This work reports that FCP1, encoding a CLE protein, negatively regulates the maintenance of the vegetative shoot apical meristem in rice. In addition, it reveals that WOX4 promotes the undifferentiated state of the meristem and is negatively regulated by FCP1.
Abstract
The shoot apical meristem is the ultimate source of the cells that constitute the entire aboveground portion of the plant body. In Arabidopsis thaliana, meristem maintenance is regulated by the negative feedback loop of WUSCHEL-CLAVATA (WUS-CLV). Although CLV-like genes, such as FLORAL ORGAN NUMBER1 (FON1) and FON2, have been shown to be involved in maintenance of the reproductive meristems in rice (Oryza sativa), current understanding of meristem maintenance remains insufficient. In this article, we demonstrate that the FON2-LIKE CLE PROTEIN1 (FCP1) and FCP2 genes encoding proteins with similar CLE domains are involved in negative regulation of meristem maintenance in the vegetative phase. In addition, we found that WUSCHEL-RELATED HOMEOBOX4 (WOX4) promotes the undifferentiated state of the meristem in rice and that WOX4 function is associated with cytokinin action. Consistent with similarities in the shoot apical meristem phenotypes caused by overexpression of FCP1 and downregulation of WOX4, expression of WOX4 was negatively regulated by FCP1 (FCP2). Thus, FCP1/2 and WOX4 are likely to be involved in maintenance of the vegetative meristem in rice.
INTRODUCTION
The aboveground aerial parts of plants are derived from the shoot apical meristem (SAM), and their development ultimately depends on SAM function. In the SAM, the stem cells divide and produce descendant cells both to self-maintain and to provide cells for the differentiation of lateral organs. The balance between self-maintenance and differentiation of the cells is essential for stem cell homeostasis in the SAM.
The genetic mechanism of this homeostasis is well understood in Arabidopsis thaliana (reviewed in Barton, 2010; Ha et al., 2010; Aichinger et al., 2012). Stem cell identity is positively regulated by WUSCHEL (WUS) protein, the expression of which is negatively regulated by the CLAVATA (CLV) signaling pathway, consisting of a peptide signal molecule processed from CLV3 protein and its receptors, such as CLV1 (Mayer et al., 1998; Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000; Reddy and Meyerowitz, 2005; Yadav et al., 2010). In turn, WUS promotes expression of CLV3, which is localized in the stem cell region. This negative feedback loop of WUS-CLV3 in the stem cell niche is the framework of SAM maintenance in Arabidopsis.
WUS encodes a type of homeobox protein, classified into the WOX protein family (Mayer et al., 1998). CLV3 encodes a small protein that has a conserved CLE domain (Fletcher et al., 1999). CLV3 is expressed in the apical domain of the central region of the meristem, whereas WUS is expressed in the region underneath the CLV3-expressing domain, called the organizing center (OC). After processing and chemical modifications, the peptide corresponding to the CLE domain acts as a signal molecule through three kinds of receptor complexes consisting of CLV1, CORYNE-CLV2, and RECEPTOR-LIKE PROTEIN KINASE2 (Clark et al., 1997; Kondo et al., 2006; Müller et al., 2008; Ogawa et al., 2008; Ohyama et al., 2009; Kinoshita et al., 2010). The signal is transmitted to the WUS gene via phosphorylation/dephosphorylation and unknown mechanisms (Williams et al., 1997; Song et al., 2006; Betsuyaku et al., 2011). The WUS protein moves from the OC to the stem cell region and directly binds the CLV3 promoter to activate expression of CLV3 (Yadav et al., 2011).
A phytohormone, cytokinin, is a key factor in stem cell homeostasis. WUS expression is positively regulated by cytokinin; in turn, WUS promotes cytokinin action by repressing the genes encoding negative regulators of cytokinin signaling, such as ARABIDOPSIS RESPONSE REGULATOR7 (ARR7) and ARR15 (Leibfried et al., 2005; Gordon et al., 2009). An active form of cytokinin produced by epidermal cells of the SAM acts as a positional cue to promote WUS expression together with ARABIDOPSIS HISTIDINE KINASE4, a cytokinin receptor, the expression domain of which is superimposed on the OC (Chickarmane et al., 2012). Thus, robust stem cell homeostasis can be achieved by a negative/positive feedback loop between CLV3/cytokinin and WUS.
In contrast with Arabidopsis, our understanding of stem cell maintenance is insufficient in other plants. Nevertheless, the genetic mechanism underlying the negative regulation of stem cell proliferation is likely to be conserved in both rice (Oryza sativa) and maize (Zea mays). Mutations in the genes FLORAL ORGAN NUMBER (FON) in rice and fasciated ear2 (fea2) and thick tassel dwarf1 (td1) in maize cause an enlargement of the size of either the flower or inflorescence meristem, resulting in an increase in floral organ numbers or thickening of inflorescences (Taguchi-Shiobara et al., 2001; Suzaki et al., 2004; Bommert et al., 2005). Rice FON1 and maize td1 encode an Leucine-rich repeat–like receptor kinase that is closely related to Arabidopsis CLV1, and fea2 encodes a CLV2-like Leucine-rich repeat protein. Rice FON2/FON4 encodes a small secreted protein with a CLE domain related to that of CLV3 and is expressed in the apical region of the meristem in a CLV3-like pattern (Chu et al., 2006; Suzaki et al., 2006). Because no effect is observed when FON2 is overexpressed in the fon1 mutant, the FON2 signal may be mediated through the FON1 receptor (Suzaki et al., 2006). In addition, it has been shown that FON2 SPARE1 (FOS1), encoding another CLE protein, redundantly regulates stem cell maintenance with FON2 in indica rice and wild species in the Oryza genus (japonica rice has a mutation in FOS1) (Suzaki et al., 2009). Thus, the CLV-like signaling pathway is likely to be involved in meristem maintenance in grasses. In addition to this pathway, cytokinin plays an important role in rice because loss of function of LONELY GUY, which encodes an enzyme responsible for conversion of the cytokinin precursor into active forms, results in failed maintenance of the floral meristem (Kurakawa et al., 2007).
In contrast with the reproductive phase, however, mutations in these genes do not bring about obvious defects in the vegetative phase in either rice or maize, suggesting that other genes are required for maintenance of the vegetative meristems. Putative candidates for these genes are FON2-like CLE PROTEIN1 (FCP1) and FCP2 in rice. Both FCP1 and FCP2 encode proteins with a CLE domain, very similar to that of FON2 (Suzaki et al., 2008). Regenerating shoots overexpressing FCP1 fail to maintain the SAM. This effect is also observed in the fon1 mutant background, suggesting that FCP1 may act through a receptor different from FON1. These findings suggest that at least two independent pathways are involved in the negative regulation of meristem maintenance in rice, depending on the type of the meristem (Suzaki et al., 2008, 2009). This situation contrasts with that in Arabidopsis, where a single CLV pathway regulates all types of meristem. In addition, the gene that promotes stem cell maintenance has not yet been identified in plants other than Arabidopsis.
Class 1 Knotted-like homeobox (KNOX) genes, such as Arabidopsis SHOOT MERISTEMLESS (STM), rice ORYZA SATIVA HOMEOBOX1 (OSH1), and maize Knotted1 (Kn1), promote the undifferentiated state of the meristem (Long et al., 1996; Sato et al., 1996; Vollbrecht et al., 2000; Tsuda et al., 2011). Loss-of-function mutants of STM and Kn1 fail to form or maintain the meristem during embryogenesis (Long et al., 1996; Vollbrecht et al., 2000). In the rice osh1 mutant, the SAM is formed but cannot be maintained after germination. In the double mutant of osh1 and d6, which encodes the related homeobox protein OSH15, the SAM is not established, suggesting that both genes are responsible for formation or maintenance of the SAM in the embryo (Tsuda et al., 2011). KNOX genes promote cytokinin accumulation by upregulating the expression of genes for its biosynthesis in both Arabidopsis and rice (Jasinski et al., 2005; Yanai et al., 2005; Sakamoto et al., 2006).
WUSCHEL-RELATED HOMEOBOX (WOX) genes are involved in various aspects of development in Arabidopsis, such as maintenance of the root apical meristem, embryogenesis, and vascular development (Haecker et al., 2004; Wu et al., 2005; Sarkar et al., 2007; Breuninger et al., 2008; Hirakawa et al., 2010; Ji et al., 2010; Suer et al., 2011). Among these genes, WOX4 promotes proliferation of stem cells in the procambium/cambium in Arabidopsis, and WOX4 itself is upregulated in response to TDIF (for tracheary element differentiation inhibitory factor) mediated by TDIF RECEPTOR/PHLOEM INTERCALATED WITH XYLEM (TDR/PXY) (Hirakawa et al., 2008, 2010; Etchells and Turner, 2010; Ji et al., 2010; Suer et al., 2011). TDIF is a CLE peptide processed from proteins encoded by the CLE41 and CLE44 genes (Ito et al., 2006). In rice, WOX genes are involved in maintenance of the root apical meristem and crown root development (Kamiya et al., 2003; Zhao et al., 2009). Although the expression patterns of other WOX genes have been reported, current understanding of the function of WOX genes is insufficient in rice (Nardmann and Werr, 2006; Dai et al., 2007).
In this work, we first confirmed that FCP1 is a negative regulator of meristem maintenance using inducible RNA silencing and overexpression experiments. Second, we revealed that rice WOX4 acts positively on the indeterminacy of the meristem and is associated with cytokinin function. Third, the expression of WOX4 was found to be negatively regulated by FCP1. Both induced overexpression of FCP1 and induced RNA silencing of WOX4 resulted in a reduction in the expression of OSH1 and FON2, suggesting that WOX4 is responsible for meristem maintenance by promoting the undifferentiated indeterminate state and by preventing premature differentiation.
RESULTS
FCP1 Overexpression Inhibits Meristem Maintenance
To examine the effect of constitutive expression of FCP1, we first introduced a construct that induces constitutive expression of FCP1 (pACT1-FCP1) into rice calli derived from the scutellum and then regenerated shoots. The resulting shoots showed abnormal morphology and lacked a normal SAM structure (Figures 1C and 1D). Although this observation suggested that FCP1 overexpression inhibits maintenance of the SAM, the detailed function of FCP1 could not be determined from the analysis of regenerated shoots.
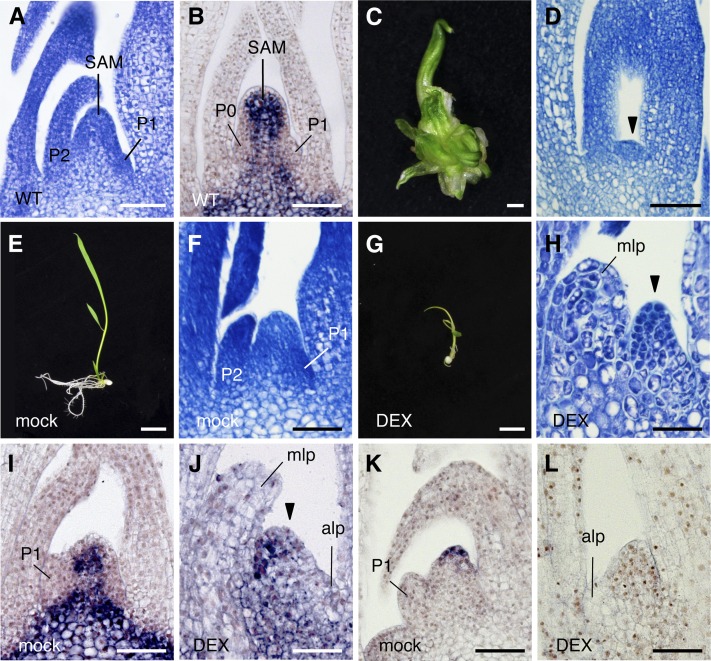
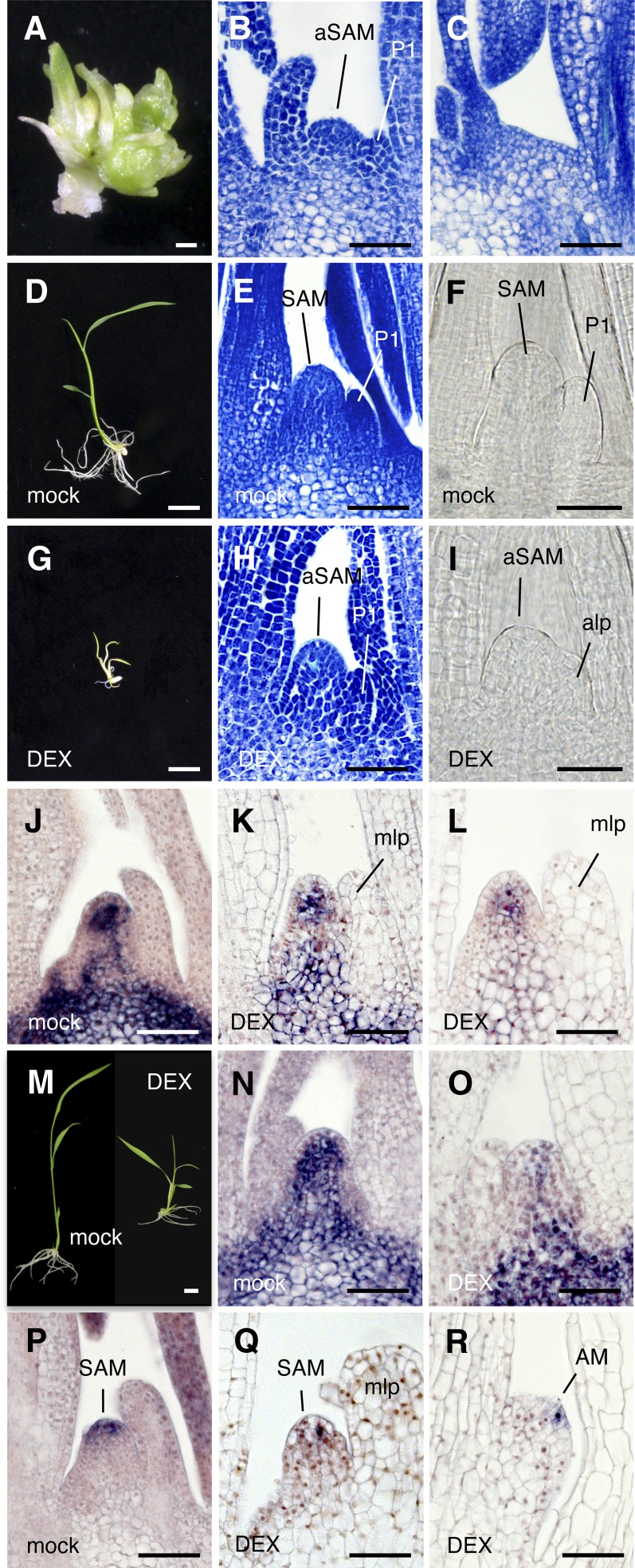
Figure 1.
Effects of Constitutive and Inducible Overexpression of FCP1.
(A) and (B) Shoot apex (A) and in situ hybridization showing spatial expression pattern of OSH1 (B) in a wild-type seedling grown in soil 30 d after germination.
(C) and (D) Generating shoot (C) with pACT1-FCP1 and its shoot apex (D). Arrowhead indicates a terminated meristem (D).
(E) and (F) Transgenic shoot with pACT1-GVG>FCP1 (without DEX).
(G) and (H) Transgenic shoot with pACT1-GVG>FCP1 (10 µM DEX). Arrowhead indicates an abnormal meristem.
(I) and (J) Spatial expression pattern of OSH1 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1. Mock (I); 50 µM DEX (J). Arrowhead indicates an abnormal meristem.
(K) and (L) Spatial expression pattern of FON2 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1. Mock (K); 50 µM DEX (L). