Diatom cell division is controlled by light. In this work, the diatom-specific cyclin dsCYC2 is identified as a rate-limiting factor that controls the onset of the cell cycle in response to blue light. Strikingly, dsCYC2 expression is under the direct control of an aureochrome blue light receptor.
Abstract
Cell division in photosynthetic organisms is tightly regulated by light. Although the light dependency of the onset of the cell cycle has been well characterized in various phototrophs, little is known about the cellular signaling cascades connecting light perception to cell cycle activation and progression. Here, we demonstrate that diatom-specific cyclin 2 (dsCYC2) in Phaeodactylum tricornutum displays a transcriptional peak within 15 min after light exposure, long before the onset of cell division. The product of dsCYC2 binds to the cyclin-dependent kinase CDKA1 and can complement G1 cyclin-deficient yeast. Consistent with the role of dsCYC2 in controlling a G1-to-S light-dependent cell cycle checkpoint, dsCYC2 silencing decreases the rate of cell division in diatoms exposed to light-dark cycles but not to constant light. Transcriptional induction of dsCYC2 is triggered by blue light in a fluence rate-dependent manner. Consistent with this, dsCYC2 is a transcriptional target of the blue light sensor AUREOCHROME1a, which functions synergistically with the basic leucine zipper (bZIP) transcription factor bZIP10 to induce dsCYC2 transcription. The functional characterization of a cyclin whose transcription is controlled by light and whose activity connects light signaling to cell cycle progression contributes significantly to our understanding of the molecular mechanisms underlying light-dependent cell cycle onset in diatoms.
INTRODUCTION
In eukaryotes, the presence of various cell cycle checkpoints ensures that the genetic information in a cell is inherited correctly by inhibiting the replication and distribution of incomplete or damaged chromosomes to the daughter cells. The major cell cycle checkpoints occur during the onset of DNA replication (G1-to-S transition) and mitosis (G2-to-M transition). During the mid-to-late G1 phase, most organisms exhibit a commitment point, before which a number of intra- and extracellular conditions must be fulfilled (Hartwell et al., 1974; Pardee, 1974; Spudich and Sager, 1980; Moulager et al., 2010). Beyond this commitment point, cells complete their cell cycle and become independent of mitogenic stimuli, such as growth factors or nutrients and, in the case of phototrophs, light. In Chlamydomonas reinhardtii, the commitment point has been shown to be preceded by a primary arrest point in G1 at which cell cycle progression becomes light dependent (Spudich and Sager, 1980).
Despite the fact that light plays a key role in the growth of photoautotrophic organisms, as demonstrated by the light-driven expression of various cell cycle genes (Bisova et al., 2005; Moulager et al., 2007, 2010; López-Juez et al., 2008; Huysman et al., 2010; Moriyama et al., 2010), little is known about the cellular signaling mechanisms that connect light perception with the activation of the cell cycle machinery in the nucleus, which includes cyclin-dependent kinases (CDKs) and their interaction partners, the cyclins (CYCs) (Morgan, 1997; Inzé and De Veylder, 2006). In the green alga Ostreococcus tauri, cyclin A plays an important role during S phase entry. This gene, the first cell cycle gene to be transcribed in the organism after dawn, is translated in a cyclic adenosine monophosphate (cAMP)-dependent manner only when cells have acquired adequate levels of light energy, thereby reflecting the metabolic state of the cells (Moulager et al., 2010). In the red alga Cyanidioschyzon merolae, the inhibition of cyclin 1 degradation through a tetrapyrrole-mediated signaling pathway during the shift from dark to light was shown to be crucial for connecting organellar and nuclear DNA replication (Kobayashi et al., 2011).
Here, we studied the molecular regulation of the light-dependent checkpoint in diatoms. Diatoms are unicellular algae that dominate primary production in many aquatic ecosystems and are responsible for about 20% of global photosynthetic carbon fixation (Van den Hoek et al., 1995; Field et al., 1998; Mann, 1999). Diatoms can grow and photosynthesize over a wide range of different light intensities and wavelengths (Holdsworth, 1985; Mercado et al., 2004), and they possess specific light sensing and acclimation strategies (Nymark et al., 2009; Bailleul et al., 2010; Park et al., 2010; Zhu and Green, 2010; Lepetit et al., 2012). Light quality and intensity not only determine the photosynthetic capacity of diatoms, but it also affects different cellular processes, including motility, sexual reproduction, and cell division (Brzezinski et al., 1990; Chen et al., 2004; Cohn et al., 2004; McLachlan et al., 2009; Mouget et al., 2009). Analogous to C. reinhardtii, the cell cycle of diatoms consists of light-dependent and -independent segments (Vaulot et al., 1986). The two major diatom groups, the centrics and the pennates (Kooistra et al., 2003; Sims et al., 2006), appear to have evolved different light-sensitive phases during their mitotic cell cycles. Flow cytometric analyses of dark-adapted cells have shown that in centric species, two light-sensitive stages are present during their cell cycle, namely, the G1 and G2/M phases (Olson et al., 1986; Vaulot et al., 1986; Brzezinski et al., 1990). Some pennate species have been reported to show a similar G1 and G2/M arrest, as reported for Cylindrotheca fusiformis (Brzezinski et al., 1990), while others display only a G1 arrest, as in Phaeodactylum tricornutum (Huysman et al., 2010) and Seminavis robusta (Gillard et al., 2008). For those species with only a light-dependent segment at the G1 phase, the immediate release of dark-arrested cells has proven to be a useful characteristic to synchronize and study the cell division process (Gillard et al., 2008; Huysman et al., 2010).
Although the light dependency of the diatom cell cycle was demonstrated more than 20 years ago (Olson et al., 1986; Vaulot et al., 1986; Brzezinski et al., 1990), to date, nothing is known about the molecular regulators that control the light-dependent cell cycle checkpoints in diatoms. In a previous study, we identified many members of the cyclin gene family in the pennate diatom P. tricornutum and the centric Thalassiosira pseudonana and described a class of diatom-specific cyclins involved in environmental signaling (Huysman et al., 2010). One of the most strongly and earliest expressed genes during the switch from dark to light in synchronized cells is the diatom-specific cyclin2 (dsCYC2), hinting at a role for this cyclin in cell cycle activation after dark arrest. To address this hypothesis, we studied the role of dsCYC2 at the light-dependent G1 checkpoint and investigated the light-dependent transcriptional regulation of this gene in P. tricornutum.
RESULTS
Light-Dependent Transcriptional and Translational Control of dsCYC2
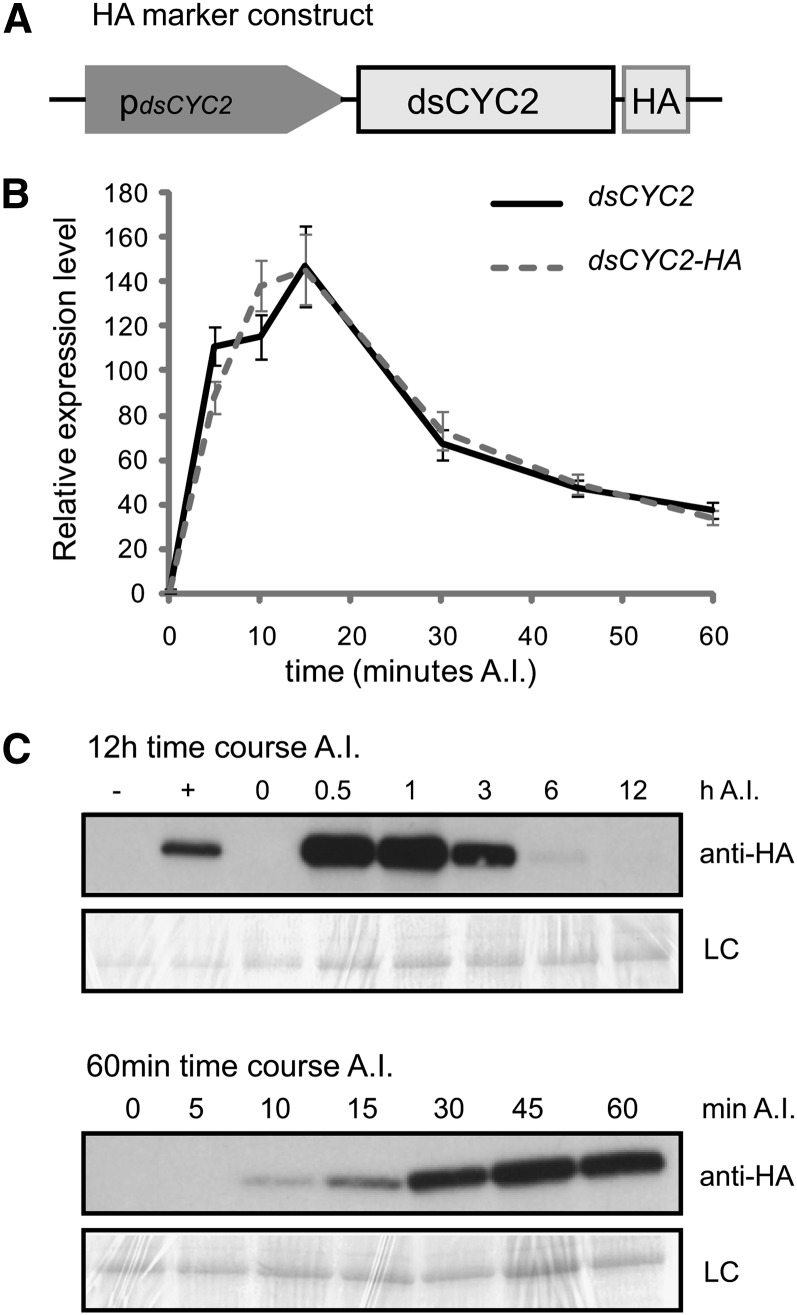
As previously shown, the transcript level of dsCYC2 changes abruptly upon exposure of dark-grown P. tricornutum cells to light (Huysman et al., 2010). To document the kinetics of dsCYC2 transcript and protein abundance upon illumination, we generated a transgenic marker line that expressed the full-length dsCYC2 open reading frame (ORF) C-terminally fused to a hemagglutinin (HA) tag under the control of the dsCYC2 promoter (pdsCYC2), which we will refer to as the HA marker line (Figure 1A). To determine the kinetics of dsCYC2 transcript levels after light exposure, we conducted a finely resolved sampling experiment during the first hour after illumination of dark-arrested cells. To this end, cells were grown exponentially under a 12-h-light/12-h-dark (12L/12D) regime and then transferred to the dark for a prolonged period (24 h) that, due to a light-dependent segment within the G1 phase, enriches cultures for G1 phase cells (Brzezinski et al., 1990; Huysman et al., 2010). When returned to light, cells progress synchronously through the cell cycle starting from the G1 phase (Huysman et al., 2010). After illumination, samples were taken at 0, 5, 10, 15, 30, 45, and 60 min for real-time quantitative PCR to monitor dsCYC2 transcript levels. An initial increase in transcript levels was observed after only 5 min of illumination, reaching a peak at 15 min, followed by a rapid decrease of the dsCYC2 mRNA levels (Figure 1B). Protein gel blot analysis over a 12-h time course showed that dsCYC2-HA protein was undetectable immediately after illumination but reached high levels at 30 to 60 min, decreasing gradually thereafter to become undetectable by 12 h (Figure 1C). Protein analysis during the first hour after illumination showed increasing levels of dsCYC2-HA starting from 10 min until 60 min after light exposure (Figure 1C), although the transcript levels were markedly lower at the later time point (Figure 1B). These data show that upon illumination, dsCYC2 transcript levels instantly reach a peak within 10 to 15 min, followed by a translational peak 30 to 60 min after light exposure.
