. subtilis wild-type and bshA mutant cells harvested before (co) and at different times after NaOCl (A) or diamide stress (C) were subjected to nonreducing SDS-PAGE and BSH-specific immunoblot analysis. (B) IAM-alkylated protein extracts of B. subtilis wild-type cells exposed to 80 μM NaOCl in the BMM and to 4 mM NaOCl in an LB medium were analyzed by BSH-specific immunoblot analysis. MetE is only expressed in BMM and identified as *MetE-SSB. (D) IAM-alkylated protein extracts of B. subtilis (B. sub), B. pumilus (B. pum), B. amyloliquefaciens (B. amy), B. megaterium (B. meg), and S. carnosus (S. carn) harvested before (co) and 10 min after NaOCl stress were analyzed by BSH-specific immunoblot analysis. In all Bacillus strains, the methionine synthase MetE is the most abundant S-bacillithiolated protein as indicated by the asterisks. BMM, Belitsky minimal medium; BSH, bacillithiol; IAM, iodoacetamide; LB, Luria Broth; PAGE, polyacrylamide gel electrophoresis; SDS, sodium dodecyl sulfate.
Furthermore, BSH immunoblots indicate that MetE is also the major BSH-modified protein in diamide-treated cells (Fig. 1C). In addition, many other BSH-modified proteins are visible after diamide stress, indicating that S-bacillithiolation is also a significant thiol modification in diamide-treated cells.
Next, we analyzed the S-bacillithiolome of NaOCl-treated cells from B. amyloliquefaciens, B. pumilus, B. megaterium, and S. carnosus (Fig. 1D). The protein extracts of all NaOCl-treated Bacillus strains showed MetE as the most abundant protein-SSB, indicating that S-bacillithiolation of MetE is a common protection mechanism against oxidative stress among Bacillus species. In B. megaterium, MetE-SSB was observed at a higher molecular weight and confirmed by LC-MS/MS analysis (Supplementary Table S2). In S. carnosus grown in LB medium, various protein-SSBs were visible, indicating that NaOCl also causes S-bacillithiolations in Staphylococcus. Because MetE synthesis is strongly repressed in LB medium, MetE-SSB was not detected in S. carnosus using BSH-specific immunoblots.
Identification of proteins with S-bacillithiolations in NaOCl-treated cells of B. subtilis, B. amyloliquefaciens, B. pumilus, B. megaterium, and S. carnosus using redox proteomics and shotgun-LC-MS/MS analyses
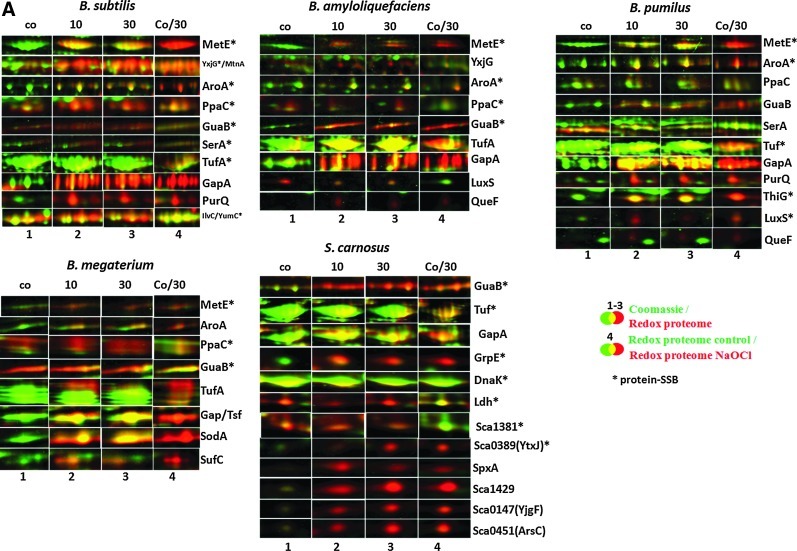
Previously, we have used redox proteomics based on the reduction of disulfides and labeling of released thiols with the fluorescence dye BODIPY FL C1-IA to identify proteins oxidized by NaOCl stress in B. subtilis (7). We applied our redox proteome approach for comparison of B. subtilis, B. amyloliquefaciens, B. megaterium, B. pumilus, and S. carnosus to identify proteins with disulfides after NaOCl stress, including those with potential SSB sites. The BODIPY-fluorescence image was overlayed with the Coomassie-stained protein amount image, and the oxidation ratios were calculated versus protein quantities (Fig. 2A, B and Supplementary Figs. S3–S7 and Table 1). The reversibly oxidized proteins were identified using MALDI-TOF/TOF mass spectrometry analysis (Supplementary Table S1), and the oxidation ratios were used for hierarchical cluster analyses (Supplementary Figs. S3–S7).
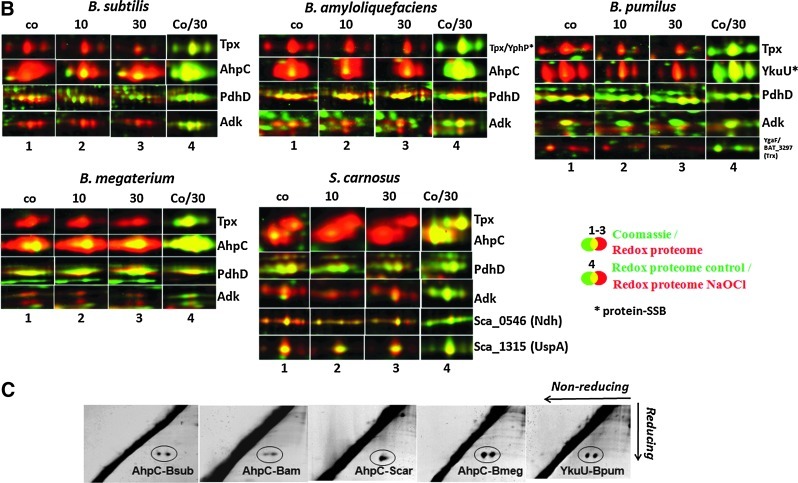
FIG. 2.
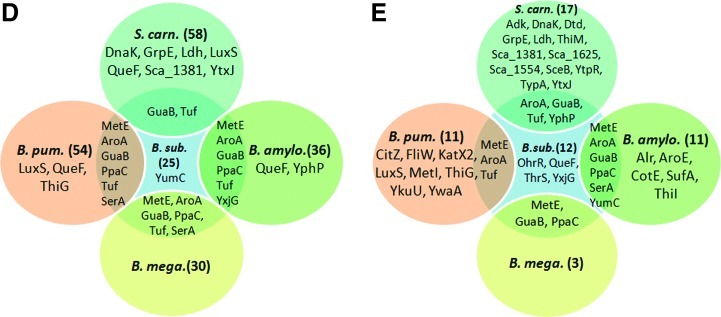
Summary of NaOCl-sensitive proteins (A) and constitutively oxidized proteins (B) in the thiol redox proteomes of four different Bacillus strains and S. carnosus including unique and conserved S-bacillithiolated proteins (D) that were identified by LC-MS/MS analysis (E). (A) Close-ups of conserved NaOCl-sensitive proteins including S-bacillithiolated proteins identified in the redox proteomes of B. subtilis, B. amyloliquefaciens, B. pumilus, B. megaterium, and S. carnosus. Reversibly oxidized proteins in the redox proteome after NaOCl treatment are shown that have been identified with specific and conserved SSB sites (*) using LC-MS/MS analysis. Overlay images represent the redox proteome (red) compared to the Coomassie-stained protein amount image (green) at control conditions (co) and 10 and 30 min after NaOCl exposure (1–3). Column 4 shows the overlay redox proteome control versus redox proteome 30-min NaOCl stress. The identified proteins and their fluorescence/protein quantity ratios are listed in Table 1, and the complete redox proteomes of all strains are given in Supplementary Figures S3–S7. (B) Examples of conserved redox-controlled proteins that are oxidized under control conditions to intermolecular or intramolecular disulfides representing thiol-dependent peroxiredoxins (AhpC, YkuU, and YgaF) and thiol peroxidases (Tpx), PdhD, and Adk. Overlay images represent the redox proteome (red) compared to the Coomassie-stained protein amount image (green) of B. subtilis, B. amyloliquefaciens, B. pumilus, B. megaterium, and S. carnosus at control conditions (co) and 10 and 30 min after NaOCl exposure (1–3). Column 4 shows the overlay redox proteome control versus redox proteome 30-min NaOCl stress. The identified proteins and their fluorescence/protein quantity ratios are listed in Table 1, and the complete redox proteomes of all strains are listed in Supplementary Figures S3–S7. (C) Diagonal nonreducing/reducing SDS-PAGE confirmed intermolecular disulfides for AhpC homologs in B. subtilis, B. amyloliquefaciens, B.megaterium, S. carnosus and for YkuU in B. pumilus. The IAM-alkylated proteins extracts (100 μg) of NaOCl-treated cells of all strains were separated by 2D nonreducing/reducing diagonal SDS-PAGE analysis as described previously (41). The AhpC homologs are encircled in the diagonal assays of B. subtilis (AhpC-Bsub), B. amyloliquefaciens (AhpC-Bam), S. carnosus (AhpC-Scar), B. megaterium (AhpC-Bmeg), and B. pumilus (YkuU-Bpum). (D) Unique and conserved S-bacillithiolated proteins identified in the redox proteomes of different Bacillus strains and S. carnosus. The number of the identified reversibly oxidized proteins in the redox proteomes of the different strains is shown in brackets. Unique and conserved proteins with identified SSB sites are listed by names that are oxidized in the redox proteome. (E) Unique and conserved S-bacillithiolated proteins identified using LC-MS/MS analysis in different Bacillus strains and S. carnosus. The number of proteins with identified SSB sites after NaOCl stress using LC-MS/MS analysis in the different Bacillus strains and S. carnosus is shown in brackets. Unique and conserved proteins with SSB sites are listed by names. LC, liquid chromatography; MS/MS, tandem mass spectrometry.
Table 1.
