The xylem in Arabidopsis roots develops as a single row of cells neighboring the undifferentiated procambium. This work determined that two closely related AHL3 and AHL4 transcription factors regulate the boundaries between the xylem and procambium. AHL4 moves from the procambium to xylem in the root meristem, likely as a heteromeric complex with AHL3.
Abstract
The xylem and phloem, major conducting and supporting tissues in vascular plants, are established by cell division and cell-type specification in the procambium/cambium. The organization of the xylem, phloem, and procambium/cambium is tightly controlled. However, the underlying regulatory mechanisms remain largely unknown. In this study, we report the discovery of two transcription factors, AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEIN 3 (AHL3) and AHL4, which regulate vascular tissue boundaries in Arabidopsis thaliana roots. In either of the knockout mutants of AHL3 and AHL4, encoding closely related AT-hook transcription factors, a misspecification of tissue boundaries between the xylem and procambium occurred and ectopic xylem developed in the procambium domain. In plants, specific types of transcription factors can serve as direct intercellular signals by moving from one cell to another, playing crucial roles in tissue patterning. Adding to this paradigm, AHL4 moves actively from the procambium to xylem in the root meristem to regulate the tissue boundaries. When the intercellular movement of AHL4 was impaired, AHL4 could not complement the xylem phenotype in the ahl4. Furthermore, AHL4 revealed unique characteristics in that it interacts with AHL3 in vivo and that this interaction facilitates their intercellular trafficking. Taken together, this study uncovered a novel mechanism in vascular tissue patterning that requires the intercellular trafficking of two interacting transcription factors.
INTRODUCTION
Morphogenesis in multicellular organisms is a tightly regulated process. Unlike animals growing from a body plan established during embryogenesis, plants grow by forming new organs throughout their life span. In this process, plants largely rely on positional information rather than lineage to control pattern formation in a temporal and spatial manner (reviewed in Scheres and Benfey, 1999; De Smet and Beeckman, 2011). Since plant cells are confined by rigid cell walls, intercellular communications within a long or short distance play crucial roles in providing developmental cues. Communications between neighboring cells in particular contribute to specifying cell types and defining boundaries between them. Many forms of molecules, including proteins, RNAs, and small molecules, serve as signals that mediate intercellular communications (reviewed in Kurata et al., 2005a; Hirakawa et al., 2011; Van Norman et al., 2011).
Vascular tissues serve as a major conductive and supporting system in vascular plants. The xylem and phloem in the vascular system are generated from procambium and cambium, stem cell populations specialized in the formation and growth of vascular tissues. The organization of vascular tissues in the root tends to be unique in each species. Vascular tissues in the Arabidopsis thaliana roots, for example, are always bisymmetrically organized. A single-cell-wide xylem axis in the stele develops two xylem vessel types: the protoxylem in the periphery of the xylem axis and the metaxylem in the center. Two phloem poles are localized perpendicular to the xylem axis, and procambium cells are between the xylem and phloem (Figure 1A). During vascular development, the xylem and phloem differentiate while the procambium/cambium remains undifferentiated. This vascular cell-type patterning is quite robust and, thus, likely requires regulatory programs that define cell type boundaries.
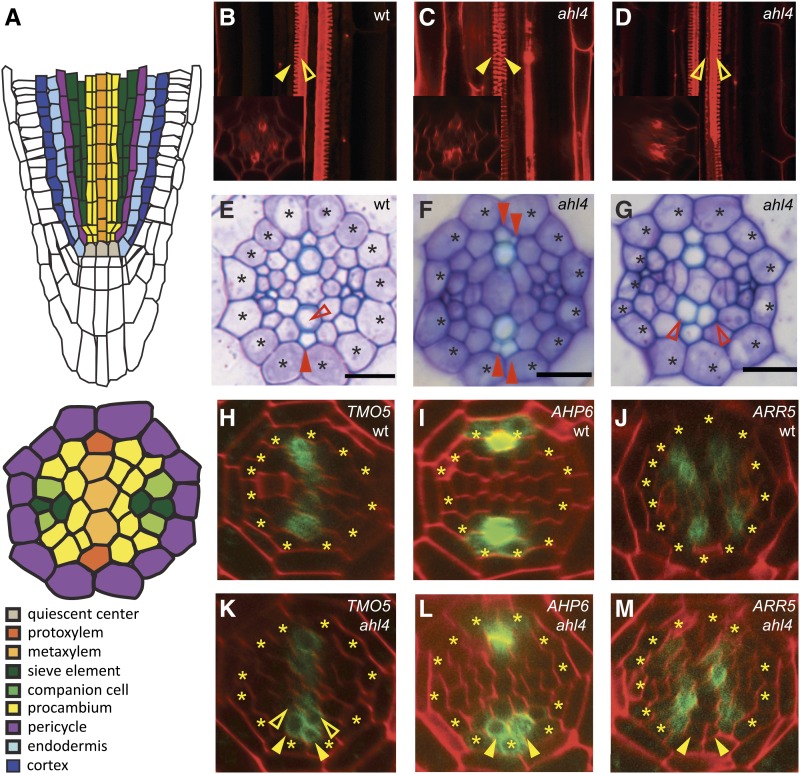
Figure 1.
Vascular Patterning in ahl4-1.
(A) Schematic representations of the Arabidopsis root meristem (longitudinal) and stele (transverse).
(B) to (G) Comparison of vascular patterns visualized by fuchsin-stained xylem ([B] to [D]) and toluidine blue–stained root section ([E] to [G]) between the wild type (wt) ([B] and [E]) and ahl4-1 ([C], [D], [F], and [G]).
(H) to (M) Marker analysis using ProTMO5 (TARGET OF MONOPTEROS 5):erGFP ([H] and [K]), ProAHP6 ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6:erGFP ([I] and [L]), and ProARR5 ARABIDOPSIS RESPONSE REGULATOR 5:erGFP ([J] and [M]) to compare the xylem precursor domain between the wild type ([H] to [J]) and ahl4-1 ([K] to [M]).
Asterisks, pericycle position; closed arrowheads, protoxylem; open arrowheads, metaxylem. Bars = 10 µm.
Several transcription factors regulate the formation of the xylem and phloem in Arabidopsis (Bonke et al., 2003; Yamaguchi et al., 2008; Carlsbecker et al., 2010; Yamaguchi et al., 2010a, 2010b). ALTERED PHLOEM DEVELOPMENT (APL), a MYB coiled-coil transcription factor, is required for phloem formation (Bonke et al., 2003). Multiple transcription factors have been identified to regulate xylem development (Yamaguchi et al., 2008, 2010a, 2010b; Carlsbecker et al., 2010). To establish the polarity and boundaries between the xylem and phloem, cell-to-cell communications are indispensible. PHLOEM INTERCALATED WITH XYLEM (PXY), CLV1-like Leucine-rich repeat–receptor-like kinases, and CLE41/44, CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE) members are key regulators in this (Fisher and Turner, 2007; Hirakawa et al., 2008). CLE41/44 peptides generated from the phloem move to the cambium cells where they bind to PXY (Hirakawa et al., 2008; Etchells and Turner, 2010; Hirakawa et al., 2010). The CLE/PXY complex in the cambium cells then activates as yet unknown signals to maintain the boundary between the cambium and xylem. Such regulation by CLEs and PXY has been observed only in the mature root and hypocotyls, not in the root meristem.
The AT-hook is a small DNA binding protein motif that is frequently associated with modulating chromatin architecture to coregulate transcription. The AT-hook motif, characterized by a highly conserved tripeptide of Gly-Arg-Pro, exists as single or multiple copies in a wide range of organisms (Aravind and Landsman, 1998). Mammals encode nonhistone chromatin-associated proteins called high mobility group (HMG) proteins, which share unique structural characteristics, including long AT-rich 3′ untranslated regions and C-terminal regions enriched with negatively charged amino acid residues. The HMG families are composed of architectural transcription factors that regulate the expression of numerous genes in vivo. One of the subfamilies, HMGA, is characterized by the AT-hook domain (reviewed in Reeves, 2001; Reeves and Beckerbauer, 2001). Another HMG subfamily member, HMGB1, is a nuclear transcription factor that is released into the extracellular matrix, acting as an alarmin (endogenous molecules that are released during immune responses). It contains two 80–amino acid HMG-1 boxes that regulate the nonspecific binding of HMGB1 to the minor grooves in DNA. Notably, HMGB1 is both released passively during cellular necrosis and secreted actively by immune cells to recruit and activate antigen-presenting cells, thereby enhancing immune responses (Chen et al., 2004; Huang et al., 2007; Yang et al., 2010).
In plants, AT-hook family proteins have evolved in a unique way by harboring both an AT-hook domain and an uncharacterized plant and prokaryotes conserved (PPC) domain. Although the PPC domain is also found in prokaryotic proteins, these do not contain the AT-hook motif (Fujimoto et al., 2004). Plant-specific AT-hook members have been shown to be involved in diverse processes, such as hypocotyl elongation, flower development, gibberellin biosynthesis, leaf senescence, and stem cell niche specification (Lim et al., 2007; Matsushita et al., 2007; Street et al., 2008; Ng et al., 2009; Gallavotti et al., 2011). In Arabidopsis, 29 members belong to the AT-hook family (Fujimoto et al., 2004; Matsushita et al., 2007). One of them was shown to have specific binding activity to DNA elements, which is required for the expression of a direct target gene (Matsushita et al., 2007). AT-hook members were also shown to bind the matrix attachment regions in the nuclei (Morisawa et al., 2000; Fujimoto et al., 2004; Lim et al., 2007; Ng and Ito, 2010).
In this study, we present the discovery of two closely related AT-hook family members, AHL3 and AHL4, which interact in vivo and regulate boundaries between the procambium and xylem in the Arabidopsis root meristem. Intriguingly, AHL3 and AHL4 proteins move between cells. Our investigation suggests that the cell-to-cell communication mediated by mobile AHL3 and AHL4 is critical for establishing the boundary between the procambium and xylem.