Figure 1.
Light-Dependent Transcription and Translation of dsCYC2.
(A) Schematic representation of the HA marker construct.
(B) Transcript levels of dsCYC2 (solid line) and HA-tagged dsCYC2 (dashed line) during a 60-min time course after illumination (A.I.) of 24-h dark-adapted HA marker cells. Values were normalized against those obtained for histone H4 and then rescaled to the gene expression levels at 0 min after illumination (=1). Error bars represent se of three technical replicates.
(C) dsCYC2-HA protein levels during a 12-h (top panel) and 60-min (bottom panel) time course after illumination of 24-h dark-adapted HA marker cells. −, Negative control (wild-type 4 h light); +, positive control (HA 4 h light). LC, loading control by Coomassie blue staining.
dsCYC2 Interacts with CDKA1 but Not CDKA2
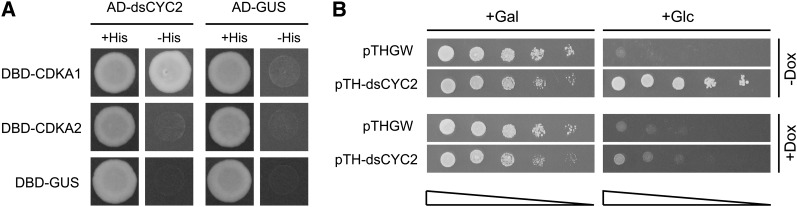
To explore the role of dsCYC2 during the cell cycle, we tested whether dsCYC2 can bind to the most conserved CDKs of P. tricornutum. To this end, we performed a yeast two-hybrid (Y2H) interaction assay in which P. tricornutum CDKA1, a G1/S-regulated cyclin-dependent kinase (CDK) containing the amino acid PSTAIRE motif, and CDKA2, a mitotically expressed CDK containing a PSTALRE motif (Huysman et al., 2010), were used as bait and dsCYC2 as prey. Growth on selective His-lacking medium was observed for the combination of dsCYC2 with CDKA1, but not with CDKA2 or any of the controls (Figure 2A). Complex formation between dsCYC2 and CDKA1 is supported by their coexpression at the G1-to-S transition in synchronized cells (Huysman et al., 2010).
Figure 2.
dsCYC2 Functions as a G1-Cyclin.
(A) Interaction of dsCYC2 with CDKA1. Yeast PJ694-α cells were cotransformed with bait and prey plasmid as indicated. Cotransformation was analyzed on medium lacking Leu and Trp (+His). Cotransformants were tested for their ability to activate the His marker gene by assessing yeast growth on medium lacking Leu, Trp, and His (-His). Constructs containing β-glucuronidase (GUS) were used as negative controls. For each combination, three independent colonies were screened, one of which is shown.
(B) Complementation of G1 cyclin–deficient yeast by dsCYC2. BF305-15d-21 cells were transformed with pTHGW (vector control) or pTH-dsCYC2. Yeast cells were serially diluted and spotted onto SD-Ura plates containing Gal (+Gal) or Glc (+Glc). When Glc was the sole carbon source, control cells were not able to grow because of the lack of G1 cyclin expression, while cells that expressed dsCYC2 overcame this phenotype. When dsCYC2 expression was repressed by the addition of doxycycline (+Dox), the complementation was lost.
Complementation of a Conditional G1 Cyclin-Deficient Yeast Mutant by Expression of dsCYC2
The interaction of dsCYC2 with CDKA1, and its peak in abundance during the early cell cycle, suggested that dsCYC2 might encode a G1-specific cyclin controlling the G1/S transition. Therefore, we examined whether dsCYC2 is able to functionally substitute for yeast G1 cyclins using a complementation assay in the yeast strain BF305-15d-21. BF305-15d-21 cells contain mutations in the endogenous cyclin1 (CLN1) and cyclin2 (CLN2) genes and express cyclin3 (CLN3) from a Gal-inducible promoter (Xiong et al., 1991). Hence, these cells are able to divide only in the presence of Gal. On Glc-containing medium, CLN3 expression is repressed and cells arrest at a regulatory transition point in the G1 phase. BF305-15d-21 cells were transformed with the pTH-dsCYC2 vector, containing the dsCYC2 ORF under control of a doxycycline-repressible promoter. Cells containing pTH-dsCYC2 were able to resume division in the presence of Glc (Figure 2B). When dsCYC2 expression was repressed by doxycycline, complementation did not occur (Figure 2B), confirming that the complementation was linked to dsCYC2 expression. These results demonstrate that dsCYC2 encodes a functional cyclin that is able to complement a G1 cyclin–deficient yeast strain.
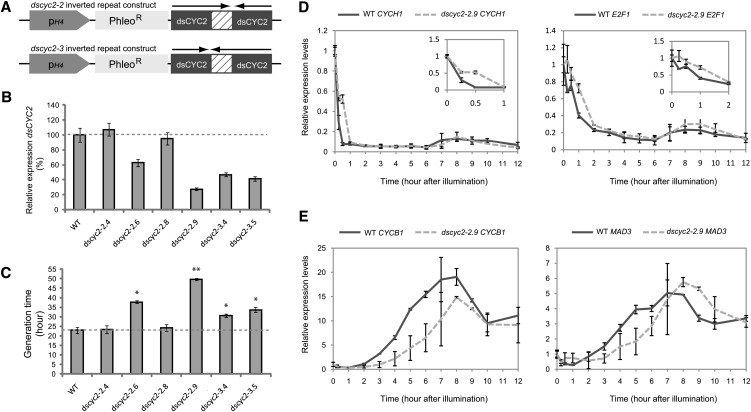
Silencing dsCYC2 Slows Cell Cycle Progression by Prolonging the Light-Dependent G1 Phase
The early light-dependent transcription of dsCYC2 suggests that its gene product plays a role in the reactivation of cell division upon illumination. To test this hypothesis, P. tricornutum dsCYC2 knockdown lines were generated by introducing a hairpin construct under control of the constitutive histone H4 promoter (De Riso et al., 2009) targeting the N-terminal region of dsCYC2 (Figure 3A). Silencing was evaluated at 15 min after illumination by comparing dsCYC2 transcript levels in wild-type cells and six independent transgenic lines harboring the RNA interference constructs. Two lines (dscyc2-2.4 and dscyc2-2.8) showed no silencing of dsCYC2, while four other lines (dscyc2-2.6, dscyc2-2.9, dscyc2-3.4, and dscyc2-3.5) showed a 40 to 75% reduction in transcript level compared with wild-type cells (Figure 3B). To test whether dsCYC2 silencing had an effect on cell cycle progression, growth rate analysis was performed on wild-type and dsCYC2 knockdown lines grown under a 12L/12D regime. No effect was observed for the nonsilenced internal control lines (dscyc2-2.4 and dscyc2-2.8) (Figure 3C). By contrast, all knockdown lines (dscyc2-2.6, dscyc2-2.9, dscyc2-3.4, and dscyc2-3.5) showed a significant increase in generation time compared with wild-type cells (Figure 3C), indicating that dsCYC2 is crucial for proper cell cycle progression.
Figure 3.
Effect of dsCYC2 Silencing on Cell Cycle Progression.
(A) Schematic representation of the dsCYC2 inverted repeat constructs used for silencing analysis. In the dscyc2-2 construct, the large fragment is positioned first, followed by the small fragment. In the dscyc2-3 construct, the small fragment is followed by the large fragment (arrows).
(B) Real-time quantitative PCR analysis of dsCYC2 transcript levels in wild-type (WT) and silenced lines. Cells were dark adapted for 24 h, and transcript levels were measured 15 min after light exposure. Transcript levels of wild-type cells were set at 100%.
(C) Generation times of wild-type and dsCYC2 silenced lines grown at 100 µE 12L/12D cycles. Error bars (in (B) and (C) represent sd of the mean of three independent experiments. *P < 0.005; **P < 0.001 (two-tailed Student’s t test).
(D) and (E) Transcript expression profiles of G1 marker genes (D) and mitotic markers (E) during a synchronized time course in wild-type and dscyc2-2.9 knockdown cells. Error bars represent se of two biological replicates.
Expression analysis of different cell cycle marker genes during the light-dependent cell cycle reentry of 24-h dark-arrested wild-type and dscyc2-2.9 cells indicated that silencing of dsCYC2 results in the attenuation of G1 progression upon light exposure. Expression of the early cell cycle marker genes cyclin H1 (CYCH1) and the transcription factor E2F1 was extended in the silenced versus wild-type cells (Figure 3D), indicating that cells with lower dsCYC2 expression levels spend more time in the G1 phase and are delayed in the onset of the cell cycle upon illumination. Also, the timing of expression of the G2/M markers CYCB1 and MAD3 (Figure 3E) was clearly delayed in the dscyc2-2.9 cells compared with wild-type cells. Thus, in addition to having an effect on cell cycle initiation at the G1 checkpoint after dark arrest, the absence of dsCYC2 expression also affects the timing of all downstream cell cycle transitions. The possibility that dsCYC2 silencing affected transcription in general or produced a general stress response could be excluded, as several miscellaneous genes that were examined showed no differential expression in wild-type versus silenced lines (see Supplemental Figure 1 online).
If dsCYC2 acts primarily at the light-dependent G1 checkpoint, no growth defects would be expected in dscyc2 cells that do not experience a dark arrest. Therefore, we monitored the growth rates of wild-type and dscyc2-2.9 cells grown under constant light conditions. Because cells grow faster and reach the stationary phase earlier in constant light compared with 12L/12D cycles, this experiment was performed at lower light intensities (50 µE) to enable the detection of the exponential phase in the growth curves. While a clear reduction of cell growth rate was observed in dscyc2-2.9 compared with wild-type cells grown in 12L/12D (Figure 3C), no significant difference in growth rate was observed when cells were grown in constant light (Table 1). However, when cells grown under continuous light were moved to 12L/12D conditions, the cells regained the growth phenotype within 3 d (Table 1). Since P. tricornutum cells are only light dependent at the G1 phase, these data suggest a primary role for dsCYC2 at the G1 phase.
Table 1. Generation Times (h) of Wild-Type (WT) and dscyc2-2.9 Cells Grown in Constant White Light at 50 µE or Shifted to a 12L/12D Cycle.
| Strain | LL | LD (Early, Days 1 and 2) | LD (Late, Days 3 to 5) |
|---|---|---|---|
| WT | 29.5 ± 3.4 | 29.2 ± 0.0 | 42.3 ± 3.9 |
| dscyc2-2.9 | 30.1 ± 1.5 | 31.2 ± 0.3 | 59.1 ± 1.0 |
LD, light-dark cycle; LL, constant white light.
Wavelength and Fluence Rate Dependency of dsCYC2 Transcription
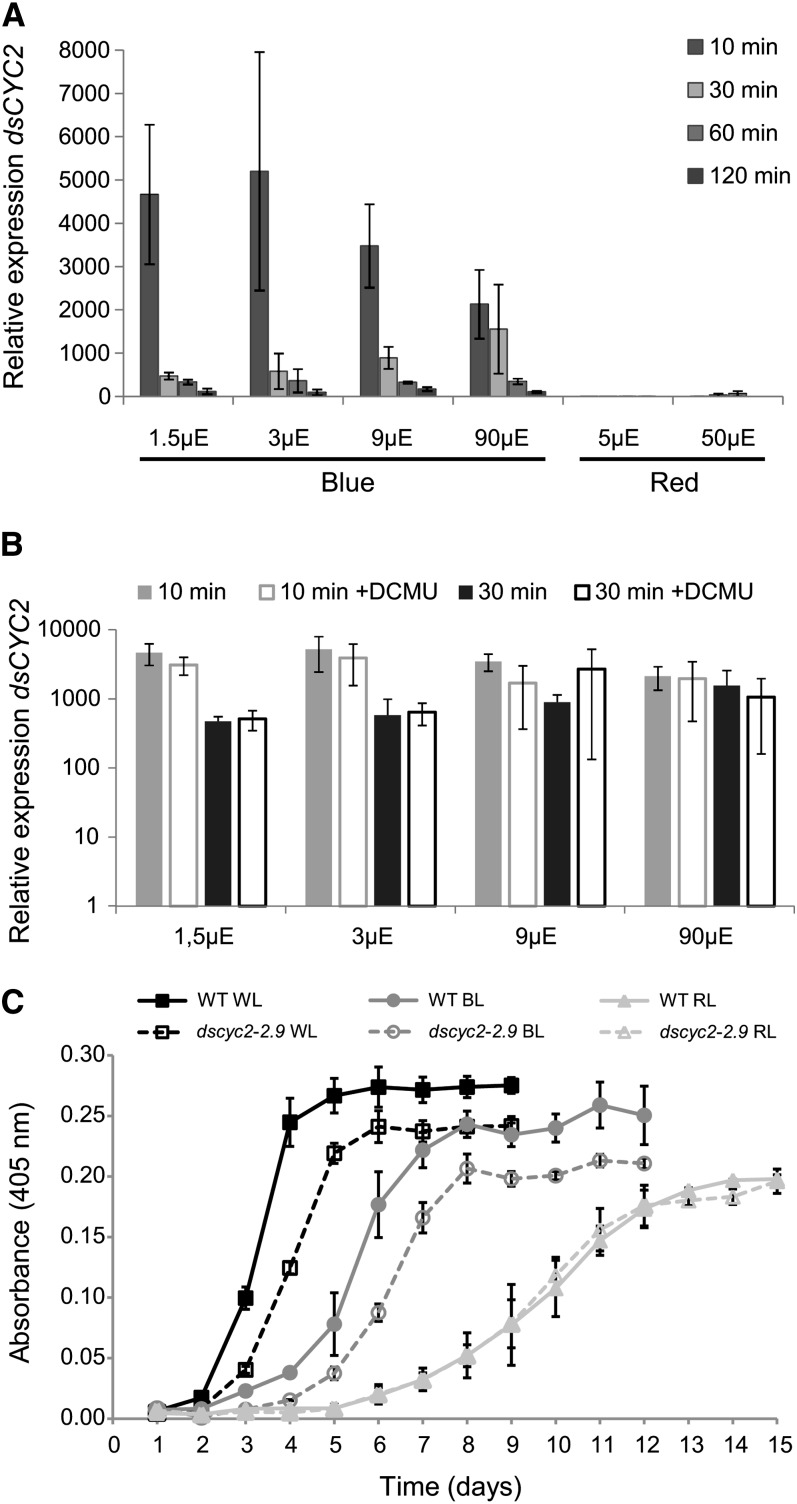
To determine whether the light-regulated induction of dsCYC2 transcripts is photoreceptor mediated, transcript levels were examined under different light conditions, including blue and red light at different fluence rates. As observed for white light, dark-adapted cells that were shifted to blue light showed an increase of dsCYC2 transcript levels 10 min after light exposure, with a stronger effect at lower light intensities (Figure 4A). Although dsCYC2 induction was lower under 90 µE blue light, the transcript levels remained high for a longer period of time compared with lower light intensities. In contrast with blue light, exposing dark-adapted cells to red light did not result in major changes in dsCYC2 transcript levels at either low or higher light intensities (Figure 4A).
Figure 4.
Blue Light Photoreceptor-Mediated Control of dsCYC2 Induction.
(A) Wavelength and fluence rate dependency of dsCYC2 induction. Wild-type cultures were dark incubated for 60 h and switched to blue or red light at different light intensities, as indicated. Relative mRNA levels of dsCYC2 at 10, 30, 60, and 120 min after light exposure are shown. Relative levels were normalized to histone H4 levels and rescaled to the expression level in dark-incubated cells (=1).
(B) Effect of DCMU on dsCYC2 induction. Log scale representation of the relative mRNA levels of dsCYC2 at 10 and 30 min after blue light exposure at different light intensities in the absence or presence of DCMU. In (A) and (B), error bars represent se of two biological replicates.
(C) Growth curves of wild-type (WT) and dscyc2-2.9 cells grown in white (WL), blue (BL), and red (RL) light adjusted to equal values of photosynthetically absorbed radiation. Error bars represent sd of three biological replicates.
To determine whether dsCYC2 induction is solely photoreceptor mediated or, to some extent, also controlled by photosynthesis-mediated metabolic changes, the effect of the addition of DCMU during the light period was tested (Figure 4B). DCMU is a specific inhibitor of noncyclic photosynthetic electron transport (PET) and blocks the transfer of electrons from photosystem II to the plastoquinone pool. The addition of DCMU prior to blue light exposure had no effect on the induction of dsCYC2 at 10 or 30 min after illumination (Figure 4B), suggesting that dsCYC2 induction is photoreceptor mediated and not dependent on PET.
To assess the effects of light color and intensity on diatom growth and the role of dsCYC2 under these conditions, the growth rates of wild-type and dsCYC2 knockdown cells exposed to a 12L/12D photoperiod of white, blue, or red light adjusted to equal values of photosynthetically absorbed radiation (QPhar) were determined. Because dsCYC2 is only induced when blue light is present, no difference in growth rate was expected to occur under red light conditions. Indeed, while dsCYC2 knockdown cells grew more slowly than wild-type cells under white and blue light, no difference was observed between the control and transgenic cultures under red light (Figure 4C). These results support the role of blue light-induced dsCYC2 expression during the cell cycle in P. tricornutum cells. In general, cells grown in red light showed a lower growth rate than those grown in blue or white light, highlighting the importance of blue light for diatom growth.
Regulation of the dsCYC2 Promoter by Light
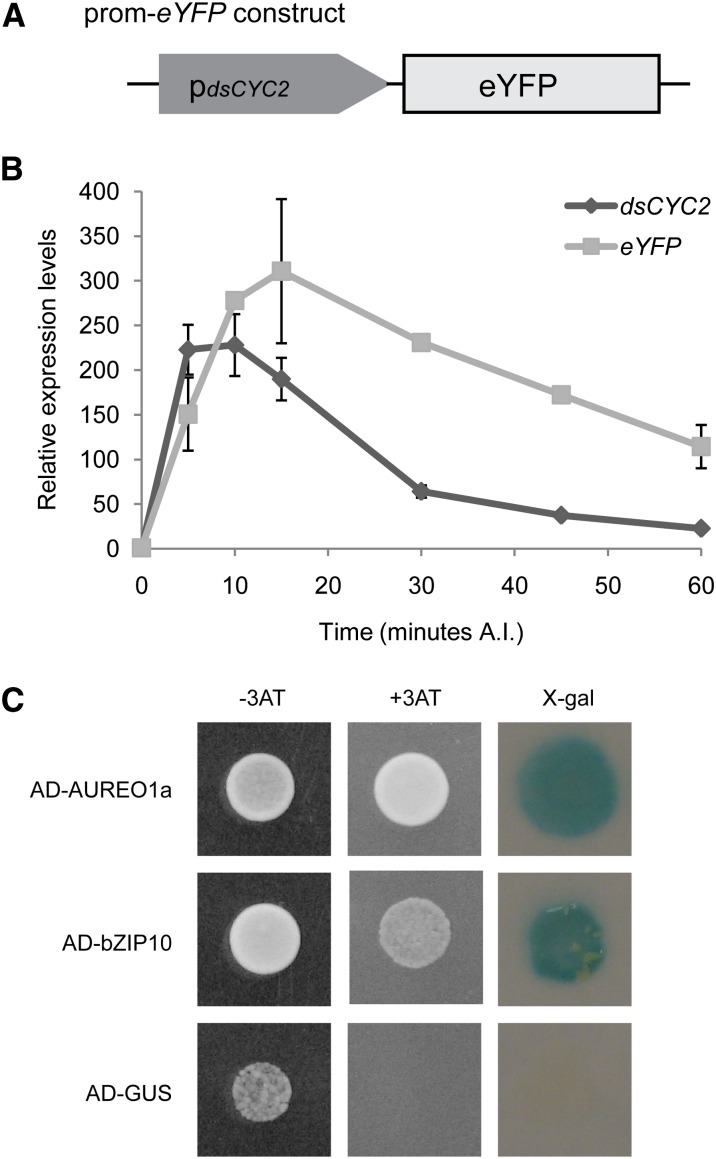
The observation that dsCYC2 transcript levels were markedly affected by light suggests that either light has a direct effect on dsCYC2 transcript stability or there is a yet undefined light-dependent signaling pathway that targets the dsCYC2 promoter sequence. In order to distinguish between these possibilities, we analyzed the short-term expression kinetics of a reporter gene placed under control of the dsCYC2 promoter during the light period following dark incubation. To construct this reporter fusion, we combined the 1018-bp region upstream of the translational start of dsCYC2 (pdsCYC2) and the coding region of enhanced yellow fluorescent protein (eYFP) (Figure 5A). Similar to dsCYC2 transcript levels, eYFP transcript levels were induced shortly after light exposure and dropped again to basal levels after longer periods of illumination (Figure 5B). The slight delay in the decrease of eYFP transcript compared with the kinetics of the dsCYC2 transcript (Figure 5B) is likely due to the higher intrinsic stability of eYFP versus dsCYC2 mRNA. Nevertheless, the overall parallel kinetics of the endogenous dsCYC2 transcript and the eYFP reporter transcript over time suggest that changes in dsCYC2 mRNA are primarily a consequence of changes in promoter activity rather than transcript stability.
Figure 5.
Regulation of the dsCYC2 Promoter by Light.
(A) Schematic representation of the prom-eYFP marker (right) constructs.
(B) Transcript levels of dsCYC2 and eYFP during a 60-min time course after illumination (A.I.) of 24-h dark-adapted pdsCYC2-eYFP cells. Values were normalized against H4 expression levels and rescaled to the levels at 0 min after illumination (= 1). Error bars represent se of two biological replicates.
(C) Y1H protein-DNA interaction assay. Interactions are positive when HIS3 (growth on 3-aminotriazole–containing medium (+3AT) and LacZ (X-Gal turns blue) expression is induced. Constructs containing GUS were used as negative controls. For each combination, three independent colonies were screened, one of which is shown.
AUREOCHROME1a and bZIP10 Associate with the dsCYC2 Promoter
To identify transcription factors that can bind to and regulate the dsCYC2 promoter, a genome-wide yeast one-hybrid (Y1H) cDNA library screen was conducted. To this end, a pdsCYC2 reporter yeast strain was generated harboring the dsCYC2 promoter upstream of the HIS3 and the LacZ reporter genes, and this strain was transformed with a yeast-compatible P. tricornutum cDNA library. The screen yielded two predicted basic leucine zipper (bZIP) transcription factors, AUREOCHROME1a (AUREO1a) and bZIP10. AUREO1a is a putative blue light photoreceptor that contains an N-terminal bZIP domain responsible for DNA binding and dimer formation and a C-terminal LOV (for light, oxygen, voltage) domain responsible for light sensing (Takahashi et al., 2007; Depauw et al., 2012). bZIP10 is a classical bZIP transcription factor (Rayko et al., 2010). Retransformation of AUREO1a and bZIP10 in the Y1H reporter strain confirmed their binding to the dsCYC2 promoter, as indicated by auxotrophic growth on selective medium and the expression of the LacZ gene (Figure 5C).
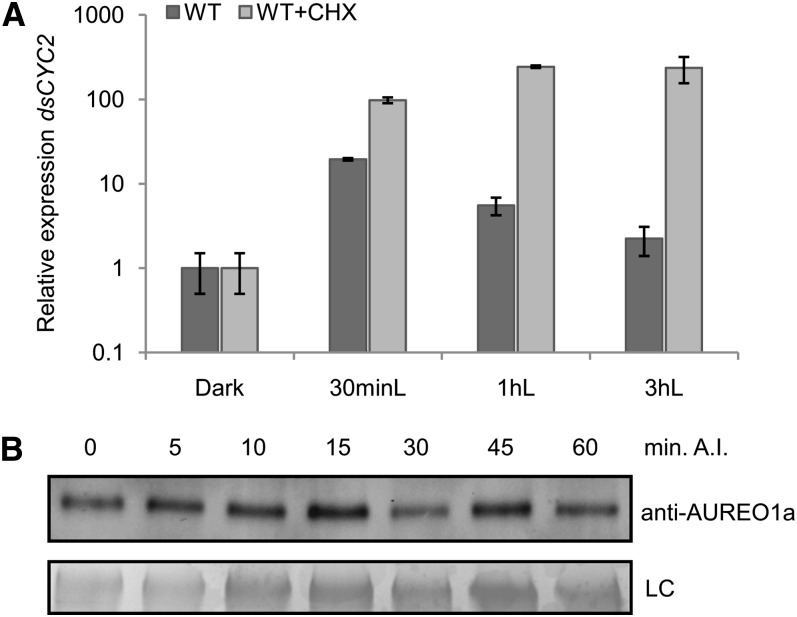
Posttranslational Control of dsCYC2 Induction
Light-dependent transcriptional induction of dsCYC2 through the activation of the LOV domain of AUREO1a would be expected to occur without the need for de novo protein synthesis. To test this hypothesis, dsCYC2 transcription was measured in wild-type cells treated with cycloheximide (CHX), an inhibitor of eukaryotic translation, just before illumination. As predicted, CHX treatment did not impair the light-dependent induction of dsCYC2 (Figure 6A). Surprisingly, in contrast with the control cultures that showed a decrease of dsCYC2 transcript levels following the initial transcriptional peak during the first hours after light exposure, transcripts accumulated to high levels in the CHX-treated cultures (Figure 6A), suggesting that upon illumination, a repressor is produced de novo that specifically targets the promoter activity of dsCYC2 to repress its expression. The addition of CHX during the dark period did not alter dsCYC2 transcript levels (see Supplemental Figure 2 online), indicating that the effect of CHX was specific to light exposure. Together, these data corroborate the hypothesis of induction of dsCYC2 transcription through activation of a photoreceptor, such as AUREO1a. Moreover, protein gel blot analysis demonstrated constitutive levels of AUREO1a during the switch from dark to light (Figure 6B), which suggests posttranslational activation of AUREO1a upon light exposure.
Figure 6.
Posttranslational Regulation of dsCYC2 Induction upon Light Exposure.
(A) Wild-type (WT) cultures were synchronized by 24-h dark treatment (Dark) and then exposed to light for 0.5 (30 minL), 1 (1 hL), or 3 h (3 hL) in the absence (dark gray) or presence (light gray) of 2 μg/mL CHX. Relative expression levels of dsCYC2 are shown. Values were normalized against H4 expression levels and then rescaled to the gene expression levels of the dark sample (=1). Error bars represent se of two independent experiments.
(B) AUREO1a protein levels during a 60-min time course after illumination (A.I.) of 24-h dark-adapted HA marker cells. LC, loading control by Coomassie blue staining.
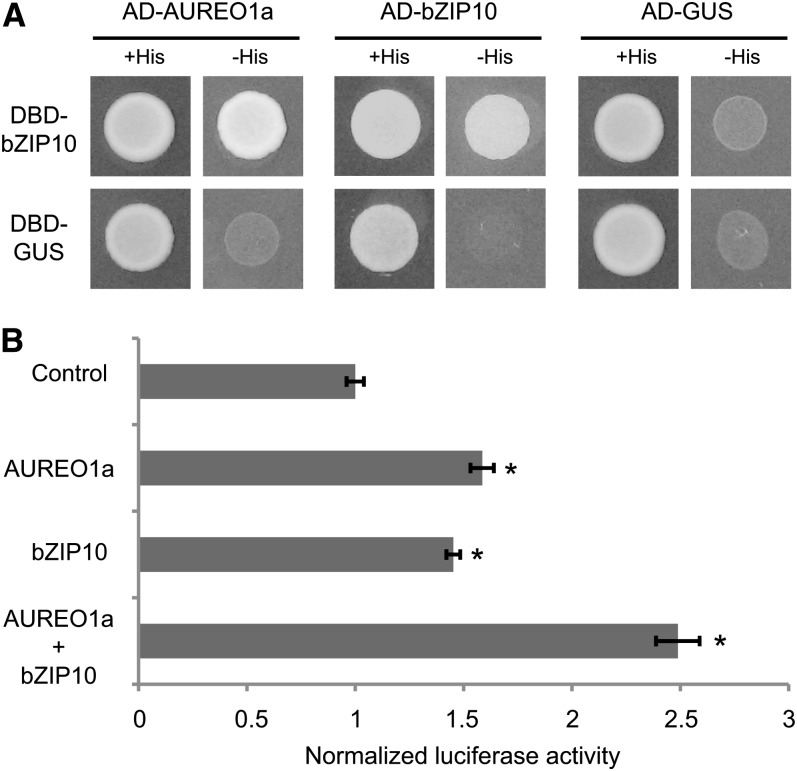
Activation of the dsCYC2 Promoter by AUREO1a and bZIP10
Because bZIP proteins are known to function as homo- or heterodimers (Schütze et al., 2008), a Y2H interaction assay was performed to test whether AUREO1a and bZIP10 can interact with themselves or with each other. In this test, when AUREO1a was used as a bait, there were high levels of self-activation, precluding any conclusions about interactions. However, when bZIP10 was used as a bait, an interaction was found to occur with both bZIP10 and AUREO1a, as indicated by auxotrophic growth on His-lacking medium (Figure 7A).
Figure 7.
Activation of the dsCYC2 Promoter by AUREO1a and bZIP10.
(A) Y2H protein–protein interaction assay. Yeast cells were cotransformed with bait and prey plasmid as indicated. Cotransformation was analyzed on medium lacking Leu and Trp (+His). Cotransformants were tested for their ability to activate the His marker gene by assessing yeast growth on medium lacking Leu, Trp, and His (-His). Constructs containing GUS were used as negative controls. For each combination, three independent colonies were screened, one of which is shown.
(B) Protoplast transactivation assay using pdsCYC2:fLUC as reporter, p35S:rLUC as normalization, and p35S:AUREO1a and p35S:bZIP10 as effector constructs. Luciferase activity of the control was arbitrarily set to 1. Error bars represent se of three biological replicates (*P ≤ 0.05, two-sided t test).
To assess the effect of AUREO1a and bZIP10 on dsCYC2 promoter activity, a transient activity assay was performed. AUREO1a and bZIP10 effector plasmids were transiently transformed either alone or together, along with a pdsCYC2:fLUC reporter construct, into tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) protoplast cells. When provided alone, both AUREO1a and bZIP10 slightly activated the dsCYC2 promoter (Figure 7B). However, the activation effect was significantly increased when both effector plasmids were coexpressed (Figure 7B). These data suggest that AUREO1a and bZIP10 function in a synergistic manner to activate the dsCYC2 promoter in response to light.
DISCUSSION
dsCYC2 Functions at the Light-Dependent G1 Checkpoint
For any photosynthetic organism, including diatoms, light is an extremely important factor that influences growth. Because diatoms can grow over a wide range of light intensities and wavelengths, these organisms are believed to have developed specific photoacclimation and photoadaptation mechanisms (Huisman et al., 2004; Lavaud et al., 2004; Lavaud et al., 2007). As with most other phytoplankton species, the timing of diatom cell division can be entrained by alternating periods of light and dark, implying that the cell cycle consists of light-dependent and -independent segments (Vaulot et al., 1986). Accordingly, both by light limitation and deprivation experiments, light-controlled restriction points have been identified in several diatom species, either during the G1 phase or during both the G1 and G2/M phases of the cell cycle (Olson et al., 1986; Vaulot et al., 1986; Brzezinski et al., 1990; Gillard et al., 2008; Huysman et al., 2010).
Previous work highlighted the role of dsCYCs in linking diverse environmental conditions to the cell cycle in diatoms (Bowler et al., 2008; Huysman et al., 2010). Here, we functionally characterized dsCYC2 as a crucial regulator of cell cycle onset after a period of darkness in P. tricornutum. Upon light exposure, dsCYC2 mRNA levels increase within minutes, followed by the induction of dsCYC2 protein. The specific expression of dsCYC2 during the G1 phase and its ability to complement G1 cyclin–deficient yeast cells suggest that dsCYC2 operates early in the cell cycle. A role for dsCYC2 in cell cycle entry is supported by the observation that lower dsCYC2 levels following light exposure prolong the G1-to-S phase transition, as shown by the delayed and altered transcript levels of the G1 markers. Also G2/M marker genes displayed a delayed expression of about 1 to 2 h compared with wild-type cells. Thus, although dsCYC2 likely acts primarily at the point of cell cycle onset, it appears that its effects go well beyond the early time points. From growth rate analysis, we determined that dscyc2-2.9 cells have a generation time almost double that of wild-type cells; thus, we would have expected to observe a longer mitotic delay. Most likely this difference can be explained by the observation that ∼10 to 15% of the dscyc2-2.9 cells are not cycling but appear to be arrested at the S phase, as observed from DNA abundance measurements (see Supplemental Figure 3 online). Therefore, the observed longer generation time most likely results from the cumulative effect of a slower cell cycle progression at the G1 phase of the cycling cells and an S phase arrest of a subset of the cells. The increase in S phase cells suggests that a prime action of dsCYC2 is to activate the CDK/cyclin complexes that are required for DNA replication, a function similar to that of the G1-specific CLN1/2 and cyclin E genes in budding yeast and mammalian somatic cells, respectively (Morgan, 2007). Although dsCYC2 protein can be detected during the S phase, it is unlikely that dsCYC2 controls DNA replication itself, as no difference in generation time was observed between control and dsCYC2 silenced lines under constant light conditions. Together with the observation that dsCYC2 levels only peak at the G1/S transition under dark/light cycles, these data suggest that dsCYC2 functions to relieve light-dependent G1 arrest, rather than regulating DNA replication.
In yeast, appropriate cell growth and metabolic status trigger G1 progression by activating CLN3, followed by CLN1 and CLN2, in association with CDC28 to finally activate the G1/S transcription factor SBF/MBF and the transcription of S phase genes (Tyers et al., 1993; reviewed in Mendenhall and Hodge, 1998). In animals and plants, D-type cyclins are stimulated by serum growth factors and hormones or Suc, respectively. D-type cyclins associate with CDKs and phosphorylate retinoblastoma (Rb) protein, leading to the release and activation of E2F transcription factors and the G1-to-S phase transition (reviewed in Oakenfull et al., 2002). Overexpression or silencing of these G1 cyclins has been reported to exhibit various effects on the G1 phase duration and overall cell cycle length, depending on the type of cyclin and cells (Quelle et al., 1993; Resnitzky et al., 1994; Sherr, 1995; Menges et al., 2006). Both CLN1-3 and D-type cyclins are characterized by PEST sequences that render the proteins unstable and confer rapid turnover (Rechsteiner and Rogers, 1996; Renaudin et al., 1996; Mendenhall and Hodge, 1998). Furthermore, plant and animal D-type cyclins, as well as the Ostreococcus cyclin A, possess an LxCxE amino acid motif at their N-terminal region that is responsible for their interaction with the Rb protein (Dowdy et al., 1993; Renaudin et al., 1996; Moulager et al., 2010). None of these motifs can be recognized in the dsCYC2 sequence, suggesting that dsCYC2 turnover is regulated by alternative mechanisms and that the protein probably does not interact directly with the P. tricornutum Rb orthologous protein. Alternatively, it is possible that dsCYC2 expression results in the transcription or activation of other G1 cyclins that regulate the Rb protein in P. tricornutum. Because diatom cell cycle progression depends not only on light, but also on other environmental factors, such as nutrient availability, it is to be expected that multiple cyclins are involved in G1 control, representing a complex integrative fine-tuning network of different signaling pathways. The presence of a critical molecule, such as dsCYC2, that rapidly coordinates the activation of the cell cycle machinery upon changing light conditions is thus of major importance for diatoms living in highly variable environments and potentially allows them to pace their cell division rate to the prevailing light conditions.
Blue Light–Dependent Induction of dsCYC2
Promoter-reporter analysis suggests that transcriptional regulation of dsCYC2 occurs through its promoter sequence. Screening for interactors of the dsCYC2 promoter yielded two transcription factors belonging to the bZIP transcription factor family: AUREO1a and bZIP10. Of particular interest is AUREO1a, which belongs to the AUREOCHROME family of blue light photoreceptors in photosynthetic stramenopiles (Takahashi et al., 2007; Ishikawa et al., 2009). AUREOCHROMES typically possess an N-terminal bZIP domain expected to be involved in dimerization and DNA binding and a C-terminal LOV domain thought to act as a photosensor (Takahashi et al., 2007; Toyooka et al., 2011). Absorption of blue light by flavin mononucleotide (FMN) attached to the LOV domain induces covalent adduct formation between FMN and a conserved Cys residue in the LOV domain. P. tricornutum encodes four AUREOCHROME-like proteins (Rayko et al., 2010; Depauw et al., 2012), but only AUREO1a seems to be involved in dsCYC2 regulation. In Vaucheria frigida, the bZIP domain of AUREO1 was found to recognize the bZIP binding site TGACGT (Jakoby et al., 2002; Takahashi et al., 2007). Interestingly, the promoter of dsCYC2 contains three of these sites (see Supplemental Figure 4 online), rendering them putative regulatory cis-acting elements. The role of AUREO1a in dsCYC2 induction is further supported by the specific transcription of dsCYC2 by blue light, but not red light. Treatment of cells with CHX or the redox inhibitor DCMU had no effect on dsCYC2 induction, indicating that no de novo protein synthesis or PET is required, reinforcing the hypothesis of direct photoreceptor-mediated regulation of dsCYC2 induction by AUREO1a. The specific response of dsCYC2 to low fluence rate blue light through AUREO1a signaling could be of particular significance to diatoms as blue light (350 to 500 nm) is the most prevalent color of light below the surface of oceanic waters (MacIntyre et al., 2000); hence, efficient blue light sensing and signaling mechanisms are expected to play a crucial role in the control of diatom growth.
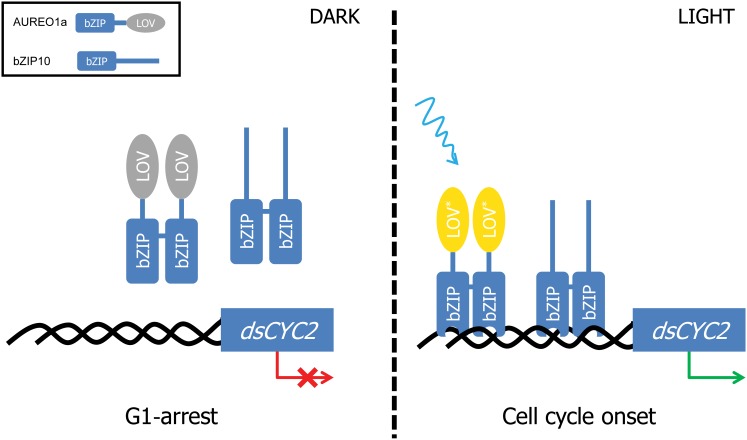
Various LOV domain–signaling mechanisms have been described for different plant, algal, and bacterial proteins, such as light-induced unfolding, rotation, dimerization, and/or DNA binding of the effector domain (reviewed in Herrou and Crosson, 2011). Here, we have shown that AUREO1a and bZIP10 can form heterodimers and that bZIP10 is able to form homodimers. Either protein could activate the dsCYC2 promoter in the BY-2 protoplast system, but activation was enhanced when both proteins were coexpressed. Based on these findings, different models of dsCYC2 regulation can be envisioned. First, upon blue light exposure, AUREO1a and bZIP10 might form heterodimers and as such bind and activate the regulatory sites present in the dsCYC2 promoter. However, previous reports have suggested that the V. frigida AUREO1 LOV domain has a dimeric nature (Mitra et al., 2012) and that two LOV domains would be needed to activate AUREO1 (Toyooka et al., 2011). Therefore, it seems plausible that upon illumination, homodimers of AUREO1a and bZIP10 would occupy different regulatory sites within the dsCYC2 promoter and act synergistically to activate it (Figure 8). How bZIP10 activates transcription of dsCYC2 remains unknown, but possible mechanisms include nuclear translocation or posttranslational modifications upon light exposure that result in the modulation of the DNA binding activity or activation potential, as described for other bZIP proteins (Jakoby et al., 2002). Further investigations are needed to uncover the precise mechanism of dsCYC2 activation by AUREO1a and bZIP10.
Figure 8.
Hypothetical Model of the Light-Dependent Regulation of dsCYC2 and Cell Cycle Onset in P. tricornutum.
Upon light exposure, the LOV domain of AUREO1a is changed from the dark state (gray) into the light state (yellow) through cysteinyl-FMN adduct formation. This induces a conformational change in the homodimer protein complex, resulting in the binding of the bZIP domains to the promoter of dsCYC2. Binding of both AUREO1a and bZIP10 homodimers to different regulatory elements in the dsCYC2 promoter results in the synergistic activation of dsCYC2 and leads to the onset of the cell cycle.
Interestingly, inhibition of protein synthesis at the dark-to-light transition delays the decrease of dsCYC2 transcript levels in the light and results in the accumulation of higher transcript levels after illumination, suggesting that upon light exposure, a repressor of dsCYC2 transcription is generated. Such a repressor might interfere with DNA binding of the activators, either directly through, for example, occupation and repression of the regulatory sites, or indirectly by interfering with the dimerization or DNA binding properties of the activators through, for example, posttranslational modifications (Schütze et al., 2008). Future work will focus on identifying the repressor(s) and their mode of regulation.
In conclusion, we identified two bZIP transcription factors that are likely to be involved in the blue light–dependent transcription of a cyclin gene that regulates the onset of the cell cycle in diatoms after a period of darkness. The involvement of aureochromes in blue light–mediated branching and sex organ development have previously been described (Takahashi et al., 2007). This study identifies a possible role for AUREO1a and its target gene dsCYC2 during the cell cycle. The dsCYC2 gene appears to be conserved in other pennate diatom species, including Fragillariopsis cyclindrus (http://genome.jgi-psf.org/Fracy1/Fracy1.home.html, Fracy1_253344) and Pseudo-Nitzschia multiseries (http://genome.jgi.doe.gov/Psemu1/Psemu1.home.html, Psemu1_301178), but no clear homolog was found in the centric T. pseudonana (Huysman et al., 2010). However, because of the high number of dsCYCs in diatom species (Huysman et al., 2010) and the presence of AUREO1a in both pennates and centrics (Depauw et al., 2012), the mechanism of light-dependent cell cycle activation through AUREO1a-mediated induction of a cyclin gene is most likely conserved in diatoms.
METHODS
Diatom Culture Conditions
Phaeodactylum tricornutum (Pt1 8.6; accession numbers CCAP 1055/1 and CCMP2561) cells were grown in f/2 medium without silica (f/2-Si) (Guillard, 1975) made from filtered and autoclaved sea water collected from the North Sea (Belgium) or artificial sea water medium (Vartanian et al., 2009). Cultures were cultivated at 18 to 20°C under a 12L/12D regime using 70 to 100 μmol photons m−2 s−1 white light (Radium NL 36W/840 Spectrolux plus, cool white). Liquid cultures were shaken at 100 rpm. For expression studies under blue and red light conditions, blue light (380 to 450 nm) was generated using neon lamps (Osram L36W/67, Lumilux Bleu), while red light (620 to 720 nm with a peak intensity of 670 nm) was generated using an LED source (Flight II DC Red + Black; Quantum Devices), and different light intensities were obtained using neutral density filters.
Vector Cloning and Biolistic Transformation
The 1018-bp promoter sequence alone, the promoter and full-length gene sequence, or the gene sequence alone of dsCYC2 of P. tricornutum was amplified with gene-specific primers (see Supplemental Table 1 online), cloned in the pDONR221 or pENTR-D-TOPO vector (Invitrogen), and subsequently recombined in a P. tricornutum destination vector (pDEST) by attL × attR recombination (Invitrogen) (Siaut et al., 2007). The dsCYC2 promoter sequence was recombined in pDEST-C-EYFP for C-terminal fusion to construct the prom-eYFP reporter line. The promoter and gene constructs were recombined in pDEST-C-HA to construct the HA marker line. Both plasmids were subsequently digested with SacII and NotI (Promega) to remove the fcpB promoter sequence. The digested product was treated with T4 DNA polymerase in the presence of 10 mM deoxynucleotide triphosphate to produce blunt ends and then ligated using T4 DNA ligase according to the manufacturer’s instructions (Promega). For the creation of dsCYC2 inverted repeat silencing constructs, a 167-bp fragment (corresponding to the dsCYC2 gene sequence from 13 to 179 bp) and a 301-bp fragment (corresponding to the gene sequence from 13 to 313 bp) were amplified from the dsCYC2 cDNA with the primers dsCYC2f1_Fw (containing a EcoRI site) and dsCYC2f1_Rv (containing a XbaI site), and dsCYC2f1_Fw and dsCYC2f2_Rv (containing a XbaI site) (see Supplemental Table 1 online), respectively. The fragments were digested with EcoRI and XbaI (Promega) and ligated in sense and antisense orientations to the EcoRI site of the linearized hir-PtGUS vector (De Riso et al., 2009).
Constructs were introduced into P. tricornutum by microparticle bombardment as previously described (Falciatore et al., 1999). The prom-eYFP reporter plasmid and the HA marker plasmid were each cotransformed with the pAF6 plasmid to confer resistance to phleomycin (Falciatore et al., 1999). Individual phleomycin-resistant colonies were both restreaked on f/2-Si agar plates and grown in liquid f/2-Si medium without antibiotics for further analysis.
Real-Time Quantitative PCR
For RNA extraction, 5 × 107 cells were collected by fast filtration, and filters with cell pellets were fast frozen in liquid nitrogen and stored at −70°C. Cell lysis and RNA extraction were performed using TriReagent (Molecular Research Center) according to the manufacturer’s instructions. Contaminating genomic DNA was removed by DNaseI treatment (GE Healthcare), and RNA was purified by ammonium acetate precipitation. RNA concentration and purity were assessed by spectrophotometry. Total RNA was reverse transcribed using iScript reverse transcriptase (Bio-Rad) or a Quantitect reverse transcription kit (Qiagen) according to the manufacturer’s instructions. Finally, an equivalent of 5 or 10 ng of reverse-transcribed RNA (cDNA) was used as template in each quantitative PCR reaction.
Samples in triplicate were amplified on the Lightcycler 480 platform (Roche) or the CFX96 Real-Time PCR detection system (Bio-Rad) with Lightcycler 480 SYBR Green I Master mix (Roche Applied Science) in the presence of 0.5 μM gene-specific primers (dsCYC2_Fw, 5′-CTATCATCGCACTCGTCATCAAC-3′, and dsCYC2_Rv, 5′-TGTCCACCAAAGCCTCCAAAC-3′; dsCYC2-HA_Fw, 5′-TCGCTCCTCTGGTGGAA-3′, and dsCYC2-HA_Rv, 5′-GTCGTAGGGGTAGGCGTAGT-3′; for other primer sequences, see Siaut et al., 2007 and Huysman et al., 2010). The cycling conditions were 10 min polymerase activation at 95°C and 45 cycles at 95°C for 10 s, 58°C for 15 s, and 72°C for 15 s. Amplicon dissociation curves were recorded after cycle 45 by heating from 65 to 95°C. Data were analyzed using the ΔCt (cycle threshold) relative quantification method using qBase (Hellemans et al., 2007), with the stably expressed histone H4 used as a normalization gene (Siaut et al., 2007).
Protein Gel Blot Analysis
For protein extraction, 5 × 107 cells were collected by fast filtration, and filters with cell pellets were fast frozen in liquid nitrogen and stored at −70°C. Proteins were extracted by adding 200 μL Laëmli buffer containing Complete Protease Inhibitor Cocktail (Roche) to the frozen cells and vortexing at high speed until the cells were lysed. Cell lysates were incubated for 15 min on ice and centrifuged at 13,000 rpm for 15 min at 4°C to remove insoluble material. Protein concentrations were determined by the Bio-Rad Protein Assay (Bio-Rad) based on the method of Bradford (1976). Equal amounts of protein extracts were resolved on 12% SDS-PAGE gels and transferred to nitrocellulose membranes (Millipore) using the wet-blot method. The dsCYC2-HA fusion protein was detected by incubating proteins transferred to nitrocellulose membranes for 1 h with a 1:500 dilution of anti-HA primary antibody (Roche) at room temperature, followed by 1 h incubation in a 1:10,000 dilution of horseradish peroxidase anti-rat secondary antibody (Abcam) at room temperature. AUREO1a protein was detected by incubating proteins transferred to nitrocellulose membranes for 1 h with a 1:1000 dilution of a specific anti-AUREO1a primary antibody at room temperature, followed by 1 h incubation in a 1:10,000 dilution of horseradish peroxidase anti-rabbit secondary antibody (GE Healthcare) at room temperature. Signals were visualized using the Western Lightning detection kit (Thermo Scientific Pierce) according to the manufacturer’s instructions.
Y2H Analysis
Y2H bait and prey plasmids were generated through recombinational Gateway cloning (Invitrogen). The full-length ORFs of the P. tricornutum dsCYC2, CDKA1, and CDKA2 genes were amplified from cDNA using gene-specific primers (see Supplemental Table 1 online), cloned in the pENTR-D-TOPO vector (Invitrogen), and subsequently recombined in the pDEST22 and pDEST32 vectors (Invitrogen) by attL × attR recombination, resulting in translational fusions between the proteins and the GAL4 transcriptional activator and DNA binding domains, respectively. AUREO1a and bZIP10 cDNAs were derived from plasmid extraction (Zymoprep; Zymo Research) from positive colonies of a Y1H library screen (see below) and recombined in the pDEST22 and pDEST32 vectors by Gateway recombination. Bait and prey plasmids were cotransformed in the yeast strain PJ694-α by the LiAc method (Gietz et al., 1992). Cotransformed yeast cells were selected on synthetic defined (SD) medium plates lacking Leu and Trp. Interaction between the introduced proteins was scored on SD plates lacking Leu, Trp, and His.
Yeast Complementation
The full-length dsCYC2 cDNA was cloned into the yeast tetracycline-repressible vector pTHGW (Peres et al., 2007) by LR cloning. The resulting plasmid, pTH-dsCYC2 (or pTHGW as a control), was transformed into the G1-deficient yeast strain BF305-15d-21 (MATa leu2-3, 112his3-11, 15ura3-52 trp1 ade1 met14; arg5,6 GAL1-CLN3 HIS3::cln 1 TRP1::cln2) by the LiAc method (Gietz et al., 1992). Complementation was assayed on Gal-containing SD medium in the presence or absence of 20 µg/mL doxycycline (Sigma-Aldrich).
Y1H Analysis
For the Y1H library screen, the dsCYC2 promoter sequence (1018 bp upstream of ATG) was cloned in the pMW#2 and pMW#3 destination vectors (Deplancke et al., 2006), yielding HIS3 and LacZ reporter constructs, respectively. The Y1H bait strain was generated as previously described (Deplancke et al., 2004, 2006). Subsequently, the Y1H bait strain was transformed with 50 µg of prey plasmids derived from a custom-made P. tricornutum Y2H cDNA library (Invitrogen) according to the Yeast Protocol Handbook (Clontech), and yeast cells that hosted a successful interaction were selected on selective SD medium lacking His, Ura, and Trp containing 25 mM of 3-amino-1,2,4-triazole and retested using a direct Y1H test.
Growth Analyses
To monitor growth rates of wild-type and dscyc2 cells, cells were grown at 12L/12D (100 µE of white light) in a 24-well plate (Falcon), in a total volume of 1 mL, over a time period of 11 d. Absorbances of the cultures were measured at 405 nm using the VICTOR3 Multilabel Plate Reader (Perkin-Elmer) each day in the morning. The growth curves of triplicate cultures were LN(2) transformed, and mean generation times were calculated by determination of the derivative of the values between the points of maximal slope (exponential growth phase).
To determine the growth rates of wild-type and dscyc2-2.9 cells under constant light conditions, batch cultures were grown under continuous illumination of white light for at least 2 weeks. At the beginning of the experiment, cells were diluted with fresh F/2 medium without silica to the same absorbance at 405 nm (0.025), and cells were either placed under constant light conditions or transferred to 12L/12D conditions at the same light intensity (50 µE). Generation times were calculated as described above.
To monitor growth curves under different light quality conditions, wild-type and dscyc2-2.9 batch cultures were cultivated at 12L/12D at 20°C in air-lifted 100-mL test tubes. Flora light-emitting diode panels (CLF Plant Climatics) were used for illumination with monochromatic blue light and red light at wavelengths of 469 nm ± 10 nm and 659 nm ± 11 nm, respectively. White fluorescence tubes (18W/865; Osram) provided illumination with white light. The spectral composition of the light sources was recorded with a spectroradiometer (Tristan). The relative absorption of incident light varied for the different light sources. Therefore, the incident light intensity was adjusted to either 72 µmol photons m−2 s−1 blue light, 120 µmol photons m−2 s−1 white light, or 123 µmol photons m−2 s−1 red light. This resulted in similar values of 30 µmol absorbed photons m−2 s−1 photosynthetically absorbed radiation (QPhar) at a culture density of 2 µg chlorophyll a mL−1 as calculated according to Gilbert et al. (2000). All cultures were inoculated with 50,000 cells mL−1. The in vivo absorption at 405 nm was recorded with a spectrophotometer (Specord M500; Zeiss).
nCounter Analysis
RNA levels were measured using the Nanostring nCounter analysis system (Nanostring Technologies) by the VIB Nucleomics Core Facility as previously described (Geiss et al., 2008). An overview of the nCounter probe pairs used in this study is shown in Supplemental Table 2 online. All probes were screened against the P. tricornutum annotated transcript database from the Department of Energy Joint Genome Initiative for potential cross-hybridization. Total RNA extract (100 ng) from two biological replicates for both wild-type and dscyc2-2.9 cells was used for hybridization, and all genes were measured simultaneously using multiplexed reactions. After a first normalization against the internal spike-in controls, genes were normalized against the four reference genes EF1a, histone H4, RPS, and UBI-4. Fold induction calculations for wild-type and dscyc2-2.9 cells values were divided by the value at the 0-h time point.
Inhibitor Studies
To determine the effect of PET inhibition, DCMU (Sigma-Aldrich) was dissolved in ethanol to a stock concentration of 100 mM and delivered to the cells 10 min before the onset of light treatment at a final concentration of 20 µM. Identical volumes of ethanol were added to the controls and had no effect on transcript expression.
To determine the effect of inhibition of protein translation on dsCYC2 transcription, cells were treated with or without CHX (Duchefa Biochemie) at a final concentration of 2 µg/mL, 5 min before the onset of light treatment.
Transient Reporter Assays
The dsCYC2 promoter sequence was cloned simultaneously with the fLUC sequence in the pm42GW7,3 destination vector (Karimi et al., 2007) by multisite Gateway cloning (Invitrogen). To generate the effector constructs, the cDNA clones of AUREO1a and bZIP10 were recombined in the p2GW7 destination vector by Gateway cloning, containing the cauliflower mosaic virus 35S promoter. Both reporter and effector plasmids were used to transfect tobacco (Nicotiana tabacum) BY-2 protoplasts using the polyethylene glycol/Ca2+ method as described by De Sutter et al. (2005). Luciferase measurements were performed using the Dual-luciferase Reporter 1000 Assay System (Promega) according to the manufacturer’s instructions and as previously described (De Sutter et al., 2005).
Accession Numbers
Sequence data from this article can be found in the P. tricornutum genome sequence database through the Joint Genome Initiative portal (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html) under the following accession numbers: dsCYC2, Phatr2_34956; CDKA1, Phatr2_20262; CDKA2, Phatr2_51279; AUREO1a, Phatr2_49116; and bZIP10, Phatr2_43744. Sequence data from other genes discussed in this article can be found in the EMBL/GenBank data libraries under the accessions numbers listed in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Silencing of dsCYC2 Does Not Result in a General Stress Response.
Supplemental Figure 2. Effect of CHX on dsCYC2 Transcript Expression in Dark and Light.
Supplemental Figure 3. S-Phase Distribution in Wild-Type (Solid Line) versus dscyc2-2.9 (Dashed Line) Cells during a Synchronized Time Course.
Supplemental Figure 4. Mapping of TGACGT Sites in the dsCYC2 Promoter Sequence.
Supplemental Table 1. Overview of the Cloning Primers.
Supplemental Table 2. Overview of the nCounter Code Set Probe Pairs.
Acknowledgments
We thank Frederik Coppens, Joke Allemeersch, and Rudy Van Eijsden for technical advice and assistance, Jonas Van Hove, Leila Tirichine, and Frauke Depauw for practical assistance, and Martine De Cock and Annick Bleys for help in preparing the article. We thank Bruce Futcher and Jim Murray for providing the BF305-15d-21 yeast strain. M.J.J.H. and M.M. thank the Agency for Innovation by Science and Technology in Flanders (IWT-Vlaanderen) for a predoctoral fellowship. This work was supported by a grant of the Research Foundation Flanders (G.0288.13). M.J.J.H. acknowledges the Federation of European Biochemical Societies (FEBS) organization for a short-term fellowship to visit A.F.’s lab at Université Pierre et Marie Curie and the European Molecular Biology Organization (EMBO) organization for a short-term fellowship (ASTF 93-2011) to visit C.B.’s lab at Institut de Biologie de l'Ecole Normale Supérieure (IBENS) Paris (France). C.B. acknowledges support from the Agence Nationale de Recherche. P.G.K. is grateful for financial support by the Deutsche Forschungsgemeinschaft (research group 1261, project 8) and the Universität Konstanz. C.W. and B.S. acknowledge the support from the Deutsche Forschungsgemeinschaft Grant FOR 1261 (Wi 764/19). A.F., A.E.F., and M.J.J.H. acknowledge support by the Human Frontier Science Program Young Investigator Grant (RGY0082/2010) and the Action Thématique et Incitative sur Programme award (2009) from Centre National de la Recherche Scientifique.
AUTHOR CONTRIBUTIONS
M.J.J.H., A.E.F., B.S.C., D.I., C.B., P.G.K., C.W., A.F., W.V., and L.D.V. conceived and designed the research. M.J.J.H., A.E.F., M.M., B.S.C., R.V., H.V.d.D., and M.S. performed the experiments. M.J.J.H., W.V., and L.D.V. analyzed the data and wrote the article. All authors read, revised, and approved the article.
Glossary
- CDK
cyclin-dependent kinase
- ORF
open reading frame
- HA
hemagglutinin
- 12L/12D
12-h-light/12-h-dark
- Y2H
yeast two-hybrid
- PET
photosynthetic electron transport
- Y1H
yeast one-hybrid
- CHX
cycloheximide
- BY-2
Bright Yellow-2
- FMN
flavin mononucleotide
- SD
synthetic defined
- GUS
β-glucuronidase
References
- Bailleul B., Rogato A., de Martino A., Coesel S., Cardol P., Bowler C., Falciatore A., Finazzi G. (2010). An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc. Natl. Acad. Sci. USA 107: 18214–18219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisova K., Krylov D.M., Umen J.G. (2005). Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 137: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244 [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brzezinski M.A., Olson R.J., Chisholm S.W. (1990). Silicon availability and cell-cycle progression in marine diatoms. Mar. Ecol. Prog. Ser. 67: 83–96 [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Cohn S.A., Bahena M., Davis J.T., Ragland R.L., Rauschenberg C.D., Smith B.J. (2004). Characterisation of the diatom photophobic response to high irradiance. Diatom Res. 19: 167–179 [Google Scholar]
- Depauw F.A., Rogato A., Ribera d’Alcalá M., Falciatore A. (2012). Exploring the molecular basis of responses to light in marine diatoms. J. Exp. Bot. 63: 1575–1591 [DOI] [PubMed] [Google Scholar]
- Deplancke B., Dupuy D., Vidal M., Walhout A.J. (2004). A Gateway-compatible yeast one-hybrid system. Genome Res. 14: 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke, B., Vermeirssen, V., Arda, H.E., Martinez, N.J., and Walhout, A.J. (2006). Gateway-compatible yeast one-hybrid screens. CSH Protoc. 2006: 5. [DOI] [PubMed]
- De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. (2009). Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sutter V., Vanderhaeghen R., Tilleman S., Lammertyn F., Vanhoutte I., Karimi M., Inzé D., Goossens A., Hilson P. (2005). Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J. 44: 1065–1076 [DOI] [PubMed] [Google Scholar]
- Dowdy S.F., Hinds P.W., Louie K., Reed S.I., Arnold A., Weinberg R.A. (1993). Physical interaction of the retinoblastoma protein with human D cyclins. Cell 73: 499–511 [DOI] [PubMed] [Google Scholar]
- Falciatore A., Casotti R., Leblanc C., Abrescia C., Bowler C. (1999). Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. (NY) 1: 239–251 [DOI] [PubMed] [Google Scholar]
- Field C.B., Behrenfeld M.J., Randerson J.T., Falkowski P. (1998). Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281: 237–240 [DOI] [PubMed] [Google Scholar]
- Geiss G.K., et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26: 317–325 [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R.A., Schiestl R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M., Wilhelm C., Richter M. (2000). Bio-optical modelling of oxygen evolution using in vivo fluorescence: Comparison of measured and calculated photosynthesis/irradiance (P-I) curves in four representative phytoplankton species. J. Plant Physiol. 157: 307–314 [Google Scholar]
- Gillard J., et al. (2008). Physiological and transcriptomic evidence for a close coupling between chloroplast ontogeny and cell cycle progression in the pennate diatom Seminavis robusta. Plant Physiol. 148: 1394–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard, R.R.L. (1975). Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals, W.L. Smith and M.H. Canley, eds (New York: Plenum Press), pp. 29–60. [Google Scholar]
- Hartwell L.H., Culotti J., Pringle J.R., Reid B.J. (1974). Genetic control of the cell division cycle in yeast. Science 183: 46–51 [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J., Crosson S. (2011). Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 9: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth E.S. (1985). Effect of growth factors and light quality on the growth, pigmentation and photosynthesis of two diatoms, Thalassiosira gravida and Phaeodactylum tricornutum. Mar. Biol. 86: 253–262 [Google Scholar]
- Huisman J., Sharples J., Stroom J.M., Visser P.M., Kardinaal W.E.A., Verspagen J.M.H., Sommeijer B. (2004). Changes in turbulent mixing shift competition for light between phytoplankton species. Ecology 85: 2960–2970 [Google Scholar]
- Huysman M.J.J., Martens C., Vandepoele K., Gillard J., Rayko E., Heijde M., Bowler C., Inzé D., Van de Peer Y., De Veylder L., Vyverman W. (2010). Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling. Genome Biol. 11: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Ishikawa M., Takahashi F., Nozaki H., Nagasato C., Motomura T., Kataoka H. (2009). Distribution and phylogeny of the blue light receptors aureochromes in eukaryotes. Planta 230: 543–552 [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis Trends Plant Sci. 7: 106–111. [DOI] [PubMed]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant Gateway vectors. Plant Physiol. 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Imamura S., Hanaoka M., Tanaka K. (2011). A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat. Cell Biol. 13: 483–487 [DOI] [PubMed] [Google Scholar]
- Kooistra W.H., De Stefano M., Mann D.G., Medlin L.K. (2003). The phylogeny of the diatoms. Prog. Mol. Subcell. Biol. 33: 59–97 [DOI] [PubMed] [Google Scholar]
- Lavaud J., Rousseau B., Etienne A.-L. (2004). General features of photoprotection by energy dissipation in planktonic diatoms (Bacillariophyceae). J. Phycol. 40: 130–137 [Google Scholar]
- Lavaud J., Strzepek R.F., Kroth P.G. (2007). Photoprotection capacity differs among diatoms: Possible consequences on the spatial distribution of diatoms related to fluctuations in the underwater light climate. Limnol. Oceanogr. 52: 1188–1194 [Google Scholar]
- Lepetit B., Goss R., Jakob T., Wilhelm C. (2012). Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth. Res. 111: 245–257 [DOI] [PubMed] [Google Scholar]
- López-Juez E., Dillon E., Magyar Z., Khan S., Hazeldine S., de Jager S.M., Murray J.A.H., Beemster G.T.S., Bögre L., Shanahan H. (2008). Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre H.L., Kana T.M., Geider R.J. (2000). The effect of water motion on short-term rates of photosynthesis by marine phytoplankton. Trends Plant Sci. 5: 12–17 [DOI] [PubMed] [Google Scholar]
- Mann D.G. (1999). The species concept in diatoms. Phycologia 38: 437–495 [Google Scholar]
- McLachlan D.H., Brownlee C., Taylor A.R., Geider R.J., Underwood G.J.C. (2009). Light-induced motile responses of the estuarine benthic diatoms Navicula perminuta and Cylindrotheca closterium (Bacillariophyceae). J. Phycol. 45: 592–599 [DOI] [PubMed] [Google Scholar]
- Mendenhall M.D., Hodge A.E. (1998). Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., Samland A.K., Planchais S., Murray J.A. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado J.M., Sánchez-Saavedra M.P., Correa-Reyes G., Lubián L., Montero O., Figueroa F.L. (2004). Blue light effect on growth, light absorption characteristics and photosynthesis of five benthic diatom strains. Aquat. Bot. 78: 265–277 [Google Scholar]
- Mitra D., Yang X., Moffat K. (2012). Crystal structures of Aureochrome1 LOV suggest new design strategies for optogenetics. Structure 20: 698–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Morgan, D.O. (2007). The Cell Cycle: Principles of Control. (London: New Science Press). [Google Scholar]
- Moriyama T., Terasawa K., Sekine K., Toyoshima M., Koike M., Fujiwara M., Sato N. (2010). Characterization of cell-cycle-driven and light-driven gene expression in a synchronous culture system in the unicellular rhodophyte Cyanidioschyzon merolae. Microbiology 156: 1730–1737 [DOI] [PubMed] [Google Scholar]
- Mouget J.-L., Gastineau R., Davidovich O., Gaudin P., Davidovich N.A. (2009). Light is a key factor in triggering sexual reproduction in the pennate diatom Haslea ostrearia. FEMS Microbiol. Ecol. 69: 194–201 [DOI] [PubMed] [Google Scholar]
- Moulager M., Corellou F., Vergé V., Escande M.-L., Bouget F.-Y. (2010). Integration of light signals by the retinoblastoma pathway in the control of S phase entry in the picophytoplanktonic cell Ostreococcus. PLoS Genet. 6: e1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulager M., Monnier A., Jesson B., Bouvet R., Mosser J., Schwartz C., Garnier L., Corellou F., Bouget F.-Y. (2007). Light-dependent regulation of cell division in Ostreococcus: Evidence for a major transcriptional input. Plant Physiol. 144: 1360–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M., Valle K.C., Brembu T., Hancke K., Winge P., Andresen K., Johnsen G., Bones A.M. (2009). An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 4: e7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull E.A., Riou-Khamlichi C., Murray J.A.H. (2002). Plant D-type cyclins and the control of G1 progression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson R.J., Vaulot D., Chisholm S.W. (1986). Effects of environmental stresses on the cell cycle of two marine phytoplankton species. Plant Physiol. 80: 918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee A.B. (1974). A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA 71: 1286–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Jung G., Hwang Y.S., Jin E. (2010). Dynamic response of the transcriptome of a psychrophilic diatom, Chaetoceros neogracile, to high irradiance. Planta 231: 349–360 [DOI] [PubMed] [Google Scholar]
- Peres A., et al. (2007). Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J. Biol. Chem. 282: 25588–25596 [DOI] [PubMed] [Google Scholar]
- Quelle D.E., Ashmun R.A., Shurtleff S.A., Kato J.Y., Bar-Sagi D., Roussel M.F., Sherr C.J. (1993). Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 7: 1559–1571 [DOI] [PubMed] [Google Scholar]
- Rayko E., Maumus F., Maheswari U., Jabbari K., Bowler C. (2010). Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 188: 52–66 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Rogers S.W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21: 267–271 [PubMed] [Google Scholar]
- Renaudin J.-P., et al. (1996). Plant cyclins: A unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol. Biol. 32: 1003–1018 [DOI] [PubMed] [Google Scholar]
- Resnitzky D., Gossen M., Bujard H., Reed S.I. (1994). Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14: 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K., Harter K., Chaban C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13: 247–255 [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (1995). D-type cyclins. Trends Biochem. Sci. 20: 187–190 [DOI] [PubMed] [Google Scholar]
- Siaut M., Heijde M., Mangogna M., Montsant A., Coesel S., Allen A., Manfredonia A., Falciatore A., Bowler C. (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406: 23–35 [DOI] [PubMed] [Google Scholar]
- Sims P.A., Mann D.G., Medlin L.K. (2006). Evolution of the diatoms: Insights from fossil, biological and molecular data. Phycologia 45: 361–402 [Google Scholar]
- Spudich J.L., Sager R. (1980). Regulation of the Chlamydomonas cell cycle by light and dark. J. Cell Biol. 85: 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Yamagata D., Ishikawa M., Fukamatsu Y., Ogura Y., Kasahara M., Kiyosue T., Kikuyama M., Wada M., Kataoka H. (2007). AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl. Acad. Sci. USA 104: 19625–19630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka T., Hisatomi O., Takahashi F., Kataoka H., Terazima M. (2011). Photoreactions of aureochrome-1. Biophys. J. 100: 2801–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Tokiwa G., Futcher B. (1993). Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12: 1955–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hoek, C., Mann, D.G., and Jahns, H.M. (1995). Algae: An Introduction to Phycology. (Cambridge, UK: Cambridge University Press). [Google Scholar]
- Vartanian M., Desclés J., Quinet M., Douady S., Lopez P.J. (2009). Plasticity and robustness of pattern formation in the model diatom Phaeodactylum tricornutum. New Phytol. 182: 429–442 [DOI] [PubMed] [Google Scholar]
- Vaulot D., Olson R.J., Chisholm S.W. (1986). Light and dark control of the cell cycle in two marine phytoplankton species. Exp. Cell Res. 167: 38–52 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Connolly T., Futcher B., Beach D. (1991). Human D-type cyclin. Cell 65: 691–699 [DOI] [PubMed] [Google Scholar]
- Zhu S.-H., Green B.R. (2010). Photoprotection in the diatom Thalassiosira pseudonana: Role of LI818-like proteins in response to high light stress. Biochim. Biophys. Acta 1797: 1449–1457 [DOI] [PubMed] [Google Scholar]