Identification of Reversibly Oxidized Proteins in the Redox Proteome in Bacillus subtilis, B. amyloliquefaciens, B. pumilus, B. megaterium, and Staphylococcus carnosus
| |
Redox ratios BODIPY fluorescence/protein amount |
|
|
|
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
B. subtilis |
B. pumilus |
B. amyloliquefaciens |
B. megaterium |
S. carnosus |
|
|
|
|
||||||||||
| Proteins | co | 10′ | 30′ | co | 10′ | 30′ | co | 10′ | 30′ | co | 10′ | 30′ | co | 10′ | 30′ | Protein function | AC-No. (Uniprot) | Function of conserved Cys | Post-translational Cys-modifications |
| AbrB BPUM_0021 | 2.1 | 2.4 | 1.5 | Possible transcriptional regulator | A8F904 | Cys54 essential for DNA-binding activity | |||||||||||||
| AccB | 1.9 | 3.6 | 3.2 | Biotin carboxyl-carrier protein of acetyl-CoA carboxylase | P49786 | Cys117 conserved | |||||||||||||
| AccD | 0.8 | 2.5 | 2.8 | Acetyl-CoA carboxylase, carboxyl transferase, beta-subunit | B4AMC5 | Cys33,35,52,54 Zn binding | |||||||||||||
| AcnA/CitB | 1.2 | 1.2 | 1.4 | 1.3 | 1.6 | 1.5 | Aconitate hydratase (Aconitase) | D5DFZ0/B9DP54 | Cys444, 510, 513 conserved: 4Fe-4S-cluster-binding site | Homolog Cys404 oxidized by NaOCl in E. coli | |||||||||
| Adk* | 5.4 | 4.7 | 4.3 | 2.3 | 1.5 | 1.5 | 1.8 | 1.8 | 1.8 | 2.2 | 2.5 | 4.9 | 2.6 | 4.5 | 2.3 | Adenylate kinase | F4E1S4 | Cys130, 133, 150 conserved, Zn-finger motif | Cys130–133 disulfide; Cys150*-S-SB by NaOCl in S. carn |
| AhpC | 5.4 | 2.7 | 2.9 | 5.4 | 3.6 | 3.8 | 4.0 | 3.7 | 3.8 | 4.1 | 4.9 | 4.8 | Alkylhydroperoxide reductase small-subunit | A7ZAJ9 | Cys47, 166 conserved; Cys47 active site | Cys47-Cys166 form catalytic disulfide | |||
| AldA | 0.4 | 1.3 | 1.1 | Putative aldehyde dehydrogenase | B9DKD6 | Cys290-conserved active site, nucleophile | |||||||||||||
| ArcA | 0.7 | 0.9 | 0.9 | Arginine deiminase 1 | B9DKE2 | Cys401-conserved amidino-cysteine-intermediate active site | |||||||||||||
| ArcC | 2.2 | 1.8 | 1.4 | Carbamate kinase 1 | B9DJ94 | No Cys conserved | |||||||||||||
| ArgG | 0.4 | 2.4 | 1.9 | Argininosuccinate synthase | B4AMD7 | No Cys conserved | Cys187 S-SCys by diamide in Bsub | ||||||||||||
| AroA* | 0.4 | 1.1 | 1.3 | 0.5 | 1.2 | 1.6 | 0.6 | 3.4 | 1.1 | 0.7 | 1.0 | 1.8 | Bifunctional 3-deoxy-7-phosphoheptulonate synthase/chorismate mutase | A7Z7R9 | Cys126, 259 conserved | Cys126*-SSB by NaOCl in B. sub., B .pum., B. amy., S. carn. | |||
| ArsC (Sca_0451) | 1.3 | 5.4 | 6.2 | Putative ArsC/Spx family protein | B9DJH2 | Cys10, 13 conserved: form redox-active disulfide bond by oxidative stress | |||||||||||||
| BMD_4393 | 0.9 | 1.2 | 0.8 | Acetyl-CoA acetyltransferase | D5DL95 | Active-site Cys90 acetylated by AcetylCoA; Cys381 important for proton exchange | |||||||||||||
| CheW | 0.3 | 1.7 | 2.2 | Chemotaxis protein CheW | A8FDA9 | No Cys conserved | |||||||||||||
| CheY | 0.6 | 0.5 | 0.6 | Response regulator CheY | A8FD98 | No Cys conserved | |||||||||||||
| CysH | 1.6 | 1.1 | 1.6 | Phosphoadenosine phosphosulfate reductase | P94498 | Cys120, 121, 203, 206, 229 conserved | Cys239 forms mixed disulfide with Cys32 of Trx in E. coli | ||||||||||||
| CysI | 3.4 | 1.7 | 2.6 | Sulfite reductase [NADPH] hemoprotein beta-component | B4AD94 | Cys436, 442, 481, 485 conserved: 4Fe-4S-cluster-binding site | |||||||||||||
| CysJ (BAT_2248) | 1.0 | 1.3 | 0.9 | 1.4 | 1.4 | 1.6 | 2.7 | 2.0 | 2.0 | Sulfite reductase [NADPH] flavoprotein, alpha-component | B4AD95 | Cys166, 559 conserved | Cys428–432 intramolecular disulfide | ||||||
| DapA | 0.4 | 2.3 | 2.4 | Dihydrodipicolinate synthase | F4EJJ2 | No Cys conserved | |||||||||||||
| Dat | 2.0 | 1.2 | 1.3 | d-alanine aminotransferase | A7Z2X8 | Cys141 close to the active-site Lys144 | |||||||||||||
| DeoD | 4.3 | 3.7 | 4.3 | Purine nucleoside phosphorylase DeoD-type | A7Z5M8 | No Cys conserved | |||||||||||||
| DhbE | 1.7 | 1.9 | 1.5 | Putative 2,3-dihydroxybenzoate-AMP ligase DhbE | A7Z8A8 | Cys218, 324, 473 conserved | |||||||||||||
| DnaJ | 2.6 | 3.7 | 3.3 | Chaperone protein DnaJ | B9DNJ9 | Cys147, 150, 204, 207 Zn-finger1; Cys164, 167, 190, 193 Zn-finger2 | |||||||||||||
| DnaK* | 0.7 | 0.5 | 0.6 | Chaperone protein DnaK | B9DNK0 | Cys15 not conserved | Cys15*-S-SB by NaOCl in S. carn. | ||||||||||||
| Eno | 0.7 | 0.8 | 0.6 | Enolase | P37869 | No Cys conserved | |||||||||||||
| FabH | 0.5 | 1.6 | 1.5 | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | B9DIT1 | Cys112 conserved active site | |||||||||||||
| FabZ | 4.0 | 5.3 | 6.3 | (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase | B9DMG5 | Cys94, 134 conserved | |||||||||||||
| FdhL (Sca_1802) | 2.9 | 3.3 | 3.0 | Putative formate dehydrogenase | B9DLU9 | Cys37, 48, 51, 63: 2Fe-2S; Cys99, 102, 109: 4Fe-4S; Cys147, 150, 153, 157: 4Fe-4S; Cys190, 193, 196, 200: 4Fe-4S; Cys264, 267, 271, 299: 4Fe-4S | |||||||||||||
| FolE2 | 1.0 | 2.3 | 2.7 | GTP cyclohydrolase | B9DKT0 | Cys176 conserved, catalytically important site, ligand for Zn2+ | |||||||||||||
| FusA | 0.5 | 0.7 | 0.5 | 1.0 | 0.6 | 0.7 | Elongation factor G | D5D9Q2 | Cys258, 389 conserved | ||||||||||
| GapA | 0.8 | 2.9 | 3.6 | 0.5 | 2.3 | 1.6 | 0.4 | 2.4 | 2.8 | 0.4 | 1.0 | 1.2 | 0.7 | 0.9 | 0.9 | Glyceraldehyde-3-phosphate dehydrogenase | F4E0S2 | Cys152 conserved active site | Cys152–156 intramolecular disulfide |
| GlmS | 0.3 | 0.7 | 0.6 | Putative glucosamine-fructose-6-phosphate aminotransferase | B9DMA1 | Cys2 catalytic site, forms γ-glutamyl-thioester intermediate during Gln hydrolysis | |||||||||||||
| GlnA | 1.6 | 2.4 | 2.2 | Glutamine synthetase | B9DPA6 | Cys96 conserved | |||||||||||||
| GltB | 5.7 | 5.6 | 4.0 | Glutamate synthase (NADPH) small subunit | A8FE17 | Cys46, 49, 54, 67, 106, 110, 114, 307 conserved: Two 4Fe-4S cluster | |||||||||||||
| GltX | 1.8 | 1.8 | 2.6 | 2.3 | 1.4 | 1.7 | 0.4 | 0.7 | 0.8 | Glutamyl-tRNA synthetase | A7Z0L4 | Cys108, 110 conserved, form Zn binding site | |||||||
| GlyA | 0.7 | 0.8 | 0.4 | Serine hydroxymethyl transferase | A8FIC1 | Cys64 conserved | Cys66 oxidized by NaOCl in E. coli | ||||||||||||
| GlyQS | 0.8 | 0.7 | 0.8 | Glycyl-tRNA synthetase | B9DNL1 | Cys230, 328, 344, 422 conserved, close to ATP binding sites | |||||||||||||
| GrpE* | 0.2 | 2.6 | 2.8 | HSP-70 cofactor protein GrpE | B9DNK1 | No Cys conserved | Cys95*-S-SB by NaOCl in S. carn. | ||||||||||||
| GuaA | 2.7 | 1.8 | 1.3 | 1.2 | 0.7 | 0.9 | GMP synthase [glutamine-hydrolyzing] | A8FAH5 | Cys85 conserved active site, nucleophilic thiolate | ||||||||||
| GuaB* | 4.9 | 3.6 | 3.8 | 1.1 | 2.2 | 2.1 | 0.8 | 2.2 | 2.5 | 1.4 | 2.0 | 1.7 | 1.3 | 3.8 | 3.6 | Inosine-5′-monophosphate dehydrogenase | F4EQ59 | Cys308 conserved- active site, forms thioimidate intermediate | Cys308*-S-SB by NaOCl in B. sub, B. amy., B. meg. |
| GuaC | 3.4 | 5.2 | 4.6 | GMP reductase | B9DP67 | Cys174 conserved active site, forms thioimidate intermediate | |||||||||||||
| Hit | 4.7 | 3.1 | 3.5 | Cell cycle regulation histidine triad (Hit) protein | G0IF34 | No Cys conserved | |||||||||||||
| Hom (BAT_0329) | 0.6 | 2.5 | 3.4 | Homoserine dehydrogenase | B4AG18 | No Cys conserved | |||||||||||||
| Icd | 0.4 | 1.4 | 1.3 | 0.4 | 0.6 | 0.6 | Isocitrate dehydrogenase [NADP] | B4AMB6 | Cys118, 411 conserved; Cys118 in active substrate binding site | ||||||||||
| IlvB | 1.8 | 1.6 | 1.1 | Acetolactate synthase, large subunit, biosynthetic type | D5DTM7 | No Cys conserved | |||||||||||||
| IlvC | 1.5 | 1.7 | 2.0 | 0.6 | 0.6 | 0.6 | 0.7 | 1.7 | 1.6 | 2.5 | 2.7 | 1.7 | Ketol-acid reductoisomerase | G0IJS5 | Cys199, 227 conserved | ||||
| IlvD | 2.1 | 2.5 | 1.9 | Dihydroxy-acid dehydratase | D5DER1 | Cys122, 189, 192, 195: 4Fe-4S cluster binding | |||||||||||||
| IspA | 1.9 | 1.5 | 1.3 | Major intracellular serine protease | A7Z3U2 | No Cys conserved | |||||||||||||
| IspG | 5.6 | 2.1 | 2.6 | 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | F4ESF5 | Cys268, 271, 303 conserved: 4Fe-4S-cluster-binding site | |||||||||||||
| Ldh* | 2.7 | 4.4 | 4.3 | l-lactate dehydrogenase | B9DN51 | Cys72 conserved | Cys72*-S-SB by NaOCl in S. carn. | ||||||||||||
| LeuC | 1.1 | 1.3 | 1.6 | 4.0 | 3.4 | 2.7 | 2.7 | 3.4 | 3.2 | 3-Isopropylmalate dehydratase large subunit | B4AM28 | Cys346, 406, 409 conserved: 4Fe-4S cluster binding | |||||||
| LuxS* | 0.7 | 2.1 | 4.5 | 6.9 | 2.7 | 3.2 | 0.3 | 2.9 | 2.7 | S-ribosyl homocysteine lyase (B. pumilus) (LuxS) | B4ANL1 | Cys84, 126 conserved: Cys84 essential for catalysis; Cys126 Fe-binding site | Cys41*-S-SB by NaOCl stress in B. pum. | ||||||
| LysC | 0.6 | 1.8 | 1.9 | Aspartokinase | A8FG00 | Cys169 conserved | |||||||||||||
| LysC (BAT_1429) | 0.5 | 1.4 | 1.3 | Aspartokinase | B4AM64 | No Cys conserved | |||||||||||||
| MenB | 2.2 | 1.4 | 1.9 | Dihydroxynaphtoic acid synthetase | B9DQ83 | Cys71, 128, 174, 205 conserved; Cys71 in substrate-binding site | |||||||||||||
| MetE* | 0.3 | 1.5 | 1.4 | 0.4 | 1.8 | 1.6 | 0.3 | 1.7 | 2.0 | 0.8 | 1.8 | 1.2 | Methionine synthase | A7Z3U1 | Cys647, 730 essential-Zinc binding active site | Cys730*-S-SB; Cys719*-SSB | |||
| MetK | 0.6 | 0.6 | 0.5 | 0.7 | 0.6 | 0.4 | S-Adenosylmethionine synthase | F4EA25 | Cys22, 44 conserved, near active metal (Mg2+, K+)-binding site | ||||||||||
| MsrA | 1.7 | 1.9 | 2.1 | Peptide methionine sulfoxide reductase | B9DNX6 | Cys10, 13 conserved; Cys 10 active site | Cys10-Cys13 disulfide | ||||||||||||
| MsrB | 2.6 | 2.2 | 3.0 | Peptide methionine sulfoxide reductase | B9DNY9 | Cys114-conserved nucleophile active site | |||||||||||||
| MtnA | 1.7 | 2.3 | 2.6 | Methylthioribose-1-phosphate isomerase | O31662 | Cys160 conserved, in active site, transition state stabilizer | |||||||||||||
| MtnW | 0.4 | 1.1 | 1.8 | 2,3-Diketo-5-methylthiopentyl-1-phosphate enolase | A8FCG8 | No Cys conserved | |||||||||||||
| MurA | 1.4 | 1.4 | 1.1 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | D5DM45 | Cys117-conserved proton donor active site, binds UDP-GlcNAc | |||||||||||||
| Ndh (Sca_0546) | 0.7 | 0.6 | 0.5 | 4.5 | 4.2 | 4.6 | 2.5 | 1.4 | 1.4 | NADH dehydrogenase ndh | B4AHV4 | No Cys conserved | |||||||
| NifS (BAT_1509) | 3.6 | 4.7 | 3.1 | Cysteine desulfurase | B4ALY7 | Cys325-conserved active site | |||||||||||||
| OhrA (BAT_2845) | 0.2 | 6.7 | 6.7 | Organic hydroperoxide resistance protein | B4AEW1 | Cys59-,123-conserved Cys | Catalytic Cys59–Cys123 disulfide | ||||||||||||
| PdhC (BAT_2691) | 1.2 | 0.6 | 0.8 | Dihydrolipoyl-lysine-residue acetyl-transferase component of PDH (E2) | B4AEF9 | No Cys conserved | |||||||||||||
| PdhD | 2.7 | 1.5 | 1.3 | 1.3 | 0.8 | 1.0 | 1.4 | 1.4 | 1.2 | 1.1 | 0.9 | 1.1 | 1.2 | 1.0 | 1.2 | Dihydrolipoyl dehydrogenase, E3 component of PDH complex | A7Z482 | Cys47-, 52-conserved, redox-active disulfide | Cys47–52 disulfide by NaOCl |
| PdxS | 4.2 | 4.6 | 4.6 | Pyridoxal biosynthesis lyase PdxS | B9DKX7 | Cys128, 130 conserved | |||||||||||||
| PfkA | 3.3 | 3.9 | 2.8 | 6-Phosphofructokinase 2 | D5DPB6 | Cys73, 119 conserved in Bacillus species | |||||||||||||
| PheT | 2.0 | 1.6 | 1.8 | Phenylalanyl-tRNA synthetase beta-chain | B9DPU9 | Cys65, 79, 120 conserved, in tRNA-binding domain | |||||||||||||
| PpaC* | 2.4 | 2.7 | 2.8 | 1.0 | 1.5 | 2.1 | 3.8 | 1.7 | 3.9 | 2.0 | 4.2 | 3.2 | Manganese-dependent inorganic pyrophosphatase | A7ZAR2 | Cys18, 118, 158 conserved | Cys158*-S-SB by NaOCl in B. sub. | |||
| Prs | 1.0 | 0.9 | 1.0 | 1.9 | 1.6 | 1.6 | Ribose-phosphate pyrophosphokinase | A8F918 | Cys57, 59, 251, 252 widely conserved | ||||||||||
| PtsI | 0.9 | 0.5 | 0.8 | Phosphoenolpyruvate-protein phosphotransferase | P23533 | Cys364, 503, 556 conserved; Cys503 proton donor active site | |||||||||||||
| PurB | 4.7 | 2.7 | 2.2 | Adenylosuccinate lyase | D5DWW6 | Cys31 conserved | |||||||||||||
| PurL | 1.4 | 1.5 | 1.1 | Phosphoribosylformylglycinamidine synthase 2 | A8FAM7 | Cys55, 164, 190, 259, 527 conserved | |||||||||||||
| PurQ | 0.7 | 4.8 | 4.6 | 2.0 | 3.0 | 3.8 | Phosphoribosylformylglycinamidine synthase 1 | B4AI84 | Cys13, 86 conserved, Cys86 nucleophilic active site | ||||||||||
| PyrAB | 1.4 | 1.0 | 0.9 | Carbamoyl-phosphate synthase large chain 2 | D5DJR1 | Cys34, 182, 204, 232, 281, 525, 1034 conserved | |||||||||||||
| PyrE | 0.9 | 1.3 | 0.6 | Orotate phosphoribosyltransferase | B4AE64 | No Cys conserved | |||||||||||||
| PyrG | 2.1 | 2.3 | 2.1 | CTP synthase | A7Z9T4 | Cys381 conserved active site | |||||||||||||
| PyrH | 3.3 | 2.1 | 2.3 | Putative uridylate kinase | B9DPH0 | Cys207 conserved | |||||||||||||
| QueC (Sca_0360) | 3.7 | 3.8 | 3.8 | Putative ExsB family protein (7-cyano-7-deazaguanine synthase) | B9DK41 | Cys191, 200, 203, 206 conserved Zn binding site | |||||||||||||
| QueF* | 0.5 | 1.2 | 1.9 | 1.0 | 5.0 | 4.8 | 1.4 | 5.4 | 4.9 | NADPH-dependent 7-cyano-7-deazaguanine reductase | B4AEQ3 | Cys56 in active site (binds Zn) | Cys56*-S-SB by NaOCl in B. sub. | ||||||
| Sat | 1.2 | 1.1 | 1.2 | Sulfate adenylyltransferase | B4AE60 | Cys319, 322, 331 conserved, close to active site that binds APS | |||||||||||||
| Sca_1381* | 2.9 | 2.5 | 2.5 | Putative transaldolase | B9DN38 | Cys200 conserved | Cys200*-S-SB by NaOCl in S. carn. | ||||||||||||
| Sca_1429 | 1.2 | 4.9 | 5.1 | Putative tRNA (cytidine/uridine-2′-O-)-methyltransferase | B9DMX6 | Cys23 conserved | |||||||||||||
| Sca_1464 | 1.9 | 2.3 | 2.9 | CobB/CobQ-like glutamine amidotransferase domain protein | B9DMV5 | Cys94 conserved nucleophilic active site | |||||||||||||
| Sca_2233 | 2.9 | 2.2 | 4.0 | Putative short-chain dehydrogenase | B9DJB3 | No Cys conserved | |||||||||||||
| SecA | 0.7 | 1.2 | 1.5 | Protein translocase subunit | P47994 | Cys828, 830, 839, 840 conserved Zn-binding site | |||||||||||||
| SerA* (BMD_2386) | 1.3 | 1.5 | 4.1 | 1.8 | 1.1 | 1.3 | 0.6 | 0.6 | 0.9 | Phosphoglycerate dehydrogenase | B4AKF2 | No Cys conserved | Cys410*-S-SB by NaOCl in B. sub. | ||||||
| SerS | 0.6 | 1.4 | 2.5 | Seryl-tRNA synthetase 2 | Q659K7 | Cys260, 353 conserved in ATP-binding active site | |||||||||||||
| SodA | 0.3 | 2.0 | 1.9 | Superoxide dismutase (Mn) | D5DMJ9 | Cys not conserved | |||||||||||||
| SpxA | 1.5 | 3.4 | 4.7 | Regulatory protein Spx | B9DIR2 | Cys10, 13 conserved redox-active disulfide | |||||||||||||
| Ssb | 2.7 | 3.4 | 2.5 | 2.4 | 1.7 | 1.5 | 1.8 | 2.1 | 1.8 | Single-stranded DNA-binding protein | A8FJE6 | No Cys conserved | |||||||
| SucC | 1.2 | 1.0 | 0.8 | 1.3 | 1.0 | 1.2 | Succinyl-CoA ligase [ADP-forming] subunit-beta | F4EIU4 | Cys102 in ATP-binding site, Cys193 in Mg/Mn-binding site | ||||||||||
| SufC | 0.6 | 1.3 | 2.1 | FeS assembly ATPase SufC | D5DM25 | No Cys conserved | |||||||||||||
| TenA (BAT_2971) | 1.7 | 2.1 | 2.3 | 0.9 | 1.0 | 0.9 | Tena/thi-4 family (Thiaminase II) | B4AF84 | Cys135-conserved catalytic active site | ||||||||||
| TenA-2 BPUM_3574 | 1.9 | 1.5 | 1.4 | Thiaminase | A8FJ01 | Cys135-conserved catalytic active site | |||||||||||||
| ThiC | 2.9 | 3.4 | 2.8 | Phosphomethylpyrimidine synthase | D5DA39 | Cys538, 541, 546: 4Fe-4S-S-AdoMet cluster | |||||||||||||
| ThiD | 2.1 | 2.1 | 1.7 | 0.3 | 2.3 | 2.5 | Phosphomethylpyrimidine kinase | A8FIN1 | No Cys conserved | ||||||||||
| ThiG* | 0.6 | 2.5 | 2.1 | Thiazole synthase | B4AF80 | Cys92 conserved | Cys102*-S-SB by NaOCl in B. pum. | ||||||||||||
| ThrS | 1.1 | 1.8 | 1.5 | Threonyl-tRNA synthetase | B9DNC9 | Cys336-conserved, catalytic Zn-binding site | |||||||||||||
| TpiA | 0.7 | 0.7 | 0.6 | 1.0 | 0.8 | 0.8 | 2.2 | 2.3 | 1.8 | 0.8 | 0.6 | 0.6 | Triosephosphate isomerase | B4AGH8 | Cys126 conserved, required for folding and stability | ||||
| Tpx | 7.2 | 4.6 | 4.1 | 7.0 | 3.7 | 5.8 | 8.6 | 4.2 | 3.3 | 5.0 | 6.2 | 5.6 | 4.9 | 5.0 | 5.8 | Probable thiol peroxidase | B4AME1 | Cys60, 95 conserved active site | Cys60-Cys95 intra-molecular disulfide |
| TrxABs/TrxBp | 1.5 | 3.9 | 3.6 | 6.9 | 10.3 | 7.8 | Thioredoxin BAT_3297 not conserved in B. subtilis | B4AP00 | Cys62 conserved in alkaliphilic Bacilli | ||||||||||
| Tsf | 0.5 | 0.4 | 0.3 | 0.6 | 0.5 | 0.5 | 0.4 | 1.0 | 1.2 | 0.8 | 0.5 | 0.6 | Elongation factor Ts | A7Z4S1 | Cys22 conserved | ||||
| Tuf* | 0.6 | 0.5 | 0.5 | 0.6 | 1.1 | 1.2 | 0.3 | 0.9 | 0.8 | 0.5 | 0.8 | 0.7 | 0.4 | 0.5 | 0.5 | Elongation factor Tu | A7Z0N5 | Cys83 conserved, in GTP-binding site | Cys83*-S-SB by NaOCl in B. sub, B. meg, B. pum |
| TypA | 1.1 | 0.6 | 0.6 | Putative GTP-binding protein family protein TypA | B9DQ05 | Cys400 conserved | |||||||||||||
| UspA (Sca_1315) | 1.8 | 1.3 | 1.2 | Putative universal stress protein | B9DNA5 | Cys111, 137 conserved in UspA-homologs | |||||||||||||
| YceC (TerC) | 3.1 | 3.5 | 3.8 | 1.8 | 2.2 | 2.7 | Tellurium resistance protein | A7Z138 | No Cys conserved | ||||||||||
| YceD (TerD) | 1.8 | 2.4 | 3.6 | 2.0 | 1.9 | 1.7 | Tellurium resistance protein | G0IK82 | No Cys conserved | ||||||||||
| YceE (TerE) | 1.2 | 2.0 | 2.2 | Tellurium resistance protein TerE | A7Z140 | No Cys conserved | |||||||||||||
| YdjL (BAT_1651) | 1.7 | 1.4 | 1.5 | Zinc-containing alcohol dehydrogenase | B4ALJ8 | Cys34,37 probably Zn-binding | |||||||||||||
| YerC | 9.3 | 4.2 | 4.1 | Sporulation protein YerC | A8FAP4 | Cys27, 35 conserved | |||||||||||||
| YflT (Sca_0046) | 3.2 | 4.2 | 2.3 | Putative uncharacterized protein | B9DM00 | No Cys conserved | |||||||||||||
| YgaF | 6.4 | 4.7 | 4.7 | Peroxiredoxin | A8FB93 | Cys45, 50 conserved; Cys45 active site | |||||||||||||
| YisY (Sca_0638) | 1.5 | 0.7 | 1.2 | Putative hydrolase of alpha-/beta-fold family | B9DQ91 | Cys226 conserved | |||||||||||||
| YitJ | 1.2 | 2.4 | 3.1 | Methylenetetrahydrofolate reductase | A8FBU4 | Cys200, 265, 266 conserved Zn-binding site | |||||||||||||
| YjcI (MetI)* | 1.3 | 1.3 | 1.3 | Cystathionine gamma-synthase/O-acetylhomoserine (thiol)-lyase | A7Z3I1 | Cys338 conserved | Cys369*-SSB by NaOCl in B. pum. | ||||||||||||
| YjgF (Sca_0147) | 1.2 | 5.0 | 4.6 | Putative purine regulatory protein | B9DLD4 | Cys103 conserved, in homotrimer interaction site | |||||||||||||
| YjoA | 3.8 | 2.0 | 2.6 | Uncharacterized protein YjoA | A7Z3N1 | No Cys conserved | |||||||||||||
| YkuU* | 6.9 | 4.7 | 5.6 | Peroxiredoxin YkuU | A8FCN5 | Cys52, 169 conserved; Cys52 active site | Cys52-Cys169 catalytic disulfide bond; Cys169*-SSB | ||||||||||||
| YneT | 2.2 | 1.8 | 1.7 | CoA-binding protein | A8FDR9 | No Cys conserved | |||||||||||||
| YodC | 4.1 | 2.7 | 3.1 | Nitroreductase YodC | A7Z5L7 | Cys153 conserved, close to FMN-binding site | |||||||||||||
| YphP* | 6.3 | 2.6 | 4.2 | Thioredoxin-like protein UPF0403 family protein | A7Z5T6 | Cys53, 55, 144 conserved; Cys53 redox-active site | Cys53*-SSB by NaOCl in B. sub. | ||||||||||||
| YtxJ (Sca_0389)* | 1.1 | 3.0 | 5.4 | Thioredoxin-like protein DUF2847 family protein | B9DJN8 | Cys30 conserved (TCPIS motif), Cys30 redox active Cys | Cys30*-SSB by NaOCl in S. carn. | ||||||||||||
| YvdA (Sca_1457) | 1.6 | 6.1 | 3.9 | Putative carbonic anhydrase | B9DMU8 | Cys38, 99, 103 conserved catalytic Zn-binding site | |||||||||||||
| YxjG* | 1.0 | 1.9 | 2.2 | 1.4 | 2.6 | 3.0 | Methionine synthase homolog | A7ZAB5 | Cys251, 346 homolog to Zn-binding Cys647, 730 in MetE | Cys346*-SSB by NaOCl in B. sub. | |||||||||
| YybR (HypR) | 7.4 | 6.1 | 12.0 | MarR/DUF24-family regulator; responds to disulfide stress | P37486 | Cys14 conserved in MarR/DUF24 family, redox-sensitive Cys | Cys14-Cys49' intersubunit disulfide by NaOCl | ||||||||||||
The proteins were cut from Coomassie-stained 2D-gels, tryptic in-gel-digested, and identified using MALDI-TOF-TOF MS/MS. The table lists the oxidation ratios/protein quantities of all identified reversibly oxidized proteins at control (co) and 10 and 30 min after NaOCl stress as average values of two biological replicate experiments (Exp1 and Exp2), their protein names and functions, Uniprot-accession numbers, and information about conserved Cys residues as revealed by the Conserved Domain Database CDD (33) (http://ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The Mascot search results with protein and peptide scores and the oxidation ratios/protein quantities of reversibly oxidized proteins at control (co) and 10 and 30 min after NaOCl stress of two biological replicate experiments (Exp1 and Exp2) with standard deviations are given in Supplementary Table S1. The redox ratios/protein quantities for the B. subtilis proteins were taken from the previous study (7). When a protein is identified in more than one Bacillus species and/or S. carnosus, the accession number is related to one species corresponding to the information about Cys conservation and functions. The asterisk (*) indicates that these proteins have been identified with peptide-SSB sites using LC-MS/MS analysis, and the BSH-modified Cys site is indicated in the last column. Proteins with oxidation/protein amount ratios that are more than 1.5-fold induced after NaOCl stress are highlighted in gray.
BSH, bacillithiol; Cys, cysteine; MS/MS, tandem mass spectrometry.
In total, 71 different proteins were identified in all five strains that are constitutively oxidized at either control or NaOCl stress conditions. Several conserved redox-sensitive proteins that coordinate metal ions (Zn, Fe-S-cluster) or form disulfide bonds for catalysis were identified in untreated cells, including the adenylate kinase Adk, the alkyl hydroperoxide reductase small-subunit AhpC, the 2-Cys peroxiredoxins YkuU and YgaF (in B. pumilus), the thiol peroxidase Tpx, the dihydrolipoamide dehydrogenase E3 subunit of the pyruvate dehydrogenase complex PdhD, and the sulfite reductase flavoprotein alpha-subunit CysJ (Fig. 2B). The intramolecular disulfide bonds were confirmed previously for PdhD, Adk, and CysJ in B. subtilis (7). In B. pumilus, which does not possess the AhpC-AhpF peroxiredoxin/reductase system (14), the YkuU 2-Cys peroxiredoxin is strongly oxidized similar to AhpC in other Bacillus strains, suggesting that YkuU might replace AhpC in peroxide detoxification. The intermolecular disulfides of AhpC homologs and YkuU were confirmed for all Bacillus strains and S. carnosus using diagonal nonreducing/reducing SDS–polyacrylamide gel electrophoresis (PAGE) analysis (Fig. 2C). Notably, in S. carnosus, the NADH dehydrogenase Sca_0546 and the universal stress protein UspA are among the most abundant proteins with strong oxidation ratios under control conditions (Supplementary Fig. S7).
Exposure of Bacillus strains and S. carnosus to sublethal NaOCl concentrations caused increased oxidation ratios of 57 different NaOCl-sensitive proteins (Table 1). The information about the conservations of Cys residues was obtained from the CDD database (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (33). Many NaOCl-sensitive proteins harbor active-site Cys residues that are nucleophilic, catalytically important, bind substrate intermediates or serve as Zn or Fe ligands. These NaOCl-sensitive proteins are associated with a variety of cellular functions, such as ROS scavenging, amino acid biosynthesis, fatty acid biosynthesis, purine biosynthesis, translation, and protein modification/folding factors. Examples are the methionine sulfoxide reductases (MsrA and MsrB) and two ArsC family proteins (SpxA and Sca_0451) that are known to form intramolecular disulfides by oxidative stress (7, 38) (Supplementary Fig. S7 and Fig. 2A). In B. megaterium, major NaOCl-sensitive proteins were identified as the Mn-dependent superoxide dismutase SodA and the Fe-S-cluster assembly ATPase SufC (Supplementary Fig. S6 and Fig. 2A). The SufA chaperone was identified with an SSB-site at the conserved Cys120 within the Fe-S-cluster site in B. amyloliquefaciens (Table 2). The glyceraldehyde-3-phosphate dehydrogenase (GapA) was among the most strongly de novo oxidized proteins by NaOCl stress in all redox proteomes. The redox-active intramolecular disulfide in GapA's active site has been mapped (7, 27).
Table 2.
Identification of S-Bacillithiolated Proteins in the Proteomes of B. subtilis, B. amyloliquefaciens, B. pumilus, B. megaterium, and S. carnosus Using Liquid Chromatography-MS/MS Analysis
| Accession | Protein | Function | Essential | Redox-sensing Cys | Peptide-SSB or -SSCys sequence | m/z precursor | m/z prec-134 (-malate) | Peptide neutral MW | Charge |
|---|---|---|---|---|---|---|---|---|---|
| B. subtilis | |||||||||
| O34777 | OhrR* | Organic hydroperoxide resistance repressor (control of the OhrA peroxiredoxin) | No | C15 redox sensing (cons.) | (K)LENQLC15(+BSH)FLLYASSR(E) | 684.9800 | 640.6500 | 2051.9183 | 3 |
| P80877 | MetE* | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (methionine synthase) | No | C647,730 Zn-binding active site (cons.) | (R)FWVNPDC730(+BSH)GLK(T) | 787.8297 | 720.9900 | 1573.6449 | 2 |
| (R)VPSTEEMYNIIVDALAVC719(+BSH)PTDR(F) | 944.7584 | 900.4600 | 2831.2533 | 3 | |||||
| P42318 | YxjG* | Methionine synthase homolog | No | C346 Zn-binding active site (cons.) | (R)YVSLDQLC341(+IAM)LSPQC346(+BSH)GFASTEEGNK(L) | 981.4173 | 936.8900 | 2941.2301 | 3 |
| P37487 | PpaC* | Manganese-dependent inorganic pyrophosphatase | Yes | C158 cons. | (K)SPTC158(+BSH)TDQDVAAAK(E) | 851.8443 | 785.1000 | 1701.6740 | 2 |
| P35136 | SerA* | d-3-phosphoglycerate dehydrogenase (serine biosynthesis) | No | C410 cons. in Bacillus species | (K)ISSSESGYDNC410(+BSH)ISVK(V) | 992.9086 | 926.0500 | 1983.8026 | 2 |
| P39912 | AroA | Phospho-2-dehydro-3-deoxyheptonate aldolase/chorismate mutase (chorismate biosynthesis) | No | C126 cons. | (R)FIVGPC126(+BSH)AVESYEQVAEVAAAAK(K) | 883.4080 | 839.4800 | 2647.2022 | 3 |
| P33166 | Tuf | Elongation factor Tu | Yes | C83 in GTP-binding site 82–86 (cons.) | (R)HYAHVDC83(+BSH)PGHADYVK(N) | 527.7176 | 494.4700 | 2106.8413 | 4 |
| P21879 | GuaB | Inosine-5′-monophosphate dehydrogenase (GMP biosynthesis) | Yes | C308 active site (cons.) | (K)VGIGPGSIC308(+BSH)TTR(V) | 519.5691 | 475.0800 | 1555.6854 | 3 |
| P54170 | YphP*,# | UPF0403 protein, thioredoxin-like protein | No | C53 active site (cons.) | (K)AEGTTLVVVNSVC53(+BSH)GC55(+IAM)AAGLAR(P) | 815.3758 | 771.1400 | 2443.1056 | 3 |
| O05268 | YumC | Ferredoxin—NADP reductase 2 (FAD-dependent pyridine nucleotide-disulfide oxidoreductase) | Yes | C85 probably active site (cons.) | (K)FDQTIC85(+Cys)LEQAVESVEK(Q) | 653.3026 | n.d. | 1956.8860 | 3 |
| P18255 | ThrS | Threonine—tRNA ligase 1 | No | C338 Zn-binding; C573 not cons. | (R)LQC573(+BSH)EGLR(V) | 607.7535 | 540.8300 | 1213.4925 | 2 |
| O31678 | QueF | NADPH-dependent 7-cyano-7-deazaguanine reductase (tRNA-Queuosine biosynthesis) | No | C56 active Zn-binding site (cons.) | (K)FNC49(+IAM)PEFTSLC56(+BSH)PK(T) | 613.5820 | 569.1400 | 1837.7241 | 3 |
| B. amyloliquefaciens | |||||||||
| A7Z3U1 | MetE | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (methionine synthase) | No | C647,730 Zn-binding active site (cons.) | (R)FWVNPDC730(+BSH)GLK(T) | 787.8297 | 721.0100 | 1573.6448 | 2 |
| (R)FWVNPDC730(+Cys)GLK(T) | 433.1948 | n.d. | 1296.5626 | 3 | |||||
| (R)VPATEEIYQIIDDALEVC719(+BSH)PTDR(F) | 962.7668 | 918.2900 | 2885.2787 | 3 | |||||
| A7ZAR2 | PpaC | Manganese-dependent inorganic pyrophosphatase | Yes | C158 cons. | (K)SPTC158(+Cys)TEQDIAAAK(E) | 727.3190 | n.d. | 1452.6235 | 2 |
| A7Z657 | SerA | d-3-phosphoglycerate dehydrogenase (serine biosynthesis) | No | C410 cons. | (K)ISSNESGYDNC410(+BSH)ISVK(V) | 671.2758 | 626.8900 | 2010.8054 | 3 |
| A7Z7R9 | AroA | Phospho-2-dehydro-3-deoxyheptonate aldolase/chorismatemutase (chorismate biosynthesis) | No | C126 cons. | (R)FIVGPC126(+BSH)AVESYEQVAEVAAAAK(K) | 883.4087 | n.d. | 2647.2042 | 3 |
| A7Z612 | AroE | 3-phosphoshikimate 1-carboxyvinyltransferase (chorismate biosynthesis) | No | C79 not cons. | (K)GIDALC79(+BSH)EPDSLLDVGNSGTTIR(L) | 881.4009 | 836.7600 | 2641.1808 | 3 |
| (K)GIDALC79(+Cys)EPDSLLDVGNSGTTIR(L) | 789.0375 | n.d. | 2364.0906 | 3 | |||||
| A7Z0D1 | GuaB | Inosine-5′-monophosphate dehydrogenase (GMP biosynthesis) | Yes | C308 active site (cons.) forms thioimidate intermediate | (K)VGIGPGSIC308(+BSH)TTR(V) | 778.8491 | 712.0500 | 1555.6837 | 2 |
| A7Z8C1 | YumC | Ferredoxin—NADP reductase 2 (FAD-dependent Pyridine nucleotide-disulfide oxidoreductase) | Yes | C85 probably active site (cons.) | (K)FDQTIC85(+Cys)LEQAVESVEK(Q) | 653.3023 | n.d. | 1956.8851 | 3 |
| (K)FDQTIC85(+BSH)LEQAVESVEK(Q) | 745.6595 | 701.2400 | 2233.9568 | 3 | |||||
| A7Z7N3 | ThiI | Probable tRNA sulfur transferase (YtbJ homolog) | No | C81 not cons. | (K)C81(+BSH)ESKLEDIK(K) | 730.8164 | n.d. | 1459.6183 | 2 |
| (Thiamine biosynthesis) | |||||||||
| A7Z4X4 | CotE | Spore coat protein E | No | C113 cons. | (K)VLQQPNC113(+Cys)LEVTISPNGNK(I) | 691.6771 | n.d. | 2072.0094 | 3 |
| F4EAC4 | SufA | Chaperone involved in Fe-S cluster assembly | No | C102,104,120 cons: Fe-S-cluster | (K)NAGTPEEC120(+BSH)(-) | 608.7031 | 541.5800 | 1215.3916 | 2 |
| A7Z1M1 | Alr | Alanine racemase 1 (cell wall biosynthesis) | Yes | C63 not cons. | (K)AALEAGASC63(+BSH)LAVAILDEAISLR(K) | 851.7523 | 807.5500 | 2552.2351 | 3 |
| B. pumilus | |||||||||
| B4AEV7 | MetE | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (methionine synthase) | No | C647,730 Zn-binding active site (cons.) | (R)FWVNPDC730(+BSH)GLK(T) | 787.8287 | 721.0000 | 1573.6428 | 2 |
| A8FIT1 | YwaA | Branched-chain amino-acid aminotransferase (valine, leucine, isoleucine biosynthesis) | No | C104 S-SCys by diamide in B. sub. | (R)LC104(+BSH)IPQIDTETVLEGLNELIR(I) | 889.1032 | 844.8300 | 2664.2878 | 3 |
| A8FGB2 | AroA | Phospho-2-dehydro-3-deoxyheptonate aldolase/chorismatemutase (chorismate biosynthesis) | No | C126 cons. | (R)FIVGPC126(+BSH)AVESYEQVAEVAAAAK(Q) | 883.4035 | n.d. | 2647.1887 | 3 |
| B4AN33 | Tuf | Elongation factor Tu | Yes | C83 in GTP-binding site 82–86 (cons.) | (R)HYAHVDC83(+BSH)PGHADYVK(N) | 527.7175 | 494.4800 | 2106.8408 | 4 |
| B4AF61 | MetI | Cystathionine-gamma synthase (CGS) (O-succinylhomoserine(thiol)-lyase) (homocysteine biosynthesis) | No | C335 cons. | (R)IANGVC369(+BSH)NK(L) | 607.7542 | 540.7300 | 1213.4939 | 2 |
| B4ANL1 | LuxS | S-ribosylhomocysteine lyase (homocysteine biosynthesis) | No | C84 essential for catalysis (cons.) C126 Fe-binding site (cons.) | (R)FC41(+BSH)QPNK(Q) | 566.7175 | 499.7200 | 1131.4204 | 2 |
| B4AF80 | ThiG | Thiazole synthase (Thiamine biosynthesis) | No | C92 cons. | (K)VEVIGC102(+BSH)SR(S) | 629.7681 | 562.9200 | 1257.5217 | 2 |
| B4AFT4 | KatX2 | Catalase | ? | No Cys cons. | (K)LLAIC461(+BSH)NFYR(A) | 503.5634 | 459.1000 | 1507.6684 | 3 |
| A8FCN5 | YkuU | 2-Cys peroxiredoxin | ? | C52, 169 cons.; C52 active site; C52–C169 catalytic disulfide | (R)VLQALQTGGLC169(+BSH)PANWKPGQK(T) | 835.7413 | 791.4100 | 2504.2022 | 3 |
| A8FHX4 | FliW | Flagellar assembly factor | No | C144 not cons. | (K)HLLEVASSC144(+BSH) | 677.7782 | 610.9300 | 1353.5418 | 2 |
| A8FG49 | CitZ | Citrate (Si)-synthase II (TCA cycle) | No | C195 cons. | (R)VC195(+BSH)VATLSDIYSGVTAAIGALK(G) | 816.7327 | 772.5300 | 2447.1762 | 3 |
| B. megaterium | |||||||||
| D5DIR6 | MetE | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (methionine synthase) | No | C649,732 Zn-binding active site | (R)ALQVLDPALFWINPDC732(+BSH)GLK(T) | 837.0709 | 792.9500 | 2508.1908 | 3 |
