Photoreceptors usually repress seedling hypocotyl elongation under light, whereas PHYB overexpression causes longer hypocotyls than in the wild type under far-red light. This work demonstrates that phyB forms a protein complex with SPA1 through a direct interaction to promote its nuclear accumulation, enhancing hypocotyl elongation in response to far-red light.
Abstract
Phytochrome A (phyA) is the primary photoreceptor mediating deetiolation under far-red (FR) light, whereas phyB predominantly regulates light responses in red light. SUPPRESSOR OF PHYA-105 (SPA1) forms an E3 ubiquitin ligase complex with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), which is responsible for the degradation of various photomorphogenesis-promoting factors, resulting in desensitization to light signaling. However, the role of phyB in FR light signaling and the regulatory pathway from light-activated phytochromes to the COP1-SPA1 complex are largely unknown. Here, we confirm that PHYB overexpression causes an etiolation response with reduced ELONGATED HYPOCOTYL5 (HY5) accumulation under FR light. Notably, phyB exerts its nuclear activities and promotes seedling etiolation in both the presence and absence of phyA in response to FR light. PhyB acts upstream of SPA1 and is functionally dependent on it in FR light signaling. PhyB interacts and forms a protein complex with SPA1, enhancing its nuclear accumulation under FR light. During the dark-to-FR transition, phyB is rapidly imported into the nucleus and facilitates nuclear SPA1 accumulation. These findings support the notion that phyB plays a role in repressing FR light signaling. Activity modulation of the COP1-SPA E3 complex by light-activated phytochromes is an effective and pivotal regulatory step in light signaling.
INTRODUCTION
As sessile organisms, plants have evolved a high degree of developmental plasticity to optimize their growth and reproduction. Light is one of the most important factors modulating many developmental processes of plants, from seed germination to the time of flowering (Deng and Quail, 1999; Li et al., 2011). Plants possess a series of photoreceptors that monitor light quality, quantity, and duration (Briggs and Olney, 2001; Lin, 2002; Christie, 2007; Rizzini et al., 2011). Prominent among these are the red (R)/far-red (FR) reversible photoreceptors, the phytochromes. The five distinct phytochromes in Arabidopsis thaliana, designated phytochrome A (phyA) to phyE, have unique and partially redundant or antagonistic roles in different photomorphogenic responses (Deng and Quail, 1999). PhyA is the only light-labile (type I) phytochrome in Arabidopsis, and phyB, phyC, and phyE are all light-stable (type II) phytochromes (Hirschfeld et al., 1998). PhyB, phyC, and phyE predominantly regulate light responses under R and white (W) light, and their actions exhibit the classical R/FR photoreversible effects characteristic of phytochrome function. PhyA can act in two distinct signaling modes: the FR light–dependent high-irradiance responses (FR-HIRs) and the very-low-fluence response (VLFR). Typical FR-HIRs facilitate the inhibition of hypocotyl elongation, opening of the apical hook, expansion of cotyledons, accumulation of anthocyanin, and FR preconditioned blocking of greening under continuous FR (FRc) light (Fankhauser and Casal, 2004; Bae and Choi, 2008; Li et al., 2011). The VLFR affects seed germination and presumably acts when a seedling emerges from soil and detects light for the first time (Botto et al., 1996; Yanovsky et al., 1997). In the dark, phytochromes are synthesized in Pr and are thought to mediate light-induced responses after absorption of R light and subsequent conversion into Pfr (Reed, 1999; Quail, 2002; Li et al., 2011). Pr-to-Pfr conversion is reversible, allowing the phyB to act as a molecular switch that is activated by R light and deactivated by FR light (Borthwick et al., 1952; Bae and Choi, 2008).
PhyA is believed to be the only photoreceptor that mediates FR-induced responses (Nagatani et al., 1993; Whitelam et al., 1993). Although the phyA phyB double mutant has hypocotyl elongation similar to that of the phyA mutant under FR light, it exhibits less unhooking than the phyA mutant (Reed et al., 1994; Neff and Chory, 1998). Previous studies have reported that overexpression of PHYB under FR light promotes hypocotyl elongation while reducing anthocyanin content and germination rate (Wagner et al., 1996; Short, 1999; Hennig et al., 2001). PhyB is thought to interfere with phyA function in FR light. However, overexpression of PHYB has no apparent effect on the abundance or degradation of phyA upon FR light exposure (Wagner et al., 1996; Short, 1999). Thus, phyB regulatory mechanisms and their relationship with phyA under FR light remain to be discovered.
The COP1 protein is an E3 ubiquitin ligase that functions in the nucleus and is responsible for the ubiquitination and targeted degradation of photoreceptors, including phyA and phyB (Seo et al., 2004; Jang et al., 2010). COP1 interacts physically with the SPA1 protein to form a heterocomplex that targets a group of photomorphogenesis-promoting factors, including HY5, LONG AFTER FAR-RED LIGHT1 (LAF1), and LONG HYPOCOTYL IN FAR-RED1 (HFR1), for degradation. This represses photomorphogenesis in the dark and prevents hyperphotomorphogenesis in the light (Saijo et al., 2003; Seo et al., 2003; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005a, 2005b).
Previous studies have shown that the COP1–SPA1 E3 complex acts as a link between light signals and downstream activities. Photoreceptors absorb light and generate transduced signals, either directly or through intermediates, ultimately modulating the COP1–SPA1 E3 complex (Holm and Deng, 1999; Saijo et al., 2003; Sullivan and Deng, 2003; Yang and Wang, 2006; Feng and Deng, 2007; Li et al., 2011). Photoreceptors may negatively regulate COP1 activity through direct interactions with COP1 (Yi and Deng, 2005). Overexpression of the C-terminal domain of either cryptochrome (CCT1 or CCT2) leads to a constitutive light response similar to that of the cop/det/fus mutants, suggesting that through direct protein–protein interaction, CCT can cause a conformational change in COP1 that reduces its effect on substrates, such as HY5 (Yang et al., 2000; Wang et al., 2001; Yi and Deng, 2005; Feng and Deng, 2007). Because the expression, abundance, and complex formation of COP1 do not appear to be significantly affected by light (Deng et al., 1992; McNellis et al., 1994b; Saijo et al., 2003), the activity of COP1, rather than its abundance, is likely to be light regulated. Recently, the blue (B) light receptor cry1 was shown to interact physically with SPA1 in a B light–dependent manner, and the cry1–SPA1 interaction may negatively regulate COP1, at least in part, by promoting its dissociation from SPA1 (Lian et al., 2011; Liu et al., 2011). In addition, the B light–dependent cry2–SPA1 interaction enhances that of cry2–COP1, suppressing COP1 activity (Zuo et al., 2011).
PhyA, phyB, and cry1 can inhibit the nuclear localization of COP1 under FR, R, or B light, respectively (Osterlund and Deng, 1998). Under FR light, the promotion of hypocotyl elongation by overexpression of COP1 is reduced by the phyB mutation, but the nuclear localization of COP1 is enhanced in the phyB mutant background compared with the wild-type background (Osterlund and Deng, 1998). This is consistent with the inhibition of hypocotyl elongation by phyB β-glucuronidase (GUS)-COP1 and the promotion of etiolation phenotypes by PHYB overexpression under FR light (Osterlund and Deng, 1998; Hennig et al., 2001). The relationship between light-activated phytochromes and the nuclear activity of the COP1–SPA1 E3 complex in light signaling remains to be elucidated.
In this study, we demonstrated that overexpression of PHYB causes an etiolation response with reduced nuclear HY5 accumulation under FR light. PhyB promotes hypocotyl elongation, cotyledon unfolding, and chlorophyll accumulation in both the presence and absence of phyA in response to FR light. PhyB acts upstream of SPA1 and is functionally dependent on it during FR light signaling. PhyB forms a protein complex with SPA1 via a direct interaction to promote SPA1 accumulation under FR light. During the transition from dark to FR light, both the nuclear activities of phyB and SPA1-phyB association rapidly increase, facilitating nuclear transport of SPA1. We also demonstrate that SPA1 promotes nuclear import of phyB in response to FR treatment in a feedback loop. Thus, phyB also represses seedling deetiolation under FR light. Activity modulation of the COP1-SPA E3 complex by light-activated phytochromes is an effective and pivotal regulatory step in light signaling.
RESULTS
PhyB Represses Seedling Deetiolation under FR Light
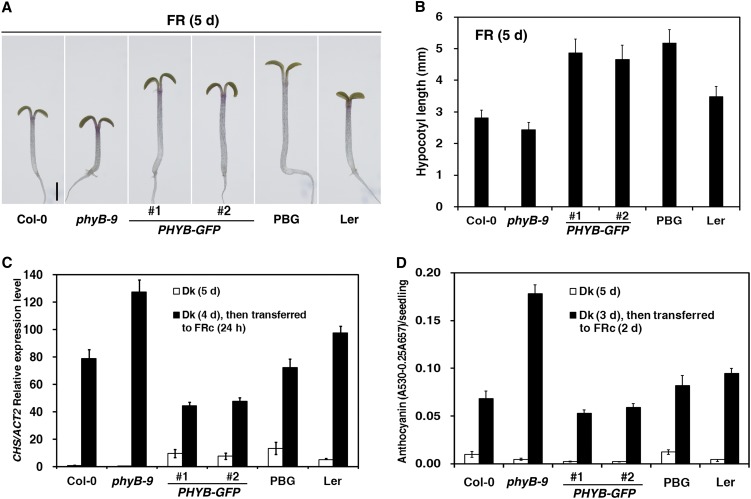
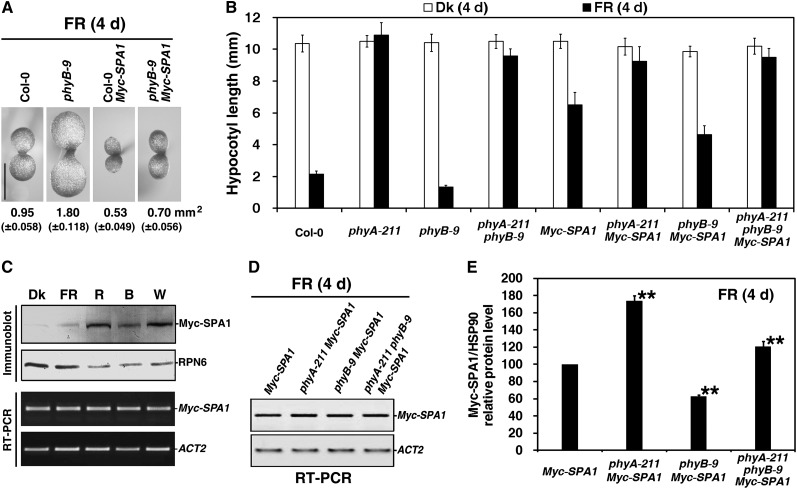
Previous studies have reported that overexpression of PHYB promotes seedling etiolation under FR light (Wagner et al., 1996; Short, 1999; Hennig et al., 2001). However, the mechanisms behind these effects of phyB in FR light are largely unknown. To investigate the role and regulatory mechanism of Arabidopsis phyB in FR light signaling, we generated transgenic plants that overexpress the full-length PHYB gene fused with green fluorescent protein (PHYB-GFP) under the control of the constitutive cauliflower mosaic virus 35S promoter. This construct was introduced into the phyB-9 mutant, a nonsense allele of PHYB (Reed et al., 1993). When first grown under R light for 5 d, the two PHYB-GFP transgenic lines possessed hypocotyls of similar lengths to those of wild-type control plants in the dark and showed enhanced seedling photomorphogenesis with short hypocotyls and large cotyledons, similar to 35S-PHYB-GFP (PBG) in the Landsberg erecta (Ler) ecotype (Yamaguchi et al., 1999), suggesting that the PHYB-GFP transgene was biologically functional (see Supplemental Figures 1A and 1B online). Next, the PHYB-GFP transgenic lines were grown in FR light for 5 d to evaluate hypocotyl elongation (Figures 1A and 1B). Seedling hypocotyl lengths in the PHYB-GFP lines and PBG were all ∼1.5 times the length of wild-type seedlings (Columbia-0 [Col-0] and Ler, respectively), whereas the hypocotyl length in the phyB-9 mutant was only 86% that of the wild type. In previous studies, overexpression of PHYB caused an approximately twofold increase in hypocotyl elongation compared with the respective wild-type backgrounds under FRc light, whereas the phyB-5 mutation significantly inhibited hypocotyl elongation (Wagner et al., 1996; Hennig et al., 2001).
Figure 1.
Arabidopsis PhyB Promotes Seedling Etiolation Responses under FR Light.
The fluence rate of the FR light was 2.5 µmol·m–2·s–1.
(A) Morphology of PHYB-GFP transgenic seedlings grown under FRc light for 5 d. #1 and #2 indicate two individual transgenic PHYB-GFP lines. The #1 line was used as the representative line for PHYB-GFP, unless otherwise indicated. PBG (Yamaguchi et al., 1999) in the Ler ecotype was used as a positive control. Bar = 1 mm.
(B) Histograms of hypocotyl lengths of PHYB-GFP transgenic seedlings grown under FRc light for 5 d (average of 40 seedlings). Error bars indicate sd.
(C) Quantitative RT-PCR analysis of CHS transcript levels in the wild type (Col-0), phyB-9, PHYB-GFP, PBG, and Ler. Seedlings were grown in the dark (Dk) for 5 d or grown in the dark for 4 d and subsequently transferred to FRc light for 24 h. Each column shows the mean relative expression of CHS/Actin2 (ACT2) of three biological repeats. Error bars indicate the sd.
(D) Measurement of anthocyanin contents of Col-0, phyB-9, PHYB-GFP, PBG, and Ler. Seedlings were grown in the dark for 5 d or grown in the dark for 3 d and subsequently transferred to FRc light for 2 d. Means and sd of three replicate experiments are shown (see Supplemental Figure 1 online).
[See online article for color version of this figure.]
To provide further molecular and biochemical evidence for the involvement of phyB in FR light signaling, we measured CHALCONE SYNTHASE (CHS; required for anthocyanin accumulation) transcript abundance and anthocyanin accumulation in PHYB transgenic lines. In response to FR light, the CHS transcript level in phyB-9 mutant seedlings was 1.6-fold higher than in the Col-0 wild-type seedlings, whereas the CHS transcript levels in the PHYB-GFP or PBG seedlings were ∼60% of wild-type levels (Figure 1C). Consequently, seedlings of the phyB-9 mutant accumulated 2.6-fold more anthocyanin than did Col-0 wild-type seedlings, whereas the PHYB-GFP or PBG seedlings accumulated ∼80% as much anthocyanin as their respective wild types (Figure 1D). ABO (35S-PHYB in the Nossen [No-0] ecotype; Wagner et al., 1991) seedlings accumulate only 60% as much anthocyanin as the wild type under FRc light (Hennig et al., 2001). Collectively, these findings suggest that phyB is involved in repressing Arabidopsis photomorphogenesis under FR light, including promoting hypocotyl elongation, affecting CHS expression and repressing anthocyanin accumulation.
The presence of sugar has been found to interfere with the inhibitory effect of phyB under FRc light (Short, 1999). Based upon that, we investigated the effects of no sugar, Suc concentration, FR light intensity, and growth period on phyB hypocotyl elongation in response to FR light. Our results suggest that phyB repression of deetiolation under FRc light occurs even in the absence of additional sugar, although the presence of sugar measurably enhances the inhibitory effect (see Supplemental Figures 2A to 2E online; see also Short, 1999; Hennig et al., 2001). We chose moderate growth conditions to perform most further experiments, including growth medium with 1% Suc, seedlings grown in FRc light for 4 to 5 d and FR light fluence rates of 2.5 and 18.1 µmol·m–2·s–1.
The Nuclear Import of PhyB Does Not Require PhyA in Response to FR Light
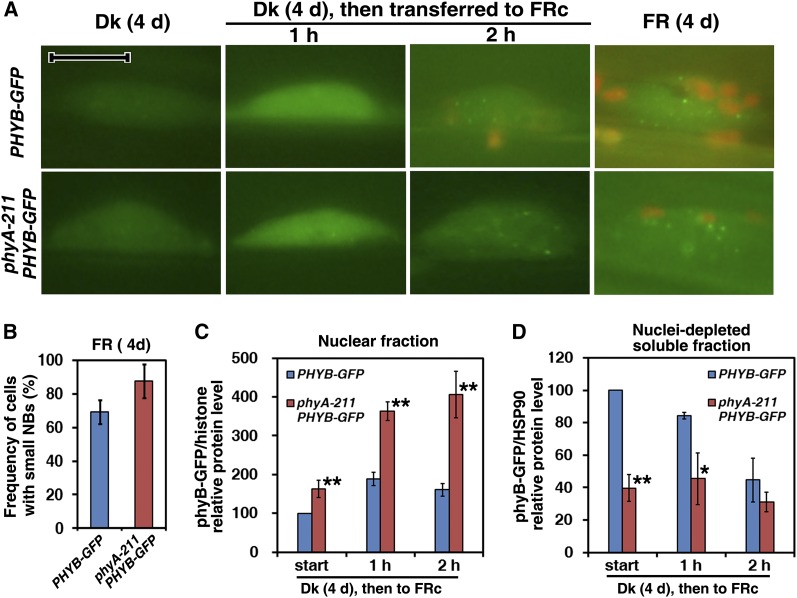
Based on fluorescence observations of phyB-GFP and phyA-GFP, the nuclear import of phyB-GFP is regulated by R light, whereas that of phyA-GFP is controlled not only by FR light but also by R light with much faster kinetics (Kircher et al., 1999, 2002; Yamaguchi et al., 1999). The high percentage of the active Pfr form of phyB in response to high steady state R light leads to large phyB-GFP nuclear bodies (NBs) (see Supplemental Figure 3A online; see also Chen et al., 2003). To determine whether FR light can activate phyB, we first checked phyB-GFP fluorescence in response to FR light treatment. In dark-grown seedlings, phyB-GFP fluorescence was very weak; 1 h of FR light exposure produced diffuse nuclear fluorescence without any NBs (Figure 2A). Following 2 h of FR exposure, or in seedlings grown under FRc light for 4 d, phyB-GFP was seen in very small, dim NBs. To confirm whether phyA was required for the nuclear activities of phyB, we also checked phyB-GFP fluorescence in a phyA-211 mutant (a nonsense allele of PHYA; Reed et al., 1994) background in response to both R and FR light treatment. Under Rc light, the phyB-GFP fluorescence exhibited large and bright NBs in both the Col-0 wild type and the phyA-211 mutant background (see Supplemental Figure 3A online). PhyB-GFP in the phyA-211 mutant background displayed almost identical fluorescence patterns to those in the wild-type background, whether in the dark or under FR light exposure. Note that the frequency of cells with small NBs in the phyA-211 mutant background was ∼25% higher than in the wild-type background when seedlings were grown under FR for 4 d (Figure 2B).
Figure 2.
The Nuclear Import of PhyB-GFP in the Wild-Type and phyA-211 Mutant Backgrounds in Response to FR Light.
Seedlings of PHYB-GFP or phyA-211 PHYB-GFP were grown in the dark (Dk) or FRc light for 4 d or in the dark for 4 d and subsequently transferred to FRc light for 1 or 2 h. The fluence rate of the FR light was 18.1 µmol·m–2·s–1.
(A) Fluorescence images of phyB-GFP in hypocotyl cell nuclei in both the wild-type and phyA-211 mutant backgrounds in response to FR light. Nuclei positions were confirmed by DAPI staining (data not shown). Bar = 10 μm.
(B) Frequencies of cells with small NBs in seedlings of PHYB-GFP and phyA-211 PHYB-GFP in FRc light. Means and sd of three replicate experiments are shown.
(C) Quantification of relative phyB-GFP/histone protein levels in the purified nuclear fraction corresponding to Supplemental Figure 3C online, indicating that nuclear import of phyB-GFP occurred in response to FR light in both of the wild-type and the phyA-211 mutant backgrounds. Means and sd of three replicate experiments are shown. Asterisks mark significant differences in relative phyB-GFP/histone protein levels between PHYB-GFP and phyA-211 PHYB-GFP according to Student’s t test (*P < 0.05; **P < 0.01).
(D) Quantification of relative phyB-GFP/HSP90 protein levels in the nuclei-depleted soluble fraction in response to FR light corresponding to Supplemental Figure 3D online. Means and sd of three replicate experiments are shown. Student’s t tests were performed as in (C) (see Supplemental Figure 3 online).
Although phyB-GFP fluorescence in dark-grown seedlings was too weak to be observed clearly, immunoblot analysis revealed a phyB-GFP signal in the nuclear fractions (see Supplemental Figures 3B and 3C online). The seedlings of PHYB-GFP or phyA-211 PHYB-GFP grown in Rc light accumulated much more nuclear phyB-GFP protein than those grown in the dark (see Supplemental Figure 3B online). To provide further evidence regarding whether the nuclear activities of phyB in response to FR light treatment required phyA, we compared nuclear accumulation of phyB-GFP in the phyA-211 mutant background with that in the wild-type background. Upon 1 h of FR light treatment, the nuclear import of phyB-GFP protein increased almost twofold compared with that in dark-grown seedlings. Unexpectedly, nuclear import of phyB-GFP protein in the phyA-211 mutant background was ∼150% higher than in the wild-type background in the dark or after FR light treatment. Over the same period, cytoplasmic levels of phyB-GFP in phyA-211 PHYB-GFP seedlings were lower than those in PHYB-GFP seedlings (Figure 2D; see Supplemental Figure 3D online). Nuclear import of endogenous phyB in the phyA-211 mutant was notably higher than that in Col-0 in the dark or after FR light treatment (see Supplemental Figure 3F online). Thus, FR exposure promoted nuclear import of phyB even in the absence of phyA.
PhyB Represses FR-HIRs in Both the Presence and Absence of PhyA
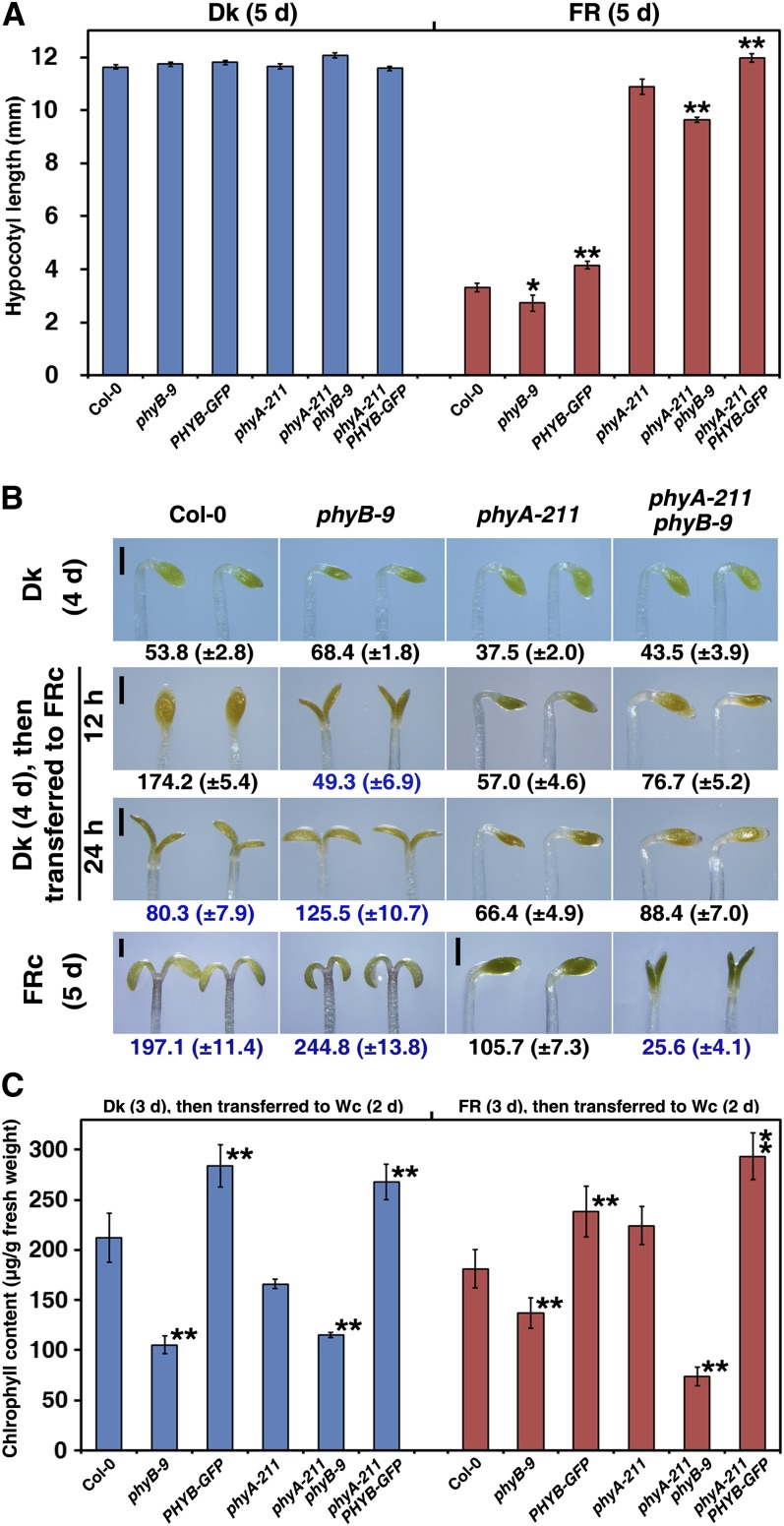
PhyA is thought to be the only active photoreceptor that promotes seedling photomorphogenesis under FR light (Nagatani et al., 1993; Whitelam et al., 1993). The first step to investigate the roles of phyB in FR light signaling is to understand the relationship between phyB and phyA. In the phyB-9 mutant or PHYB-GFP transgenic line, the hypocotyls were 18% shorter or 25% longer, respectively, than the wild-type hypocotyls when grown under FR light (Figure 3A). Then, we compared the hypocotyl lengths of the phyA-211 phyB-9 and phyA-211 PHYB-GFP double mutants with that of the phyA-211 single mutant. Although phyA-211 exhibited a dominant effect on hypocotyl elongation, the phyA-211 phyB-9 double mutant seedlings had 11% less hypocotyl elongation compared with the phyA-211 single mutant grown under FR light for 5 d. By contrast, phyA-211 PHYB-GFP double mutant seedlings had 10% greater hypocotyl elongation compared with the phyA-211 single mutant. All differences in hypocotyl lengths between phyA-211 and phyA-211 phyB-9 or phyA-211 PHYB-GFP were significant as assessed by Student’s t test.
Figure 3.
PhyB Inhibits the FR-HIRs in Both the Presence and Absence of PhyA.
(A) Histograms comparing hypocotyl lengths of the wild type (Col-0), phyB-9, PHYB-GFP, phyA-211, phyA-211 phyB-9, and phyA-211 PHYB-GFP in the dark (Dk) or under FR light (18.1 µmol·m–2·s–1) for 5 d. Hypocotyl lengths of at least 30 seedlings were determined after 5 d in each replicate. The means of five replicates are shown ± se. Asterisks mark significant differences between phyB-9 or PHYB-GFP and Col-0, phyA-211 phyB-9, or phyA-211 PHYB-GFP and phyA-211, according to Student’s t test (*P < 0.05; **P < 0.01).
(B) Apical unhooking and cotyledon unfolding of the wild type (Col-0) and phyB-9, phyA-211, and phyA-211 phyB-9 mutants. Seedlings were grown in the dark for 4 d, under high-FR light fluency (90.5 µmol·m–2·s–1) for 5 d or in the dark for 4 d and subsequently transferred to high-FR light for 12 or 24 h. Numbers in black indicate the angles between the apical hook and the hypocotyl; numbers in blue show the cotyledon unfolding angles and numbers in brackets represent the sd. Bars = 0.5 mm.
(C) PhyB inhibits FR-induced block of greening under continuous W (Wc) light. Seedlings were grown in the dark or under FR light (18.1 µmol·m–2·s–1) for 3 d and subsequently transferred to continuous W light (100.0 µmol·m–2·s–1) for 2 d. Means and sd of four replicate experiments are shown. Student’s t tests were performed as in (A).
In previous studies, the phyA phyB double mutant conferred 18 to 27% less unhooking than the phyA mutant under FRc light (Reed et al., 1994; Neff and Chory, 1998). Next, we compared apical unhooking and cotyledon unfolding between the Col-0 wild type possessing phyB-9 or phyA-211 with the phyA-211 phyB-9 double mutant. Minor differences were observed in the apical unhooking angles of seedlings grown in continuous dark for 4 d; seedlings of the phyB-9 single mutant or phyA-211 phyB-9 double mutant had larger apical unhooking angles compared with those of the Col-0 wild type or phyA-211 single mutants (Figure 3B). After growth in continuous dark for 4 d followed by high-intensity FR light (90.5 µmol·m–2·s–1) for 12 h, the Col-0 wild-type seedlings had a straight apical hook with unopened cotyledons, whereas the phyB-9 single mutant seedlings exhibited a cotyledon opening angle of 49.3 ± 6.9°. Although the cotyledons of the phyA-211 phyB-9 double mutant seedlings were unopened after 24 h of high-intensity FR treatment, their apical unhooking angles were 33% larger than those of phyA-211 single mutant seedlings. After growth in intense FRc light (90.5 µmol·m–2·s–1) for 5 d, the phyA-211 phyB-9 double mutant seedlings had open cotyledons (25.6 ± 4.1°), whereas those of the phyA-211 single mutant seedlings remained unopened.
A previous study showed that the FR block of greening is phyA dependent (Barnes et al., 1996). When grown under FR light for 3 d and under W light for an additional 2 d, either PHYB-GFP or phyA-211 PHYB-GFP seedlings had ∼30% higher chlorophyll concentrations compared with those of Col-0 wild type or phyA-211 mutants (Figure 3C). This suggests that phyB inhibited the FR-induced block of greening in subsequent W light in both wild-type and phyA-211 backgrounds. From these observations, we conclude that phyB enhanced Arabidopsis etiolation under FRc light in both the presence and absence of phyA.
PhyB Acts Upstream of SPA1 in FR Light Signaling
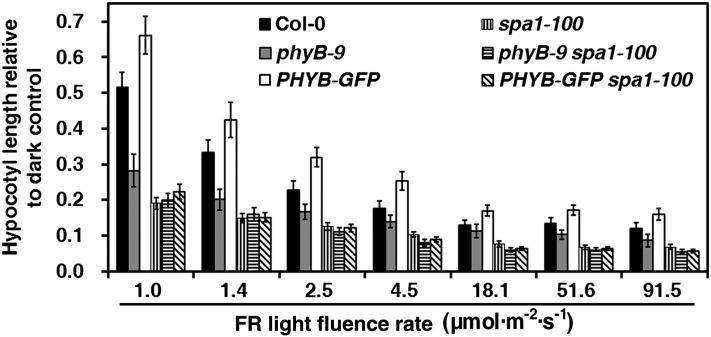
The negative effect of SPA1 was previously shown to occur early in the phyA-specific signaling pathway (Hoecker et al., 1998), which forms the E3 complex with COP1 to mediate the degradation of various photomorphogenesis-promoting factors (Saijo et al., 2003; Seo et al., 2003; Yang et al., 2005a, 2005b). Another study showed that the spa1-2 phyB-1 double mutant and the spa1-2 single mutant (Rsch-0/RLD [RLD] ecotype) exhibit similar hypocotyl elongation under FRc light (Baumgardt et al., 2002). To confirm the epistatic relationship between SPA1 and phyB in FR light signaling, we introduced phyB-9 and the PHYB-GFP transgenic line into the spa1-100 mutant via genetic crosses, constructing phyB-9 spa1-100 and PHYB-GFP spa1-100 double mutants (Figure 4). Under different FR intensities, spa1-100 completely suppressed the deetiolation and etiolation phenotypes caused by phyB-9 and PHYB-GFP, suggesting not only that phyB acts upstream of SPA1 but also that its functions are dependent on SPA1 under FR light.
Figure 4.
Epistatic Analysis of PhyB with SPA1 under FR Light.
Histograms comparing hypocotyl lengths relative to the dark control (hypocotyl length/hypocotyl length in the dark) (average of 60 seedlings) of Col-0, phyB-9, PHYB-GFP, spa1-100, phyB-9 spa1-100, and PHYB-GFP spa1-100 seedlings. Seedlings were grown under FRc light of various fluence rates for 4 d. Error bars indicate the sd.
PhyB Promotes SPA1 Accumulation under FR Light
Overexpression of Arabidopsis SPA1 (Myc-SPA1, driven by the constitutive 35S promoter in the Col-0 background) results in a hyperetiolation phenotype under R, FR, and B light conditions (Yang and Wang, 2006). To evaluate the effects of phyA and phyB on SPA1 function, we introduced the Myc-SPA1 transgene into the phyA-211 and phyB-9 single mutants and phyA-211 phyB-9 double mutant to generate double or triple homozygous plants. Overexpression of Myc-SPA1 leads to a cotyledon size much smaller than that of the Col-0 wild-type (Yang and Wang, 2006). In this study, the cotyledon size of the phyB-9 mutant was ∼1.89-fold that of Col-0 under FR illumination. In addition, the phyB-9 mutant rescued the small cotyledon size caused by Myc-SPA1 overexpression (Figure 5A). The phyA-211 Myc-SPA1 double mutant showed greater hypocotyl elongation than Myc-SPA1 (the parental line) under FR light; its hypocotyl length was 1.32-fold that of the parental line (Figure 5B). The hypocotyls of phyB-9 Myc-SPA1 double mutant seedlings were ∼30% shorter than those of the parental Myc-SPA1 line under high-FR light. Hypocotyl elongation of the phyA-211 phyB-9 Myc-SPA1 triple mutant resembled that of the phyA-211 phyB-9 double mutant, with visibly shorter hypocotyls compared with the phyA-211 single mutant. Taken together, these results suggest that both phyA and phyB are necessary for the proper functioning of SPA1 under FR light.
Figure 5.
PhyB Enhances SPA1 Accumulation under FR Light.
Seedlings were grown in the dark (Dk) or under FRc light for 4 d. The fluence rate of the FR light was 18.1 µmol·m–2·s–1, unless otherwise indicated.
(A) The phyB-9 mutant rescued the small cotyledon size caused by overexpression of Myc-SPA1 (parental line, in Col-0 background; Yang and Wang, 2006). Seedlings were grown under FRc (2.5 µmol·m–2·s–1) for 4 d. Numbers indicate the cotyledon area and numbers in parentheses represent sd. Bar = 1 mm.
(B) Histograms comparing hypocotyl lengths of the wild type (Col-0), phyA-211, phyB-9, phyA-211 phyB-9, Myc-SPA1, phyA-211 Myc-SPA1, phyB-9 Myc-SPA1, and phyA-211 phyB-9 Myc-SPA1 (average of 60) seedlings in the dark or under FR light. Error bars indicate the sd.
(C) Immunoblot and RT-PCR analyses showing that the Myc-SPA1 transgenic line accumulated different protein levels, with comparable transcript abundance, in the dark or under FRc (2.5 µmol·m–2·s–1), R light (30.0 µmol·m–2·s–1), B light (5.0 µmol·m–2·s–1), and W light (100.0 µmol·m–2·s–1). For immunoblot analysis, an anti-RPN6 (a 26S proteasome subunit) immunoblot is shown at the bottom to indicate approximately equal protein loading. For RT-PCR analysis, amplification of the ACT2 gene is shown below as a positive control.
(D) RT-PCR analysis of Myc-SPA1 transcript levels in seedlings of Myc-SPA1, phyA-211 Myc-SPA1, phyB-9 Myc-SPA1, and phyA-211 phyB-9 Myc-SPA1 grown under FR light for 4 d. RT-PCR of the ACT2 gene is shown at the bottom as a positive control.
(E) Quantification of relative Myc-SPA1/HSP90 protein levels from Myc-SPA1, phyA-211 Myc-SPA1, phyB-9 Myc-SPA1, and phyA-211 phyB-9 Myc-SPA1 corresponding to Supplemental Figure 4 online, showing that phyB enhanced Myc-SPA1 protein accumulation in the presence and absence of phyA under FR light. Error bars represent sd from triplicate experiments. Asterisks mark significant differences in relative Myc-SPA1/HSP90 protein levels from that in Myc-SPA1 according to Student’s t test (*P < 0.05; **P < 0.01).
To investigate the role of light regulation in SPA1 activities, we determined Myc-SPA1 protein accumulation in the dark and under FR, R, B, and W light conditions. Figure 5C demonstrates that SPA1 protein levels, but not transcript abundance, were tightly controlled in the dark and under FR, R, B, and W light and that all light conditions led to greater Myc-SPA1 accumulation. Elevated SPA1 abundance has been reported under FRc and Rc light conditions (Fittinghoff et al., 2006; Saijo et al., 2008). To determine whether phyA and phyB are involved in light-regulated SPA1 accumulation, Myc-SPA1 protein levels were compared in wild-type, phyA-211, phyB-9, and phyA-211 phyB-9 backgrounds under FR light. Myc-SPA1 transcript levels were comparable in the Col-0 wild type and phyA-211, phyB-9, and phyA-211 phyB-9 mutants under FR light (Figure 5D). Under FR light, Myc-SPA1 protein accumulation in the phyA-211 Myc-SPA1 or phyB-9 Myc-SPA1 double mutant seedlings was 170 or 36%, respectively, of that in seedlings of the parental line (Figure 5E; see Supplemental Figure 4 online). The Myc-SPA1 protein level in the phyA-211 phyB-9 double mutant background was intermediate to those in the phyA-211 and phyB-9 mutant backgrounds. Notably, Myc-SPA1 protein levels in the wild-type, phyA-211, phyB-9, and phyA-211 phyB-9 backgrounds were entirely consistent with their hypocotyl elongation under FR light. These data imply that both phyA and phyB play a pivotal role in the regulation of SPA1 abundance and that phyB enhances SPA1 accumulation in both the presence and absence of phyA.
PhyB Interacts with SPA1 in Vitro and in Vivo
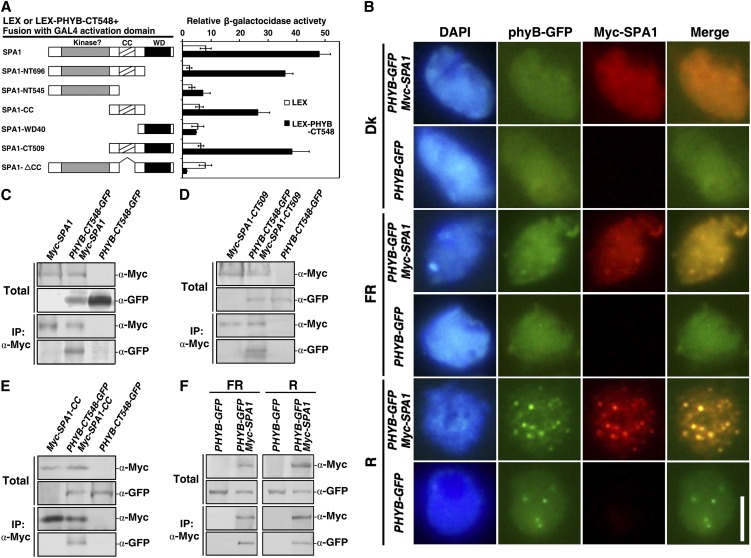
Since phyB was required for SPA1 activity under FR light, we determined whether a direct interaction between phyB and SPA1 was involved in their functional regulation. We first used yeast two-hybrid assays to determine their interaction and the domains responsible. PhyB-FL (amino acids 1 to 1172) alone activated reporter gene expression in our yeast two-hybrid system (data not shown), so phyB-CT548 (amino acids 626 to 1172) was used to test the interaction between phyB and SPA1. The full-length SPA1 protein as well as deletion derivatives, including SPA1-NT696, SPA1-CC, and SPA1-CT509, were capable of interacting with phyB-CT548. However, SPA1-NT545, SPA1-WD40, and SPA1-ΔCC were unable to interact with phyB-CT548 (Figure 6A). PhyB-NT651 (amino acids 1 to 651) and phyB-NT450 (amino acids 1 to 450) failed to interact with SPA1-FL or its deletion derivatives (data not shown). These observations suggest that the C-terminal domain of SPA1, including both coiled-coil and WD40 domains, is required for the phyB–SPA1 interaction.
Figure 6.
PhyB Interacts with SPA1 in Vitro and in Vivo.
The fluence rates of the FR and R light were 2.5 and 30.0 µmol·m–2·s–1, respectively.
(A) The phyB–SPA1 interaction was analyzed by yeast two-hybrid assay. The panel on the left illustrates the prey and bait constructs. SPA1-FL, amino acids 1 to 1029; SPA1-NT696, amino acids 1 to 696; SPA1-NT545, amino acids 1 to 545; SPA1-CC (coiled-coil domain), amino acids 521 to 696; SPA1-WD40 (WD40 repeat domain), amino acids 647 to 1029; SPA1-CT509, amino acids 521 to 1029; SPA1-ΔCC (which lacks the coiled-coil domain), amino acids 1 to 545 and 647 to 1029; phyB-CT548, amino acids 626 to 1172. The panel on the right shows the corresponding β-galactosidase activities. Values represent the mean of six individual yeast colonies, and error bars represent the sd.
(B) Colocalization of phyB and SPA1 in the nucleus of Arabidopsis cells. Nuclei were isolated from seedlings of PHYB-GFP or PHYB-GFP Myc-SPA1 grown in the dark (Dk), FR light, or R light for 4 d. PhyB-GFP fluorescence was examined as in Figure 2A. Samples were probed with anti-Myc (mouse monoclonal IgG) followed by TRITC-conjugated anti-mouse IgG (R). Images of the same cell from separate color channels were merged using Photoshop (Adobe; Merge). Bar = 5 μm.
(C) In vivo co-IP of phyB-CT548 by Myc-SPA1. N. benthamiana plants were coinfiltrated with p19 and either or both of 35S-driven Myc-SPA1 and PHYB-CT548-GFP in EHA105 and then transferred to FR light for 3 d; leaf extracts were incubated with anti-Myc–conjugated agarose under FR light. The pellet was analyzed by immunoblotting with anti-Myc and anti-GFP antibodies.
(D) and (E) In vivo co-IP of phyB-CT548 by Myc-SPA1-CT509 (D) or Myc-SPA1-CC (E). Sample preparation and co-IP were performed as in (C).
(F) In vivo co-IP of phyB-GFP by Myc-SPA1 under FR or R light. Native protein extracts were prepared from PHYB-GFP or PHYB-GFP Myc-SPA1 seedlings grown in FR light or R light for 4 d and incubated with anti-Myc–conjugated agarose under FR or R light. Co-IP was performed as in (C).
Next, we employed an immunofluorescence assay using a PHYB-GFP Myc-SPA1 double-transgenic line to determine whether phyB and SPA1 colocalized in the nuclei of Arabidopsis cells. After growth in the dark or under R or FR light for 5 d, the PHYB-GFP Myc-SPA1 seedlings were harvested and fixed, and the GFP fluorescence of phyB-GFP or Myc immunofluorescence of Myc-SPA1 was observed. In the dark, the phyB-GFP and Myc-SPA1 proteins colocalized in the nucleus with diffuse fluorescence (Figure 6B, yellow nuclear fluorescence). Under R light, they colocalized in large and bright NBs (yellow dots). By contrast, phyB-GFP and Myc-SPA1 colocalized in both NBs and exhibited diffuse fluorescence under FR conditions (small yellow dots and yellow nuclear fluorescence). Although the phyB-GFP and Myc-SPA1 proteins colocalized in the nucleus under all light conditions, their different fluorescence patterns, diffuse fluorescence, small NBs, and large NBs imply that they exist as various active forms in the dark and under FR and R light conditions.
To determine whether phyB and SPA1 colocalize in the same protein complex, we conducted in vivo coimmunoprecipitation (co-IP) assays (Liu et al., 2010). Agrobacterium tumefaciens strains EHA105 carrying pCAMBIA1302-PHYB-CT548-GFP, 35S:p19 (Liu et al., 2010), and pJIM19-Myc-SPA1 (Yang and Wang, 2006) constructs were used to infect Nicotiana benthamiana leaves. The plants were grown under FR light for 3 d, after which native protein was extracted from the leaves and subjected to co-IP using antibodies against the Myc epitope. Myc-tagged full-length SPA1, SPA1-CT509, or SPA1-CC proteins copurified with GFP-tagged phyB-CT548 (Figures 6C to 6E). Neither SPA1-NT545 nor SPA1-WD40 copurified with phyB-CT548 (data not shown). To further investigate whether the colocalization of phyB and SPA1 in the same protein complex is light dependent, we compared the interaction between SPA1 and phyB by means of in vivo co-IP assays using PHYB-GFP transgenic line and PHYB-GFP Myc-SPA1 double transgenic line seedlings grown under FR or R light. The results suggested that phyB-GFP interacted with Myc-SPA1 under both FR and R light (Figure 6F). Taken together, these findings indicate not only that phyB and SPA1 colocalize in the same protein complex under FR light, but also that under FR light, the interaction between phyB and SPA1 is important for their activities.
PhyB Enhances Nuclear Transport of SPA1 during the Transition from Dark to FR Light
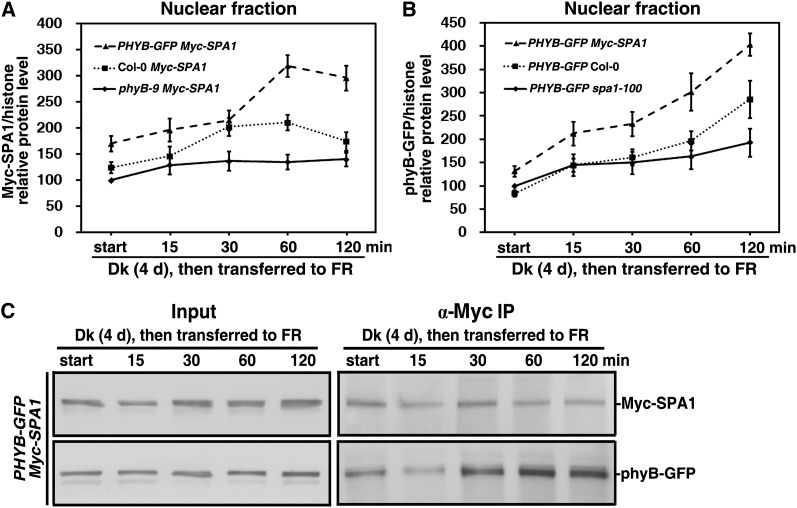
FR exposure effectively promoted nuclear accumulation of phyB (Figure 2), and phyB also enhanced SPA1 accumulation under FR light (Figure 5E; see Supplemental Figure 4 online). To test whether phyB directly promotes SPA1 nuclear transport in response to FR light, we compared the effect of different phyB levels, phyB-9, the wild type, and PHYB-GFP on nuclear transport of Myc-SPA1 during the transition from dark to FR light. In phyB-9 Myc-SPA1 double mutant seedlings, nuclear transport of Myc-SPA1 increased by 29 to 40% over that of dark-grown seedlings (Figure 7A; see Supplemental Figure 5A online). Relative nuclear Myc-SPA1 protein levels in Col-0 Myc-SPA1 seedlings were 124, 146, 202, 211, and 175% that of dark-grown phyB-9 Myc-SPA1 seedlings during the dark to FR light transition for 0, 15, 30, 60, and 120 min, respectively. Over the same time course, relative nuclear Myc-SPA1 protein levels in PHYB-GFP Myc-SPA1 seedlings were 170, 196, 215, 319, and 296% compared with those of dark-grown phyB-9 Myc-SPA1. Therefore, phyB facilitated the nuclear transport of SPA1 during the transition from dark to FR light. SPA1, which contains two putative nuclear localization sequences, strongly localizes to the nucleus in both dark- and light-treated cells (Hoecker et al., 1999). Our data demonstrated that SPA1 was involved in feedback regulation of nuclear phyB accumulation under FR light and that phyB and SPA1 displayed nuclear cotransport during the transition from dark to FR light (Figure 7B; see Supplemental Text1 and Supplemental Figure 5B online).
Figure 7.
PhyB and SPA1 Coordinately Promote Nuclear Transport of Each Other during the Transition from Dark to FR.
Seedlings were grown in the dark (Dk) for 4 d and subsequently transferred to FR light for 15, 30, 60, or 120 min. The fluence rate of the FR light was 18.1 µmol·m–2·s–1. All lines or double mutants, including Myc-SPA1, PHYB-GFP, phyB-9 Myc-SPA1, PHYB-GFP Myc-SPA1, and PHYB-GFP spa1-100, were of the Col-0 ecotype.
(A) Quantification of relative Myc-SPA1/histone protein levels corresponding to Supplemental Figure 5A online, showing that nuclear accumulation of Myc-SPA1 increased with phyB levels during the transition from dark (Dk) to FR. Error bars represent the sd from triplicate experiments.
(B) Quantification of relative phyB-GFP/histone protein levels corresponding to Supplemental Figure 5B online, showing that nuclear accumulation of phyB-GFP increased with SPA1 levels during the transition from dark to FR. Error bars represent the sd from triplicate experiments.
(C) In vivo co-IP of phyB-GFP by Myc-SPA1 showing that the strength of Myc-SPA1 and phyB-GFP binding was enhanced during the transition from dark to FR. Native protein extracts were prepared from PHYB-GFP Myc-SPA1 seedlings harvested at each time point during the transition from dark to FR and incubated with anti-Myc–conjugated agarose in the dark. Co-IP was performed as in Figure 6D.
To determine whether synchronous nuclear accumulation of both phyB and SPA1 is due to their direct interaction, we tested their binding during the dark to FR transition using an in vivo co-IP assay. At a comparable level of Myc-SPA1, the phyB-GFP levels evidently increased during the dark-to-FR transition compared with the dark control (Figure 7C). Note that nuclear accumulation of Myc-SPA1 was related to the amount bound to phyB-GFP during the dark-to-FR transition. Thus, we conclude that phyB promoted nuclear accumulation of SPA1 through their direct interaction in response to FR light.
SPA1 Is Required for COP1 Nuclear Accumulation under FR Light
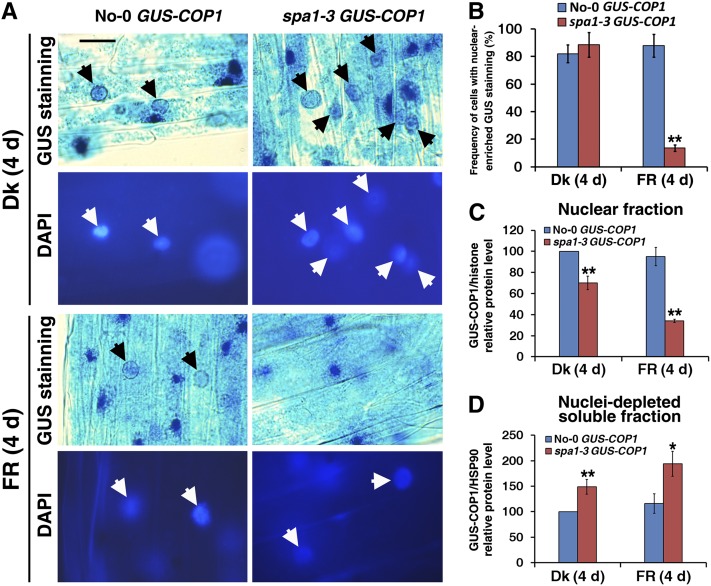
Arabidopsis COP1 acts as a repressor of seedling photomorphogenic development within the nucleus, and light inactivation of COP1 is accomplished by a reduction in its nuclear abundance (von Arnim and Deng, 1994; Subramanian et al., 2004). COP1 and SPA1 are functionally interdependent, and SPA1 enhances COP1 activation in the dark and under FR light (Saijo et al., 2003; Yang and Wang, 2006). The enhanced etiolation phenotype conferred by a 35S promoter–driven GUS-COP1 transgene is largely suppressed by the spa1-3 mutation under FR light, but not in darkness (see Supplemental Figure 6A online; see also Yang and Wang, 2006). To determine whether SPA1 is involved in the light-regulated nuclear accumulation of COP1, we compared the nuclear abundance of GUS-COP1 in the parental line (No-0 wild-type background; von Arnim and Deng, 1994) and GUS-COP1 in the spa1-3 mutant background both in the dark and under FRc light. Histochemical staining revealed that GUS-COP1 was similarly enriched in the nuclei of hypocotyl cells in dark-grown seedlings of the parental line and in the spa1-3 mutant background (Figure 8A). However, after an overnight incubation in GUS staining solution, nuclear GUS-COP1 was clearly visible in the FR light–grown seedlings of the parental line but was barely detectable in seedlings with the spa1-3 mutant background (Figure 8A; see Supplemental Figure 6B online). Under FR light, the cell frequency with nuclear-enriched GUS staining in the No-0 wild-type seedlings was 6.4-fold higher than in the spa1-3 mutant seedlings, whereas no clear differences in the rate of nuclear-enriched GUS staining were observed between the seedlings of No-0 GUS-COP1 and spa1-3 GUS-COP1 in the dark (Figure 8B). Immunoblot analysis showed that the nuclear GUS-COP1 protein level in the No-0 wild-type background was ∼1.4- or 2.8-fold higher than in the spa1-3 mutant background in the dark or under FR light (Figure 8C; see Supplemental Figure 6C online). These results matched the hypocotyl elongation and GUS nuclear-enriched staining results of No-0 GUS-COP1 and spa1-3 GUS-COP1 under FR light. The apparent reduction in the nuclear abundance of GUS-COP1 was not due to reduced GUS-COP1 protein levels in the spa1-3 mutant background, as spa1-3 mutant seedlings showed stronger overall histochemical staining for GUS-COP1 and accumulated much more cytoplasmic GUS-COP1 protein than the parental plants (Figure 8D; see Supplemental Figures 6B and 6D online). Notably, GUS-COP1 transcript levels were comparable in the No-0 wild-type and spa1-3 mutant backgrounds both in darkness and under FR light (see Supplemental Figure 6E online). These observations suggest that nuclear accumulation of GUS-COP1 was impaired in the spa1-3 mutant background under FR light with a concomitant increase in its cytoplasmic accumulation.
Figure 8.
Nuclear Accumulation of COP1 Is Defective in the spa1-3 Mutant under FR Light.
Seedlings were grown in the dark (Dk) or FR light (2.5 µmol·m–2·s–1) for 4 d.
(A) Nuclear accumulation of GUS-COP1 in the parental No-0 GUS-COP1 plants (left panels, indicated by black arrows) but not in the spa1-3 GUS-COP1 plants (right panels) under FR light. The top panels present GUS-stained images and the bottom panels display DAPI staining of the corresponding image to show the positions of the nuclei (white arrows). The unmarked blue GUS staining dots are GUS-COP1 cytoplasmic inclusion bodies. Bar = 20 μm.
(B) Difference in frequency of cells with nuclear-enriched GUS staining in No-0 GUS-COP1 and spa1-3 GUS-COP1 in the dark or under FR light.
(C) Quantification of relative GUS-COP1/histone protein levels in the purified nuclear fraction corresponding to Supplemental Figure 6C online, indicating reduced nuclear abundance of GUS-COP1 in the spa1-3 mutant background. Error bars represent sd from triplicate experiments. Asterisks mark significant differences in relative GUS-COP1/histone protein levels from No-0 GUS-COP1 according to Student’s t test (*P < 0.05; **P < 0.01).
(D) Quantification of relative GUS-COP1/HSP90 protein levels in the nuclei-depleted soluble fraction in response to FR light, corresponding to Supplemental Figure 6D online. Error bars represent sd from triplicate experiments. Student’s t tests were performed as in (C) (see Supplemental Figure 6 online).
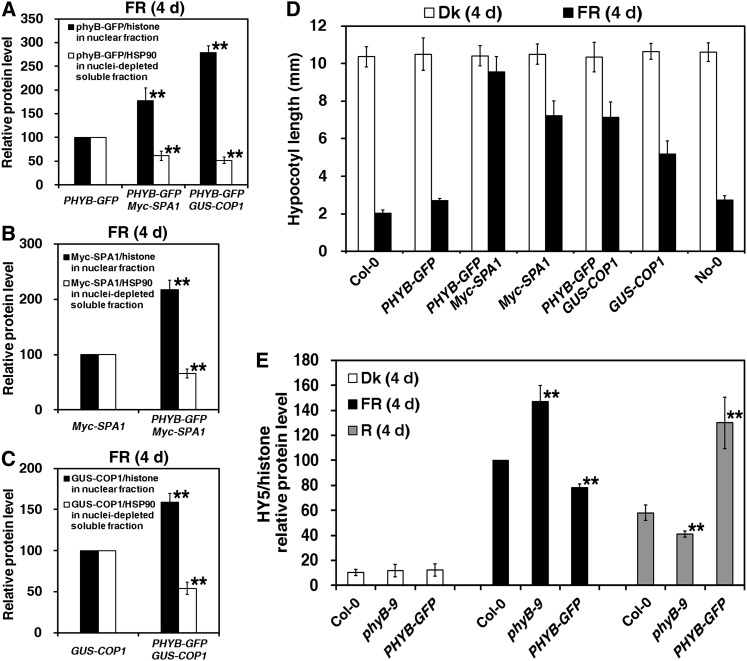
PhyB Is Necessary for the Nuclear Accumulation of Both SPA1 and COP1 under FR Light
After demonstrating the functional deficiency of SPA1 in the phyB-9 mutant in response to FR light (Figures 5B, 5E, and 7A), we tested whether PHYB overexpression enhanced the nuclear abundance of both SPA1 and COP1. Under FR light, seedlings of the PHYB-GFP Myc-SPA1 and PHYB-GFP GUS-COP1 double-transgenic lines accumulated ∼1.8- and 2.8-fold more nuclear phyB-GFP protein, respectively, compared with seedlings of their parental line (PHYB-GFP) (Figure 9A; see Supplemental Figure 7A online). The nuclear level of Myc-SPA1 in seedlings of the PHYB-GFP Myc-SPA1 double-transgenic line was more than two times higher than that in its parental line (Myc-SPA1) (Figure 9B; see Supplemental Figure 7B online). Additionally, the nuclear protein level of GUS-COP1 in seedlings of the PHYB-GFP GUS-COP1 double transgenic line was ∼59% higher than its parental line (GUS-COP1) (Figure 9C; see Supplemental Figure 7C online).
Figure 9.
Nuclear Accumulations of Both SPA1 and COP1 Are Enhanced in PHYB-GFP Background under FR Light.
Seedlings were grown in the dark (Dk), under FR light (18.1 µmol·m–2·s–1), or under R light (30.0 µmol·m–2·s–1) for 4 d.
(A) Quantification of relative phyB-GFP/histone protein levels in the purified nuclear fraction and relative phyB-GFP/HSP90 protein levels in the nuclei-depleted soluble fraction corresponding to Supplemental Figure 7A online, indicating that nuclear accumulation of phyB-GFP was enhanced in the transgenic Myc-SPA1 and GUS-COP1 backgrounds. Error bars represent sd from triplicate experiments. Asterisks mark significant differences in relative protein levels of phyB-GFP/histone in the nuclear fraction or phyB-GFP/HSP90 in the nuclei-depleted soluble fraction in the double transgenic lines of PHYB-GFP Myc-SPA1 and PHYB-GFP GUS-COP1 from PHYB-GFP (parental line) according to Student’s t test (**P < 0.01).
(B) and (C) Quantification of relative Myc-SPA1 (B) and GUS-COP1 (C) protein levels corresponding to Supplemental Figures 7B and 7C online, indicating enhanced nuclear accumulation of Myc-SPA1 and GUS-COP1 in the transgenic PHYB-GFP background. Error bars represent sd from triplicate experiments. Student’s t tests were performed as in (A).
(D) Histograms showing greater hypocotyl elongation of the PHYB-GFP Myc-SPA1 and PHYB-GFP GUS-COP1 double transgenic lines than in their parental lines (Myc-SPA1 and No-0 GUS-COP1, respectively) (at least 60 seedlings) in the dark or under FR light. Error bars indicate sd.
(E) Quantification of relative HY5/histone protein levels in purified nuclear fractions from Col-0, phyB-9, and PHYB-GFP seedlings grown in the dark or under FR or R light for 4 d, corresponding to Supplemental Figure 7E online. Error bars represent sd from triplicate experiments. Student’s t tests were performed as in (A) (see Supplemental Figure 7 online).
Although the hypocotyl lengths of Myc-SPA1 and No-0 GUS-COP1 transgenic plants were 5.2- and 1.9-times greater, respectively, than those of their wild types under FR light, the hypocotyl lengths of the PHYB-GFP Myc-SPA1 and PHYB-GFP GUS-COP1 double mutants remained 32 and 38% higher, respectively, than those of their parental lines (Myc-SPA1 and No-0 GUS-COP1) (Figure 9D). Additionally, phyB enhanced nuclear protein accumulation and hypocotyl elongation caused by Myc-SPA1 overexpression, even in the absence of additional sugar under FR light (see Supplemental Figures 8A and 8B online). Therefore, phyB promotes seedling etiolation through enhancement of nuclear abundance of the COP1-SPA1 E3 ligase complex under FR light.
Previous studies have shown that SPA1 forms an E3 complex with COP1 to mediate degradation of nuclear HY5, a basic domain/leucine zipper transcription factor that acts downstream of light signaling to promote photomorphogenesis (Osterlund et al., 2000; Saijo et al., 2003; Yang and Wang, 2006). Thus, HY5 abundance is correlated with not only the extent of photomorphogenic development but also the E3 activity of the COP1-SPA1 complex (Osterlund et al., 2000; Saijo et al., 2003). In the dark, all Col-0, phyB-9, and PHYB-GFP seedlings accumulated only almost undetectable amounts of nuclear HY5 protein (Figure 9E; see Supplemental Figure 7E online). Under Rc light, phyB promotes seedling deetiolation; HY5 abundance in the nuclei of phyB-9 mutant and PHYB-GFP transgenic line seedlings was 71 and 224%, respectively, that of wild-type seedlings. Conversely, nuclear HY5 levels in phyB-9 mutant and PHYB-GFP transgenic line seedlings were 147 and 78%, respectively, compared with those of wild-type seedlings under FRc light. Therefore, we conclude that phyB enhances seedling etiolation under FR light via promoting E3 activity of the COP1-SPA1 complex in the nucleus.
DISCUSSION
The Low Pfr:Ptot Ratio of PhyB Is Responsible for Repression of FR-HIRs
Typical FR-HIRs allow seedlings to inhibit hypocotyl elongation to expand cotyledons and to accumulate anthocyanin under FRc light (Bae and Choi, 2008; Li et al., 2011). Etiolation caused by overexpression of PHYB under FRc light was first noted by McCormac et al. (1993) and Wagner et al. (1996) and has since been explored further (Short, 1999; Casal et al., 2000; Hennig et al., 2001). Seedlings overexpressing PHYB-GFP have drastic etiolation phenotypes, with elongated hypocotyls and reduced anthocyanin accumulation under FRc light (Figures 1A to 1D; McCormac et al., 1993; Wagner et al., 1996; Short, 1999; Casal et al., 2000; Hennig et al., 2001). Conversely, the PHYB-deficient phyB-9 mutant displayed a shorter hypocotyl, increased anthocyanin accumulation (Figures 1A to 1D), greater cotyledon unfolding (Figure 3B) and larger cotyledon size (Figure 5A), and markedly increased HY5 accumulation under FRc light compared with those in the wild type (Figure 9E) (Hennig et al., 2001). These results suggest that phyB plays a repressive role in the FR-HIRs (Figure 10; Casal et al., 2000; Hennig et al., 2001).
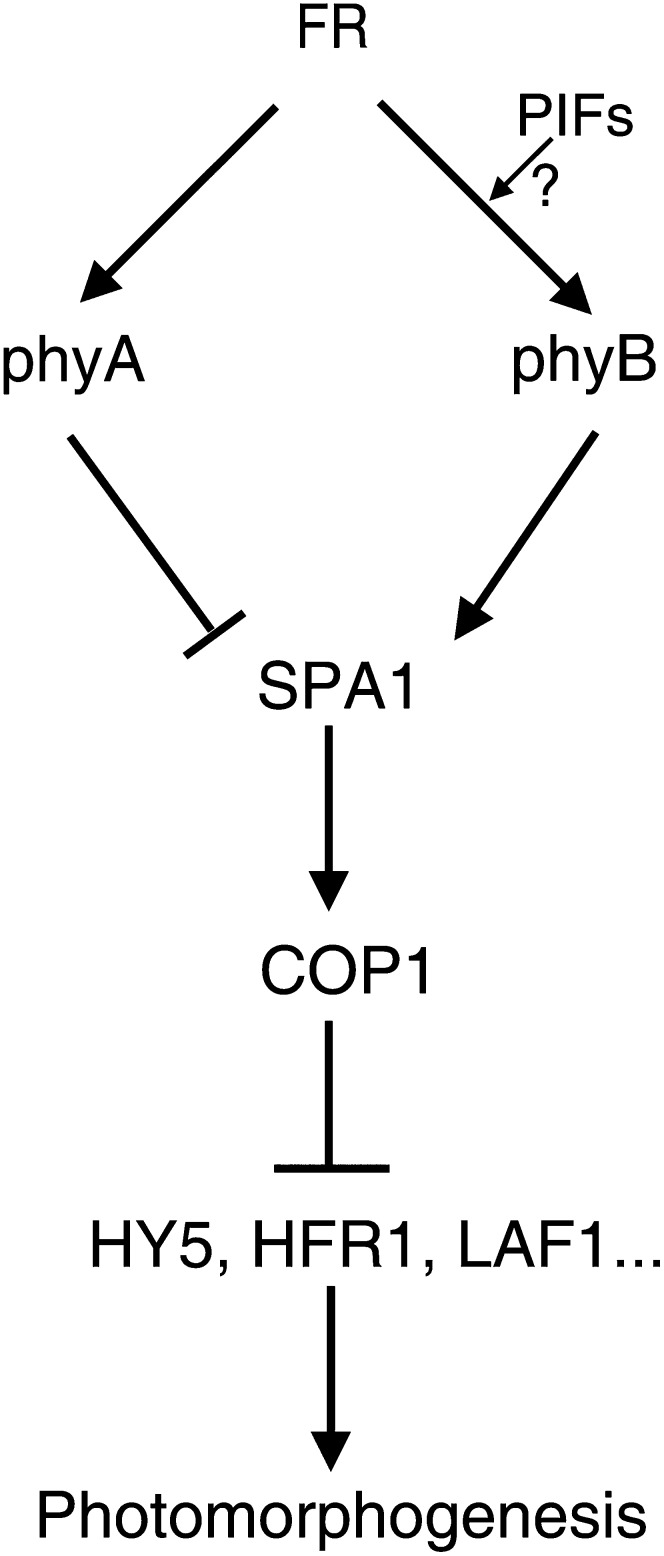
Figure 10.
Model of the Effect of PhyB and PhyA on the Nuclear Abundance of SPA1 under FR Light.
FR light irradiation not only activates nuclear import of phyB but also enhances its SPA1 binding strength to promote nuclear accumulation of SPA1. COP1 and SPA1 are functionally interdependent, and SPA1 maintains proper COP1 activation in the nucleus to promote the degradation of HY5, HFR1, LAF1, and others, resulting in seedling etiolation responses under FR light. PhyB may antagonize the phyA effect in terms of the nuclear abundance of SPA1 in response to FR light (see Supplemental Figure 9 online).
The Pfr form of phytochrome is considered the biologically active form, whereas the Pr form is biologically inactive (Quail, 2002; Bae and Choi, 2008; Li et al., 2011). The Pfr:Ptot ratio of phyA (the active Pfr form in the total phyA protein) is ∼40-fold lower under FR light than R light; however, phyA nuclear localization and phyA-mediated responses are triggered most efficiently by FR light (Rausenberger et al., 2011). Overexpression of the phyB C357S mutation, which prevents incorporation of the chromophore, had no effects on hypocotyl elongation under either R or FR light; these results suggest that photoactivation is necessary for phyB inhibition of FR-HIRs (Wagner et al., 1996; Hennig et al., 2001). Large NBs of phyB-GFP are observed when a high percentage of phyB is in the active Pfr form under R light, and a low Pfr:Ptot ratio of phyB-GFP, caused by low fluence rate of R or R:FR light schemes, is associated with smaller and fewer NBs, and even diffuse nuclear localization (Chen et al., 2003). Although we observed a high level of phyB nuclear accumulation under FRc light (Figures 2C and 7B; see Supplemental Figure 3F online), the NBs of phyB-GFP were small and dim (Figures 2A and 6B), implying that the phyB protein under FR light involves a low ratio of Pfr:Ptot rather than a high ratio.
PHYB overexpression leads to steady etiolation phenotypes under all FR intensities (Figure 4; Casal et al., 2000), whereas a low Pfr: Ptot ratio of the PHYB-GFP transgenic line under low R light does not cause a dominant-negative phenotype as with FR irradiation (Wagner and Quail, 1995; Wagner et al., 1996; Chen et al., 2003; Matsushita et al., 2003). Varying R-FR dichromatic irradiation yields three categories of Pfr:Ptot ratios of phyB: >0.3, 0.2 to 0.3, and <0.2, corresponding to enhancement, no effect, or repression of seedling deetiolation compared with the wild type, respectively (Hennig et al., 2001). These results suggest that the low Pfr: Ptot ratio of phyB caused by only FR irradiation is necessary to trigger the repression of seedling deetiolation.
Several reports suggested that Suc and light follow separate, but linked, signaling pathways (Cheng et al., 1992; Barnes et al., 1996; Dijkwel et al., 1997; Thum et al., 2003). Repression of FR-HIRs by phyB has been demonstrated to be dependent on not only the FR fluence rate but also on the availability of metabolizable sugars (see Supplemental Figures 2C and 2D online; Short, 1999). Although phyB promotion of hypocotyl elongation and SPA1 protein accumulation occur even in the absence of additional sugar (see Supplemental Figures 2A, 8A, and 8B online; Hennig et al., 2001), the presence of sugar observably interferes with the phyB inhibitory effect on seedling photomorphogenesis under FRc light (see Supplemental Figure 2C online; Short, 1999). Further studies are needed to address the interactions of carbon with light-signaling pathways.
PhyB Enhances SPA1 Accumulation to Repress FR Light Signaling in Both the Presence and Absence of PhyA
In response to FR light, phyB-GFP forms small, dim NBs and effectively accumulates in the nucleus, and the nuclear import of phyB remains stable in the phyA-211 mutant background (Figures 2A to 2C). Promotion of hypocotyl elongation, cotyledon folding, and chlorophyll accumulation by phyB in both the presence and absence of phyA suggests that the repressive effects of phyB on seedling deetiolation under FR light do not require phyA (Figures 3A to 3C). Compared with the phyA-211 single mutant, the shorter hypocotyl seen in the phyA-211 phyB-9 double mutant implies that phyA plays the dominant role under FR light and that the antagonistic effect of phyA and phyB on FR signaling does not directly affect the abundance of either protein. As PHYB overexpression has no apparent effect on the abundance or degradation of phyA under FR light, phyA and phyB may interact with a common partner (Wagner et al., 1996; Short, 1999; Casal et al., 2000). Our results demonstrated that phyB promotes SPA1 accumulation in both the presence and absence of phyA (i.e., phyB functions at the same level as phyA in the regulation of SPA1 accumulation) (Figures 5B and 5E). In fact, nuclear import of phyB-GFP or endogenous phyB in the phyA-211 mutant is higher than that in the wild-type background (Figure 2C; see Supplemental Figure 3F online). The increased SPA1 nuclear accumulation in the phyA-211 mutant (Figure 5E) may facilitate nuclear import of phyB (Figures 7B and 7C).
Phytochrome-interacting factors (PIFs) and PIF-like factors bind the Pfr form of phyB and/or phyA and function as negative regulators in the phyA and/or phyB signaling cascades (Casal et al., 2000; Zhu et al., 2000; Kim et al., 2003; Bauer et al., 2004; Oh et al., 2004; Park et al., 2004; Leivar et al., 2008; Lorrain et al., 2009). Both PIF4 and PIF5 were first identified as specific negative regulators of phyB-mediated R light signaling (Huq and Quail, 2002; Shen et al., 2007; Leivar et al., 2008; Jang et al., 2010). Recently, the pif4 pif5 double loss-of-function mutant has been shown to be hypersensitive to low FR fluence rates, without altering phyA concentration (Lorrain et al., 2009). PIF3 facilitates the nuclear import of phyB under R and FR light irradiation (Pfeiffer et al., 2012). Thus, PIFs may be involved in phyB-mediated repression of seedling deetiolation under FR light (Figure 10).
PhyB and SPA1 Coordinately Promote Nuclear Transport and Accumulation of Each Other in Response to FR Light
Phytochromes are believed to be synthesized in the cytoplasm in the dark and to rapidly translocate into the nucleus to initiate signaling events upon irradiation (Reed, 1999; Quail, 2002; Bae and Choi, 2008). The nuclear transport of phyA, but not phyB, is regulated by the transport facilitators FR ELONGATED HYPOCOTYLS1 (FHY1) and FHY1-LIKE (Hiltbrunner et al., 2005, 2006; Genoud et al., 2008). PhyB-GFP fluorescence (or GUS-phyB staining) in dark-grown seedlings was very weak (Figures 2A and 6B) (Sakamoto and Nagatani, 1996; Kircher et al., 1999); however, darkness does not completely block the nuclear import kinetics of phyB (see Supplemental Figures 3B, 3C, and 3F online). Indeed, phyB-GFP exhibits weak fluorescence in dark-grown seedlings (Figures 2A and 6B) (Yamaguchi et al., 1999; Matsushita et al., 2003; Oka et al., 2004; Hiltbrunner et al., 2006). The phyB-9 mutant enhanced apical unhooking in both the wild-type and phyA-211 mutant backgrounds in the dark (Figure 3B), and phyB regulates gene expression even in dark-grown seedlings (Mazzella et al., 2005). This implies that phyB also functions in the dark; however, both photoactivation and nuclear import are necessary and sufficient for full phyB biological function (Huq et al., 2003).
FR illumination is believed to have little effect on phyB activities compared with R light (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Kircher et al., 2002; Bauer et al., 2004). FR exposure induced phyB-GFP to form very small, dim NBs (Figure 2A), and the transition from dark to FR light clearly promoted the nuclear import of phyB. However, evidence regarding kinetics of phyB nuclear import in the dark and during the transition from dark to FR light is lacking. The phyB C-terminal domain is responsible for its nuclear import and nuclear speckle formation, whereas the N-terminal domain is functional in the nucleus, with no speckle (Quail, 1997; Kircher et al., 1999; Yamaguchi et al., 1999; Usami et al., 2007). In a model of the intracellular action of phyB, the nuclear import activity of the C-terminal domain is blocked by the N-terminal domain in the dark; upon absorption of light, structural changes in these regions prevent this interaction, and the released C-terminal region induces the nuclear import of phytochromes (Chen et al., 2005). PhyB interacts and forms a protein complex with SPA1 under FR light (Figures 6A to 6F). Both phyB (Edgerton and Jones, 1992; Quail, 1997; Sharrock and Clack, 2004; Chen et al., 2005) and SPA1 (see Supplemental Figures 9A and 9B online; Zhu et al., 2008) form homodimers in plants, so we propose a heterotetramer model to elucidate the SPA1-phyB complex under FR light (see Supplemental Figure 9C online). First, SPA1 may activate phyB through competitive interaction with the phyB C-terminal domain, thus releasing it from repressive interaction with the N-terminal domain to promote phyB nuclear import (Figures 6A and 7B). Second, SPA1, which contains two putative nuclear localization sequences, strongly localizes to the nucleus in both dark- and light-treated cells (see Supplemental Figure 9C online; Hoecker et al., 1999). Third, the phyB and SPA1 proteins colocalized in the same nuclear complex under FR light (Figures 6B and 6F). Thus, phyB and SPA1 coordinately promote nuclear transport and accumulation of each other in response to FR light (Figures 7A and 7B).
Light-Activated Photoreceptors Directly Modulate the Activities of the COP1-SPA1 Complex
The in vivo co-IP assays of phyB-CT548 (lacking the N-terminal photosensory domain) and SPA1 imply that both phyB and SPA1 reside in the same protein complex in planta irrespective of light conditions (Figures 6C to 6E). However, the Pr form, low Pfr:Ptot ratio, and high Pfr:Ptot ratio of phyB influence the fluorescence patterns of phyB-SPA1 complex in the dark and under FR and R light, respectively (Figure 6B). Furthermore, FR light irradiation not only activates nuclear import of phyB (Figures 2A, 2C, and 7B) but also enhances its SPA1 binding strength (Figure 7C), resulting in accumulation of SPA1 in the nucleus under FR light (Figure 7A). COP1 and SPA1 are functionally interdependent, and SPA1 enhances COP1 activation in the dark and under FR, R, and B light conditions (Figures 8A to 8C; Saijo et al., 2003, 2008; Seo et al., 2003; Yang and Wang, 2006). Under FR light, phyB activates or stabilizes the COP1-SPA1 E3 ligase complex by promoting SPA1 nuclear activity (Figures 5E and 7A) to enhance the degradation of HY5, HFR1, LAF1, and others, resulting in seedling etiolation responses (Figure 9E) (Seo et al., 2003; Saijo et al., 2003; Yang and Wang, 2006). We hypothesize that SPA1 is rapidly regulated by the shift in the Pfr:Pr ratio of the phytochromes in response to environmental light changes, resulting in modulation of COP1-SPA1 E3 ligase complex activity (Figure 10).
Osterlund and Deng (1998) first noted that photoreceptors play critical roles in inhibiting nuclear localization of COP1. Compared with the wild-type background, overexpression of both PHYB and PHYA inhibits the nuclear localization of GUS-COP1, resulting in repression of the elongated hypocotyl phenotype by GUS-COP1 under Rc light (Osterlund and Deng, 1998). Through direct interaction, the C-terminal domain of either cryptochrome (CCT1 and CCT2) modifies COP1 structure, leading to its inactivation (Yang et al., 2000; Wang et al., 2001; Yi and Deng, 2005; Feng and Deng, 2007). Cry1 was recently confirmed to interact with SPA1 in a B light–dependent manner to promote dissociation of SPA1 from COP1 (Lian et al., 2011; Liu et al., 2011). In addition, the B light–dependent cry2–SPA1 interaction enhances that of cry2-COP1, suppressing COP1 activity (Zuo et al., 2011). Furthermore, our data confirm that phyB promotes SPA1 nuclear accumulation in response to FR light, enhancing COP1-SPA1 E3 ligase activity (Figures 7 to 9). Therefore, activity modulation of the COP1-SPA1 E3 ligase complex by light-activated photoreceptors through direct interactions is an effective and pivotal regulatory step in light signaling (Figure 10).
METHODS
Plant Materials and Plant Growth Conditions
The phyA-211 (Reed et al., 1994), phyB-9 (Reed et al., 1993; McNellis et al., 1994a), and spa1-100 (Yang et al., 2005b) mutants and Myc-SPA1 (line B1-17) transgenic lines (Yang and Wang, 2006) are of the Col-0 ecotype. PBG (Yamaguchi et al. 1999), GUS-COP1 (von Arnim and Deng 1994), and spa1-3 (Hoecker et al., 1998) are in the Ler ecotype, the No-0 ecotype, and the RLD ecotype background, respectively. Seeds were surface sterilized and spread on Murashige and Skoog agar plates (1× Murashige and Skoog salts, 1% Suc, and 0.9% agar; Sigma-Aldrich). Growth conditions for seedlings were as described previously (Yang et al., 2005a). FR and R light was supplied using light-emitting diode light sources (model E-30LEDL3; Percival Scientific), with irradiance fluence rates of ∼2.5 and 30.0 µmol·m–2·s–1, respectively, unless otherwise indicated (model ILT1400A with sensor model SEL-033/F/W; International Light Technologies). W light was supplied using cool-white fluorescent lamps. To erase postmaturation germination problems, seeds stored for 3 to 4 months at room temperature were used for seedling hypocotyl measurements.
Plasmid Construction
A full-length PHYB cDNA fragment was generated by RT-PCR using the primer pair PHYB-1NF (5′-accatgggcATGGTTTCCGGAGTCGGGGGT-3′) with a NcoI site (underlined) and PHYB-3516NR (5′-agctagcATATGGCATCATCAGCATCATG-3′) with an NheI site (underlined); the PCR products were cloned into pMD19-T vector (TaKaRa) to generate the pMD19-PHYB clone. Then, the NcoI restriction site in PHYB was erased in the correct clone without changing the coded amino acid using a site-directed mutagenesis kit (Stratagene) following the manufacturer’s instructions, with the primer pair PHYB-mF (5′-GAGCCGGAGTCAGCaATGGGAAACTGCGGA-3′) and PHYB-mR (5′-TCCGCAGTTTCCCATtGCTGACTCCGGCTC-3′). An NcoI-NheI fragment containing the full-length PHYB coding region from pMD19-PHYB was cloned into the NcoI-SpeI sites of the binary vector pCAMBIA1302 to generate pCAMBIA1302-PHYB.
Plant Transformation and Selection of Transgenic Plants
The pCAMBIA1302-PHYB binary construct was electroporated into the Agrobacterium tumefaciens strain GV3101 and then introduced into Arabidopsis thaliana via a floral dip method (Clough and Bent, 1998). Transgenic plants were selected on germination plates containing 25 μg/mL hygromycin for PHYB-GFP. We selected ∼40 T1 transgenic lines and allowed them to self-produce T2 seeds. Phenotypic analyses were conducted using T2 plants with single T-DNA insertion and then confirmed in the T3 generation. For most experiments, homozygous T3 or T4 transgenic plants were used.
Construction of Double or Triple Mutants
The double mutants phyA-211 phyB-9, phyA-211 PHYB-GFP, phyB-9 spa1-100, PHYB-GFP spa1-100, phyA-211 Myc-SPA1, phyB-9 Myc-SPA1, phyA-211 phyB-9 Myc-SPA1, PHYB-GFP Myc-SPA1, and PHYB-GFP GUS-COP1 were derived from genetic crosses of the two respective single parental mutants (or transgenic lines). Putative double or triple mutants were selected in the F2 generation and confirmed in the F3 generation based on the mutant phenotype, or antibiotic selection markers, immunoblot, and RT-PCR analysis. A previous double mutant was included in this study: spa1-3 GUS-COP1 (Yang and Wang, 2006).
Anthocyanin Measurement
The anthocyanin measurement method was modified from Mancinelli et al. (1991) and Holm et al. (2002). Briefly, 100 seedlings from each light treatment/genotype were incubated in 300 μL methanol acidified using 1% HCl overnight, with gentle shaking in the dark. Then, 250 μL distilled water and 500 μL chloroform were added, vortexed, and spun quickly to separate anthocyanin from chlorophyll. To 350 μL of supernatant, 650 μL HCl was added. Total anthocyanin was determined by measuring the A530 and A657 of the aqueous phase spectrophotometrically. The relative amount of anthocyanin was calculated as (A530 − 0.25 A657) per seedling.
Chlorophyll Measurement
Chlorophyll measurement of seedlings was conducted according to Fankhauser and Casal (2004).
RT-PCR and Quantitative RT-PCR Analyses
For RT-PCR and quantitative RT-PCR analyses, Arabidopsis seedlings were grown under different light conditions, as indicated in the text. Total RNA was extracted using RNeasy Plant Mini Kits (Qiagen) and converted into cDNA by M-MLV reverse transcriptase (Promega). Quantitative RT-PCR analyses for light-responsive gene expression (CHS) were performed in a total volume of 20 μL according to the manual for SYBR Premix Ex Taq (TaKaRa). Three replicates were performed per sample. Quantitative PCR was performed using the Chromo4 Quantitative PCR detection system (Bio-Rad) according to the manufacturer’s instructions. Relative expression was determined after normalization against the reference gene ACTIN 2 using Opticon Monitor version 3.1 (Bio-Rad). Each column represents the mean relative expression of three biological repeats; error bars indicate the se. Primer pairs used for RT-PCR were the following: for PHYB, PHYB-3216F (5′-AAACGCGATTGTAAGTCAAGCG-3′) and PHYB-3682R (5′-CACTAGCAGTTGACAATGGTCG-3′); for CHS, CHS-F (5′-CTACTTCCG CATCACCAAC-3′) and CHS-R (5′-AGAGCAGACAACGAGGACAC-3′); for Myc-SPA1, Myc-F (5′-CGAATTCTGCAGATATCCATC-3′) and SPA1-5TR (5′-GAAGCTCAGCTCACAGGAATGTTG-3′); for Actin 2, ACT2-F (5′-GACCAGCTCTTCCATCGAGAA-3′) and ACT2-R (5′-CAAACGAGGGCTGGAACAAG-3′).
Nuclear Fractionation
Measurement of nuclear fractions was conducted using the modified method of Shen et al. (2007) and Saijo et al. (2008). Arabidopsis seedlings were grown under different light conditions for 4 d. Then, 1.5 g of fresh seedlings was homogenized quickly in 3 mL Honda buffer (2.5% Ficoll 400, 5% dextran T40, 0.4 M Suc, 25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 mM PMSF, and 1× complete protease inhibitor cocktail; Honda et al., 1966) using an ice-cold mortar and pestle, then filtered through a 62-μm-pore nylon mesh. After adding Triton X-100 to a final concentration of 0.5%, the homogenate was incubated on ice for 15 min and centrifuged at 1500g for 5 min. The supernatant fraction centrifuged at 12,000g for 10 min and was saved as the nuclei-depleted soluble fraction. The pellet was resuspended gently in 1 mL Honda buffer with 0.1% Triton X-100. Then, the solution was centrifuged at 1500g for 5 min and the pellet was resuspended in 1 mL Honda buffer, after which the preparation was centrifuged at 100g for 1 min and the supernatant transferred to a new tube. The supernatant was centrifuged at 2000g for 5 min. The pellet was resuspended in 300 μL of buffer G (1.7 M Suc, 10 mM Tris-HCl, pH 8.0, 0.15% Triton-X 100, 2 mM MgCl2, 5 mM DTT, and 1× protease inhibitor cocktail) and transferred to the top of 300 μL buffer G in a new tube. The preparation was centrifuged at 16,000g for 1 h. The pellet was then resuspended in 100 μL Honda buffer and 100 μL 2× SDS loading buffer. Anti-Histone H3 (1:10,000, ab1791; Abcam) and anti-HSP90 (1:8000, at-115; Santa Cruz Biotechnology) antibodies were used to detect nuclear and cytosolic markers, respectively.
Immunoblot Analysis
For immunoblots of Arabidopsis plant extracts, the total protein fraction was extracted with the lysis buffer: 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 25 mM β-glycerophosphate, 2 mM sodium orthovanadate, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1 mM PMSF, and 1× Complete Protease Inhibitor Cocktail (Roche). Specific anti-HY5 peptide antibodies were raised in rabbits and affinity purified against the peptides ETSGRESGSATGQE (amino acids 54 to 67) and GESQRKRGRTPAEK (amino acids 74 to 87). The specific anti-phyB peptide antibody was raised in rabbits and affinity purified against the peptide EQAQSSGTKSLRPR (phyB, amino acids 27 to 40) (HangZhou HuaAn Biotechnology). Myc-SPA1 and phyB-GFP proteins were detected with a monoclonal anti-myc antibody (clone 9E10; Santa Cruz Biotechnology) and an anti-GFP polyclonal anti-GFP antibody (P30010; Shanghai Abmart Biotech), respectively. Proteins were visualized by incubating specimens with goat anti-mouse or rabbit secondary antibodies conjugated to alkaline phosphatase (1:5000; Sigma-Aldrich) in the presence of 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium as substrates.
Quantification of immunoblots was conducted according to Saijo et al. (2008). Briefly, band intensities of Myc-SPA1, HSP90 (loading control for total lysates), or histone (loading control for nuclear fraction) were measured with ImageJ (http://rsb.info.nih.gov/ij/). Relative band intensities were then calculated using the ratio of Myc-SPA1/HSP90 or Myc-SPA1/histone for each immunoblot panel. All immunoblot experiments were repeated at least three times, essentially with the same conclusions, and representative results are shown.
Microscopy of Localization of GUS-COP1 and PhyB-GFP and Immunofluorescence Assay of Myc-SPA1
Histochemical GUS staining was used to examine GUS-COP1 localization, as described previously (von Arnim and Deng, 1994; Osterlund and Deng, 1998; Wang et al., 2009). The cells examined were from the hypocotyl region, approximately one-third of the distance from the hypocotyl/root junction to the base of the cotyledons. The seedlings were incubated in the GUS staining solution overnight. The stained seedlings were fixed for 30 min in 10% acetic acid, 3.7% formaldehyde, and 50% ethanol. Then, the seedlings were decolored through a graded ethanol series and mounted in 1 μg mL−1 4′,6-diamidino-2-phenylindole (DAPI) solution before observation. GUS-COP1 localization was examined under a BX41 fluorescence microscope (Olympus). Representative images were photographed with a digital DP20 camera system (Olympus). All images were taken from the same hypocotyl region with identical exposures.
To visualize the phyB-GFP fusion proteins, the PHYB-GFP transgenic line was mounted on slides and examined with a BX41 fluorescence microscope (Olympus). For each condition, at least 20 seedlings were observed; a representative image is presented. Representative images were prepared as GUS-COP1 localization mentioned above.
To visualize immunofluorescence of Myc-SPA1 and PHYB-GFP, Myc-SPA1 seedlings were grown under R light (30.0 µmol·m–2·s–1) or FR light (2.5 µmol·m–2·s–1) for 5 d and harvested and fixed in fixation buffer (4% formaldehyde, 10 mM Tris-HCl, pH 7.5, 10 mM EDTA, and 100 mM NaCl) (Zuo et al., 2011) under R or FR light for 15 min. Then, nuclei were isolated using ice-cold Honda buffer (Honda et al., 1966) (see also above), washed three times with PBS, and incubated with c-Myc antibody (clone 9E10; Santa Cruz Biotechnology) diluted in PBS (1:250) at 4°C for 4 h. After five washings in PBS, nuclei were incubated with tetramethylrhodamine isothiocyanate (TRITC) conjugated anti-mouse IgG (T5393, 1:200; Sigma-Aldrich) at 4°C for 4 h, followed by a further five washings in PBS. The nuclei were then diluted in PBS with DAPI (1 µg/mL). Representative Myc-SPA1 immunofluorescence images were obtained as GUS-COP1 localization mentioned above.
Yeast Two-Hybrid Analysis
The assay system and all procedures were described by Serino et al. (1999). The various LexA and GAD constructs of SPA1 and its domain deletions used in this study were as described previously (Saijo et al., 2003; Yang et al., 2005a, 2005b). The primer pair used to produce PHYB-CT548 was PHYB-CT548F (5′-ccaattgatgAACTCTAAAGTTGTG-3′) with a MfeI site (underlined) and PHYB-CT548R (5′-agtcgacCTAATATGGCATCATCAGCATCATG-3′), which contained a SalI site (underlined).
In Vivo Co-IP Assay
For the in vivo co-IP assay, PHYB-CT548 cDNA was amplified with the primer pair AtB-1873NF (5′-CCATGGGCATGAACTCTAAAGTTGTGGATG-3′) and AtB-3516NR (5′-GCTAGCATATGGCATCATCAGCATCATG-3′) and cloned into the pCAMBIA1302 binary vector to generate pCAMBIA1302-CT548-GFP. Agrobacterium strain EHA105 carrying pCAMBIA1302-PHYB-CT548-GFP, pJIM19-Myc-SPA1 (Yang and Wang, 2006), or 35S-p19 (Liu et al., 2010) constructs were plated on Luria-Bertani (LB) medium containing the appropriate selective antibiotics at 28°C. A single clone was grown in 5 mL of LB liquid selection medium for 24 h. Then, 500 μL of culture was transferred to new LB liquid selection medium with 10 mM MES, pH 5.6, and 40 μM acetosyringone and grown at 28°C in a shaker. When the bacterial growth reached an OD600 of ∼3.0, the culture was centrifuged at 2150g for 10 min. The pellet was resuspended in infiltration buffer (10 mM MES, 150 μM acetosyringone, and 10 mM MgCl2), and the bacteria were then incubated for 3 h at room temperature. The same volume of suspension carrying different constructs was coinfiltrated into a Nicotiana benthamiana leaf. After growth under FR light (2.5 µmol·m–2·s–1) for 3 d, the leaf was harvested and native extraction buffer (50 mM Tris-MES, pH 8.0, 0.5 M Suc, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, and 1× protease inhibitor cocktail) was used to lyse the sample. Then, 250 μL of extract was incubated with 20 μL anti-Myc–conjugated agarose (M20012; Shanghai Abmart Biotech) under FR light (2.5 µmol·m–2·s–1) for 8 h at 4°C. The agarose was washed twice with 200 μL native extraction buffer. The pellet was subjected to immunoblot analysis using anti-Myc and anti-GFP, as described above. PHYB-GFP or PHYB-GFP Myc-SPA1 double transgenic line seedlings were grown under FR (2.5 µmol·m–2·s–1) or R (30.0 µmol·m–2·s–1) light for 4 d. In vivo co-IP assays of phyB-GFP and Myc-SPA1 were performed as described above, except using the lysis buffer (see also above) instead of the native extraction buffer.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PHYA (At1g09570), PHYB (At2g18790), SPA1 (At2g46350), and ACTIN2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes and RT-PCR and Immunoblot Analyses of Transgenic PHYB-GFP Lines; Related to Figure 1.
Supplemental Figure 2. Evaluation of Appropriate Growth Conditions to Investigate PhyB Functions under Far-Red Light.
Supplemental Figure 3. Nuclear Import of PhyB in Both the Wild-Type and phyA-211 Mutant Backgrounds in Response to Red and Far-Red Light; Related to Figure 2.
Supplemental Figure 4. Anti-Myc Immunoblot Analyses of Myc-SPA1 in Col-0, phyA-211, phyB-9, and phyA-211 phyB-9 Backgrounds under Far-Red Light; Related to Figure 5E.
Supplemental Figure 5. Immunoblot Analyses Showing That PhyB and SPA1 Coordinately Promote Nuclear Transport of Each Other during the Transition from Dark to Far Red; Related to Figure 7.
Supplemental Figure 6. GUS-Stained Images and Immunoblot and RT-PCR Analyses of GUS-COP1 in No-0 and spa1-3 backgrounds in the Dark and under Far-Red Light; Related to Figure 8.
Supplemental Figure 7. PhyB Promotes the Stabilization of Both COP1 and SPA1 to Repress HY5 Accumulation in the Nucleus under Far-Red Light; Related to Figure 9.
Supplemental Figure 8. PhyB Regulates SPA1 Activities in the Absence of Additional Sugars under Far-Red Light; Related to Figure 9.
Supplemental Figure 9. A Heterotetramer Model of the PhyB-SPA1 Complex.
Supplemental Text 1. Evaluation of Appropriate Growth Conditions to Investigate PhyB Functions under Far-Red Light; SPA1 also Enhances Nuclear Import of PhyB in Response to Far-Red Light.
Acknowledgments
We thank Qi Xie and Akira Nagatani for providing the 35S:p19 vector and the PBG transgenic line, respectively. We thank Xing Wang Deng for providing the seed of GUS-COP1 and RPN6 antibody. We thank Xiaofeng Cao, Hongquan Yang, Wojciech Pawlowski, David Pincus, and Ruairidh Sawers for their reading of, and comments on, the article. This research was supported by funds from the Genetically Modified Organisms Breeding Major Projects of China (2011ZX08010-002), the National Natural Science Foundation of China (30871438 and 31170267), the Beijing Natural Science Foundation (5092019), and the Chongqing Natural Science Foundation (CSTC2009BA1088).
AUTHOR CONTRIBUTIONS
X.Z. and S.W. designed and performed the research. P.Z., M.S., L.S., Y.X., Z.L., Y.C., F.M., and L.Y. performed the research. X.Z., S.W., and H.W. analyzed the data. J.Y. and H.Z designed the research and wrote the article. All authors discussed the results and contributed to the article.
Glossary
- FR
far-red
- R
red
- W
white
- FR-HIR
FR light–dependent high-irradiance response
- FRc
continuous FR
- B
blue
- GFP
green fluorescent protein
- PBG
35S-PHYB-GFP
- Ler
Landsberg erecta
- Col-0
Columbia-0
- No-0
Nossen
- NB
nuclear body
- co-IP
coimmunoprecipitation
- GUS
β-glucuronidase
- DAPI
4′,6-diamidino-2-phenylindole
- LB
Luria-Bertani
References
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Barnes S.A., Nishizawa N.K., Quaggio R.B., Whitelam G.C., Chua N.H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt R.L., Oliverio K.A., Casal J.J., Hoecker U. (2002). SPA1, a component of phytochrome A signal transduction, regulates the light signaling current. Planta 215: 745–753 [DOI] [PubMed] [Google Scholar]
- Borthwick H.A., Hendricks S.B., Parker M.W., Toole E.H., Toole V.K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA 38: 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto J.F., Sanchez R.A., Whitelam G.C., Casal J.J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs W.R., Olney M.A. (2001). Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J., Yanovsky M.J., Luppi J.P. (2000). Two photobiological pathways of phytochrome A activity, only one of which shows dominant negative suppression by phytochrome B. Photochem. Photobiol. 71: 481–486 [DOI] [PubMed] [Google Scholar]
- Chen M., Schwab R., Chory J. (2003). Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl. Acad. Sci. USA 100: 14493–14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Tao Y., Lim J., Shaw A., Chory J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15: 637–642 [DOI] [PubMed] [Google Scholar]
- Cheng C.L., Acedo G.N., Cristinsin M., Conkling M.A. (1992). Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc. Natl. Acad. Sci. USA 89: 1861–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng X.W., Matsui M., Wei N., Wagner D., Chu A.M., Feldmann K.A., Quail P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G β homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Deng X.W., Quail P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10: 121–129 [DOI] [PubMed] [Google Scholar]
- Dijkwel P.P., Huijser C., Weisbeek P.J., Chua N.H., Smeekens S.C. (1997). Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9: 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Edgerton M.D., Jones A.M. (1992). Localization of protein-protein interactions between subunits of phytochrome. Plant Cell 4: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Deng X.W. (2007). The role of ubiquitin/proteasome–mediated proteolysis in photoreceptor action. In Light and Plant Development, G. C. Whitelam and K. J. Halliday, eds (Oxford: Blackwell Publishing), pp 128–154 [Google Scholar]
- Fittinghoff K., Laubinger S., Nixdorf M., Fackendahl P., Baumgardt R.L., Batschauer A., Hoecker U. (2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 47: 577–590 [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Casal J.J. (2004). Phenotypic characterization of a photomorphogenic mutant. Plant J. 39: 747–760 [DOI] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Poppe C., Sweere U., Martin A., Schäfer E. (2001). Negative interference of endogenous phytochrome B with phytochrome A function in Arabidopsis. Plant Physiol. 125: 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Hirschfeld M., Tepperman J.M., Clack T., Quail P.H., Sharrock R.A. (1998). Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U., Tepperman J.M., Quail P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499 [DOI] [PubMed] [Google Scholar]
- Hoecker U., Xu Y., Quail P.H. (1998). SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Deng X.W. (1999). Structural organization and interactions of COP1, a light-regulated developmental switch. Plant Mol. Biol. 41: 151–158 [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S.I., Hongladarom T., Laties G.G. (1966). A new isolation medium for plant organelles. J. Exp. Bot. 17: 460–472 [Google Scholar]
- Huq E., Al-Sady B., Quail P.H. (2003). Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35: 660–664 [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Muller T., Bauer D., Adám E., Schäfer E., Nagy F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schäfer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. (2002). Blue light receptors and signal transduction. Plant Cell 14(suppl): S207–S225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S., Fankhauser C. (2009). Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Mancinelli A.L., Rossi F., Moroni A. (1991). Cryptochrome, phytochrome, and anthocyanin production. Plant Physiol. 96: 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T., Mochizuki N., Nagatani A. (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Mazzella M.A., Arana M.V., Staneloni R.J., Perelman S., Rodriguez Batiller M.J., Muschietti J., Cerdán P.D., Chen K., Sánchez R.A., Zhu T., Chory J., Casal J.J. (2005). Phytochrome control of the Arabidopsis transcriptome anticipates seedling exposure to light. Plant Cell 17: 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac A.C., Wagner D., Boylan M.T., Quail P.H., Smith H. (1993). Photoresponses of transgenic Arabidopsis seedlings expressing introduced phytochrome–B encoding cDNAs: Evidence that phytochrome A and phytochrome B have distinct photoregulatory functions. Plant J. 4: 19–27 [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994a). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Deng X.W. (1994b). Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6: 1391–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A., Reed J.W., Chory J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Chory J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y., Matsushita T., Mochizuki N., Suzuki T., Tokutomi S., Nagatani A. (2004). Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell 16: 2104–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Deng X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.I., Liu J.R., Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45: 968–975 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.K., Popp C., Wüst F., Bindics J., Viczián A., Hiltbrunner A., Nagy F., Kunkel T., Schäfer E. (2012). Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (1997). An emerging molecular map of the phytochromes. Plant Cell Environ. 20: 657–665 [Google Scholar]
- Quail P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Reed J.W. (1999). Phytochromes are Pr-ipatetic kinases. Curr. Opin. Plant Biol. 2: 393–397 [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W. (2003). The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Zhu D., Li J., Rubio V., Zhou Z., Shen Y., Hoecker U., Wang H., Deng X.W. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 31: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Nagatani A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Serino G., Tsuge T., Kwok S., Matsui M., Wei N., Deng X.W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11: 1967–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R.A., Clack T. (2004). Heterodimerization of type II phytochromes in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 11500–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Short T.W. (1999). Overexpression of Arabidopsis phytochrome B inhibits phytochrome A function in the presence of sucrose. Plant Physiol. 119: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C., Kim B.H., Lyssenko N.N., Xu X., Johnson C.H., von Arnim A.G. (2004). The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: Mutational analysis by bioluminescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 101: 6798–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.A., Deng X.W. (2003). From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Thum K.E., Shasha D.E., Lejay L.V., Coruzzi G.M. (2003). Light- and carbon-signaling pathways. Modeling circuits of interactions. Plant Physiol. 132: 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T., Matsushita T., Oka Y., Mochizuki N., Nagatani A. (2007). Roles for the N- and C-terminal domains of phytochrome B in interactions between phytochrome B and cryptochrome signaling cascades. Plant Cell Physiol. 48: 424–433 [DOI] [PubMed] [Google Scholar]
- von Arnim A.G., Deng X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Wagner D., Koloszvari M., Quail P.H. (1996). Two small spatially distinct regions of phytochrome B are required for efficient signaling rates. Plant Cell 8: 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Quail P.H. (1995). Mutational analysis of phytochrome B identifies a small COOH-terminal-domain region critical for regulatory activity. Proc. Natl. Acad. Sci. USA 92: 8596–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Tepperman J.M., Quail P.H. (1991). Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell 3: 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang X., Li W., Piqueras R., Cao K., Deng X.W., Wei N. (2009). Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J. 58: 655–667 [DOI] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R., Nakamura M., Mochizuki N., Kay S.A., Nagatani A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q., Wu Y.J., Tang R.H., Liu D., Liu Y., Cashmore A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Hoecker U., Liu B., Xu L., Wang H. (2005b). Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J. 43: 131–141 [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005a). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang H. (2006). The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J. 47: 564–576 [DOI] [PubMed] [Google Scholar]
- Yanovsky M.J., Casal J.J., Luppi J.P. (1997). The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J. 12: 659–667 [DOI] [PubMed] [Google Scholar]
- Yi C., Deng X.W. (2005). COP1 - From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tepperman J.M., Fairchild C.D., Quail P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97: 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]