Abstract
RNA silencing refers to a collection of gene regulatory mechanisms that use small RNAs for sequence specific repression. These mechanisms rely on ARGONAUTE (AGO) proteins that directly bind small RNAs and thereby constitute the central component of the RNA-induced silencing complex (RISC). AGO protein function has been probed extensively by mutational analyses, particularly in plants where large allelic series of several AGO proteins have been isolated. Structures of entire human and yeast AGO proteins have only very recently been obtained, and they allow more precise analyses of functional consequences of mutations obtained by forward genetics. To a large extent, these analyses support current models of regions of particular functional importance of AGO proteins. Interestingly, they also identify previously unrecognized parts of AGO proteins with profound structural and functional importance and provide the first hints at structural elements that have important functions specific to individual AGO family members. A particularly important outcome of the analysis concerns the evidence for existence of Gly-Trp (GW) repeat interactors of AGO proteins acting in the plant microRNA pathway. The parallel analysis of AGO structures and plant AGO mutations also suggests that such interactions with GW proteins may be a determinant of whether an endonucleolytically competent RISC is formed.
ELEMENTS OF ARGONAUTE FUNCTION IN PLANT BIOLOGY
Small RNA and Argonaute: The Core of RNA Silencing
RNA silencing is a central mechanism of gene regulation in eukaryotes. It relies on small 20- to 30-nucleotide RNAs referred to as microRNAs (miRNAs), short interfering RNAs (siRNAs), and Piwi-interacting RNAs. miRNAs and siRNAs are produced as duplexes from longer double-stranded precursors by Dicer, a ribonuclease III. In a process referred to as small RNA loading, one of the strands, the guide strand, becomes stably associated with a protein of the ARGONAUTE (AGO) family, while the other strand, the passenger strand, is degraded. Piwi-interacting RNAs are found only in animals and are produced from longer single-stranded RNA precursors through the action of an animal-specific subclass of AGO proteins called Piwi.
The AGO-small RNA complex constitutes the core of the RNA-induced silencing complex (RISC) that uses base pairing to silence complementary mRNA at the posttranscriptional level or genomic loci producing complementary RNA at the cotranscriptional level (Carthew and Sontheimer, 2009). Posttranscriptional silencing can occur via translational repression, often coupled to mRNA decay, or via endonucleolytic cleavage (slicing) catalyzed by the AGO protein (Huntzinger and Izaurralde, 2011). Cotranscriptional silencing, on the other hand, involves repressive chromatin modifications and, at least in plants, DNA methylation (Haag and Pikaard, 2011). The sequence specificity of any RNA silencing reaction is conferred by the guide RNA, while the precise nature of silencing depends on the properties of the associated AGO protein, including its ribonuclease activity, interacting proteins, and subcellular localization.
Classes of Plant Small RNAs and Functional Specialization of Plant AGO Proteins
In plants, several different classes of small RNAs with specialized biological roles have been identified. In Arabidopsis thaliana, 10 different AGO proteins mediate the effects of several distinct types of small RNAs (Vaucheret, 2008). The most abundant class consists of RNA polymerase IV- and V-dependent 24-nucleotide small interfering RNAs (p4/p5-siRNAs) that mostly derive from transposable elements and other repetitive sequences and act to silence such loci at the transcriptional level via DNA methylation and repressive chromatin modifications. These functions are performed by a phylogenetic clade comprising AGO4, AGO6, and AGO9 that all seem to be functionally similar, although they are differentially expressed (Zilberman et al., 2003; Zheng et al., 2007; Havecker et al., 2010). Some 24-nucleotide p4/p5-siRNAs associate with AGO1 (H. Wang et al., 2011), but the biological role of this association remains elusive.
The second largest class comprises miRNAs. miRNAs derive as single species from imperfectly base paired, endogenous hairpin transcripts and are of fundamental importance for posttranscriptional regulation of protein coding genes. Their activities are essential in hormone responses (particularly auxin), meristem function, and stress adaptation (Voinnet, 2009). AGO1 is the major mediator of miRNA activities (Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005), but its close homolog, AGO10, plays additional, specialized roles associated with the function of some miRNAs (Brodersen et al., 2008; Mallory et al., 2009; Ji et al., 2011; Zhu et al., 2011). Functionally relevant associations of miRNAs with AGO2 (Maunoury and Vaucheret, 2011; Zhang et al., 2011) or AGO7 (see below) occasionally occur.
A third class comprises trans-acting siRNAs. Their biogenesis is initiated by miRNA-guided cleavage of a single-stranded, often noncoding, transcript. A cleavage fragment is then converted into double-stranded RNA by an RNA-dependent RNA polymerase and diced to yield a population of trans-acting siRNAs that in turn regulate gene expression in association with AGO1 (Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005). The production of the deeply conserved TAS3 ta-siRNAs that are involved in specification of leaf polarity via control of auxin response factors constitutes a particular case. Their biogenesis requires a specialized AGO protein, AGO7, that associates almost exclusively with miR390 to guide cleavage of TAS3 precursor RNA (Montgomery et al., 2008).
A fourth major class consists of exogenous siRNAs (i.e., siRNAs that are produced from extragenomic sources). Viral siRNAs are by far the most important group of exogenous siRNAs. Indeed, RNA silencing constitutes the first line of antiviral immunity in plants, but other pathogens that use nucleic acids directly in their virulence strategies can also be sources of siRNAs (Dunoyer et al., 2006; Ding and Voinnet, 2007). Viral siRNAs associate with AGO1 and AGO2 that are major effectors in antiviral immunity (Azevedo et al., 2010; Harvey et al., 2011; Jaubert et al., 2011; X.B. Wang et al., 2011).
In addition to these major classes of small RNAs, a number of other endogenous small RNAs have been discovered. They include siRNAs derived from endogenous hairpins (often referred to as endo-siRNAs), siRNAs derived from convergent, overlapping transcripts (natural antisense transcript siRNAs), and siRNAs induced in close proximity to DNA double-strand breaks (double-strand break–induced RNAs [diRNAs]) (Borsani et al., 2005; Lindow et al., 2007; Wei et al., 2012). endo-siRNAs typically accumulate as AGO1-associated 21- to 22-nucleotide species and AGO4-associated 24-nucleotide species (Dunoyer et al., 2010), while diRNAs associate specifically with AGO2 that is required for repair via homologous recombination (Wei et al., 2012).
AGO STRUCTURE/FUNCTION RELATIONSHIPS
Core Properties of AGO Proteins
To carry out their diverse functions in RNA silencing reactions, AGO proteins have three key biochemical properties. First, they all bind small RNAs tightly to facilitate base pairing with complementary target RNAs (Hammond et al., 2001). Second, many AGO proteins possess an RNaseH-like endonuclease activity, the slicer activity (Song et al., 2004), that serves at least three purposes: (1) It contributes to silencing by cleavage of target RNA provided that guide and target RNAs are base paired around the middle of the duplex (Hutvágner and Zamore, 2002; Llave et al., 2002); (2) it can amplify silencing responses by production of cleaved RNA fragments that are substrates for RNA-dependent RNA polymerases and hence become sources of new waves of secondary siRNAs analogous to tasiRNA biogenesis (Irvine et al., 2006; Qi et al., 2006); and (3) it facilitates strand separation of fully complementary small RNA duplexes by passenger strand cleavage (Matranga et al., 2005). Third, AGO proteins serve as platforms onto which silencing cofactors can bind. It is a recurrent principle in many plant, animal, and fungal AGOs that they bind to proteins containing Gly-Trp (GW) dipeptides (El-Shami et al., 2007; Till et al., 2007). These GW proteins are often essential cofactors in the RNA silencing reaction. Two prominent examples are provided by the protein GW182 in animals and the large subunit of the plant-specific RNA polymerase V. In the miRNA pathway in animals, AGO proteins associate with members of the GW182 protein family (Meister et al., 2005). These proteins are sufficient to confer silencing when tethered to an mRNA and act, among other mechanisms, to recruit one of the major deadenylase complexes to targeted mRNAs for initiation of mRNA decay (Lazzaretti et al., 2009; Braun et al., 2011). In plants, AGO4 binds directly to GW repeats in the large subunit of RNA polymerase V, a plant-specific RNA polymerase required for AGO4-dependent chromatin silencing of repeats downstream of siRNA production (El-Shami et al., 2007).
Domain Organization of AGO Proteins
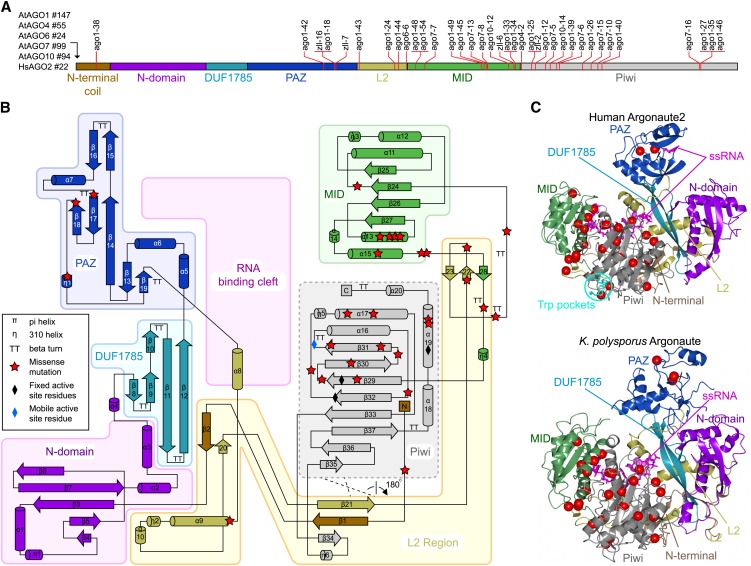
The core biochemical properties reside in four domains common to all AGO proteins: an N-terminal domain, a Piwi-Argonaute-Zwille (PAZ) domain, a domain in the middle of the primary structure (MID), and a C-terminal (PIWI) domain. In addition, the PAZ and MID domains are connected by a large piece of intervening sequence referred to as linker 2 (L2; Figure 1A). The N-terminal domain can be divided into three elements of primary structure, designated N-terminal coil, N domain, and domain of unknown function 1785 (DUF1785), previously referred to as linker 1.
Figure 1.
Structural Mapping of AGO Missense Mutations.
(A) Schematic diagram of common elements of the primary structure of the AGO protein family depicting the location of known plant missense alleles. The length of N-terminal extensions in each AGO protein is shown to the left of the diagram.
(B) Topology diagram of the human Ago2 structure colored according to domains and structural regions. The RNA binding cleft is located between one lobe containing the N, DUF1785, and PAZ domains and a second lobe containing the MID and Piwi domains. The L2 connects the two lobes and forms secondary structure elements with several domains and the structured N terminus. The plant ago missense mutations are marked with red stars.
(C) The tertiary structures of human Ago2 and K. polysporus Ago as determined by crystallography. Note the overall structural similarity between human and yeast Ago, except in the PAZ domain that exists in closed (human Ago2) and open (K. polysporus Ago) conformations. The positions of plant ago missense mutations are marked with red spheres. ssRNA, single-stranded RNA.
Small RNA binding involves the MID and PAZ domains, such that the 5′phosphate is bound by the MID domain and the 3′end is bound by the PAZ domain (Ma et al., 2004, 2005). The slicer activity resides in the Piwi domain that adopts an RNaseH-like fold. Three active-site residues (Asp-Asp-His) placed in analogous positions in human and archaeal AGO are required for slicer activity in human, plant, and fungal AGOs (Liu et al., 2004; Song et al., 2004; Baumberger and Baulcombe, 2005; Irvine et al., 2006). Interactions with GW repeat proteins are not understood in detail, but mutations that disrupt such interactions have been identified in the Piwi domain, suggesting that it is also the domain responsible for GW interactions (Till et al., 2007).
Mutational Analyses of Plant AGOs
The functions of Arabidopsis AGO proteins have been studied extensively using genetic approaches. In addition to screens aimed at identifying genes required for different silencing phenomena, several genes encoding AGO proteins (AGO1, AGO7, and AGO10) were originally identified in developmental screens, and the name of the protein family indeed derives from the tentacle-shaped leaves of Arabidopsis ago1 knockout mutants that produce seedlings resembling small squids (Argonautus) (Bohmert et al., 1998; Vaucheret, 2008). These forward genetic analyses have helped assign specific AGO proteins to distinct silencing pathways and have generated extensive series of mutant alleles defective for function. For example, 56 different mutant alleles of AGO1 have been described in the literature (see Table 1 for the 20 missense alleles; see Supplemental Table 1 online for a complete list), and they can be ordered according to their strength (i.e., the severity of their developmental defects). These mutant collections therefore constitute a precious resource of information on amino acid residues important for function of plant AGOs.
Table 1. Missense Mutations Recovered in Arabidopsis AGO Proteins by Forward Genetics.
| Allele | Mutation | Domain | Hs-AGO2 | Expected to Affect | Strength | Reference |
|---|---|---|---|---|---|---|
| AGO1 | ||||||
| ago1-12 | H765L | Piwi | His-600 | Slicing | Strong | Kidner and Martienssen (2004) |
| ago1-18 | P493S | PAZ | Pro-340 | PAZ fold | Strong | Sorin et al. (2005) |
| ago1-24 | L573F | L2 | Ile-418 | MID-Piwi interface | Null | Fagard et al. (2000) |
| ago1-25 | G760S | Piwi | Gly-595 | Piwi fold | Weak | Morel et al. (2002) |
| ago1-26 | P840S | Piwi | Pro-661 | Putative Trp pocket | Medium | Morel et al. (2002) |
| ago1-27 | A994V | Piwi | Ala-813 | MID-Piwi interface | Weak | Morel et al. (2002) |
| ago1-33 | G735R | MID | Gly-573 | MID-Piwi interface | Weak | Sorin et al. (2005) |
| ago1-34 | G735R | MID | Gly-573 | MID-Piwi interface | Weak | Sorin et al. (2005) |
| ago1-35 | A994V | Piwi | Ala-813 | MID-Piwi interface | Weak | Sorin et al. (2005) |
| ago1-38 | G186R | N coil | Gly-32 | N coil | Weak | Gregory et al. (2008) |
| ago1-39 | G793E | Piwi | Ala-627 | MID-Piwi interface | Very weak | This work |
| ago1-40 | A865V | Piwi | Ala-686 | Unclear | Very weak | Mallory et al. (2009) |
| ago1-42 | P481T | PAZ | Pro-326 | PAZ fold | Strong | This work |
| ago1-43 | P527L | L2 | Pro-373/Ala-372* | N- and C-lobe interface | Medium | This work |
| ago1-44 | G579E | L2 | Gly-422 | MID-Piwi interface | Medium | This work |
| ago1-45 | R696H | MID | Arg-534 | Unclear | Weak | Smith et al. (2009) |
| ago1-46 | A994T | Piwi | Ala-813 | MID-Piwi interface | Weak | Smith et al. (2009) |
| ago1-48 | G601N | MID | Gly-445 | MID-L2 interface | Medium | This work |
| ago1-49 | G692N | MID | Ala-530 | 5′RNA binding | Medium | This work |
| ago1-54 | G601D | MID | Gly-445 | MID-L2 interface | Medium | This work |
| AGO7 | ||||||
| ago7-5 | A750T | Piwi | Ala-613 | Piwi fold | Cuperus et al. (2010) | |
| ago7-6 | S769F | Piwi | Val-631 | Unclear, Piwi fold? | Cuperus et al. (2010) | |
| ago7-7 | G583E | MID | Ala-457 | MID fold | Cuperus et al. (2010) | |
| ago7-8 | S668L | MID | Gly-536 | MID fold | Cuperus et al. (2010) | |
| ago7-10 | L818F | Piwi | Leu-680 | N coil | Cuperus et al. (2010) | |
| ago7-13 | E669K | MID | Asp-537 | 5′phosphate binding | Cuperus et al. (2010) | |
| ago7-15 | S810N | Piwi | Ser-672 | Unclear, target RNA binding? | Cuperus et al. (2010 ) | |
| ago7-16 | R944K | Piwi | Arg-812 | 5′ phosphate binding | Cuperus et al. (2010) | |
| AGO10 | ||||||
| zll-2 | G707D | Piwi | Gly-595 | Piwi fold | Strong | Moussian et al. (1998) |
| zll-6 | G681R | MID | Gly-572 | Unclear, MID fold? | Strong | Moussian et al. (1998) |
| zll-7 | E445A | PAZ | Glu-342 | PAZ fold | Strong | Moussian et al. (1998) |
| zll-16 | P443S | PAZ | Pro-340 | PAZ fold | Strong | Moussian et al. (1998) |
| ago10-12 | L674F | MID | Leu-565 | MID-Piwi interface | Weak | Ji et al. (2011) |
| ago10-14 | D731N | Piwi | Asp-619 | Putative Trp pocket | Weak | Ji et al. (2011) |
| AGO4 | ||||||
| ago4-2 | E641K | Piwi | Gln-581* | Unclear | Agorio and Vera (2007) | |
| AGO6 | ||||||
| ago6-6 | G455R | L2 | Gly-433 | MID-Piwi interface | Eun et al. (2011) |
For all mutations, the location, predicted biochemical or structural effect, and phenotypic effect are indicated. Amino acid residues marked with an asterisk show ambiguous alignments with human Ago2. For AGO10, allele strength is ecotype dependent. The classifications given here apply to ecotype Landsberg erecta (Mallory et al., 2009). Allele strengths cannot be clearly assigned for ago7, ago4 and ago6. A list of all published Arabidopsis ago mutations obtained by forward genetics is given in Supplemental Table 1 online.
The mutational data set is biased toward amino acids encoded by GC-rich codons, however. This is because most genetic screens have used the guanine O6-alkylating agent ethyl methanesulfonate (EMS) as mutagen for induction of point mutations, and guanine O6-alkylation results almost exclusively in GC-AT transitions. Thus, of 20 independently isolated ago1 point mutant alleles, 18 (90%) affect the four residues (20%) encoded by GC-rich codons (Gly, Ala, Pro, and Arg). Although EMS-based screening is close to exhaustive based on the recovery of several independent, yet identical alleles, the failure to recover mutants in certain regions of AGO proteins does not necessarily exclude their functional importance; the codons for important amino acid residues may simply be immune to EMS mutagenesis. Notwithstanding this caveat, the collection of ago mutant alleles constitutes an invaluable toolbox for a range of genetic and biochemical approaches to further our understanding of RNA silencing mechanisms. For example, specific ago alleles with defects in interaction surfaces could be used to identify components of AGO-containing complexes important for their function using genetics and would be useful tools to validate the importance of interactors identified by biochemical means. But how can the correct alleles be chosen for such analyses? All such experiments have been hampered by our complete lack of understanding of the specific molecular defect in any missense mutant of plant AGO proteins, despite detailed analyses of their phenotypic effects. In this article, we use the recently published eukaryotic AGO structures as models for plant AGOs to give more precise interpretations of the information hidden in the mutant AGO alleles recovered in multiple screens over the last 15 years of Arabidopsis research. We also include a previously published but uncharacterized allelic series of ago1 isolated by continued screening for mutants defective in posttranscriptional silencing of β-glucuronidase or nitrate reductase transgenes (Morel et al., 2002; Jauvion et al., 2010). While many mutations are likely to affect folding or structural integrity of parts of the protein, a distinct set of mutations with more well-defined functional defects can also be identified. These include mutations that affect core biochemical activities as well as uncharacterized, surface-exposed patches likely to constitute sites of protein–protein interaction. The analyses also reveal regions of AGO proteins of unexpected structural importance.
EUKARYOTIC AGO STRUCTURES
Eukaryotic AGO Structures as Models for Plant AGO Proteins
Until very recently, all structural work on AGO proteins has focused on either isolated fragments of eukaryotic AGO proteins or on full-length proteins from thermophilic bacteria or archaea (Faehnle and Joshua-Tor, 2010). The thermophile AGO proteins have no known endogenous function and have fundamentally different biochemical properties because they use guide DNA strands. In addition, their sequence identity to eukaryotic AGO proteins is barely detectable by standard BLAST searches. Thus, although important lessons have been learned from the structural studies of these proteins, it has been unclear how similar eukaryotic AGO proteins involved in RNA silencing really are. Therefore, it is a considerable breakthrough that three recent articles now describe high-resolution crystal structures of small RNA–bound human Ago2 and budding yeast (Kluyveromyces polysporus) Ago (Elkayam et al., 2012; Nakanishi et al., 2012; Schirle and MacRae, 2012). Perhaps surprisingly, the eukaryotic AGO structures are remarkably similar to the distantly related thermophile AGO, although there are differences in the overall topology of the molecules that may be important for function.
A structural comparison between the still distantly related human and yeast AGO shows that each of the domains adopts close to identical structures. For the PAZ, MID, and Piwi domains, the root mean square deviation values of the Cα atoms are between 1.3 and 1.9 Å. The same range of values is obtained when comparing the crystal structures of the Arabidopsis AGO1, AGO2, and AGO5 MID domains (Frank et al., 2012). Given this high structural similarity and that Arabidopsis AGOs, in particular the miRNA-related AGOs, AGO1 and AGO10, are considerably more closely related in sequence to human Ago2 than is the budding yeast Ago (43 and 42% versus 24% identity), we think it safe to assume that human Ago2 is a good structural model for plant AGO proteins.
In the following, we use the new eukaryotic AGO structures to analyze the structural and functional consequences of missense mutations known to produce defective AGO proteins. Throughout the article, we show parts of the human Ago2 structure (Protein Data Bank entries 4EI3 and 4F3T) and a homology model of plant proteins to illustrate how given mutant alleles exhibit specific biochemical defects.
Overall Structural Description of AGO
As described previously for thermophile AGO proteins, eukaryotic AGOs adopt a two-lobed structure with a central cleft lined by positively charged amino acid residues that allow binding of the negatively charged small RNA (Figures 1B and 1C). The MID-Piwi domains constitute one lobe, while the N domain on the other side of the cleft together with the DUF1785 region and the PAZ domain constitute the other lobe. The linker, L2, connects the PAZ and MID domains. It is a remarkable part of the protein because it stretches more than 200 Å across the N, DUF1785, Piwi, and MID domains and contributes substantially to the stabilization of the overall structure.
A simple mapping of the positions of all identified missense mutations in plant AGO proteins provides a first rough idea of regions sensitive to structural change and of functionally important sites. Thus, mutations cluster particularly in the MID/Piwi lobe, some are found in the PAZ domain and in L2, whereas the N and DUF1785 domains are nearly devoid of deleterious mutations (Figure 1C).
Small RNA binding involves at least the PAZ, Piwi, and MID domains. The structures provide clear views of the 5′phosphate bound by the MID domain, the 5′end of the RNA guide bound by the Piwi domain, and the 3′end bound by the PAZ domain. The negatively charged sugar-phosphate backbone of the small RNA runs through the entire protein along the positively charged central cleft, although not all nucleotides are structured and thereby visible in the crystal structures. Well-defined electron density can be observed for the first seven nucleotides of the small RNA guide primarily bound by the Piwi domain.
The anchoring of the sugar-phosphate backbone of RNA to the MID and Piwi domains allows stabilization of the bases. The Watson-Crick edges (Leontis and Westhof, 2001) of nucleotides 2 to 7 (the “seed”) are solvent exposed and hence poised for base pairing with target RNAs. However, despite their solvent exposure, the seed nucleotides are not readily accessible in the crystallized conformation. This suggests that considerable conformational change is required for these nucleotides to seed (i.e., initiate) sRNA-target RNA base pairing. Finally, a comparison between the human and yeast structures shows that the PAZ domain can undergo a conformational change between a more closed and a more open position (Figure 1C). Only in the more closed position is the 3′RNA end bound to the PAZ domain.
The N-Terminal Coil: An Unrecognized Unit of AGO Proteins
The eukaryotic structures reveal that the division of AGO proteins into N, PAZ, MID, and Piwi domains based on primary structure does not fully account for their spatial organization. Interestingly, the N-terminal extreme of the N domain adopts a well-defined coil-like structure that is tucked in between residues of the Piwi domain. The coil extends more than 36 Å across the MID/Piwi lobe on the border to the N/PAZ lobe. After a 90° turn, the N-terminal extends into a β-strand that interacts with elements from the Piwi, DUF1785, and L2 domains, before it ends in the globular N domain.
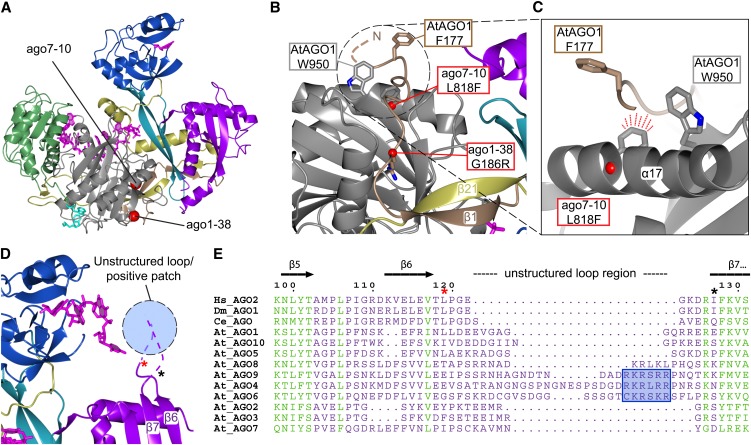
Mutations recovered in plant AGOs suggest that this structured N-terminal coil has important functions. The weak ago1-38 mutant changes an invariant Gly to Arg, which is expected to dislocate the N-terminal coil and distort its fixation (Figures 2A and 2B). Similarly, the Phe replacing a Leu in ago7-10 is predicted to clash with the very end of the structured N-terminal coil, as it points out toward the coil from the Piwi domain (Figures 2A and 2B).
Figure 2.
The Structurally Fixed N-Terminal Coil and the N Domain.
(A) Ribbon diagram of human Ago2 pointing out the locations of the two missense mutations ago7-10 and ago1-38.
(B) The structured N terminus (brown) is tightly interacting with the Piwi domain (gray) as an extended coil-like structure, and the β1-strand is fixed between β21 (L2; gold) and β34 (Piwi; not seen). The G186R mutation in ago1-38 (red sphere; modeled) is expected to locally dislocate the N-terminal coil since the Arg side chain is predicted to heavily clash with the Piwi domain. The L818F mutation in ago7-10 located in the α17-helix is likewise expected to dislocate the extended N-terminal coil.
(C) Close-up view of the α17-helix containing the modeled L818F ago7-10 mutation. The bulkier Phe is predicted to clash with the peptide backbone of the N-terminal region. The side chains of two conserved hydrophobic and solvent exposed residues, At-AGO1 F177 and At-AGO1 W950, are presented.
(D) The close proximity between the 3′end of the guide strand and the unstructured region connecting β6-β7 is displayed in the human Ago2 structure loaded with miR-20a (Protein Data Bank entry 4F3T). The first and last residues in the unstructured loop are indicated with asterisks matching those in (E).
(E) The primary sequence alignment of human (Hs), Drosophila (Dm), C. elegans (Ce), and Arabidopsis AGOs shows how the loop region connecting β6-β7 varies in length and sequence. A conserved positively charged patch in AGO4, 6, and 9 (blue box) is suggested to interact with the 3′end of the guide strand.
What might be the function of this fixed coil and the surrounding structural elements? A simple possibility would be that it stabilizes the interface between the two lobes. Although the tight interaction with the Piwi domain may well contribute to stability, we consider it unlikely that a distortion of the very tip of the N terminus could have an impact on the overall structure. The function may therefore be found elsewhere. Membrane association is emerging as a general property of animal and plant AGO proteins (including AGO1 and AGO7), and ago1-38 was reported to exhibit reduced membrane association (Cikaluk et al., 1999; Gibbings et al., 2009; Lee et al., 2009; Brodersen et al., 2012 ; Jouannet et al., 2012). Since membrane association was proposed to depend on other interacting proteins, the region may serve as a patch for protein–protein interaction that enables membrane recruitment. In this regard, it is interesting to note that Trp-769 and Phe-23 in human Ago2 (Trp-950 and Phe-177 in Arabidopsis AGO1), located in the same area, project their hydrophobic side chains into the solvent as if to create an interface for protein binding (Figures 2B and 2C). The perfect conservation of these residues in several AGO proteins that associate with miRNAs (Drosophila melanogaster Ago1, Caenorhabditis elegans ALG-1 and ALG-2, and Arabidopsis AGO1 and AGO10) suggests functional importance (see Supplemental Figure 1 online). These residues are therefore obvious targets of mutagenesis to test the importance of the patch in membrane recruitment.
The N/DUF1785/PAZ Lobe: Getting the Right Size of RNA Loaded and Bound?
The functions of the N and DUF1785 domains are unclear, although in vitro biochemical assays using human Ago2 point to a role in strand separation during the loading process (Kwak and Tomari, 2012). No clues to their function are discernible from Arabidopsis forward genetics, since none of the 38 point mutants recovered in plant AGOs affects the N-DUF1785 domains. Structurally, the N/DUF1785/PAZ lobe is more extended and likely more flexible than the MID/Piwi lobe. DUF1785 consists of a centrally positioned five-stranded β-sheet that connects the globular part of the N domain and the PAZ domain (Figure 3A).
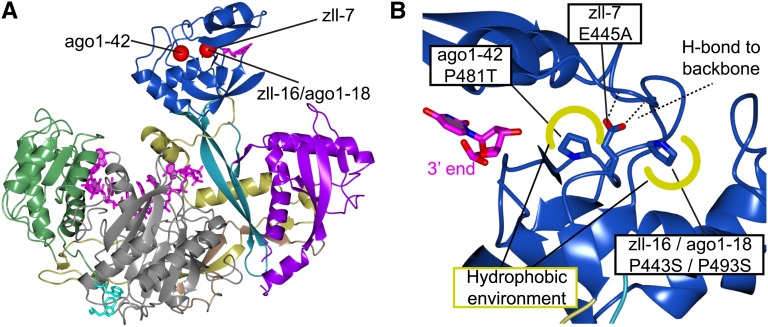
Figure 3.
Structural Integrity of the PAZ Domain Is Important for Function.
(A) Ribbon diagram of human Ago2 pointing out the locations of three plant missense mutations in the PAZ domain.
(B) Three mutations in zll-16/ago1-18 and ago1-42 change two conserved Pro residues constituting important hydrophobic interactions (illustrated in yellow) needed for proper folding of the domain. The zll-7 mutation E445A disrupts H-bonds to two backbone amides also resulting in destabilization of the PAZ domain.
The length and structural flexibility of two β-strands in DUF1785 allow the PAZ domain to be freely accessible and to bend by at least 20° as suggested by conformational differences between human and yeast AGOs. Another flexible region connects β6 and β7 (Figure 2D) and is protruding from the globular N domain toward the PAZ domain. Its spatial position suggests an involvement in coordinating the least structured part of the 3′guide strand. In this respect, it is interesting to note that AGO4, 6, and 9 harbor an insertion of four to five positively charged residues in this region that is absent in other plant AGOs (Figure 2E). Because AGO4, 6, and 9 bind 24-nucleotide rather than 21-nucleotide small RNAs, this positively charged patch could act to coordinate additional nucleotides and hence may be a determinant of size preferences of AGO proteins.
Therefore, it is possible that the N/DUF1785 domains mainly provide a flexible support for RNA binding and for exposure of the PAZ domain. Such a role might explain its robustness to point mutations compared with the more sensitive RNA and protein binding interfaces in the rigid MID-Piwi lobe. The role in supporting the PAZ domain may reconcile two recent reports that point to the importance of either N/DUF1785 or PAZ domains in strand separation during loading of human Ago2 (Gu et al., 2012; Kwak and Tomari, 2012). Since PAZ domain mutants had a much more profound effect on strand separation than N/DUF1785 mutants (Gu et al., 2012), it is possible that strand separation defects in N/DUF1785 mutants arise as an indirect consequence of reduced support of the PAZ domain. The functional importance of the PAZ domain is supported by four mutations in AGO1 and AGO10 (Table 1, Figure 3). These mutations do not seem to disrupt small RNA binding specifically but are likely to affect the fold of the entire PAZ domain (Figure 3B). Although binding of the 3′end of the small RNA is an important function of the PAZ domain, other functions can be envisaged. Indeed, we can speculate that the exposure and flexibility of the PAZ domain may be important in RNA loading, target recognition, or both (Bouasker and Simard, 2009).
L2: The Backbone of Argonaute
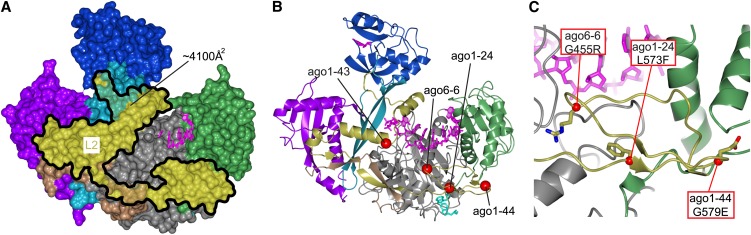
The L2 part has several structural contributions and interacts with N, DUF1785, Piwi, and MID domains, as well as with the RNA. The total interface area between L2 and the surrounding protein reaches a remarkably high 4100 Å2 (Figure 4A). Close to the PAZ domain, L2 adopts a coil conformation. This region contains conserved positively charged residues likely to be involved in RNA binding. Parallel to the DUF1785 region, an α-helix is present that interacts with the Piwi-bound RNA and is responsible for a kink between the nucleotide bases 6 and 7.
Figure 4.
L2 Is a Significant Structural Unit.
(A) Surface representation of the human Ago2 highlighting the large footprint of L2. Note that all domains are interacting with L2.
(B) Structural overview of the missense mutations in L2 with orientation as in (A). The ago1-43 missense mutation is likely to reduce the stability of the interface between the N- and C-lobes.
(C) View of three modeled L2 missense mutations located between the MID (green) and Piwi (gray) domains. The G455R (ago6-6) and L573F (ago1-24) could have an indirect effect on the RNA binding, whereas the destabilization caused by the G579E mutation in ago1-44 is less clear.
The L2 interactions stabilize both the interface between the N and DUF1785 domains and, in particular, between the MID and Piwi domains. These latter interactions are important as shown by three mutations in plant AGOs. Two mutations, ago1-44 and ago6-6, change structurally important Gly residues involved in β-turns (Figures 4B and 4C). For example, the intermediate-strength ago1-44 mutation changes a Gly to a Glu in a position of the structure where the dihedral angles only allow a Gly. The third, and most remarkable mutation, is the L573F change in ago1-24. This mutation produces a null phenotype, demonstrating that even seemingly mild changes in this domain can result in complete loss of function, probably as a consequence of lost structural integrity of L2. This finding is unexpected as the sequence of this part of L2 is not particularly conserved in plants. It does, however, clearly fill out a space between the MID and Piwi domains close to the bound RNA, and it provides stabilization of a coil in the Piwi domain that is directly involved in coordination of the 5′end of the RNA.
The MID-Piwi Interface and 5′End Binding of Small RNA
The MID/Piwi lobe binds the structurally well-defined 5′end of the small RNA, in contrast with the N/DUF1785/PAZ lobe that harbors its flexible part. This fundamental property of AGO-small RNA complexes was also observed in structures of DNA-bound thermophile AGO complexes (Wang et al., 2008). It explains a wealth of computational and biochemical evidence, especially from animals, pointing to the 5′end of the small RNA as being of particular importance for target interaction.
The vast majority of the 38 missense mutations summarized here are located in the MID and Piwi domains (Figures 1B and 1C) probably in part as a consequence of their importance in binding the small RNA in a well-defined conformation that allows target interaction. The two globular domains are highly conserved and are held together by an extensive network of contacts. In addition, the low root mean square deviation values between the Cα atoms of homologous AGO structures indicate that the orientation of the two domains is fixed (human and Kluyveromyces polysporus, 1.54 Å/380 Cα). This appears to be of fundamental functional importance because of the conspicuous density of mutations, 13 in total, on or in close proximity to the interface between the two domains (Figure 5A). Thus, disruption or even slight distortion of the MID–Piwi interaction seems to profoundly affect function. A plausible explanation for this is that small RNA binding requires specific spacing between the domains as well as a series of specific interactions to conserved side chains that are very sensitive to regional mutations.
Figure 5.
The MID and Piwi Domains Are Highly Sensitive to Mutations.
(A) Structural overview highlighting the density of missense mutations on or in close proximity to the MID-Piwi interface. A close-up view of the region shows the location of the 13 missense mutations in this region.
(B) Presentation of mutations directly involved in the 5′RNA binding. Conserved residues are part of a H-bond network involved in 5′phosphate coordination.
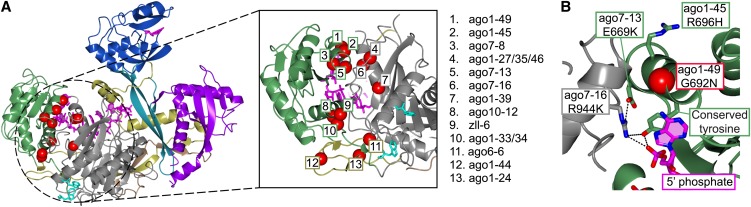
Some of the mutations in plant AGOs directly illustrate the importance of coordination of the 5′phosphate of the small RNA in the MID domain. The ago7-16 mutation introduces a seemingly conservative change of a deeply conserved Arg residue to a Lys (R944K). However, more careful analysis shows that this Arg, in spite of its location in the Piwi domain, is directly involved in coordination of the 5′phosphate. To be positioned close to the 5′phosphate, the guanidinium group of the Arg requires coordination by hydrogen bonding to a Glu located in the MID domain (Figure 5B). Such coordination cannot be performed by a Lys. Interestingly, the ago7-13 mutation changes the very same Glu that allows Arg-944 to contact the 5′phosphate, confirming that this part of the structure is needed for proper 5′phosphate binding (Figure 5B).
AGO proteins are well known to exhibit selective small RNA binding depending on the identity of the 5′nucleobase of the small RNA. For example, AGO1 exhibits a strong preference for 5′uridine, while AGO2 and AGO4 associate mainly with small RNAs starting with a 5′adenosine (Mi et al., 2008). Recent structures of plant and animal MID domains have identified the so-called nucleotide specificity loop in the MID domain that makes contacts to the 5′nucleobase (Frank et al., 2010, 2012). No mutations have been recovered in the nucleotide specificity loop, but the ago1-49 mutation illustrates the importance of binding of the 5′nucleotide base. This strong loss-of-function mutation results in a Gly-to-Asn change. Although the Gly is not directly associated with small RNA binding, its preceding residue, a strongly conserved Tyr, is stacking with the 5′uridine of the small RNA and coordinates the 5′phosphate by its hydroxyl group. The strong phenotype of ago1-49 can therefore be explained by a structural change in the local environment disturbing 5′nucleotide binding.
New Twists on Interactions with GW Proteins
The structural work on thermophile AGO proteins cannot be used as a model for understanding interactions with GW proteins because such interactions have only been described for eukaryotic AGO proteins. While none of the reported eukaryotic AGO structures provide direct views of complexes with GW proteins, the work of Schirle and MacRae (2012) on human Ago2 provides interesting clues to the molecular basis for these interactions. They succeeded in growing crystals only in the presence of 100 mM phenol and noticed that two well-defined phenol molecules could be found in two hydrophobic pockets in the surface of the Piwi domain far away from the bound small RNA. Structures with free Trp bound showed the indole ring inserted into the hydrophobic pockets, suggesting that they are the binding sites for GW repeat proteins (Schirle and MacRae, 2012). Human Ago2 mutations that abolish GW interactions were previously found to cluster around the 5′phosphate binding site in the MID domain (Till et al., 2007). However, reinspection of these mutational data in light of the structure of human Ago2 shows that nearly as many disruptive mutations are concentrated around the Trp binding pockets, including a conserved Lys whose side chain makes up one side of a pocket. Loss of GW interaction in the MID domain mutants may well be an indirect consequence of loss of small RNA binding, since recent proteomics studies show that human Ago2 interacts with only partly overlapping sets of proteins depending on its small RNA loading, and one GW182 family member is bound exclusively by Ago-miRNA complexes (Frohn et al., 2012). In addition, a Drosophila Ago1 mutant with reduced GW182 interaction, yet intact miRNA binding, is located in the putative GW binding pocket (Eulalio et al., 2009).
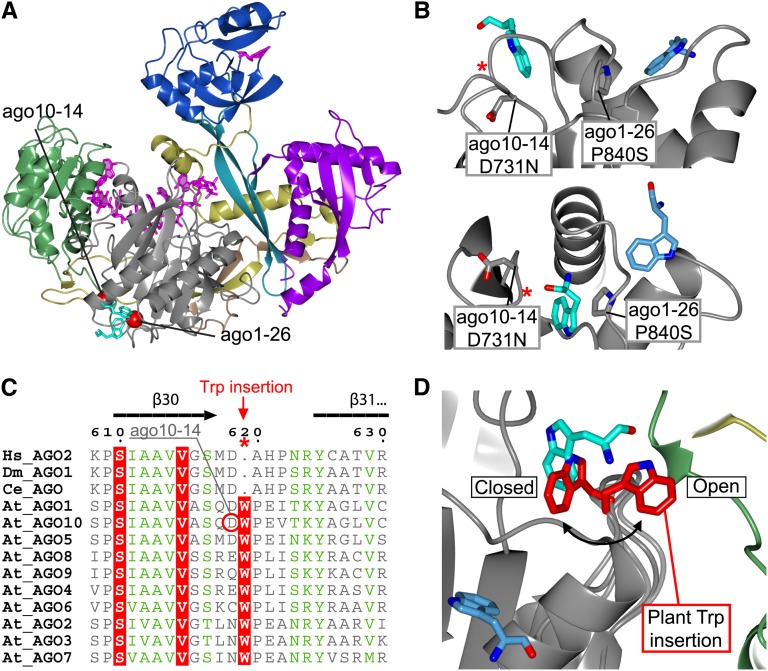
Do we learn anything new about the functions of plant AGO proteins from the identification of these putative GW binding sites? Alignment of sequences surrounding the two Trp binding pockets in human Ago2 suggests that at least one binding pocket is well conserved in AGO1 and AGO10 (see Supplemental Figure 1 online). The functional importance of this binding site is supported by the ago1-26 allele that has a Ser in place of a conserved Pro residue (Pro-840 in At-AGO1 and Pro-661 in Hs-Ago2). This Pro lines the binding pocket and its replacement by Ser is predicted to decrease hydrophobicity and dislocate conserved residues (Leu-650, Lys-660, and Tyr-698 in Hs-Ago2) in the Trp binding environment (Figures 6A and 6B). Therefore, the ago1-26 allele indirectly provides evidence that one or more so far elusive GW interaction partners are required for silencing by miRNAs and siRNAs in association with AGO1.
Figure 6.
Hs-Ago2 Trp Binding Pockets Are Likely to Be Conserved in Plants.
(A) Structural overview of Hs-Ago2 pointing out the close proximity between the ago10-14 and ago1-26 mutations and the ligand-bound Trp residues (cyan), suggesting a functional connection to the Trp binding sites.
(B) Top: Side view of the two Trp binding pockets. Pro-840 mutated in ago1-26 constitutes parts of one of the pockets (Trp; light blue), but the mutation could interfere with both pockets due to its central location (cyan and light blue). The ago10-14 D731N mutation shows how changing the nonconserved environment involved in Trp binding in Hs-Ago2 has an effect in plants. Bottom: Different view of top panel. The red asterisks indicate the location of the Trp insertion pointed out in (C).
(C) The primary sequence alignment of Human (Hs), Drosophila (Dm), C. elegans (Ce), and Arabidopsis AGOs reveals insertion of a conserved Trp in the loop connecting β30-β31 in plant AGOs.
(D) Backbone modeling of the At-AGO1 loop connecting β30-β31. The model shows the conformation of the plant-specific Trp (red) inserted at the Hs-Ago2 Trp binding site (cyan). The model suggests two possible conformations: one excluding additional Trp binding (closed) and one allowing binding of hydrophobic elements from an external source (open).
The existence of the second Trp binding pocket is less clear based on sequence conservation between human and plant AGOs alone (see Supplemental Figure 1 online). Evidence for its existence in plant AGOs is provided by the ago10-14 allele. This mutation affects a conserved Asp that engages in hydrogen bonding with two backbone amide groups to stabilize a turn in the Piwi domain necessary for formation of the pocket (Figures 6A and 6B). This observation is of particular importance for models of AGO10 function. AGO10 has been proposed to function as a competitive inhibitor of miR165/166-AGO1 complexes through specific, efficient loading of this miRNA but poor silencing activity of the AGO10-miR166 complex (Zhu et al., 2011). In this model, loss of the shoot apical meristem observed in ago10 loss-of-function mutants is a consequence of exaggerated silencing by AGO1-miR166 of a family of transcription factors that are required for meristem maintenance. There is good evidence for this model: Shoot apical meristem phenotypes of ago10 can be suppressed by inhibition of miR166 activity, and specific features in pre-miR166 have been identified that dictate preferential loading of miR166 into AGO10 (Zhu et al., 2011). However, more classical silencing functions of AGO10 in association with other miRNAs have also been proposed (Brodersen et al., 2008; Mallory et al., 2009; Ji et al., 2011). From a molecular perspective, the competitive inhibitor mode of AGO10 function would only require miR166 binding, while the silencing function would require the full range of biochemical properties of AGO proteins. Since the Trp binding pockets are situated far away from the small RNA interaction surface in the Piwi domain, one may predict that mutations in the Trp pockets should affect only the silencing mode and not the competitive inhibitor mode of AGO10 function. This condition is met, since the ago10-14 allele does not exhibit the shoot apical meristem defects of ago10 null mutants (Y.J. Kim and X. Chen, unpublished data; Ji et al. 2011). Thus, the structural analysis of ago10 mutants is consistent with both proposed modes of AGO10 function and suggests that silencing by AGO10 involves one or more GW proteins yet to be identified. It further indicates that ago10-14 could be particularly useful for exploring such GW interactions, as could ago1-26 in the case of AGO1.
The Missing GW Interactors: A Case of Autoinhibition?
Despite serious attempts in several laboratories, no plant GW protein acting in the miRNA pathway with AGO1 or AGO10 has yet been purified, and it has only very recently been shown that the helicase SDE3 uses GW motifs to interact with AGO1 in transposon regulation (Garcia et al., 2012). Genetic evidence implicates the GW-containing protein SUO in repression of some miRNA targets, but it is unclear whether SUO interacts physically with AGO1 and whether it requires GW dipeptides for function (Yang et al., 2012). By contrast, GW proteins readily copurify with AGO proteins from animal cells. This is all the more puzzling given the mutational evidence for the importance of intact Trp binding pockets in the plant AGOs.
Inspection of the alignment of human and plant AGOs in the region surrounding the least conserved binding pocket reveals an interesting clue to the solution of this problem: The plant proteins all contain an insertion of a Trp residue in the loop atop the binding pocket (Figure 6C; see Supplemental Figure 1 online). Flexible backbone modeling of this loop shows two preferred scenarios regarding this inserted Trp: Either the indole side chain is inserted into the pocket (closed conformation), or it projects outwards into the solvent (open conformation) (Figure 6D). This predicts that although interactions with GW proteins are possible in the open conformation, the affinity may be much reduced compared with the situation in animal AGOs. This could explain why such interactors are easily lost in affinity purifications of AGO proteins and immediately suggests strategies to circumvent this problem.
What might have favored the appearance of such an autoinhibition mechanism in plant AGO proteins? One possibility is protection from viruses, since AGO proteins, as key antiviral host factors, may be extremely vulnerable if simple GW repeats are all it takes to evolve a strong interactor. It is also possible that high-affinity AGO-GW interactions would compromise specificity of RISC formation with the different AGO proteins, necessitating evolution of a mechanism to reduce spurious AGO-GW interactions. However, the most intriguing possibility relates to a new model for the constitution of the active site for slicing that we discuss below.
The Mechanism of Slicing: Four Is More Dynamic Than Three
One of the most important new observations emanating from the eukaryotic AGO structures concerns the catalytic mechanism of slicing. Based on the thermophile AGO structures and the mutational evidence from a range of eukaryotic AGOs, it has been generally accepted that the active site of AGO proteins is composed of a catalytic triad, typically two Asp residues and a His residue or three Asp residues. Nakanishi et al. (2012) now point out that the eukaryotic AGO structures strongly suggest the existence of a catalytic tetrad, rather than a triad. Mutational evidence in yeast Ago confirmed that a fourth residue, an invariant Glu corresponding to Glu-637 in human Ago2, is indeed essential for catalysis. The authors further suggested that this fourth residue, positioned in a loop, is recruited to the active site only when needed for slicing. In both the yeast and human structures, the Glu-637 loop and Glu-637 itself is part of an extensive hydrogen bond network that may guide its active site recruitment (Nakanishi et al., 2012).
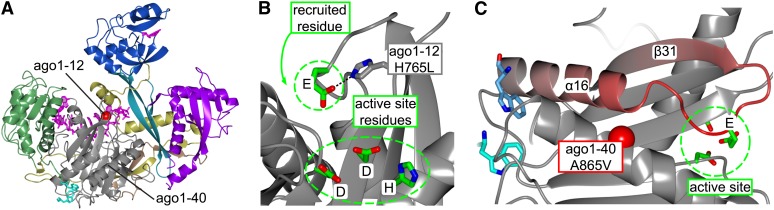
Inspection of sequence alignments and the mutational data on Arabidopsis AGO1 provide evidence that the active site in plant AGOs is similarly composed of such a catalytic tetrad. The catalytic Glu residue and its hydrogen bond partners are conserved, and one of the strongest missense mutations in AGO1 results in the His765Leu change. The corresponding His-600 residue in human Ago2 directly coordinates the catalytic Glu (Figures 7A and 7B), and mutation of this His residue in budding yeast Ago was shown in biochemical assays to decrease slicer activity by approximately sevenfold (Nakanishi et al., 2012). Although evidence for the catalytic tetrad model is solid, the present structural data do not reveal exactly how the catalytic Glu residue contributes to catalysis. In the crystallized conformation with guide strand bound and not target strand, the carboxylate group is fully occupied by hydrogen bond and salt bridge partners. Several rearrangements must therefore take place before it can participate in catalysis.
Figure 7.
The ago1-12 Allele Provides Evidence for a Catalytic Tetrad in Plants.
(A) Overview showing the location of the ago1-12 mutation.
(B) Close-up view of the Hs-Ago2 active site region showing the catalytic tetrad (EDDH). The ago1-12 H765L mutation disrupts the H-bond to the deeply conserved Glu (Glu-736 in Hs-Ago2), with the active site residue suggested to be recruited for slicing.
(C) The proximity of the Trp binding sites and the E736 loop (in red). Interactors bound to the Trp binding pockets could reach the E736 loop and thereby modulate its active site recruitment. Note that A865V missense mutation in ago1-40 is located in this area expected to be included in the GW interaction surface.
The model of a dynamic constitution of the active site opens some interesting venues for investigation of plant small RNA biology. In plants, miRNAs and siRNAs are well known to exhibit extended complementarity to their targets, and evidence for slicing of targets can generally be found. Nonetheless, miRNAs and siRNAs can also regulate their targets via translational repression, raising the obvious question of how slicing is avoided in these cases. No convincing solution has been proposed, and it is indeed a genuinely difficult problem: Biochemical evidence clearly shows that the guide RNA-AGO complex constitutes a minimal, slicer-competent RISC (Rivas et al., 2005), implying that cofactors must be required to inhibit this activity, if translational repression rather than mRNA cleavage is the result of a RISC–mRNA interaction with a highly base-paired guide RNA-target RNA duplex. On the other hand, the structural studies on thermophile, and now also eukaryotic AGOs, indicate that the canonical catalytic triad would be difficult to modulate by protein–protein interaction: These three residues are placed in the most rigid and conserved part of the Piwi domain (Figure 1B), are facing the RNA binding cleft, and are out of reach for protein interaction partners.
The model of recruitment of a fourth active site residue offers an attractive solution, since it predicts that slicing should be inhibited if recruitment is prevented. The eukaryotic AGO structures suggest that such modulation of active-site recruitment is possible. The loop containing the catalytic Glu-637 residue connects β-strand 31 and α-helix 16 that follows directly after the two GW binding pockets (Figure 1B). In line with this, the Trp residues bound to human Ago2 are oriented with the Cα-CO bond pointing toward the Glu-637 loop. Although the Piwi domain is unlikely to undergo major structural changes, it is plausible that movements in this loop can be influenced by GW proteins docking to the nearby pockets (Figure 7C).
The possibility that GW proteins could regulate slicer activity suggests interesting explanations for why plant AGOs, contrary to animal AGOs, harbor the putative autoinhibitory Trp residue. Slicer inhibition probably must be tightly regulated and should only occur in the presence of sufficiently high concentrations of specific partners. Such switch-like decision making would be facilitated by the proposed autoinhibition mechanism.
We also note that a viral suppressor of RNA silencing, P38 from turnip crinkle virus, is known to require a GW motif for suppressor activity and AGO1 interaction (Azevedo et al., 2010). Slicer activity is believed to be important in antiviral defense and is therefore an obvious target of viral virulence factors. Intriguingly, association of siRNAs, but not miRNAs, with AGO1 is inhibited in the presence of P38 (Schott et al., 2012). This could well be a consequence of slicer inhibition because only the duplex intermediate of siRNAs is perfectly self-complementary. Evidence from plant and animal systems shows that loading of such siRNAs requires slicer activity for passenger strand cleavage, contrary to miRNAs (Matranga et al., 2005; Iki et al., 2010).
Changing Unknown Unknowns to Known Unknowns
Although impressive progress has been made in understanding small RNA and AGO protein biology, much remains to be learned. The analysis of structure and point mutations presented here shows how mutations can directly interfere with some of the core functions of AGO proteins or affect structural integrity. Yet, a small set of mutations cannot reasonably be explained based on structures and known biochemical properties. Therefore, these mutations are likely to define AGO protein functions that remain to be understood. The most obvious examples are lesions in surface-exposed residues without direct contacts to important secondary structure elements and include the mutations ago1-45, ago1-40, ago4-2, and ago7-15.
The weak ago1-45 R696H mutation is located in α-helix 13 in the Piwi domain, a region that also harbors three additional mutant alleles: ago7-13, ago1-49, and ago7-8 (Figure 1B). This α-helix is involved in RNA binding and participates in the MID–Piwi domain interaction. These functions are probably affected in ago7-13, ago1-49, and ago7-8. However, the side chain of Arg-696 is solvent exposed and does not seem to have any significant intramolecular interactions that can be hampered by the mutation. The structure of the isolated Arabidopsis AGO1 MID domain is consistent with this conclusion (Frank et al., 2012). This argues against structural differences between Hs-Ago2 and At-AGO1 as the cause for difficulty in functional assignment of Arg-696, conserved in most plant AGOs and proven by genetics to have important function.
The ago1-40 A865V mutation, located in α17, is partly surface exposed and cannot be expected to have a major effect on the fold. Accordingly, this mutation produces a very weak phenotype. Its location between the Trp binding pockets and the catalytic Glu loop raises the possibility that it may be linked to GW interactors.
The ago4-2 allele has a Glu-Lys mutation at a freely accessible position in an apparently unconserved loop region connecting the MID and Piwi domains. Remarkably, the ago4-2 mutation is dominant negative (Agorio and Vera, 2007). Taken together, these structural and genetic properties of the ago4-2 mutation strongly suggest that this loop engages in protein–protein interactions essential for AGO4 function. The lack of conservation of the loop and its likely participation in protein–protein interactions is interesting because it suggests that this loop may be a starting point to understand the properties that make AGO family members different and enable participation in specialized RNA silencing pathways.
Finally, the ago7-15 mutation S810N is interesting. This residue is completely conserved, and it is positioned in the Piwi domain facing the cleft between the N/DUF1785/PAZ- and MID-Piwi lobes. Therefore, it is possible that this residue is involved in RNA target binding. While such a function would be common to all AGO proteins, and therefore consistent with the complete conservation of Ser-810, even weak reductions of this function would be particularly deleterious to AGO7. This is because AGO7 is strictly required for biogenesis TAS3 tasiRNAs not only as a result of TAS3 precursor transcript cleavage, but also because it stably associates with the precursor transcript in a noncleaving mode upstream of the cleavage site (Montgomery et al., 2008). The molecular basis of this noncleaving mode of AGO7 function is not understood, and ago7-15 is a candidate for a mutation affecting this function, perhaps via decreased target RNA binding.
Concluding Remarks
The determination of structures of small RNA-bound entire eukaryotic AGO proteins is an achievement of key importance to the understanding of RNA silencing. It offers precise physico-chemical explanations to many biochemical and genetic observations of AGO function, as shown here with a systematic analysis of functional consequences of ago mutations recovered in Arabidopsis genetic screens. This enables the selection of specific mutant alleles defective in defined AGO functions in future genetic and biochemical studies. The parallel analysis of structural and genetic data also provides a solid platform on which to construct novel hypotheses for future research. Indeed, these functional and structural assignments will be useful for design of genetic and biochemical approaches that use the existing mutant alleles to explore aspects of AGO function unknown at present. We can therefore now look forward to a substantially accelerated understanding of how the AGO family exerts its key regulatory functions in plant biology.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of Plant, Yeast, and Animal AGOs.
Supplemental Table 1. Mutant Alleles of Arabidopsis AGO Genes Recovered by Forward Genetics.
Acknowledgments
We thank Dominique Lauressergues for sequencing novel ago1 alleles, and Laura Arribas for sequence information on ago1-22. This work was supported by grants to P.B. from the Lundbeck Foundation and from the Novo Nordisk Foundation.
AUTHOR CONTRIBUTIONS:
C.P. and P.B. designed the study. C.P. analyzed structures. H.V. isolated and characterized novel ago1 mutant alleles. P.B. and H.V. assembled information on published mutant ago alleles. P.B. and C.P. wrote the article.
References
- Agorio A., Vera P. (2007). ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Azevedo J., Garcia D., Pontier D., Ohnesorge S., Yu A., Garcia S., Braun L., Bergdoll M., Hakimi M.A., Lagrange T., Voinnet O. (2010). Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17: 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouasker S., Simard M.J. (2009). Structural biology: Tracing Argonaute binding. Nature 461: 743–744 [DOI] [PubMed] [Google Scholar]
- Braun J.E., Huntzinger E., Fauser M., Izaurralde E. (2011). GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Schaller H., Khafif M., Schott G., Bendahmane A., Voinnet O. (2012). Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikaluk D.E., Tahbaz N., Hendricks L.C., DiMattia G.E., Hansen D., Pilgrim D., Hobman T.C. (1999). GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell 10: 3357–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Montgomery T.A., Fahlgren N., Burke R.T., Townsend T., Sullivan C.M., Carrington J.C. (2010). Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.W., Voinnet O. (2007). Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Brosnan C.A., Schott G., Wang Y., Jay F., Alioua A., Himber C., Voinnet O. (2010). An endogenous, systemic RNAi pathway in plants. EMBO J. 29: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunoyer P., Himber C., Voinnet O. (2006). Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 38: 258–263 [DOI] [PubMed] [Google Scholar]
- Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L. (2012). The structure of human argonaute-2 in complex with miR-20a. Cell 150: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. (2007). Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21: 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Helms S., Fritzsch C., Fauser M., Izaurralde E. (2009). A C-terminal silencing domain in GW182 is essential for miRNA function. RNA 15: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun C., Lorkovic Z.J., Naumann U., Long Q., Havecker E.R., Simon S.A., Meyers B.C., Matzke A.J., Matzke M. (2011). AGO6 functions in RNA-mediated transcriptional gene silencing in shoot and root meristems in Arabidopsis thaliana. PLoS ONE 6: e25730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle C.R., Joshua-Tor L. (2010). Argonaute MID domain takes centre stage. EMBO Rep. 11: 564–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Boutet S., Morel J.-B., Bellini C., Vaucheret H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97: 11650–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F., Hauver J., Sonenberg N., Nagar B. (2012). Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 31: 3588–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F., Sonenberg N., Nagar B. (2010). Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465: 818–822 [DOI] [PubMed] [Google Scholar]
- Frohn A., Eberl H.C., Stöhr J., Glasmacher E., Rüdel S., Heissmeyer V., Mann M., Meister G. (2012). Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol. Cell. Proteomics 11: 1442–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Garcia, S., Pontier, D., Marchais, A., Renou, J.P., Lagrange, T., and Voinnet, O. (2012). Ago hook and RNA helicase motifs underpin dual roles for SDE3 in antiviral defense and silencing of nonconserved intergenic regions. Mol. Cell 48: 109–120. [DOI] [PubMed]
- Gibbings D.J., Ciaudo C., Erhardt M., Voinnet O. (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Gu S., Jin L., Huang Y., Zhang F., Kay M.A. (2012). Slicing-independent RISC activation requires the argonaute PAZ domain. Curr. Biol. 22: 1536–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J.R., Pikaard C.S. (2011). Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492 [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Harvey J.J., Lewsey M.G., Patel K., Westwood J., Heimstädt S., Carr J.P., Baulcombe D.C. (2011). An antiviral defense role of AGO2 in plants. PLoS ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E., Izaurralde E. (2011). Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Hutvágner G., Zamore P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. (2010). In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Irvine D.V., Zaratiegui M., Tolia N.H., Goto D.B., Chitwood D.H., Vaughn M.W., Joshua-Tor L., Martienssen R.A. (2006). Argonaute slicing is required for heterochromatic silencing and spreading. Science 313: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Jaubert M., Bhattacharjee S., Mello A.F., Perry K.L., Moffett P. (2011). ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156: 1556–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauvion V., Elmayan T., Vaucheret H. (2010). The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 22: 2697–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., et al. (2011). ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V., Moreno A.B., Elmayan T., Vaucheret H., Crespi M.D., Maizel A. (2012). Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 31: 1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C.A., Martienssen R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kwak P.B., Tomari Y. (2012). The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 19: 145–151 [DOI] [PubMed] [Google Scholar]
- Lazzaretti D., Tournier I., Izaurralde E. (2009). The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 15: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., et al. (2009). Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 11: 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N.B., Westhof E. (2001). Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow M., Jacobsen A., Nygaard S., Mang Y., Krogh A. (2007). Intragenomic matching reveals a huge potential for miRNA-mediated regulation in plants. PLoS Comput. Biol. 3: e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Ma J.B., Ye K., Patel D.J. (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J. (2005). Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Hinze A., Tucker M.R., Bouché N., Gasciolli V., Elmayan T., Lauressergues D., Jauvion V., Vaucheret H., Laux T. (2009). Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 5: e1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. (2005). Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Maunoury N., Vaucheret H. (2011). AGO1 and AGO2 act redundantly in miR408-mediated Plantacyanin regulation. PLoS ONE 6: e28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Lührmann R., Tuschl T. (2005). Identification of novel argonaute-associated proteins. Curr. Biol. 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Morel J.-B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B., Schoof H., Haecker A., Jürgens G., Laux T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17: 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J. (2012). Structure of yeast Argonaute with guide RNA. Nature 486: 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Denli A.M., Hannon G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qi Y., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Schirle N.T., MacRae I.J. (2012). The crystal structure of human Argonaute2. Science 336: 1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott G., Mari-Ordonez A., Himber C., Alioua A., Voinnet O., Dunoyer P. (2012). Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J. 31: 2553–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.R., Willmann M.R., Wu G., Berardini T.Z., Möller B., Weijers D., Poethig R.S. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5424–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Sorin C., Bussell J.D., Camus I., Ljung K., Kowalczyk M., Geiss G., McKhann H., Garcion C., Vaucheret H., Sandberg G., Bellini C. (2005). Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17: 1343–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. (2007). A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 14: 897–903 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P., Bartel D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang X., Liu J., Kiba T., Woo J., Ojo T., Hafner M., Tuschl T., Chua N.H., Wang X.J. (2011). Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 67: 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.X., Gasciolli V., Vaucheret H., Ding S.W. (2011). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J. (2008). Structure of the guide-strand-containing argonaute silencing complex. Nature 456: 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y. (2012). A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112 [DOI] [PubMed] [Google Scholar]
- Yang L., Wu G., Poethig R.S. (2012). Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao H., Gao S., Wang W.C., Katiyar-Agarwal S., Huang H.D., Raikhel N., Jin H. (2011). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(∗)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 42: 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Zhu J., Kapoor A., Zhu J.K. (2007). Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Hu F., Wang R., Zhou X., Sze S.H., Liou L.W., Barefoot A., Dickman M., Zhang X. (2011). Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]