| D5DER3 | PpaC | Manganese-dependent inorganic pyrophosphatase | Yes | C158 cons. | (K)SPTC158(+BSH)TDQDVAAAK(E) | 568.2314 | 523.7300 | 1701.6723 | 3 |
| (K)SPTC158(+Cys)TDQDVAAAK(E) | 713.3030 | n.d. | 1424.5915 | 2 | |||||
| D5D924 | GuaB | Inosine-5′-monophosphate dehydrogenase (GMP biosynthesis) | Yes | C308 active site, forms thioimidate intermediate (cons.) | (K)VGIGPGSIC308(+BSH)TTR(V) | 778.8499 | 712.1000 | 1555.6853 | 2 |
| S. carnosus | |||||||||
| B9DN54 | AroA | Phospho-2-dehydro-3-deoxyheptonate aldolase/chorismate mutase (chorismate biosynthesis) | No | C83, 125 cons; C83 catalytic site; C125 Fe-binding site | (K)SFIFGPC125(+BSH)SVESQEQVDK(V) | 765.9924 | 721.8800 | 2294.9553 | 3 |
| B9DKV8 | Tuf | Elongation factor Tu | No | C82 in GTP-binding site 81–85 | (R)HYAHVDC82(+BSH)PGHADYVK(N) | 527.7177 | 494.5300 | 2106.8417 | 4 |
| B9DM03 | GuaB | Inosine-5′-monophosphate dehydrogenase, (GMP biosynthesis) | No | C307 active site, forms thioimidate intermediate (cons.) | (K)VGIGPGSIC307(+BSH)TTR(I) | 519.5693 | 475.0700 | 1555.6862 | 3 |
| B9DNY0 | YphP | UPF0403 protein (thioredoxin-like protein) | No | C54 active site (CxC-motiv) cons. | (K)NVGKDETTFVVINSTC54(+BSH)GC56(+IAM)AAGLAR(P) | 720.5766 | 687.4300 | 2878.2773 | 4 |
| B9DJN8 | YtxJ | DUF2847 protein (thioredoxin-like protein) | No | C30 active site (TCPI-motiv) cons. | (K)HSNTC30(+BSH)PISANAYDQFNK(F) | 769.3155 | 724.9500 | 2304.9246 | 3 |
| B9DNK0 | DnaK | Chaperone protein | No | No Cys cons. | (K)IIGIDLGTTNSC15(+BSH)VAVLEGDEPK(V) | 880.7465 | 836.5500 | 2639.2176 | 3 |
| B9DNK1 | GrpE | Chaperone protein | No | No Cys cons. | (K)TYQAQC95(+BSH)VLTDILPTIDNIER(A) | 901.4220 | 856.9600 | 2701.2442 | 3 |
| B9DN51 | Ldh | l-lactate dehydrogenase | No | C72 cons. | (K)AGSYEDC72(+BSH)SDADLVVITAGAPQKPGETR(L) | 787.3511 | 754.4000 | 3145.3684 | 4 |
| Q9RGS6 | ThiM | Hydroxyethylthiazole kinase (thiamine biosynthesis) | No | C258 not cons. | (R)IDSDAVAENC258(+BSH)NLEEVK(-) | 715.6331 | 671.2800 | 2143.8775 | 3 |
| B9DMD5 | Sca_1625 | Putative aldehyde dehydrogenase family protein (similar to GbsA betaine aldehyde dehydrogenase) | ? | C279 active site (cons.) | (K)VYNNTGQVC279(+BSH)TAGTR(T) | 627.2645 | 582.8600 | 1878.7716 | 3 |
| (K)VYNNTGQVC279(+Cys)TAGTR(T) | 534.9047 | n.d. | 1601.6922 | 3 | |||||
| B9DN59 | YtpR | Similar to Phe-tRNA synthetase (YtpR homolog) | No | C126,167 cons. | (K)VNVGNEELQIVC126(+BSH)GAPNVEAGQK(V) | 888.7393 | 844.1200 | 2663.1961 | 3 |
| (K)VNVGNEELQIVC126(+Cys)GAPNVEAGQK(V) | 796.3812 | n.d. | 2386.1218 | 3 | |||||
| B9DNG1 | Dtd | d-tyrosyl-tRNA(Tyr) deacylase (YrvI homolog) | No | No Cys cons. | (R)LYEAFNEALC113(+Cys)QYGVEVK(T) | 698.9884 | n.d. | 2093.9434 | 3 |
| B9DLW2 | SceB | SceB precursor (SsaA homolog) | ? | C166 cons | (R)TSSGANYYTAGQC166(+BSH)TYYAFDR(A) | 879.0096 | 834.6200 | 2634.0069 | 3 |
| (R)TSSGANYYTAGQC166(+Cys)TYYAFDR(A) | 786.6523 | n.d. | 2356.9352 | 3 | |||||
| B9DN38 | Sca_1381 | Putative transaldolase | ? | C200 cons. | (R)ELLNVIQADEIGADIITC200(+BSH)PSGVISK(I) | 998.8192 | 954.2100 | 2993.4358 | 3 |
| B9DQ05 | TypA | Putative GTP-binding protein family protein (YlaG homolog) | No | C408 not cons. | (R)VQC408(+BSH)EVPQENAGAVIESLGQR(K) | 841.7123 | n.d. | 2522.1150 | 3 |
| B9DML0 | Sca_1554 | Putative ABC transporter ATP-binding protein | ? | No Cys cons. | (R)QIENC583(+IAM)EAEIEAC590(+BSH)EQK(I) | 730.6216 | 686.2800 | 2188.8431 | 3 |
| B9DM56 | Adk | Adenylate kinase | Yes | C130, 133, 150 cons., Zn-finger motif | (K)VDGVC150(+BSH)DLDGGK(L) | 491.8620 | 447.3800 | 1472.5642 | 3 |
Cytoplasmic protein extracts of B. subtilis, B. amyloliquefaciens, B. pumilus, B. megaterium, and S. carnosus were prepared in an urea-IAM-buffer, tryptic in-gel-digested, and analyzed in a LTQ Orbitrap-Velos™ mass spectrometer as described in the Materials and Methods section. Peptides with S-bacillithiolations were identified by the additional mass of 396 Da at Cys residues and the diagnostic neutral malate loss fragment ions that appeared as abundant ions in the fragment ion MS/MS spectra. Peptides with S-cysteinylation were identified with a mass difference of 119 Da at Cys residues. The table includes the Uniprot accession numbers and protein functions as derived from the UniprotKB database (http://uniprot.org/) and information about conserved Cys residues and Cys functions as revealed by the Conserved Domain Database CDD (34) (http://ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The table also includes the m/z of the precursor ions, m/z of the precursor malate loss ions (−134 Da), and the neutral molecular mass of the BSH-modified peptide and peptide charges. The Xcorr, ΔCn scores, and mass deviations of the S-bacillithiolated peptides and complete collision-induced diffraction (CID) MS/MS spectrum of all peptides and the b and y fragment ion series are given in Supplementary Figures S8 and S9 as follows: OhrR (S8A), MetE (S8B), YxjG (S8C), PpaC (S8D), SerA (S8E), AroA (S8H), Tuf (S8I), GuaB (S8J), YphP (S8F), YtxJ (S8G), YumC (S8K), ThrS (S8L), QueF (S8M), YwaA (S8N), MetI (S8O), LuxS (S8P), ThiG (S8U), KatX2 (S8S), YkuU (S8T), FliW (S8Q), CitZ (S8R), AroE (S8V), CotE (S8W), SufA (S8X), Alr (S8Y), ThiI (S8Z), DnaK (S9A), GrpE (S9B), Ldh (S9C), ThiM (S9D), Sca_1625 (S9E), YtpR (S9F), Dtd (S9G), SceB (S9H), Sca_1381 (S9I), TypA (S9J), Sca_1554 (S9K), and Adk (S9L). * indicates that these proteins with SSB sites have been identified in the previous study in B. subtilis (7) and are listed here for comparison. Bold texts indicate conserved S-bacillithiolated proteins that are identified in more than two of the four Bacillus species and S. carnosus in this study. #The YphP protein was identified in untreated and NaOCl-treated B. subtilis cells. ? indicates that it is unknown if this protein is essential. n.d. indicates that the malate-loss precursor ion was not determined.
IAM, iodoacetamide; LTQ, linear-trap quadrupole; MW, molecular weight.
We were interested to identify proteins with NaOCl-sensitive thiols that form SSB sites in Bacillus spp. and S. carnosus. Tryptic digests of protein extracts from control and NaOCl-treated cells were analyzed for S-bacillithiolations using Orbitrap-Velos LC-MS/MS analysis. The CID MS/MS spectra of the BSH-modified peptides show characteristic and abundant malate loss precursor ions (precursor-134 Da) that were used as diagnostic fragment ions to search for BSH-modified peptides and labeled in the MS/MS spectra (Table 2, Supplementary Tables S2 and Supplementary Figs. S8 and S9) (7). In our previous study, we found S-bacillithiolation sites for the thioredoxin-like protein YphP in B. subtilis under control conditions (7), but no further SSB sites were identified in Bacillus species and S. carnosus. In response to NaOCl stress, we could identify in total 54 proteins with characteristic SSB sites, including 29 unique proteins and eight conserved proteins with identical SSB sites in two or more Bacillus strains and S. carnosus. The eight proteins with conserved SSB sites include the methionine synthase (MetE), the inorganic pyrophosphatase (PpaC), the 3-D-phosphoglycerate dehydrogenase (SerA), the bifunctional 3-deoxy-7-phosphoheptulonate synthase/chorismate mutase (AroA), the translation elongation factor EF-Tu (TufA), the inosine 5′-monophosphate (IMP) dehydrogenase (GuaB), the thioredoxin-like protein (YphP), and the ferredoxin–NADP+ oxidoreductase (YumC) (Fig. 2A, E and Supplementary Figs. S8 and S9).
The methionine synthase MetE was oxidized most strongly and specifically by NaOCl stress in the redox proteome of all Bacillus strains (Fig. 2A and Supplementary Figs. S3–S7), and the conserved Cys730-SSB and Cys719-SSB sites were mapped by LC-MS/MS analyses (Supplementary Fig. S8). The YxjG methionine synthase paralog was only oxidized by NaOCl stress in B. subtilis and B. amyloliquefaciens (Supplementary Figs. S3 and S4). Further, NaOCl-sensitive enzymes with SSB sites at nonconserved Cys369 and Cys41, respectively, include the cystathionine-gamma synthase (MetI) and S-ribosylhomocysteine lyase (LuxS) of B. pumilus involved in homocysteine biosynthesis (Supplementary Figs. S8 and S9). The conserved proteins SerA with Cys410-SSB and PpaC with Cys158-SSB modifications (7) are constitutively oxidized in the redox proteomes of all strains, suggesting that basal S-thiolation is observed for selected proteins during aerobic growth (Fig. 2A and Supplementary Figs. S3–S7). The AroA protein was strongly oxidized in all redox proteomes by NaOCl, and the conserved Cys126-SSB site was identified by LC-MS/MS analysis (Fig. 2A and Supplementary Figs. S3–S7 and Table 2). AroA functions in the first step of chorismate synthesis catalyzing the production of 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP), and thus its S-bacillithiolation could regulate the biosynthesis of aromatic amino acids (Fig. 3). In B. amyloliquefaciens, the AroE protein that functions in the later steps of chorismate biosynthesis was also identified with a Cys79-SSB site. The essential GuaB protein that functions in guanosine 5′-phosphate biosynthesis showed increased oxidation ratios by NaOCl stress in most Bacillus spp., and the active-site Cys308 was identified as a conserved SSB site (Figs. 2A and 3 and Supplementary Figs. S3–S7 and Table 2). Enzymes for thiamine biosynthesis also harbor NaOCl-sensitive thiols, such as the thiazole synthase ThiG in B. pumilus that was identified with a Cys102-SSB site. The hydroxyethylthiazole kinase ThiM of S. carnosus was identified with a Cys258-SSB modification (Figs. 2A and 3 and Supplementary Figs. S3–S7 and Table 2). The NADPH-dependent 7-cyano-7-deazaguanine reductase QueF was identified with an SSB site at the active site Zn-binding Cys55, and was NaOCl-oxidized in the redox proteomes of B. amyloliquefaciens and B. pumilus. QueF is involved in the biosynthesis of queuosine, a hypermodified base found in the wobble positions of tRNAs (25). Additional proteins with increased oxidation ratios and identified SSB sites include the translation elongation factor EF-Tu, aminoacyl-tRNA synthetases (ThrS, YtpR, and TypA), and the DnaK and GrpE chaperones (Fig. 2A, Tables 1 and 2). Oxidation of DnaK and GrpE was only observed in the S. carnosus redox proteome, and the SSB sites were mapped at Cys15 and Cys95, respectively (Fig. 2A and Supplementary Figs. S3–S7 and Table 2).
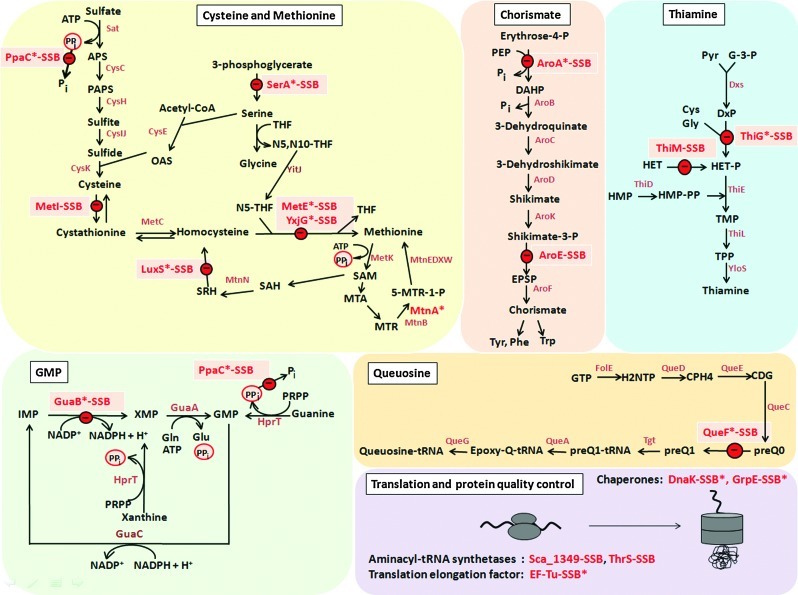
FIG. 3.
Metabolic enzymes with SSB sites were identified within the biosynthetic pathways for methionine, Cys, chorismate, thiamine, GMP, and queuosine. The translation and protein quality control machinery is another target of S-bacillithiolation. The schematics of the metabolic pathways refer to the annotated genes of B. subtilis modified from the SubtiPathways database (http://subtiwiki.uni-goettingen.de/subtipathways.html), and possible inhibitory steps in these pathways are shown by the identified S-bacillithiolated proteins SerA, PpaC, MetE, YxjG, MetI, LuxS (Met, Cys biosynthesis); AroA, AroE (Chorismate biosynthesis); ThiG, ThiM (Thiamine biosynthesis); GuaB and QueF (GMP, queuosine biosynthesis). The schematic for queuosine biosynthesis is adapted from (37), and the Met and Cys biosynthesis pathway is from (48). Cys, cysteine; GMP, guanosine 5′-phosphate. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
Among the redox and antioxidant proteins, the thioredoxin-like proteins YphP and YtxJ (Sca_0389) were identified with SSB sites at their active-site Cys53 and Cys30, respectively, and oxidized in the redox proteome of S. carnosus by NaOCl stress (Fig. 2A and Supplementary Figs. S8 and S9). The ferredoxin–NADP+ oxidoreductase YumC (23) was identified with an SSB modification at the active-site Cys85.
In S. carnosus, the lactate dehydrogenase Ldh and the putative transaldolase Sca_1381 are constitutively oxidized in the redox proteome, and the Cys72-SSB and Cys200-SSB sites, respectively, were identified by LC-MS/MS (Fig. 2A; Table 2). The aldehyde dehydrogenase AldA and the arginine deiminase ArcA are NaOCl-sensitive proteins in the redox proteome of S. carnosus that also possess active-site Cys residues.
NaOCl stress caused a decreased BSH/BSSB redox ratio in B. subtilis as shown by thiol metabolomics
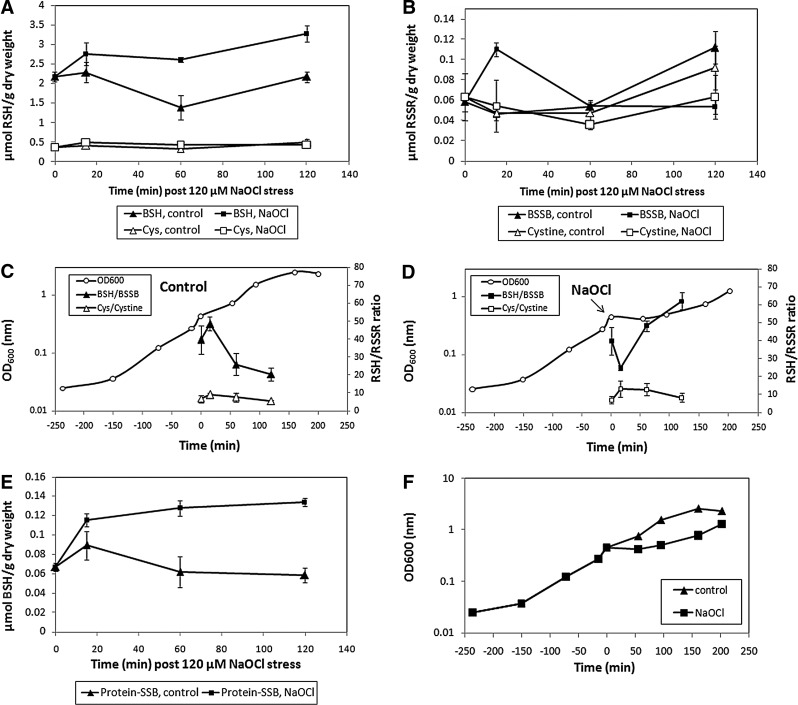
In B. subtilis, grown in an LB medium, similar amounts of BSH (0.6–1 μmol/g dry weight [DW]) and Cys (0.6 μmol/g DW) have been quantified previously (12, 40). The redox ratios were calculated previously as ∼400:1 for BSH/BSSB and ∼120:1 for Cys/Cystine in LB-grown B. subtilis cells (40). We quantified the change of the BSH/BSSB and Cys/Cystine redox ratios in B. subtilis cells grown in the BMM after NaOCl stress (Fig. 4 and Supplementary Fig. S10). The BSH level was 2.2 μmol/g DW in growing cells (OD500=0.4) and increased to 2.7 μmol/g DW directly after NaOCl stress and to 3.2 μmol/g DW during postexponential growth (Fig. 4A). BSH synthesis is about two-fold increased by NaOCl stress compared to control cells. The BSSB level was 0.06 μmol/g DW in growing cells, and NaOCl exposure resulted in a rapid two-fold increase to 0.11 μmol/g DW after 15 min (Fig. 4B). The BSSB level decreased within 60 min after NaOCl stress to control levels. The BSH/BSSB redox ratio was calculated as 40–50:1 during exponential growth, dropped two-fold shortly after NaOCl exposure, and recovered to 50–60:1 within 2 h (Fig. 4C, D). Approximately 3% (0.06 μmol/g DW) of the total cellular BSH content was bound as protein-SSB during growth and increased two-fold after exposure to 120 μM NaOCl (Fig. 4E). The BSH level is four- to eight-fold higher than Cys, and the levels of Cys (0.4 μmol/g DW), Cystine (0.05 μmol/g DW), and the Cys/Cystine redox ratio (∼10:1) were not affected by NaOCl stress. These data confirm that BSH is used as major redox buffer and oxidized to BSSB by NaOCl stress, causing a decreased BSH/BSSB redox ratio and S-bacillithiolation in B. subtilis cells.
FIG. 4.
The BSH/BSSB redox ratio and BSH protein-mixed disulfides are changed in B. subtilis after NaOCl exposure. B. subtilis cells grown in BMM were exposed to 120 μM NaOCl stress at OD600=0.4 and harvested immediately before (0 min) and 15, 60, and 120 min post-NaOCl stress. The reduced thiol redox buffer (RSH) including BSH and Cys (A), the oxidized thiol redox buffer (RSSR) including BSSB and Cystine (B), and protein BSH contents (E) were measured as described in the Materials and Methods section. The RSH/RSSR ratios are calculated for control cells (C) and NaOCl-treated cells (D) in relation to the growth curve. The time point of NaOCl exposure is indicated by an arrow. The combined growth curves of untreated and NaOCl-treated cells are also shown for comparison (F). Experiments were performed in triplicate, and the error bars are given as standard error of the means values. Representing peaks for BSH, Cys, BSSB, Cystine and protein BSH are shown in the high performance liquid chromatography chromatograms in Supplementary Figure S10. BSSB, oxidized bacillithiol disulfide.
Discussion
Previously, we have shown that BSH forms mixed protein disulfides (S-bacillithiolations) with essential catalytic Cys residues of enzymes involved in the methionine biosynthesis pathway and with the redox-sensing OhrR repressor after NaOCl stress in B. subtilis (7). Thus, S-bacillithiolation acts as a redox-switch under hypochlorite stress to induce methionine starvation and up-regulation of the OhrA peroxiredoxin for detoxification. In this work, S-bacillithiolation sites in 54 proteins have been identified among four different industrially important Bacillus species and S. carnosus after exposure to NaOCl. We applied our previous LC-MS/MS analysis to directly identify S-bacillithiolation sites and proved the MS/MS spectra by their characteristic malate loss precursor ions. The number of S-bacillithiolated proteins is lower than that of glutathionylated proteins identified in eukaryotes. This discrepancy could be due to the labile nature of the Cys-GlcN-Mal moiety that limits its detection, and also because the amount of BSH is much lower (2–3 μmol/g DW) compared to GSH in E. coli (19 μmol/g DW) or mycothiol (MSH) in Mycobacterium smegmatis (40 μmol/g DW) (40). Thus, selective thiol-trapping and enrichment tools need to be developed to quantify S-bacillithiolated proteins more comprehensively. To identify S-glutathionylated proteins in the proteome of eukaryotic organisms, redox proteomic approaches have been developed that use glutaredoxin (Grx) for specific de-glutathionylation of cell extracts, followed by biotin-N-ethylmaleimide (NEM) alkylation and affinity purification (e.g., the biotin-switch assay) (21, 28, 30, 31). By analogy, large-scale identification and quantification of BSH-mixed protein disulfides would require selective reduction by bacilliredoxins (Brxs), followed by alkylation and purification protocols. We identified possible candidates for putative Brxs (YphP and YtxJ) that were S-bacillithiolated at their active-site Cys residues. Current attempts are directed to develop Brx-based redox proteomics methods to further characterize and quantify the S-bacillithiolome, and to analyze the substrate specificities of these putative Brxs.
The fluorescence-based redox proteomics approach was used to quantify all oxidized proteins, and many conserved S-bacillithiolated proteins show increased oxidation ratios upon NaOCl exposure (e.g., MetE, YxjG, AroA, and ThiG). However, some conserved S-bacillithiolated proteins like TufA, GuaB, PpaC, and SerA did not show increased oxidation ratios by NaOCl stress in the redox proteomes of most Bacillus spp., indicating that S-bacillithiolation also occurs under nonstress aerobic growth conditions. This confirms results of S-glutathionylations in malaria parasites, human, and yeast cells, where the basal level S-thiolation occurs even under nonstress conditions as a result of aerobic growth (21, 24, 30). In Plasmodium falciparum, more than 400 S-glutathionylated proteins were identified in nonstressed cells (21). The authors further demonstrated in vitro that the activities of only a few identified metabolic enzymes were inhibited by S-glutathionylation. The physiological role and verification of the predicted basal level S-bacillithiolation of essential proteins in our study require further detailed investigations, since we could not identify S-bacillithiolations of these proteins in untreated cells using LC-MS/MS analysis. Overall, our combined redox proteomics and LC-MS/MS analyses showed a similar composition of the S-bacillithiolome compared to the S-glutathionylomes described for eukaryotes in oxidatively stressed cells (8, 31). The S-bacillithiolome contains mainly biosynthetic enzymes for amino acids, cofactors, nucleotides, as well as translation factors, chaperones, and redox and antioxidant proteins. Most proteins are enzymes involved in amino acid biosynthesis pathways, such as methionine, Cys (MetE, YxjG, MetI, LuxS, and PpaC), serine (SerA), aromatic amino acids (AroA and AroE), branched chain amino acids (YwaA), GTP synthesis (GuaB), queousine biosynthesis (QueF), and thiamine biosynthesis (ThiG, ThiI, and ThiM). We further identified S-bacillithiolated proteins involved in translation, such as elongation factor EF-Tu and aminoacyl-tRNA-synthetases; the heat-specific chaperones (DnaK and GrpE); and redox and antioxidant proteins, including thioredoxin-like proteins (YphP and YtxJ), the ferredoxin-NADP+ oxidoreductase (YumC), and peroxiredoxins (YkuU), which could function in the de-bacillithiolation process or require BSH for their regeneration.
The identified S-bacillithiolated proteins include eight conserved proteins identified in B. subtilis (MetE, GuaB, PpaC, SerA, AroA, Tuf, YphP, and YumC), which could be confirmed as S-bacillithiolated proteins in other species (Fig. 2D, E). Most conserved S-bacillithiolated proteins are abundant proteins oxidized in the redox proteomes of Bacillus and Staphylococcus species (e.g., MetE, Tuf, GuaB, PpaC, SerA, and AroA) and have conserved SSB peptides (Fig. 2A and Table 2). In addition, we detected 29 unique proteins that were S-bacillithiolated only in one Bacillus species or S. carnosus. The most unique proteins with SSB sites were identified in S. carnosus (12 SSB sites) and B. pumilus (8 SSB sites). These proteins are less abundant in the gel-based proteome and were only detected by LC-MS/MS analysis. For example, ThiM and ThiG proteins were found as specific targets for S-bacillithiolation in S. carnosus and B. pumilus, respectively. Novel S-bacillithiolated proteins were mapped in S. carnosus as the putative aldehyde dehydrogenase (Sca_1625), the transaldolase (Sca_1381), and the lactate dehydrogenases (Ldh), as well as the SceB precursor. These proteins are not conserved in Bacillus species, but homologs are present in the pathogen S. aureus. Interestingly, the chaperones GrpE and DnaK were S-bacillithiolated in S. carnosus, but Cys residues are absent in the GrpE and DnaK homologs of Bacillus species. The detection of some unique SSB-peptides could be also due to better peptide ionization properties of the tryptic peptides of less-conserved proteins.
Using BSH-specific immunoblots, we could demonstrate that MetE is the most abundantly S-bacillithiolated protein in all Bacillus species. MetE is most strongly oxidized by NaOCl stress in the redox proteome of B. subtilis wild-type and bshA mutant cells and is S-cysteinylated in the bshA mutant (7). Similar MetE oxidation ratios were calculated after 30 and 60 min post NaOCl stress in the wild-type and bshA mutant cells, but reduction of MetE took significant longer in the bshA mutant (Supplementary Fig. S11). Similarly, AroA was S-bacillithiolated in the wild-type cells and displays similar oxidation ratios in the wild-type and bshA mutant cells within the first hour, but is more slowly reduced in the bshA mutant. This may suggest that S-cysteinylation can fully compensate for S-bacillithiolation in the absence of BSH, but the reduction of these mixed disulfides occurs with different efficiencies.
The physiological importance of MetE inhibition by S-thiolation has previously been demonstrated. Methionine auxotrophy is one common phenotype in bacteria resulting from S-thiolation of MetE in B. subtilis and E. coli in response to oxidative stress (7, 19). In all Bacillus species, the conserved, active-site Cys730 and the nonconserved Cys719 were identified as SSB sites for MetE. The observed S-bacillithiolation of LuxS and MetI could be required to inhibit homocysteine biosynthesis after NaOCl stress, since homocysteine is toxic and accumulates when Met biosynthesis is inhibited, due to MetE-SSB modification (Fig. 3).
Among the conserved S-bacillithiolated proteins, the chorismate synthase AroA was identified, which catalyzes the synthesis of DAHP. Notably, the DAHP synthase Aro4p of yeast cells was found as a target for the Grx2 and showed increased thiol redox ratios at Cys76 and Cys244 in the grx2 mutant (35). The essential IMP dehydrogenase GuaB and the pyrophosphatase PpaC have been previously found to be S-glutathionylated in human T-lymphocytes and endothelial cells under diamide stress conditions (11, 30).
An important finding of this study is the identification of S-bacillithiolated proteins involved in translation and protein quality control, such as elongation factor EF-Tu and the DnaK and GrpE chaperones. The heat-specific Hsp70 and Hsp90 chaperones are well represented in the S-glutathionylomes of human and yeast cells (11, 45). The elongation factor TufA promotes GTP-dependent binding of aminoacyl-tRNAs to the A-sites of the ribosome, but also has chaperone functions (4, 47, 49). The TufA protein was oxidized at Cys256 by NaOCl stress in E. coli, and a tufA mutant was highly sensitive to NaOCl (27). In the Bacillus spp. and S. carnosus TufA proteins, we identified the conserved Cys82-SSB modification in the GTP-binding site. Since TufA is essential in B. subtilis, its S-thiolation in oxidatively stressed cells could cause inhibition of protein synthesis during oxidative stress as previously shown in yeast cells (45). Thus, the fact that amino acid biosynthetic enzymes and translation proteins dominate in the conserved S-bacillithiolome suggests that S-bacillithiolation could control protein synthesis during oxidative stress. The strong repression of the stringent controlled RelA regulon, including genes for ribosomal proteins, by NaOCl stress in our previous microarray analysis further indicates a decreased protein synthesis that could be caused by S-bacillithiolation (7). The activation of the stringent response by ROS and RNS has also been shown in Salmonella (2, 17). Specifically, the stringent response regulatory DnaK suppressor protein (DksA) was shown to be play a role in controlling GSH biosynthesis and other metabolic pathways associated with the generation of reducing power under oxidative stress conditions (17).
The next question is how the reversibility of this redox mechanism (protein de-bacillithiolation) might be mediated. Redoxins catalyzing the reversible S-thiolation/reduction of S-glutathionylated proteins include Grxs, thioredoxins, glutathione S-transferases, and GSH disulfide reductases that were identified in large-scale redox proteomics studies (21). The mechanisms of protein de-glutathionylation by mono- and dithiol Grx have been studied in plants and mammalian systems (13, 52). In both mechanisms, the nucleophilic active-site Cys first attacks the GSH-mixed protein disulfide, resulting in substrate de-glutathionylation and formation of a Grx-SSG intermediate (52). In plant dithiol Grx, this Grx-SSB intermediate is then attacked by a second reactive Cys not present in the CXXC motif that leads to Grx intramolecular disulfide formation. This Grx disulfide is finally resolved by an NADPH-dependent ferredoxin:thioredoxin reductase. In the monothiol mechanism, the Grx-SSG intermediate is regenerated by GSH that is oxidized to GSSG and requires the GSH reductase to restore the reduced GSH pool (52). The thioredoxin-like proteins YphP and YtxJ could possibly function as putative Brxs. YphP is a DUF1094 family protein whose previously solved crystal structure adopts a thioredoxin-fold, has a conserved C53XC55 motif (with a reduction potential of −130 mV), and was demonstrated to display some weak disulfide isomerase activity (10). Under NaOCl stress, YphP is S-bacillithiolated at its more solvent-exposed active-site Cys53. The DUF2847 family Brx protein YtxJ is another thioredoxin-like protein with a conserved TCPIS motif reminiscent of the same redox-active motif found in many monothiol Grxs (18). YtxJ was identified in S. carnosus, S-bacillithiolated in its TCPIS motif. The detection of S-bacillithiolated YphP and YtxJ during NaOCl stress could represent the trapping of transient intermediates of Brx-mediated de-bacillithiolation pathways. The distribution of YphP and YtxJ homologs among bacteria encoding BSH biosynthesis genes (12) further suggests their roles as Brx candidates. Our current experiments are directed to further investigate the functions of these putative Brx proteins in the de-bacillithiolation pathway.
We further analyzed whether the BSH/BSSB redox ratio is affected by NaOCl stress that could contribute to S-bacillithiolation of proteins. In E. coli cells, a four-fold decrease in the GSH/GSSG redox ratio from 370:1 to 87:1 was observed in menadione-treated cells (46). The two-fold increase of BSSB post NaOCl stress is comparable to the increase of GSSG in E. coli cells exposed to menadione (46). The increased GSSG levels result in a decreased GSH/GSSG ratio that could contribute to protein S-glutathionylation spontaneously via thiol–disulfide exchange between GSSG and protein thiols (13). However, this thiol–disulfide exchange is kinetically too slow and very unlikely in vivo, since it would require a drop in the GSH:GSSG ratio from 100:1 to 1:1 for 50% conversion of protein thiols to protein GSH-mixed disulfides (13, 52). In our studies, the BSH:BSSB redox ratio dropped two-fold, confirming that BSH is oxidized to BSSB by NaOCl. This BSH:BSSB ratio of 25:1 is unlikely to be sufficient for spontaneous formation of protein-SSB via a direct thiol–disulfide exchange reaction between protein-SH and BSSB. Instead, S-bacillithiolation first requires oxidation of reactive Cys residues to activated thiol derivatives, such as sulfenic acids (9, 15). NaOCl leads to chlorination of Cys residues to unstable sulfenylchloride intermediates that rapidly react further to form disulfides (50). Thus, we suggest that sulfenylchloride formation at reactive Cys residues by NaOCl and the decreased BSH/BSSB redox ratio contribute to the mechanism of S-bacillithiolation in Firmicutes bacteria.
Finally, the question arises if hypochlorite is a physiologically relevant oxidant that mediates protein S-bacillithiolation in Bacillus and Staphylococcus species. Hypochlorite is encountered by soil bacteria and pathogenic Bacillus and Staphylococcus species, since it is present in household bleach and other disinfectants, and it is also produced by activated macrophages during the infection process by the enzyme myeloperoxidase. The observed S-bacillithiolations in soil-dwelling Bacillus species after NaOCl stress might mimic the protective response to toxic ROS, such as hydroxyl radicals, which are generated during respiration, explaining the conservation of some of the protein targets across different species.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains used were B. subtilis wild-type strains 168 (trpC2) and CU1065 (trpC2 pheA1) and bshA-mutant strain HB11002 (CU1065 trpC2 and bshA::mlsr) (12), B. amyloliquefaciens FZB42 (5), B. pumilus SBUG1799, B. megaterium SBUG1152 (36), and S. carnosus TM300 (43). Cultivation was performed as detailed in the Supplementary Materials and Methods section.
Thiol redox proteome analysis and protein identification using MALDI-TOF-TOF MS/MS
The thiol redox proteome analysis was performed as described (7). Cells were harvested at control and NaOCl stress conditions, sonicated, alkylated in 8 M urea/1% chaps/100 mM iodoacetamide (IAM), and acetone-precipitated. The pellet was resolved in an urea/chaps buffer without IAM, and 200 μg of the protein extract was reduced with 10 mM Tris-(2-carboxyethyl)-phosphine and labeled with BODIPY FL C1-IA [N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)-methyl)-iodoacetamide] (Invitrogen, Eugene, OR). The fluorescence-labeled protein extract was separated using 2D PAGE, scanned for BODIPY fluorescence, and stained with Coomassie as described (7). Quantitative image analysis was performed with DECODON Delta 2D software (www.decodon.com). The oxidation ratios were calculated as fluorescence/protein amount ratios and are listed in Supplementary Table S1. The log2 oxidation ratios were used for hierarchical clustering analysis using Multi-Experiment Viewer (MeV v.4.8) software (http://mev.tm4.org/) (32). Tryptic digestion of proteins, peptide spotting, and MALDI-TOF-TOF measurement were performed as described in the Supplementary Materials and Methods section.
Linear-trap quadrupole–Orbitrap Velos mass spectrometry
Bacterial cells were sonicated in an urea/chaps alkylation buffer with 100 mM IAM and separated by 15% nonreducing SDS-PAGE. In-gel tryptic digests and Orbitrap Velos LC-MS/MS analysis were performed as described in the Supplementary Materials and Methods section. S-bacillithiolated peptides were identified by searching all MS/MS spectra in .dta format against B. subtilis 168, B. megaterium, B. amyloliquefaciens FZB42, B. pumilus, and S. carnosus TM300 target-decoy protein sequence databases extracted from UniprotKB release 12.7 using Sorcerer™-SEQUEST® according to the parameters and SEQUEST filter criteria as described in the Supplementary Materials and Methods section.
Preparation of keyhole limpet hemocyanin-BSH for generation of BSH-antibodies
Two milligrams of synthetic BSH (44) was conjugated to 10 mg keyhole limpet hemocyanin (KLH) using the Maleimide-Activated KLH Conjugation Kit (Sigma) according to the details described in the Supplementary Materials and Methods section.
Nonreducing BSH-immunoblot analysis and nonreducing/reducing diagonal SDS-PAGE
Polyclonal BSH antisera were used at 1:500 dilution, and 25 μg IAM-alkylated protein extracts was used for nonreducing 15% SDS-PAGE and immunoblot analysis as described (6). Nonreducing/reducing diagonal SDS-PAGE analysis was performed as previously described (6).
Quantification of reduced thiol redox buffer (RSH), oxidized thiol redox buffer (RSSR), and protein BSH
B. subtilis CU1065 was grown in BMM to an OD600 of 0.44 and harvested before and after NaOCl stress for subsequent RSH, RSSR, and protein RSH quantification as described in the Supplementary Materials and Methods section.
Supplementary Material
Abbreviations Used
- APS
adenosine-5′-phosphosulfate
- BMM
Belitsky minimal medium
- Brx
bacilliredoxin
- BSH
bacillithiol
- BSSB
oxidized bacillithiol disulfide
- CDG
7-carboxy-7-deazaguanine
- CPH4
6-carboxy-5,6,7,8-tetrahydropterin
- Cys
cysteine
- DAHP
3-deoxy-d-arabino-heptulosonate 7-phosphate
- DW
dry weight
- DxP
1-deoxy-d-xylulose 5-phosphate
- EPSP
5-enolpyruvylshikimate-3-phosphate
- GMP
guanosine 5′-phosphate
- Grx
glutaredoxin
- GSH
glutathione
- GSSG
oxidized glutathione disulfide
- GST
glutathione S-transferase
- H2NTP
7,8-dihydroneopterin triphosphate
- HET-P
4-methyl-5-(beta-hydroxyethyl)thiazole monophosphate
- HMP
2-methyl-4-amino-5-hydroxymethyl pyrimidine
- HMP-PP
2-methyl-4-amino-5-hydroxymethyl pyrimidine pyrophosphate
- IAM
iodoacetamide
- IMP
inosine 5′-phosphate
- KLH
keyhole limpet hemocyanin
- LB
Luria broth
- LC
liquid chromatography
- LMW
low molecular weight
- LTQ
linear-trap quadrupole
- Met
methionine
- MS/MS
tandem mass spectrometry
- MW
molecular weight
- N5,N10-THF
5,10-methylenetetrahydrofolate
- N5-THF
5-methyltetrahydrofolate
- OAS
O-acetylserine
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- PEP
phosphoenol pyruvate
- PAGE
polyacrylamide gel electrophoresis
- PPi
inorganic pyrophosphate
- PPP
pentose phosphate pathway
- preQ0
7-cyano-7-deazaguanine
- preQ1
7-aminomethyl-7-deazaguanine
- protein-SSB
BSH protein-mixed disulfide
- PRPP
5-phospho-alpha-d-ribose 1-diphosphate
- Pyr
pyruvate
- Q
queuosine
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RSH
reduced thiol redox buffer
- RSSR
oxidized thiol redox buffer
- SAM
S-adenosyl methionine
- SDS
sodium dodecyl sulfate
- THF
tetrahydrofolate
- TMP
thiamine monophosphate
- TPP
thiamine pyrophosphate
- XMP
xanthosine 5′-phosphate
Acknowledgments
We thank the Decodon Company for support with Decodon Delta 2D software, Frieder Schauer for providing strains B. pumilus SBUG1799 and B. megaterium SBUG1152, and Dana Clausen for excellent technical assistance. We are very grateful to Ahmed Gaballa and John D. Helmann for discussion of the results before publication and for providing the B. subtilis bshA mutant. This work was supported by grants from the Deutsche Forschungsgemeinschaft (AN746/2-1 and AN746/3-1) to H.A., by project TRIG A from the University of Science, Hanoi National University to T.T.T.H., and by a grant from the Biotechnology and Biological Science Research Council (BB/H013504/1) to C.J.H.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Antelmann H. Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourret TJ. Porwollik S. McClelland M. Zhao R. Greco T. Ischiropoulos H. Vazquez-Torres A. Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One. 2008;3:e1833. doi: 10.1371/journal.pone.0001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandes N. Schmitt S. Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldas TD. El Yaagoubi A. Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–11482. doi: 10.1074/jbc.273.19.11478. [DOI] [PubMed] [Google Scholar]
- 5.Chen XH. Koumoutsi A. Scholz R. Eisenreich A. Schneider K. Heinemeyer I. Morgenstern B. Voss B. Hess WR. Reva O. Junge H. Voigt B. Jungblut PR. Vater J. Sussmuth R. Liesegang H. Strittmatter A. Gottschalk G. Borriss R. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- 6.Chi BK. Albrecht D. Gronau K. Becher D. Hecker M. Antelmann H. The redox-sensing regulator YodB senses quinones and diamide via a thiol-disulfide switch in Bacillus subtilis. Proteomics. 2010;10:3155–3164. doi: 10.1002/pmic.201000230. [DOI] [PubMed] [Google Scholar]
- 7.Chi BK. Gronau K. Mader U. Hessling B. Becher D. Antelmann H. S-Bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.009506. 009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalle-Donne I. Rossi R. Colombo G. Giustarini D. Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Davies MJ. Myeloperoxidase-derived oxidation: mechanisms of biological damage and its prevention. J Clin Biochem Nutr. 2011;48:8–19. doi: 10.3164/jcbn.11-006FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derewenda U. Boczek T. Gorres KL. Yu M. Hung LW. Cooper D. Joachimiak A. Raines RT. Derewenda ZS. Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry. 2009;48:8664–8671. doi: 10.1021/bi900437z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fratelli M. Demol H. Puype M. Casagrande S. Eberini I. Salmona M. Bonetto V. Mengozzi M. Duffieux F. Miclet E. Bachi A. Vandekerckhove J. Gianazza E. Ghezzi P. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc Natl Acad Sci U S A. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaballa A. Newton GL. Antelmann H. Parsonage D. Upton H. Rawat M. Claiborne A. Fahey RC. Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A. 2010;107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallogly MM. Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Gioia J. Yerrapragada S. Qin X. Jiang H. Igboeli OC. Muzny D. Dugan-Rocha S. Ding Y. Hawes A. Liu W. Perez L. Kovar C. Dinh H. Lee S. Nazareth L. Blyth P. Holder M. Buhay C. Tirumalai MR. Liu Y. Dasgupta I. Bokhetache L. Fujita M. Karouia F. Eswara Moorthy P. Siefert J. Uzman A. Buzumbo P. Verma A. Zwiya H. McWilliams BD. Olowu A. Clinkenbeard KD. Newcombe D. Golebiewski L. Petrosino JF. Nicholson WL. Fox GE. Venkateswaran K. Highlander SK. Weinstock GM. Paradoxical DNA repair and peroxide resistance gene conservation in Bacillus pumilus SAFR-032. PLoS One. 2007;2:e928. doi: 10.1371/journal.pone.0000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins CL. Pattison DI. Davies MJ. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids. 2003;25:259–274. doi: 10.1007/s00726-003-0016-x. [DOI] [PubMed] [Google Scholar]
- 16.Helmann JD. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid Redox Signal. 2011;15:123–133. doi: 10.1089/ars.2010.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henard CA. Bourret TJ. Song M. Vazquez-Torres A. Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J Biol Chem. 2010;285:36785–36793. doi: 10.1074/jbc.M110.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero E. de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hondorp ER. Matthews RG. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2004;2:e336. doi: 10.1371/journal.pbio.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jothivasan VK. Hamilton CJ. Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat Prod Rep. 2008;25:1091–1117. doi: 10.1039/b616489g. [DOI] [PubMed] [Google Scholar]
- 21.Kehr S. Jortzik E. Delahunty C. Yates JR., 3rd Rahlfs S. Becker K. Protein S-glutathionylation in malaria parasites. Antioxid Redox Signal. 2011;15:2855–2865. doi: 10.1089/ars.2011.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SO. Merchant K. Nudelman R. Beyer WF., Jr. Keng T. DeAngelo J. Hausladen A. Stamler JS. OxyR: a molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 23.Komori H. Seo D. Sakurai T. Higuchi Y. Crystallization and preliminary X-ray studies of ferredoxin-NADP+ oxidoreductase encoded by Bacillus subtilis yumC. Acta Crystallogr F Struct Biol Cryst Commun. 2010;66:301–303. doi: 10.1107/S1744309110000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Moan N. Clement G. Le Maout S. Tacnet F. Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 25.Lee BW. Van Lanen SG. Iwata-Reuyl D. Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in queuosine biosynthesis. Biochemistry. 2007;46:12844–12854. doi: 10.1021/bi701265r. [DOI] [PubMed] [Google Scholar]
- 26.Lee JW. Soonsanga S. Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leichert LI. Gehrke F. Gudiseva HV. Blackwell T. Ilbert M. Walker AK. Strahler JR. Andrews PC. Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leonard SE. Carroll KS. Chemical ‘omics' approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Lillig CH. Potamitou A. Schwenn JD. Vlamis-Gardikas A. Holmgren A. Redox regulation of 3′-phosphoadenylylsulfate reductase from Escherichia coli by glutathione and glutaredoxins. J Biol Chem. 2003;278:22325–22330. doi: 10.1074/jbc.M302304200. [DOI] [PubMed] [Google Scholar]
- 30.Lind C. Gerdes R. Hamnell Y. Schuppe-Koistinen I. von Lowenhielm HB. Holmgren A. Cotgreave IA. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl M. Mata-Cabana A. Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal. 2011;14:2581–2642. doi: 10.1089/ars.2010.3551. [DOI] [PubMed] [Google Scholar]
- 32.Mar JC. Wells CA. Quackenbush J. Defining an informativeness metric for clustering gene expression data. Bioinformatics. 2011;27:1094–1100. doi: 10.1093/bioinformatics/btr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchler-Bauer A. Lu S. Anderson JB. Chitsaz F. Derbyshire MK. DeWeese-Scott C. Fong JH. Geer LY. Geer RC. Gonzales NR. Gwadz M. Hurwitz DI. Jackson JD. Ke Z. Lanczycki CJ. Lu F. Marchler GH. Mullokandov M. Omelchenko MV. Robertson CL. Song JS. Thanki N. Yamashita RA. Zhang D. Zhang N. Zheng C. Bryant SH. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masip L. Veeravalli K. Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 35.McDonagh B. Padilla CA. Pedrajas JR. Barcena JA. Biosynthetic and iron metabolism is regulated by thiol proteome changes dependent on glutaredoxin-2 and mitochondrial peroxiredoxin-1 in Saccharomyces cerevisiae. J Biol Chem. 2011;286:15565–15576. doi: 10.1074/jbc.M110.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikolasch A. Niedermeyer TH. Lalk M. Witt S. Seefeldt S. Hammer E. Schauer F. Gesell Salazar M. Hessel S. Julich WD. Lindequist U. Novel cephalosporins synthesized by amination of 2,5-dihydroxybenzoic acid derivatives using fungal laccases II. Chem Pharm Bull (Tokyo) 2007;55:412–416. doi: 10.1248/cpb.55.412. [DOI] [PubMed] [Google Scholar]
- 37.Miles ZD. McCarty RM. Molnar G. Bandarian V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci U S A. 2011;108:7368–7372. doi: 10.1073/pnas.1018636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakano S. Erwin KN. Ralle M. Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol. 2005;55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- 39.Newton GL. Buchmeier N. Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton GL. Rawat M. La Clair JJ. Jothivasan VK. Budiarto T. Hamilton CJ. Claiborne A. Helmann JD. Fahey RC. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palm GJ. Khanh Chi B. Waack P. Gronau K. Becher D. Albrecht D. Hinrichs W. Read RJ. Antelmann H. Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR. Nucleic Acids Res. 2012;40:4178–4192. doi: 10.1093/nar/gkr1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pattison DI. Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 43.Rosenstein R. Nerz C. Biswas L. Resch A. Raddatz G. Schuster SC. Gotz F. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl Environ Microbiol. 2009;75:811–822. doi: 10.1128/AEM.01982-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma SV. Jothivasan VK. Newton GL. Upton H. Wakabayashi JI. Kane MG. Roberts AA. Rawat M. La Clair JJ. Hamilton CJ. Chemical and chemoenzymatic syntheses of bacillithiol: a unique low-molecular-weight thiol amongst low G+C Gram-positive bacteria. Angew Chem Int Ed Engl. 2011;50:7101–7104. doi: 10.1002/anie.201100196. [DOI] [PubMed] [Google Scholar]
- 45.Shenton D. Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smirnova GV. Muzyka NG. Glukhovchenko MN. Oktyabrsky ON. Effects of menadione and hydrogen peroxide on glutathione status in growing Escherichia coli. Free Radic Biol Med. 2000;28:1009–1016. doi: 10.1016/s0891-5849(99)00256-7. [DOI] [PubMed] [Google Scholar]
- 47.Thompson RC. Dix DB. Karim AM. The reaction of ribosomes with elongation factor Tu.GTP complexes. Aminoacyl-tRNA-independent reactions in the elongation cycle determine the accuracy of protein synthesis. J Biol Chem. 1986;261:4868–4874. [PubMed] [Google Scholar]
- 48.Tomsic J. McDaniel BA. Grundy FJ. Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wholey WY. Jakob U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol Microbiol. 2012;83:981–991. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winterbourn CC. Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Xiong Y. Uys JD. Tew KD. Townsend DM. S-glutathionylation: from molecular mechanisms to health outcomes. Antioxid Redox Signal. 2011;15:233–270. doi: 10.1089/ars.2010.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaffagnini M. Bedhomme M. Lemaire SD. Trost P. The emerging roles of protein glutathionylation in chloroplasts. Plant Sci. 2012;185–186:86–96. doi: 10.1016/j.plantsci.2012.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.