RESULTS
AHL4 Regulates Boundaries between the Xylem and Procambium
Xylem precursors in the root are established in the meristem close to the underlying quiescent center (QC) (Mähönen et al., 2000). In Arabidopsis, five or six xylem precursor cells form a single row of xylem axis, among which the periphery and center differentiate into protoxylem and metaxylem vessels, respectively (Figures 1A, 1B, and 1E). To identify transcription factors potentially involved in this xylem patterning, we surveyed and selected transcription factors that are enriched in the xylem precursor cells using high-resolution microarray-based gene expression data available for most cell types in the Arabidopsis root (Brady et al., 2007). Fifteen transcription factors were chosen for further characterization based on the selection criteria of at least twofold enrichment in the xylem precursor cell type over other root cell types with corrected P values < 0.001 (see Supplemental Figure 1A online). To identify the transcription factors impaired in xylem patterning, T-DNA insertion lines of each candidate were screened for abnormal xylem phenotypes. Insertion lines associated with genes for three transcription factors (i.e., AT1G29950, AT4G25320, and AT5G51590) exhibited abnormal xylem phenotypes. Among these, AT5G51590 (AHL4) and AT4G25320 (AHL3) encode proteins that belong to the AT-hook family (SALK_124619, ahl4-1; and FLAG_445H04, ahl3-1 hereafter). T-DNA insertions disrupted the third exon of AHL4 in ahl4-1 and the first exon of AHL3 in ahl3-1, respectively (see Supplemental Figure 2A online). Real-time RT-PCR analysis showed that AHL4 expression was reduced by 99% in the ahl4-1 compared with the wild-type root and that AHL3 expression was reduced by 60% in the ahl3-1 (see Supplemental Figure 2B online). Since these data indicated that ahl4-1 is likely a null mutant, whereas ahl3-1 is not, we first characterized ahl4-1 for its vascular tissue patterning in the root.
In ahl4-1, we observed additional strands of protoxylem (Figures 1C and 1F; closed arrowheads) and metaxylem (Figures 1D and 1G; open arrowheads). This extra protoxylem phenotype was detected in ∼75% of 30 individual ahl4-1 plants examined (see Supplemental Figure 3 online; P value for λ2 test = 1.59547E-48). Here, we used the protoxylem phenotype to further dissect AHL4 function. Our histological analysis did not indicate any obvious defect in the phloem patterning of ahl4-1 (Figures 1E to 1G). Extra xylem vessel formation occurred in the maturation zone of the ahl4-1 root. However, cell type–specific root expression data indicated that AHL4 is expressed in the root meristem before vascular cells begin differentiation. To identify the role of AHL4 in the root meristem, we introduced several vascular cell type–specific markers and compared their expression patterns in ahl4-1 to those in the wild-type root meristem. First, we investigated the impact of AHL4 on xylem domain specification using ProTMO5:erGFP (endoplasmic reticulum–localized green fluorescent protein), which is specifically expressed in the xylem precursor cells (Figure 1H) (Lee et al., 2006). In contrast with the wild type, where ProTMO5:erGFP expression is restricted to a single row of xylem precursors (Figure 1H), the expression of ProTMO5:erGFP in ahl4-1 expanded to the adjacent cells in the procambium domain (Figure 1K; see Supplemental Figure 4 online). This expression pattern is consistent with the formation of extra xylem strands in ahl4-1. Another marker, ProAHP6:erGFP, is expressed in a protoxylem precursor and the two neighboring pericycle cells (Figure 1I) (Mähönen et al., 2006). Consistent with TMO5, the expression domain of ProAHP6:erGFP also expanded in ahl4-1, being detected in the cells which would belong to procambium in the wild type (Figure 1L). These xylem marker analyses suggested that the proper formation of boundaries between the procambium and xylem might require AHL4. To further test this idea, we checked the status of procambium in ahl4-1 by introducing ProARR5:erGFP, which precisely marks procambium domains that neighbor xylem precursors in the root meristem (Figure 1J) (Lee et al., 2006). As expected, ProARR5:erGFP in ahl4-1 was absent in the cells where xylem markers had expanded (Figure 1M, arrowheads).
Next, to test whether phloem development is affected, we introduced ProAPL:erGFP, which is expressed in the developing protophloem sieve element and then switches to the companion cells and metaphloem sieve element, into ahl4-1 (Bonke et al., 2003). However, as indicated by our histological analysis, no obvious changes in ProAPL:erGFP expression were detected in ahl4-1 (see Supplemental Figure 5 online). Note also that the overall root growth in ahl4-1 was normal.
Collectively, our results support the conclusion that AHL4 is specifically involved in defining the boundary between the xylem and procambium.
AHL4 Promoter Expression Is Specific to Procambium but Its Translational Domain Is Expanded
To gain insight into the spatio-temporal regulation of xylem development by AHL4, we generated transcriptional and translational fusion lines of AHL4. For this, we produced constructs driving the expression of erGFP or β-glucuronidase (GUS) and the coding region of AHL4 fused to free GFP (AHL4-GFP), respectively, under the 2-kb-long upstream intergenic region of AHL4. When ProAHL4:AHL4-GFP was introduced into ahl4-1, all six independent transgenic lines complemented its mutant phenotype, negating the formation of ectopic xylem strands (Figures 2A and 2B; see Supplemental Table 1 online). This result indicates that the AHL4-GFP fusion protein is fully functional and that the selected promoter is sufficient for AHL4 activity. We next analyzed the spatial domain where AHL4-GFP is expressed. Consistent with its regulation of the boundaries between the xylem and procambium in the root meristem, AHL4-GFP was enriched in the stele of a root meristem (Figure 2C). The expression level of transcriptional erGFP was very weak (see Supplemental Figure 6A online) but mostly detected in the procambium, which was different from the expression domain detected by ProAHL4:AHL4-GFP. We further confirmed this discrepancy between transcriptional and protein expression domains using the transcriptional GUS line. Contrary to the ubiquitous distribution of AHL4-GFP throughout the stele cells of the root meristem, the GUS expression was specific to the procambium (Figure 2D). AHL4 was originally selected as a gene enriched in the xylem precursor; however, ProAHL4:GUS expression suggested that the AHL4 transcriptional domain is specific to the procambium. This is likely because the selection for xylem precursor enriched genes was made in the absence of procambium profiling data from the current root expression map.
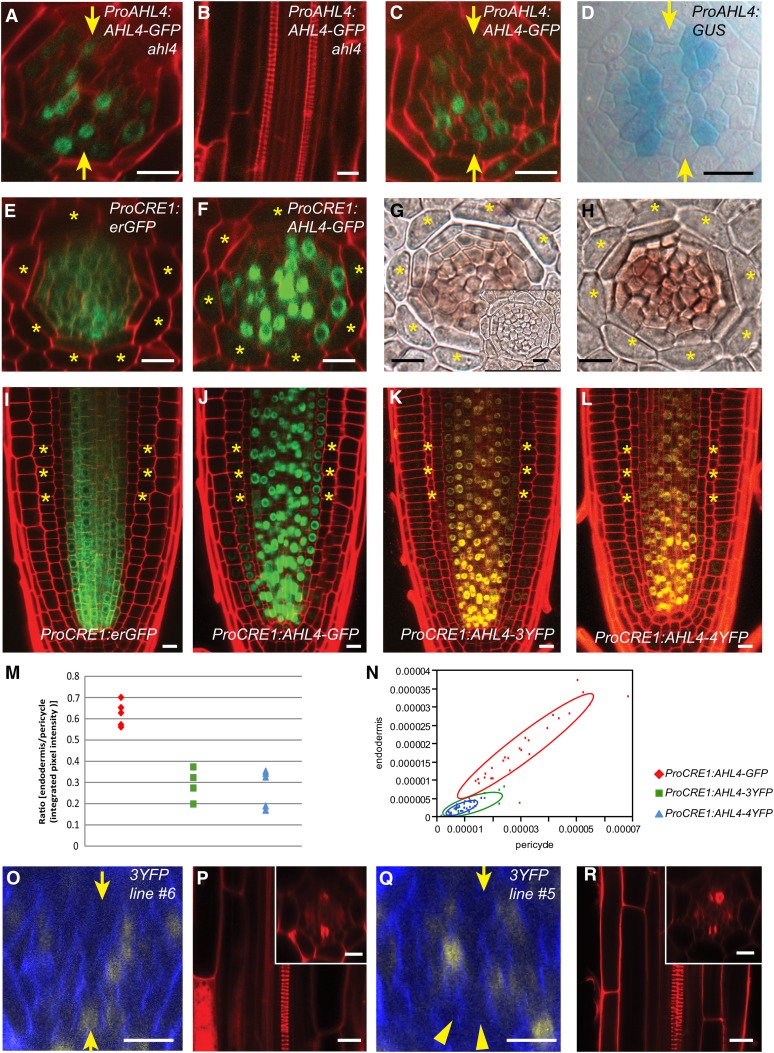
Figure 2.
Movement of AHL4 Is Crucial for Establishing the Boundaries between the Xylem and Procambium.
(A) to (C) ProAHL4:AHL4:GFP can be found in both the procambium and xylem in ahl4-1 (A) and the wild type (C). ProAHL4:AHL4:GFP; ahl4-1 recovers the ectopic xylem phenotype in ahl4-1 (B).
(D) ProAHL4:GUS expression is specific to the procambium.
(E) to (N) ProCRE1:erGFP is expressed in the stele ([E] and [I]), while ProCRE1:AHL4:GFP is also found outside the stele ([F] and [J]). In situ hybridization with an antisense GFP probe suggests that mRNA distributions in ProCRE1:erGFP (G) and ProCRE1:AHL4:GFP (H) are similar (inset in [G] shows a sense GFP probe). ProCRE1:AHL4:3xYFP (K) and ProCRE1:AHL4:4xYFP (L) exhibit reduced AHL4 mobility, as supported by quantification of the movement ([M] and [N]).
(O) to (R) An independent line of ProAHL4:AHL4:3xYFP; ahl4-1 with AHL4 moving into xylem precursors (O) is capable of rescuing the xylem phenotype (P), while another line of ProAHL4:AHL4:3xYFP; ahl4-1 with retarded AHL4 mobility (Q) cannot rescue the xylem phenotype (R).
Asterisk, cortex position; arrowheads, protoxylem; arrows, xylem axis.
AHL4 Protein Moves between Cells
The spatial expansion of the AHL4 protein domain from the transcriptional domain indicated that AHL4 proteins or RNAs might move between cells. To examine this further, we analyzed the spatial distribution of AHL4 proteins expressed under the stele-specific CYTOKININ RESPONSE 1 (CRE1) promoter (Figures 2E and 2I) (Mähönen et al., 2000). Consistent with observations made with transcriptional and translational fusion lines, AHL4-GFP signals expanded outside the stele and reached the lateral root cap cells in the meristem region (Figures 2F and 2J). In the meristematic zone, we always detected AHL4-GFP in the endodermis, where CRE1 is not normally expressed.
Though most of the mobile transcription factors have been shown to move as proteins, there are cases when mRNAs move between cells (Lucas et al., 1995; Ruiz-Medrano et al., 1999). To test the latter possibility for AHL4, we examined the mRNA domains of AHL4:GFP in ProCRE1:AHL4:GFP transgenic plants by performing RNA in situ hybridization. An antisense GFP probe successfully detected AHL4:GFP mRNA only in the stele cells (Figure 2H), which was consistent with the control experiment employing ProCRE1:erGFP (Figure 2G) (Mähönen et al., 2000). Based on the results from expression analyses of proteins and mRNAs of AHL4:GFP in the ProCRE1:AHL4:GFP transgenic plants, we concluded that the AHL4 protein, rather than its transcript acts non-cell-autonomously.
Movement of AHL4 Is Critical for the Boundary between the Xylem and Procambium
Numerous studies have shown that many mobile proteins move from one cell to another through plasmodesmata (PD) (Lucas et al., 1995; Zambryski and Crawford, 2000; Wu et al., 2003; Kurata et al., 2005b; Schlereth et al., 2010; Tsukagoshi et al., 2010). Given that the size exclusion limit of PD in the root meristem is suggested to be around 60 kD (Crawford and Zambryski, 2001; Rim et al., 2011), AHL4-GFP (72 kD) might move between cells in a targeted manner. To gain a better understanding of AHL4 movement, the coding region of AHL4 was fused with tandem yellow fluorescent proteins (YFPs; 3x or 4xYFP) driven by the CRE1 promoter and introduced into an ahl4-1 mutant. The extra xylem phenotype associated with ahl4-1 was complemented in all nine transgenic lines expressing ProCRE1:AHL4:3x/4xYFPs (Table 1). This result shows that fusion proteins between AHL4 and tandem YFPs are also fully functional. We then analyzed their cell-to-cell movement capacity relative to ProCRE1:AHL4:GFP(1x). Triple and quadruple YFP have large molecular masses (81 and 108 kD, respectively) and therefore can significantly reduce the protein movement between cells (Crawford and Zambryski, 2001; Kurata et al., 2005b; Tsukagoshi et al., 2010). Our confocal analysis suggested that adding tandem YFPs significantly interferes with the movement of AHL4 but does not abolish the movement activity completely (Figure 2K and 2L).
Table 1. Movement Comparison between AHL4-GFP and AHL4-3x/4xYFP.
| Genotype | Percentage (n) of Independent Lines Exhibiting GFP (YFP) Signals in the Endodermis | Total No. of Lines Examined (n) | Percentage (n) of Individual Plants Exhibiting GFP (YFP) Signals in the Endodermis | Percentage (n) of Individual Plants Rescued the Ectopic Xylem Phenotype | Total No. of Individual Plants Scored (n) |
|---|---|---|---|---|---|
| ProCRE1:AHL4:GFP; wild type | 100 (5) | 5 | 100 (25) | N/A | 25 |
| ProCRE1:AHL4:3xYFP; ahl4-1 | 75 (3) | 4 | 60 (12) | 100 (20) | 20 |
| ProCRE1:AHL4:4xYFP; ahl4-1 | 60 (3) | 5 | 52 (13) | 100 (25) | 25 |
For ProCRE1:AHL4:GFP; wild type and ProCRE1:AHL4:4xYFP; ahl4-1, five independent lines were characterized. For ProCRE1:AHL4:3YFP; ahl4-1, four independent lines were characterized. For each independent line, five individual plants were examined for the movement. N/A, not applicable.
Next, we calculated how frequently movement occurred for the AHL4-GFP and AHL4-tandem-YFP proteins. AHL4-GFP movement from the stele to endodermis was detected in all five independent lines, accounting for a 100% rate. By contrast, only 75 and 60% of the transgenic lines expressing AHL4-3x and 4xYFP, respectively, exhibited protein trafficking from the stele to endodermis (Table 1). We then quantified the movement of proteins in the same transgenic lines by measuring and comparing fluorescence signals in the pericycle cells and the neighboring endodermal cells (for further details about the imaging and calculation, see Supplemental Figure 7 online and Methods). The average fluorescence intensity of AHL4-GFP in the endodermis was 62% (±5.7% sd) of the fluorescence measured in the adjacent pericycle. By contrast, the signal intensities measured for AHL4-3xYFPs and AHL4-4xYFP were 29% ± 7.4% (sd) and 27% ± 8.9% (sd), respectively, of the adjacent pericycle (Figure 2M). The levels of AHL4-GFP and AHL4-tandem-YFP were similar in the pericycle, indicating that the low endodermis to pericycle fluorescence ratio measured for AHL4-tandem-YFP is not the result of decreased expression (Figure 2N). These results indicate that AHL4 has a strong tendency to move between cells, which cannot be easily blocked by increasing its protein size to a great extent.
To gain insight into the non-cell-autonomous function of AHL4 in developmental control, we expressed AHL4 fused with 3xYFPs under the AHL4 promoter in ahl4-1. Similar to what was observed in ProCRE1:AHL4:3xYFP plants, fusing 3xYFP to AHL4 restricted its movement only partially (see Supplemental Table 1 online). Next, to test whether the movement of AHL4 into the xylem precursors contributes to the complementation of the extra protoxylem phenotype, we measured the frequencies of the events detecting AHL4-3x/4xYFP signals in the xylem precursors and associated normal xylem phenotypes using 10 individual progenies each from six independent transgenic lines. Unlike ProAHL4:AHL4:GFP, which rescued the xylem phenotype of ahl4-1 in 100% of plants examined, ProAHL4:AHL4:3xYFP rescued the xylem phenotype only in ∼50% (see Supplemental Table 1 and Supplemental Figure 3 online). In seedlings where AHL4-3xYFP was detected in the xylem precursors, all 31 exhibited a normal xylem phenotype (Figures 2O and 2P). By contrast, in the absence of AHL4-3xYFP in the xylem precursors, the extra xylem phenotype was not rescued in any case (29/29) (Figures 2Q and 2R). Such a strong correlation between AHL4 movement to the xylem precursors and xylem phenotype indicates that the movement of AHL4 from procambial cells to xylem precursors is required to define and maintain the boundary between the xylem and procambium.
AHL3 Functions in a Complementary Manner to AHL4
A previous phylogenetic analysis suggested that AHL3 is the most closely related homolog of AHL4 in Arabidopsis (Fujimoto et al., 2004). Given that AHL3 and AHL4 share 69% amino acid similarity and that both play a role in xylem patterning, they might function in a partially redundant manner. However, the ahl3-1 ahl4-1 double mutant did not show any additional xylem phenotype to what was observed in each single mutant (Figure 3A). Real-time RT-PCR showed that the expression level of AHL4 was not affected in the ahl3-1 background or vice versa (see Supplemental Figure 2B online). We confirmed this again by generating and phenotyping artificial microRNA lines that simultaneously knocked down AHL3 and AHL4. These plants exhibited a similar phenotype to the single and double mutants (see Supplemental Figures 8C to 8F online). In addition, ProAHL4:AHL4:GFP introduced into ahl3-1 was not able to rescue the extra xylem phenotype despite the clear presence of AHL4-GFP protein in the xylem precursors (Figures 3B and 3C). Collectively, these results suggest that AHL3 and AHL4 function in a complementary manner (see Discussion).
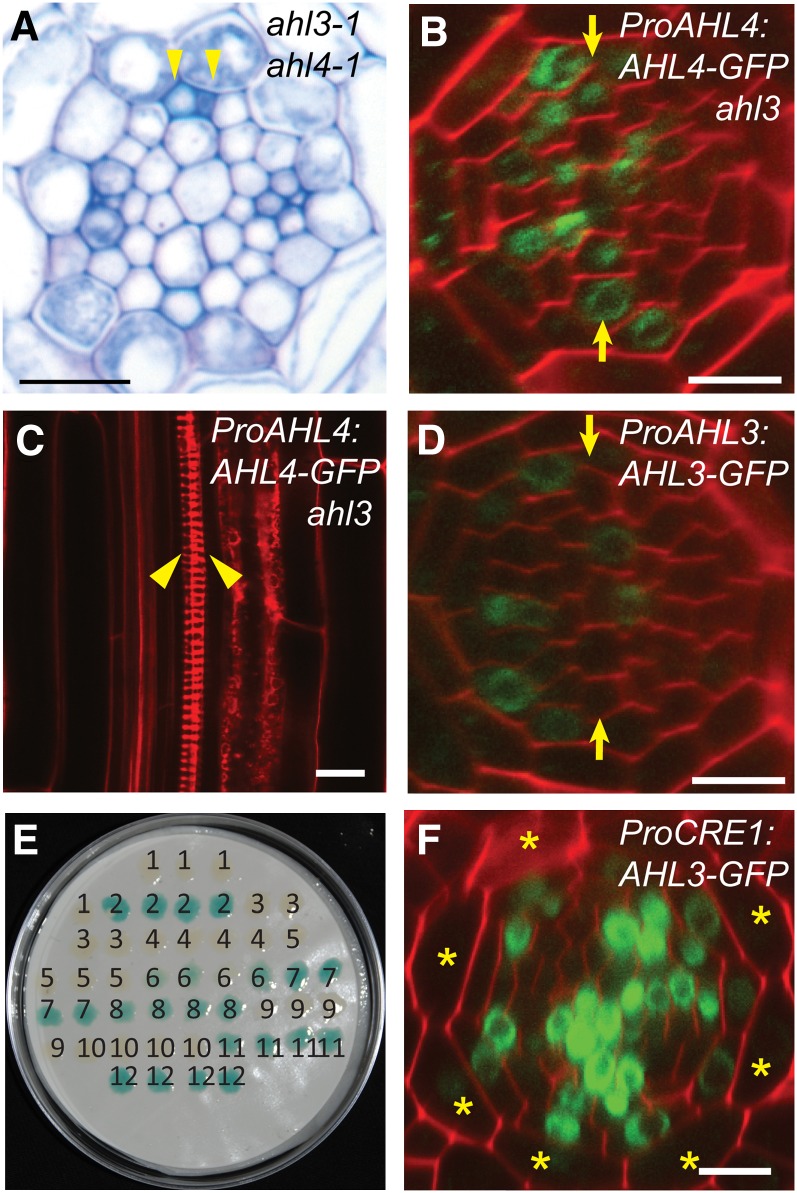
Figure 3.
AHL3/4 Proteins Interact with Each Other and Together Influence Xylem Patterning.
(A) ahl3-1 ahl4-1 forms ectopic xylem strands as shown by toluidine blue staining.
(B) and (C) ProAHL4:AHL4:GFP in ahl3-1. AHL4-GFP is observed in the xylem precursors (B), yet the ectopic xylem formation cannot be rescued (C).
(D) Confocal cross-section image of a root expressing ProAHL3:AHL3:GFP in the wild type.
(E) Yeast two-hybrid assay demonstrating the direct interaction between AHL3/4 proteins and the self-interaction of AHL3 and AHL4 (1, BD + AD; 2, BD-krev + AD-RalGDS-wt; 3, BD-krev + AD-RalGDS-m1; 4, BD-krev + AD-RalGDS-m2; 5, BD + AD-AHL4; 6, BD-AHL4 + AD; 7, BD-AHL4 + AD-AHL3; 8, BD-AHL4 + AD-AHL4; 9, BD + AD-AHL3; 10, BD-AHL3 + AD; 11, BD-AHL3 + AD-AHL4; 12, BD-AHL3 + AD-AHL3).
(F) Confocal cross-section image of a root expressing ProCRE1:AHL3:GFP in the wild type demonstrates the movement of AHL3-GFP outside the stele.
Asterisks, cortex position; yellow arrows, xylem axis; arrowhead, protoxylem. Bars = 10 µm.
For AHL3 to function complementarily to AHL4, it is likely to be present in the same spatial domain as AHL4 (see Supplemental Figure 1 online). Indeed, AHL3-GFP proteins expressed under the AHL3 promoter in the wild-type background were observed in the nuclei of both procambium and xylem precursors of the root meristem, strengthening the notion that they function in a complementary manner (Figure 3D). However, GUS expressed under the same AHL3 promoter was mainly restricted to the endodermis in the late elongation zone and onwards (see Supplemental Figures 6B and 6C online), revealing different distribution patterns from the AHL3-GFP. Thus, AHL3 proteins, which are generated in the elongation and maturation zones of a root, seem to move down to the root meristem through the stele.
AHL3 and AHL4 Interact in Vivo and Function as a Heteroprotein Complex
Next, we investigated whether AHL3 and AHL4 form a functional protein complex by performing yeast two-hybrid assays. This experiment showed that they undergo not only homomeric but also heteromeric interactions in yeast (Figure 3E). To demonstrate their interaction directly in vivo, we next performed fluorescence resonance energy transfer (FRET) analysis using an enhanced green fluorescent protein (EGFP) and mCherry pair by employing acceptor photobleaching. This method is useful for determining the extent of FRET by monitoring an increase in donor (e.g., EGFP) fluorescence after acceptor (e.g., mCherry) photobleaching (Day et al., 2001; Karpova et al., 2003; Tramier et al., 2006). First, an EGFP and mCherry pair was used as the FRET negative control by coexpressing them in Arabidopsis leaf epidermal cells via biolistic DNA delivery. The nuclei of the target cells were selected as the region of interest (ROI) to monitor the FRET. Multichannel images collected before bleaching showed the colocalization of EGFP and mCherry clearly, with similar signal intensities in the nucleus (Figure 4A). When the acceptor was bleached in this experiment, a 75% reduction in mCherry emission was observed. However, this caused a negligible change in the signal intensity of the donor EGFP (Figures 4A and 4B), confirming that no FRET occurs between the free EGFP and mCherry even though they are colocalized. Additional controls with combinatorial fluorescent pairs (i.e., EGFP/AHL4-mCherry and AHL3-EGFP/mCherry) also confirmed the absence of FRET (Figures 4C and 4D).
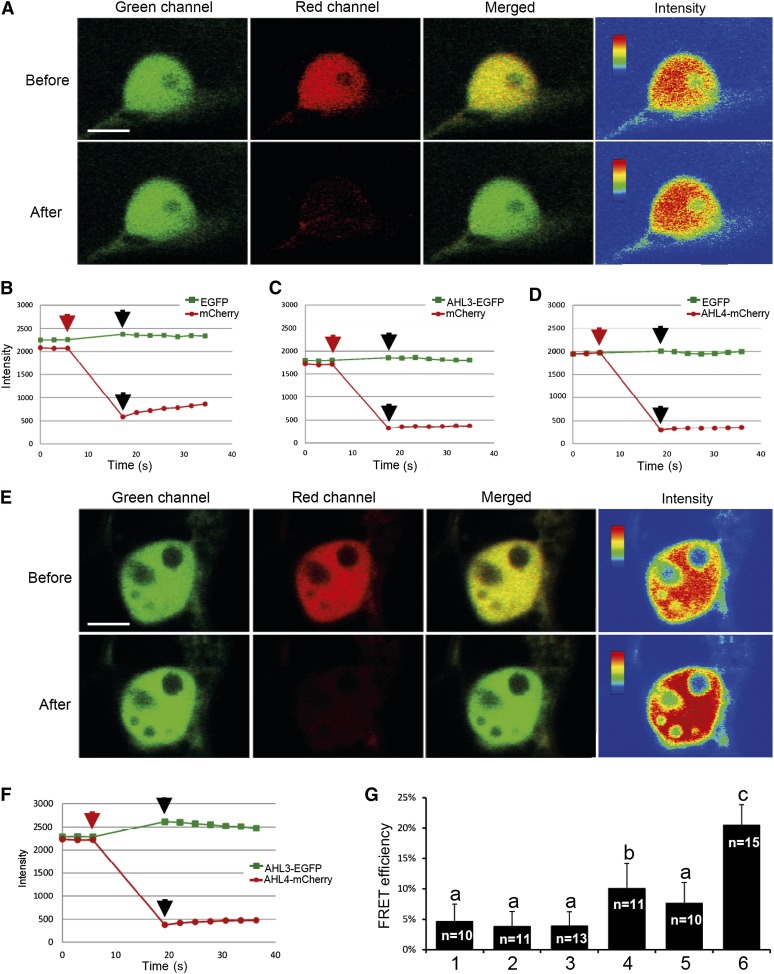
Figure 4.
The in Vivo Interaction between AHL4 and AHL3 as Determined by FRET Analysis.
(A) Single-scan confocal images taken before and after acceptor photobleaching of a representative example cell coexpressing EGFP and mCherry. The nucleus was selected as the ROI for photobleaching in this experiment. Fluorescence signals of EGFP and mCherry are presented in green, red, and merged channels. For the qualitative detection of FRET, EGFP fluorescence images before and after photobleaching are provided as pseudo-colored intensity maps.
(B) to (D) Quantitative changes in fluorescence intensities of the target nucleus labeled with EGFP and mCherry (B), AHL3-EGFP and mCherry (C), and EGFP and AHL4mCherry (D). Red arrows indicate prebleaching points, and black arrows indicate postbleaching points. The time lapse between the two points (marked by red and black arrows) in each experiment indicates the length of bleaching (10 s). Images shown in (A) represent the pre- and postbleaching points marked in (B).
(E) and (F) Single-scan confocal images taken before and after acceptor photobleaching of a representative cell coexpressing AHL3-EGFP and AHL4-mCherry (E) and quantification of the fluorescence intensities (F).
(G) Quantification of FRET efficiencies from all experiments: 1, EGFP and mCherry; 2, AHL3-EGFP and mCherry; 3, EGFP and AHL4-mCherry; 4, AHL3-EGFP and AHL3-mCherry; 5, AHL4-EGFP and AHL4-mCherry; and 6, AHL3-EGFP and AHL4-mCherry pairs. n, number of total repeats from three independent experiments. Statistical significance (levels not connected by same letter) was determined using analysis of variance test (P value < 1E-20). Bars indicate sd.
We then examined the existence of FRET between AHL3-EGFP as donor and AHL4-mCherry as acceptor under the same experimental conditions and parameters used for acceptor photobleaching of the negative controls. To monitor the FRET of this pair, the nuclei of target cells coexpressing both proteins were selected once more. Bleaching of the acceptor resulted in ∼80% loss of the AHL4-mCherry signal in a typical experiment as shown in Figures 4E and 4F. In contrast with the negative controls, however, the acceptor bleaching led to a significant gain in the AHL3-EGFP signal by ∼13% without background subtraction. For quantitative analysis, three independent FRET experiments for all combinations were subsequently performed. The result showed that the mean FRET efficiency between AHL3-EGFP and AHL4-mCherry was 20%, which was statistically significant compared with the mean values, 3 to 5%, of the negative controls (Figure 4G). This result provides solid evidence that the heteromeric complexes can form in planta. What about homomeric complexes, as suggested by the yeast two-hybrid experiment? Intriguingly, the FRET efficiency of homomeric AHL4 formation was statistically not different from the negative controls. By contrast, the FRET efficiency of homomeric AHL3 was slightly above the threshold level but still significantly lower than that of the AHL3/4 heteromeric complex (Figure 4G). Based on these FRET analyses, the heteromeric complex is likely the more prevalent form than the homomeric complex in planta. These FRET experiments firmly establish in vivo interaction between AHL4 and AHL3 and that this interaction is necessary for successful boundary formation between the xylem and procambium (see Discussion).
AHL4 and AHL3 Move Cell to Cell When Coexpressed in Leaf Epidermal Cells
Cell-to-cell movement of some transcription factors is influenced by their physical interaction with other transcription factors (Cui et al., 2007; Balkunde et al., 2011). Having established in vivo interaction between AHL3 and AHL4, we asked how their interaction relates to the cell-to-cell movement. To this end, we employed transient expression assays using Arabidopsis leaf tissues. AHL4 and AHL3 coding regions were fused to mCherry and EGFP, respectively, and biolistically bombarded into leaves detached from 3-week-old Arabidopsis Columbia-0 (Col-0). Control bombardment using vector plasmids expressing free EGFP (pcGC) or mCherry (pdCC) resulted in cell-to-cell movement in ∼30 to 50% of transfected cells (Table 2), ranging from limited (single cell layer) to extensive (more than two cell layers) movement, as has been observed previously under similar experimental conditions (Lee et al., 2011). AHL4-mCherry or AHL3-EGFP alone resulted in single epidermal cell expression only, revealing no movement activity. The total number of transfected cells examined was 144 and 187 each (Table 2). This result is not surprising, given that the intercellular movement of non-cell-autonomous transcription factors functioning in roots, such as SHORTROOT (SHR), is influenced by a cell type–specific factor, SCARECROW, for example (Sena et al., 2004; Cui et al., 2007). Expression data from the eFP Browser database indicate that neither AHL3 nor AHL4 is significantly expressed in the examined leaf developmental stage (Winter et al., 2007; see Supplemental Figure 1B online).
Table 2. Quantification of Cell-to-Cell Protein Trafficking.
| Proteins | Total No. of Transfected Cellsa | Total No. of Trafficking Eventsb | Trafficking Frequency (%) |
|---|---|---|---|
| Single expression | |||
| EGFP | 397 | 210 | 53 |
| mCherry | 257 | 94 | 37 |
| AHL4-mCherry | 144 | 0 | 0 |
| AHL3-EGFP | 187 | 0 | 0 |
| Coexpression | |||
| EGFP + mCherry | 370 | 178 (EGFP) | 48 |
| 178 (mCherry) | 48 | ||
| AHL4-mCherry + EGFP | 176 | 0 (AHL4-mCherry) | 0 |
| 91 (EGFP) | 52 | ||
| AHL3-EGFP + mCherry | 244 | 0 (AHL3-EGFP) | 0 |
| 82 (mCherry) | 34 | ||
| AHL4-mCherry + AHL3-EGFP | 239 | 6 (AHL4-mCherry) | 3 |
| 6 (AHL3-EGFP) | 3 |
Independent transfection events were scored by examining fluorescent epidermal cells under a confocal microscope at 48 h postbombardment using 3-week-old Arabidopsis Col-0 leaves. Results from three independent repeats were combined.
Detection of fluorescent signals within the cells surrounding a single transfected target cell was scored as one trafficking event regardless of the extent of movement.
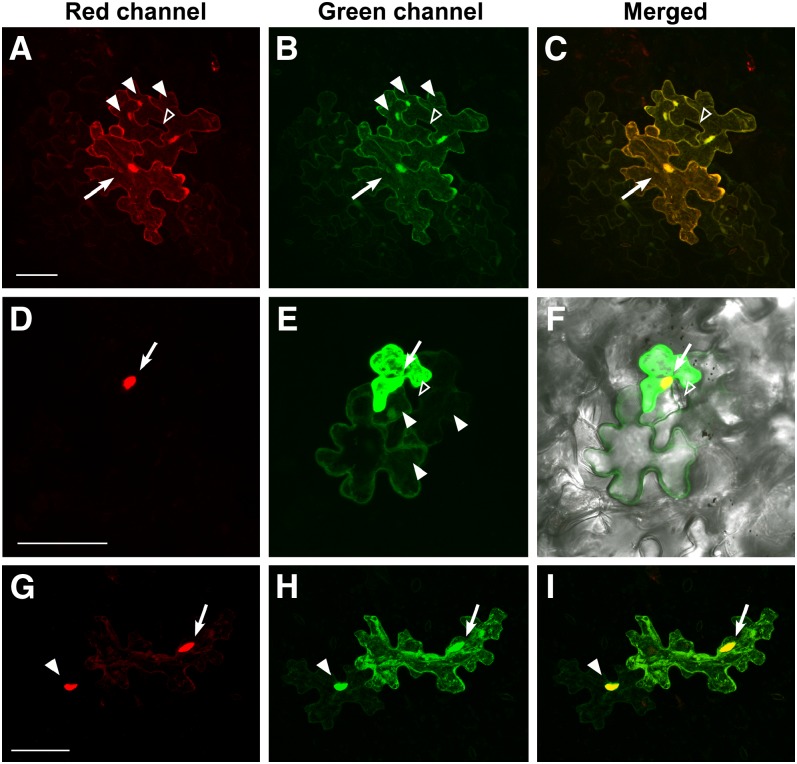
Next, we examined the intercellular movement of AHL4 in the presence of AHL3. As controls for dual-protein trafficking, we performed a combinatorial experiment in which AHL4-mCherry was cobombarded with pdGC for EGFP and AHL3-EGFP with pdCC for mCherry. These combinations showed that although free EGFP or mCherry moved out of the cotransfected target cell at a normal frequency, AHL4-mCherry or AHL3-EGFP did not (Figures 5A to 5F). This result is consistent with the lack of movement of AHL4 or AHL3 when expressed alone. However, to our surprise, when AHL4 and AHL3 were coexpressed, their movement into a neighboring epidermal cell was observed (Figures 5G to 5I). Interestingly, the fluorescent signals associated with both proteins were dispersed throughout the cytoplasm in addition to strong nuclear expression within the target cells, while accumulating disproportionately in the nucleus within the neighboring cell into which they moved. Although the occurrence of this movement was observed at a low frequency (3%) and the extent of movement was limited to a single neighboring cell, this movement was reproducible and consistent in multiple repeats (Table 2). This result does not address whether these proteins move as a complex per se. However, it supports the notion that coexpression facilitates their movement.
Figure 5.
AHL4 and AHL3 Move Together in Leaf Epidermal Cells.
Three-dimensional maximum intensity projection of serial z-sections capturing epidermal cells that coexpress fluorescent protein pairs from the transfected target cells (arrow) and the neighboring cells the fluorescent proteins moved into. Bars = 50 µm.
(A) to (C) Extensive diffusion of free mCherry (A) and EGFP (B) into the cytoplasms and nuclei (filled arrowhead) of adjacent cells. Note the lack of the fluorescent signals in mature guard cells (unfilled arrowhead).
(D) to (F) Trafficking of EGFP (filled arrowhead) (E) from the cotransfected cell (arrow) with AHL4-mCherry (D). Note the lack of EGFP diffusion into the guard cells (open arrowhead).
(G) to (I) Cotrafficking of AHL4-mCherry (G) and AHL3-EGFP (H) from the target cell (arrow) into a neighboring cell (closed arrowhead). Note the strong nuclear accumulation of both proteins.
Having found that AHL3 cotrafficks with AHL4 in leaf epidermis, we queried whether AHL3 is also capable of moving between root cells. Consistent with the bombardment assay, we observed GFP signals within the endodermis of the transgenic plants that expressed ProCRE1:AHL3:GFP in the wild-type background (Figure 3F). We also investigated whether the intercellular trafficking of AHL4 is facilitated by AHL3 as shown in the leaves, by examining AHL4-GFP localization in the ahl3-1 roots expressing ProAHL4:AHL4:GFP (Figure 3B). However, we did not find any significant reduction in the frequencies of xylem localization of AHL4-GFP in the ahl3-1. This result suggests that AHL4 movement is not fully dependent on AHL3, AHL4 may move on its own in the root, or another member of the AHL family may substitute for AHL3 in promoting AHL4 movement. Given that AHL4 requires AHL3 for activating downstream components, such a heteromeric complex would be functionally critical regardless of its involvement in the cell-to-cell mobility.
DISCUSSION
AHL3 and AHL4 Regulate the Formation of Tissue Boundary between the Procambium and Xylem in Arabidopsis Roots
In our study, we determined that the two novel AT-hook family proteins AHL3 and AHL4 regulate the establishment of the tissue boundary between the xylem and procambium. Disrupting the functions of either AHL3 or AHL4 caused the formation of additional proto- and metaxylem strands. The formation of extra xylem strands likely resulted from either of the following two events. First, it could have been from the abnormal cell division that triggered the formation of extra xylem precursor cells. Second, it could have been from the breakage of boundaries between cell types, which led to the misspecification of cell fates. We favor the latter scenario based on the analysis of the cell marker ProTMO5:erGFP in ahl4 roots presented in this study. The expression of this cell marker indicates that cell fate determination into the procambium or xylem in ahl4 roots is complete at a very early stage, being only one or two cells apart from the QC (see Supplemental Figure 4 online). Additionally, ProTMO5:erGFP expression expands as soon as the xylem axis becomes discernible immediately above the QC. Complementary to TMO5, ProARR5:erGFP expression was no longer detected in those cells (Figures 1K and 1M). If abnormal proliferation activity in the xylem precursor cells was attributable to the extra xylem formation, we should have detected xylem precursors being split in ahl4-1, but this was not the case. Therefore, we propose that AHL4 contributes to correct vascular patterning by defining the boundaries between procambium and xylem and not by inducing the cell division of xylem precursors.
For AHL4 to define the tissue boundary, its closest homolog, AHL3, is required. For example, reducing AHL3 expression results in an extra protoxylem phenotype similar to ahl4-1. The double mutant ahl3-1ahl4-1 phenocopies ahl3-1 and ahl4-1 single mutants, indicating that AHL3 and AHL4 function in the same pathway. Furthermore, our FRET analysis strongly supports that AHL3 and AHL4 directly interact in vivo. A recent study showed that a monocotyledon-specific AT-hook member in maize (Zea mays), BARREN STALK FASTIGIATED1, forms both hetero- and homodimers with other putative AT-hook DNA binding proteins in yeast (Gallavotti et al., 2011). It might be a common characteristic of AT-hook proteins that they function as heterocomplexes. In this regard, an important future investigation will be to delineate the relationship between the type of complex they form in vivo and their specific biological function.
Surprisingly, the transcriptional fusion line expressing ProAHL3:GUS revealed that AHL3 promoter activity is limited to the maturation zone of the root (see Supplemental Figure 6B online). Given that AHL3-GFP in ProAHL3:AHL3-GFP was detected in the procambium and xylem precursor cells of the meristematic zone, the ProAHL3:GUS data suggest the intriguing possibility that AHL3 produced in the maturation zone somehow has to travel to the root tip where it functions to regulate the boundaries. Dissecting the mechanism underlying this multilayered, non-cell-autonomous signaling pathway will be a challenging but worthwhile endeavor.
AHL3/4 Comovement, a Novel Characteristic of the AT-Hook Transcription Factors?
Consistent with the previously proposed roles associated with nuclear functions for AT-hook members, AHL3/4-GFP fusion proteins are selectively localized to the nuclei. Our investigation further shows that AHL4 proteins actively move between cells, likely through PD. When we increased the size of AHL4-YFP fusion proteins to 154 kD by expressing ProCRE1:AHL4:4xYFP in ahl4, we could still observe the movement of AHL4-YFP outside the stele domain. However, unlike the other known non-cell-autonomous proteins, CAPRICE (Kurata et al., 2005b) and SHR (Sena et al., 2004; Gallagher and Benfey, 2005; Gallagher and Benfey, 2009), AHL4 movement does not seem to be directional. In our analysis, AHL4 moved not only between the vascular cell types but also outside the vascular cylinder. Regardless, its movement from the procambium to the xylem precursor is crucial for regulating the boundary between the xylem and procambium. This argument is supported by our analysis of AHL4:3xYFP expressed under the AHL4 promoter in ahl4.
When ProAHL4:AHL4-GFP was introduced into ahl4, AHL4-GFP was also found in the xylem precursor and the extra xylem phenotype in ahl4 was complemented at 100%. On the contrary, AHL4-3xYFP in the xylem precursors was observed less frequently than AHL4-GFP. As a result, the frequency of the phenotypic complementation of ahl4 expressing ProAHL4:AHL4-3xYFP was significantly lower than the one expressing ProAHL4:AHL4-GFP. AHL4-3xYFP is fully functional given that it complements the ahl4 mutant phenotype when expressed under the CRE1 promoter. Thus, the decrease in the intercellular movement of AHL4 is likely responsible for the reduction in phenotypic complementation.
Thus far, AHL3 and AHL4 are the only AT-hook family members that are reported to move between cells. Further analysis on the domains of AHL3/4 proteins will help improve our understanding of the mechanisms underlying the intercellular movement of these proteins and establish whether the non-cell-autonomous activity is a shared feature of the AT-hook family members. As shown by studies of KNOTTED1 (KN1) and SHR, one of the mechanisms plants use to regulate the movement of non-cell-autonomous transcription factors involves the coupling of nuclear localization and intercellular trafficking (Lucas et al., 1995; Kim et al., 2005; Gallagher and Benfey, 2009; reviewed in Lee et al., 2010). Impairment of nuclear localization of KN1 and SHR abolished their movement between cells. The PPC domain has been reported to be important for the nuclear localization of AHL1, another AT-hook member in Arabidopsis (Fujimoto et al., 2004). It would be interesting to learn whether the PPC domain in AHL3 and AHL4 would provide a similar function. Analyzing the PPC domains of AHL members may also prove insightful in addressing whether the movement is a shared characteristic in the family and whether the nuclear localization is coupled to their non-cell-autonomous activity.
Intriguingly, our transient protein movement analysis in the Arabidopsis leaf suggests that the coexpression of AHL3 and AHL4 in epidermal cells facilitates their intercellular movement. The number of intercellular movements observed was low at only 3%. However, this is significant because the movement events were observed only when AHL3 and AHL4 were coexpressed. This suggests that the heteromeric complex formed between AHL3 and AHL4 could be more movement competent than the homomeric complex at least in leaf cells. Despite this observation in leaves, AHL4 movement was normal in ahl3 roots. We speculate that this apparent discrepancy could be explained if an additional AHL isoform(s), expressed preferentially in the root, can complement ahl3 for the AHL4 movement. Consistent with this notion, the relative expression of other AHL isoforms is much higher in roots than in leaves, similar to the expression patterns of AHL3 and AHL4 (see Supplemental Figure 1 online). Moreover, AHL4 can form a heteromeric complex not only with AHL3 but also with AHL1 (and perhaps other AHLs) in yeast two-hybrid assays. However, the heteromeric complex formed between AHL4 and AHL1 or other isoforms unlikely complements AHL4/AHL3 function in determining the boundary between the procambium and protoxylem.
Our current working model is that a specific heteromeric complex formation is required for their non-cell-autonomous activity and nuclear function. Our analysis of ProAHL3:GUS suggests that AHL3 might be predominantly expressed in the elongation and maturation zone in the root (see Supplemental Figure 6B online). Together with the AHL3 protein expression pattern observed in ProAHL3:AHL3:GFP, we propose that AHL3 moves from the upper part of the root to the root tip, where it interacts with AHL4 to maintain the boundaries. If this is correct, the intercellular movement of AHL3 is unlikely to require AHL4. Further in-depth studies would be necessary before we fully comprehend the mechanisms by which AHL3 and AHL4 function non-cell-autonomously during boundary formation.
As for the movement of AHL4, we speculate that this could be facilitated by the presence of AHL3. However, given that AHL4 can move in ahl3-1, AHL3 might not be the only factor that affects AHL4 movement, as discussed above. For example, it is conceivable that additional AT-hook members, such as AHL1 and AHL6, may also be involved in facilitating the cell-to-cell movement of AHL4 in roots. AHL1 and 6 are not only closely related to AHL3 and 4 but are also highly expressed in the roots (see Supplemental Figure 1 online). Notably, our preliminary data suggested that AHL1 indeed interacts with AHL4, pointing to the possibility that AHL1 and AHL6 might also interact with AHL3 and AHL4 to facilitate cooperative movement. However, it is most probable that these complexes, although mobility competent, are not functional in the boundary formation between the procambium and xylem and that this function requires an AHL3/4 heteromeric complex. Future investigations on additional AHL members and their functional relationships may open the door to the discovery of novel mechanisms by which combinatorial nuclear factors are recruited as mobile cellular signaling molecules to determine specific developmental processes.
AHL3/4 and Hormonal Signaling in Xylem Patterning
In the ahl3 or ahl4 mutant, the formation of extra xylem strands occurs not only to the protoxylem, but also to the metaxylem. This phenotype is a novel phenotype, as there is no prior report of single mutants that affect tissue boundaries for both the protoxylem and metaxylem in the root. The vascular pattern in the root meristem is established long before the vascular cell types become morphologically distinctive. It has been reported that high levels of cytokinin and auxin are distributed in the root meristem in a mutually exclusive manner. Such a distribution pattern was suggested to contribute to the delineation of the procambium and protoxylem domains (Bishopp et al., 2011a). A high level of auxin in the protoxylem precursor cells was found to promote the expression of AHP6 (Bishopp et al., 2011a). AHP6, a cytokinin signaling inhibitor, in turn suppresses the cytokinin signal to establish the protoxylem precursor cell domain in the root meristem (Mähönen et al., 2006). High levels of cytokinin in the procambium facilitate this process by regulating auxin efflux carriers that channel auxin maxima to the protoxylem precursors and thereby maintain procambium cell identity. Therefore, disrupting the balance between cytokinin and auxin levels alters the formation of protoxylem strands, and cytokinin and auxin antagonize each other to define the boundaries between the procambium and protoxylem (Bishopp et al., 2011a, 2011b).
TMO5 is a direct target gene of MONOPTEROS/AUXIN RESPONSE FACTOR5 (Schlereth et al., 2010). ProTMO5:erGFP in the ahl4 also exhibits the expansion of its expression. Complementary to these observations, ARR5, an A-type Arabidopsis response regulator induced by high cytokinin, retracts its expression from the extra xylem domains in ahl4 (Figure 1M) (D’Agostino et al., 2000; Bishopp et al., 2011a). We also observed the expansion of ProAHP6:erGFP in ahl4, which is highly similar to what happens in the cytokinin signaling mutants (Figure 1L). These results indicate that loss of AHL4 function affects both the auxin and cytokinin domains in the vascular tissue. We thus asked whether AHL4 is involved in the regulation of cytokinin signaling. In wild-type seedlings, exogenous cytokinin treatment inhibits protoxylem formation, whereas in cytokinin signaling mutants, it does not (Mähönen et al., 2006; Yokoyama et al., 2007; Argyros et al., 2008). To test the possibility that AHL4 is a part of cytokinin signaling, ahl4-1 seedlings were treated with exogenous cytokinin. This experiment showed that ahl4-1 seedlings were as responsive to cytokinin as the wild-type seedlings, failing to form the protoxylem (see Supplemental Figures 9A to 9F online). We also investigated whether AHL4 expression is affected by high levels of cytokinin. When we treated roots expressing ProAHL4:GUS with exogenous cytokinin, we observed dramatic expansion of GUS expression domains to the elongation and maturation zones. We also found a change of GUS expression from the boundary cells between the xylem and phloem to the boundary cells neighboring metaxylem as well as the metaxylem precursors (see Supplemental Figures 6D to 6H online). Despite the dramatic response of AHL4 expression to the exogenous cytokinin, we did not find changes in the spatial distribution of AHL4 proteins in the ProAHL4:AHL4:GFP transgenic roots that were treated with cytokinin (see Supplemental Figure 9G online). Thus, AHL4 does not seem to mediate changes in protoxylem domains under high cytokinin. However, these data do not rule out the possibility that expansion of the protoxylem domain in the cytokinin receptor mutants, such as cre1 ahk3, might result from the suppression of AHL4. Taken together, we speculate that AHL3/4 regulates tissue boundaries in parallel to, or partially overlapping with, the cytokinin pathway.
Concluding Remarks
Approximately 25% of transcription factors in Arabidopsis move from one cell to another (Lee et al., 2006). Several studies have shown their involvement in the cell-type specification; however, their involvement in the boundary formation between cell types has not been demonstrated until now. Xylem vessels generated from the procambium/cambium are critical for distributing water and minerals throughout plant bodies. The xylem also constitutes a significant portion of plant biomasses. Densities and numbers of xylem vessels in stems change depending on the water availability. Considering that the root is an entry point for water uptake from the soil, there are likely molecular mechanisms that enable control of xylem vessel formation specifically in the root in response to dynamic environmental challenges. AHL3 and AHL4, together with cytokinin signaling, might govern at least part of these processes at transcriptional and/or posttranscriptional levels. Further investigation will broaden the understanding of this underexplored area of plant biology.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used. Seeds were surface sterilized, plated (0.5× Murashige and Skoog [MS] medium with 1% Suc), and grown under a 16-h-light/8-h-dark cycle at 22 to 23°C in a plant growth chamber. ahl4-1 (SALK_124619) was obtained from the ABRC, and ahl3-1 (FLAG_445H04) was obtained from Versailles Genetics and Plant Breeding Laboratory Arabidopsis thaliana Resource Centre (INRA Versailles France). The following marker lines were characterized previously: ProTMO5:erGFP (Lee et al., 2006), ProARR5:erGFP (Lee et al., 2006), ProAHP6:erGFP (Mähönen et al., 2006), and ProCRE1:erGFP (Mähönen et al., 2000). The primers used for genotyping are listed in Supplemental Table 2 online.
Plasmid Construction
Gateway cloning technology (Invitrogen) was used for DNA manipulations. AHL4, AHL3, and CRE1 promoters were amplified from Arabidopsis Col-0 genomic DNA and cloned into pDONR P4_P1R. AHL3 and AHL4 cDNA were cloned into pDONR221. 3xYFP and 4xYFP in pDONR P2R_P3 have been previously described (Tsukagoshi et al., 2010). ProCRE1:AHL3/4:GFP, ProAHL4:erGFP, ProAHL4:AHL4:GFP, and ProAHL3:AHL3:GFP were constructed in dpGreen-Bar (Lee et al., 2006) by MultiSite Gateway LR recombination. ProCRE1:AHL4:3x/4xYFP and ProAHL4:GUS were constructed into dpGreen-BarT. A suitable target site for the microRNA targeting AHL3/4 was identified and generated by following the instructions on http://wmd3.weigelworld.org/cgi-bin/webapp.cgi, except that oligoA was modified to clone amiRNA-AHL3-4 into a pENTR/D-TOPO vector. ProCRE1:amiRNA-AHL3-4 was constructed into dpGreen-BarT by MultiSite Gateway LR recombination. All clones in the binary vector were transformed into Agrobacterium tumefaciens GV3101 with pSOUP and transformed into either the wild type or ahl4-1. Coding regions of AHL3 and AHL4 were introduced into pDEST22 and pDEST32 by Gateway LR recombination to fuse AHL3 and AHL4 with the GAL4 activation domain and the GAL4 DNA binding domain, respectively. For the coexpression and trafficking experiments, AHL4 and AHL3 were cloned into the pdGC and pdCC expression vectors that are designed to produce C-terminal fluorescent fusion proteins (Lee et al., 2005; Ben-Nissan et al., 2008). Briefly, AHL4 and AHL3 open reading frames were PCR amplified using Phusion Taq polymerase (NEB) followed by restriction enzyme double digestion with XhoI and KpnI and direct cloning into XhoI- and KpnI-digested pdGC and pdCC, respectively. The primers used for cloning are listed in Supplemental Table 2 online.
Histological Analysis
All seedling samples were collected at 6 d after germination. Confocal images were obtained using a Leica TCS SP5 laser scanning confocal microscope with the preset excitation/emission wavelengths of 488 nm/505 to 530 nm for GFP, 510 nm/525 to 560 nm for YFP, and 561 nm/591 to 635 nm for propidium iodide. For the visualization of root structure, all seedlings were stained with 2 µg/mL propidium iodide. Root transverse sections and toluidine blue staining were performed as described previously (Scheres et al., 1995). Basic fuchsin staining has been described elsewhere (Mähönen et al., 2000). GUS staining was performed as described previously (Sundaresan et al., 1995) and followed with root transverse sectioning, as described above. To analyze the response of AHL4 to cytokinin, ProAHL4:GUS or ahl4-1 seeds were plated on MS plates and grown for 3 d. Then, half of the seedlings were transferred onto MS plates containing 50 nM benzyl aminopurine. After growing seedlings for another 3 d, they were processed for GUS expression analysis or xylem phenotyping.
Confocal Imaging for Fluorescence Intensity Measurement
For quantitative analysis of AHL4 movement, GFP/YFP intensity was measured in both the endodermis and its neighboring pericycle cells in ProCRE1:AHL4:GFP and ProCRE1:AHL4:3x/4xYFP. For each genetic background, five plants from each of four or five independent lines were examined. A plant was first examined longitudinally under a ×63 objective to locate a focal plane where the adjacent endodermis and pericycle cell files were aligned in parallel (see Supplemental Figure 4A online). Then, the image was zoomed in 2.2 times to center the aforementioned endodermis and pericycle cells (see Supplemental Figures 4B and 4C online). Two sequential images were obtained on slightly different focal planes to capture the fluorescence signals within the center of the nuclei in the endodermis and pericycle, respectively. All the images were captured using the same setting. At least three pairs of nuclei aligned parallel to the endodermis and pericycle cells were measured for fluorescence signal intensity by ImageJ software, and the integrated pixel intensity was recorded for further analysis.
In Situ Hybridization
Six-day-old roots were fixed, embedded, and sectioned for in situ hybridization as previously described (Mähönen et al., 2000). We amplified 700-bp long GFP DNA templates including a T7 promoter for either sense or antisense probes, and probes were hydrolyzed to generate working probes of 150 bp long. The primers used for the riboprobes are listed in Supplemental Table 2 online.
Quantitative Real-Time RT-PCR
Two-millimeter-long sections of root tips from 6-d-old seedlings were harvested, and total RNAs were isolated with an RNeasy mini kit (Qiagen). cDNA was synthesized using a SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) as described previously (Carlsbecker et al., 2010). The primers used for gene expression level measurement are listed in Supplemental Table 2 online.
Yeast Two-Hybrid Assay
A ProQuest two-hybrid system (Invitrogen) was used for yeast two-hybrid analysis. All the procedures were performed according to the manufacturer’s standard protocol. Recombinant hybrid proteins were tested for self-activation. Plasmid DNA pairs between pEXP32-Krev1 and pEXP22-RalGDS-wt, m1, and m2 were used as controls for strong positive, weak positive, and negative interactions, respectively.
Transient Expression in Leaf Epidermal Cells
Biolistic DNA delivery for the transient expression and cell-to-cell movement assay in Arabidopsis was basically performed as previously described (Lee et al., 2011). Briefly, leaves were detached from 3-week-old Arabidopsis Col-0 and layered on an MS agar plate for bombardment. An equal amount of DNA (5 µg each) was mixed and used for gold coating for coexpression assays. The bombarded leaves were examined at 48 h for cell-to-cell movement assays. At least three independent bombardment assays were performed for each or a combination of constructs, and at least 50 to 100 cells on average were examined per bombardment.
Confocal Microscopy for Trafficking and FRET Analysis
Confocal microscopy for epidermal cell-to-cell movement was performed using an LSM 510 META scanhead on a Zeiss LSM 5 DUO confocal microscope as described by Lee et al. (2011). A series of optical sections was acquired as Z-stacks and rendered as three-dimensional maximum intensity projections with Zeiss LSM 510 AIM software (Rel. 4.2). Cells were examined with a ×40 C-Apochromat (1.2 numerical aperture) water immersion objective lens and 4× zoom.
For acceptor photobleaching FRET analysis, the following microscope settings were used: Single-labeled images were used to determine spectral bleed-through levels, and dual labeled images were acquired in fast-line switch mode to eliminate crosstalk. Images of single-labeled donor (EGFP or AHL3-EGFP) or acceptor (mCherry or AHL4-mCherry) and multilabeled donor plus acceptor were collected at the donor excitation wavelength (488 nm) with a 505- to 550-nm band-pass emission filter or acceptor excitation wavelength (561 nm) with a 575- to 615-nm band-pass emission filter. Ten multiscanned images for each experiment were collected using the same imaging conditions and parameters. mCherry was photobleached by continuously scanning ROI with the 561-nm laser line set at 100% intensity for 10 s. Multichannel images of EGFP and mCherry were then collected using the respective excitation lines. To minimize photobleaching during the imaging process, images were collected at 0.2% of the laser intensity. To ensure that imaging-associated bleaching was minimal, we monitored the level of bleaching in each experiment by collecting three EGFP/mCherry image pairs before bleaching and seven after bleaching. Data were collected from several cells and at least three independent experiments were conducted. Recovery of the donor from quenching was quantified by subtracting the donor emission after bleaching from the donor emission before bleaching. The FRET images were processed by background subtraction. Processing and analysis of FRET images, pseudocoloring of fluorescence intensities, and calculation of FRET efficiencies were performed using the software and methods of Zeiss Zen 2010 D (v7,0,0,223).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AHL1 (AT4G12080), AHL3 (AT4G25320), AHL4 (AT5G51590), and AHL6 (AT5G62260). Accession numbers for the xylem precursor-enriched transcription factors are labeled in Supplemental Figure 1A online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Patterns of AHLs and Other Xylem-Enriched Transcription Factors.
Supplemental Figure 2. Isolation of ahl4-1 and ahl3-1.
Supplemental Figure 3. Xylem Phenotyping.
Supplemental Figure 4. Xylem Development in ahl4.
Supplemental Figure 5. Phloem Development in ahl4.
Supplemental Figure 6. Transcriptional Domains of AHL3 and AHL4.
Supplemental Figure 7. Confocal Microscopy for the Quantification of AHL4 Movement, Shown in Figures 2M and 2N.
Supplemental Figure 8. Ectopic Xylem Formation in the ahl3-1 and amiRNA Lines.
Supplemental Figure 9. Sensitivity of ahl4-1 to Exogenous Cytokinin Treatment.
Supplemental Table 1. AHL4-GFP/AHL4-3xYFP Localization in the Xylem and Its Correlation to Phenotype Complementation.
Supplemental Table 2. List of Primers Used in This Article.
Acknowledgments
We thank James Eaglesham for assisting in the genotyping, and Mike Scanlon, Tom Brutnell, Susan McCouch, and the Lee lab members for commenting on this article. This article is a part of the doctoral dissertation prepared by Jing Zhou. This work was supported by the startup funds from Boyce Thompson Institute and Seoul National University to Ji-Young Lee, grants from the National Science Foundation to Ji-Young Lee (IOS-0818071) and to Jung-Youn Lee (IOS-0954931), and partially by grants from the National Center for Research Resources (5P30RR031160-03) and the National Institute of General Medical Sciences (8 P30 GM103519-03) from the National Institutes of Health to Jung-Youn Lee. We thank the Plant Cell Imaging Center at the Boyce Thompson Institute for supporting microscopy and J. Caplan at the University of Delaware for helpful discussion on FRET technique and analysis.
AUTHOR CONTRIBUTIONS
J.Z. and Ji-Young Lee designed the research. J.Z. performed research. X.W. performed the FRET and epidermal trafficking analyses. J.Z., Jung-Young Lee, and Ji-Young Lee wrote the article.
Glossary
- HMG
high mobility group
- QC
quiescent center
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- YFP
yellow fluorescent protein
- PD
plasmodesmata
- FRET
fluorescence resonance energy transfer
- EGFP
enhanced green fluorescent protein
- ROI
region of interest
- Col-0
Columbia-0
- PPC
plant and prokaryotes conserved
- MS
Murashige and Skoog
References
- Aravind L., Landsman D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26: 4413–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkunde R., Bouyer D., Hülskamp M. (2011). Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138: 5039–5048 [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G., Cui W., Kim D.J., Yang Y., Yoo B.C., Lee J.Y. (2008). Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol. 148: 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A., Help H., El-Showk S., Weijers D., Scheres B., Friml J., Benková E., Mähönen A.P., Helariutta Y. (2011a). A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21: 917–926 [DOI] [PubMed] [Google Scholar]
- Bishopp A., Lehesranta S., Vatén A., Help H., El-Showk S., Scheres B., Helariutta K., Mähönen A.P., Sakakibara H., Helariutta Y. (2011b). Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 21: 927–932 [DOI] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mähönen A.P., Hauser M.-T., Helariutta Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Ward M.F., Sama A.E., Wang H. (2004). Extracellular HMGB1 as a proinflammatory cytokine. J. Interferon Cytokine Res. 24: 329–333 [DOI] [PubMed] [Google Scholar]
- Crawford K.M., Zambryski P.C. (2001). Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 125: 1802–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- D’Agostino I.B., Deruère J., Kieber J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R.N., Periasamy A., Schaufele F. (2001). Fluorescence resonance energy transfer microscopy of localized protein interactions in the living cell nucleus. Methods 25: 4–18 [DOI] [PubMed] [Google Scholar]
- De Smet I., Beeckman T. (2011). Asymmetric cell division in land plants and algae: The driving force for differentiation. Nat. Rev. Mol. Cell Biol. 12: 177–188 [DOI] [PubMed] [Google Scholar]
- Etchells J.P., Turner S.R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Fisher K., Turner S. (2007). PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Fujimoto S., Matsunaga S., Yonemura M., Uchiyama S., Azuma T., Fukui K. (2004). Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol. Biol. 56: 225–239 [DOI] [PubMed] [Google Scholar]
- Gallagher K.L., Benfey P.N. (2005). Not just another hole in the wall: Understanding intercellular protein trafficking. Genes Dev. 19: 189–195 [DOI] [PubMed] [Google Scholar]
- Gallagher K.L., Benfey P.N. (2009). Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 57: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Malcomber S., Gaines C., Stanfield S., Whipple C., Kellogg E., Schmidt R.J. (2011). BARREN STALK FASTIGIATE1 is an AT-hook protein required for the formation of maize ears. Plant Cell 23: 1756–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2011). Establishment and maintenance of vascular cell communities through local signaling. Curr. Opin. Plant Biol. 14: 17–23 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., et al. (2007). Extracellular hmgb1 functions as an innate immune-mediator implicated in murine cardiac allograft acute rejection. Am. J. Transplant. 7: 799–808 [DOI] [PubMed] [Google Scholar]
- Karpova T.S., Baumann C.T., He L., Wu X., Grammer A., Lipsky P., Hager G.L., McNally J.G. (2003). Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209: 56–70 [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Rim Y., Wang J., Jackson D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19: 788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T., Okada K., Wada T. (2005a). Intercellular movement of transcription factors. Curr. Opin. Plant Biol. 8: 600–605 [DOI] [PubMed] [Google Scholar]
- Kurata T., et al. (2005b). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Cho S.K., Sager R. (2010). Plasmodesmata and non-cell-autonomous signaling in plants. In The Plant Plasma Membrane, 1st ed, Angus S. Murphy, Burkhard Schulz, and Wendy Peer, eds (Berlin Heidelberg:Springer), pp. 87–108
- Lee J.Y., Colinas J., Wang J.Y., Mace D., Ohler U., Benfey P.N. (2006). Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Taoka K.-i., Yoo B.-C., Ben-Nissan G., Kim D.-J., Lucas W.J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Wang X., Cui W., Sager R., Modla S., Czymmek K., Zybaliov B., van Wijk K., Zhang C., Lu H., Lakshmanan V. (2011). A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Kim Y., Breeze E., Koo J.C., Woo H.R., Ryu J.S., Park D.H., Beynon J., Tabrett A., Buchanan-Wollaston V., Nam H.G. (2007). Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52: 1140–1153 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouché-Pillon S., Jackson D.P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983 [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bishopp A., Higuchi M., Nieminen K.M., Kinoshita K., Törmäkangas K., Ikeda Y., Oka A., Kakimoto T., Helariutta Y. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A., Furumoto T., Ishida S., Takahashi Y. (2007). AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 143: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisawa G., Han-Yama A., Moda I., Tamai A., Iwabuchi M., Meshi T. (2000). AHM1, a novel type of nuclear matrix-localized, MAR binding protein with a single AT hook and a J domain-homologous region. Plant Cell 12: 1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.H., Ito T. (2010). Shedding light on the role of AT-hook/PPC domain protein in Arabidopsis thaliana. Plant Signal. Behav. 5: 200–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.H., Yu H., Ito T. (2009). AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 7: e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. (2001). Molecular biology of HMGA proteins: Hubs of nuclear function. Gene 277: 63–81 [DOI] [PubMed] [Google Scholar]
- Reeves R., Beckerbauer L. (2001). HMGI/Y proteins: Flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519: 13–29 [DOI] [PubMed] [Google Scholar]
- Rim Y., Huang L., Chu H., Han X., Cho W., Jeon C., Kim H., Hong J.-C., Lucas W., Kim J.-Y. (2011). Analysis of Arabidopsis transcription factor families revealed extensive capacity for cell-to-cell movement as well as discrete trafficking patterns. Mol. Cells 32: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cázares B., Lucas W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126: 4405–4419 [DOI] [PubMed] [Google Scholar]
- Scheres B., Benfey P.N. (1999). Asymmetric cell division in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 505–537 [DOI] [PubMed] [Google Scholar]
- Scheres B., Di Laurenzio L., Willemsen V., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E.H., Schmid M., Jürgens G., Weijers D. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Sena G., Jung J.W., Benfey P.N. (2004). A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Street I.H., Shah P.K., Smith A.M., Avery N., Neff M.M. (2008). The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 54: 1–14 [DOI] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tramier M., Zahid M., Mevel J.C., Masse M.J., Coppey-Moisan M. (2006). Sensitivity of CFP/YFP and GFP/mCherry pairs to donor photobleaching on FRET determination by fluorescence lifetime imaging microscopy in living cells. Microsc. Res. Tech. 69: 933–939 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Van Norman J.M., Breakfield N.W., Benfey P.N. (2011). Intercellular communication during plant development. Plant Cell 23: 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dinneny J.R., Crawford K.M., Rhee Y., Citovsky V., Zambryski P.C., Weigel D. (2003). Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Goué N., Igarashi H., Ohtani M., Nakano Y., Mortimer J.C., Nishikubo N., Kubo M., Katayama Y., Kakegawa K., Dupree P., Demura T. (2010b). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153: 906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Kubo M., Fukuda H., Demura T. (2008). Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Ohtani M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., Demura T. (2010a). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Tewary P., de la Rosa G., Wei F., Oppenheim J.J. (2010). The alarmin functions of high-mobility group proteins. Biochim. Biophys. Acta Mech. 1799: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A., Yamashino T., Amano Y.-I., Tajima Y., Imamura A., Sakakibara H., Mizuno T. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48: 84–96 [DOI] [PubMed] [Google Scholar]
- Zambryski P., Crawford K. (2000). Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 16: 393–421 [DOI] [PubMed] [Google Scholar]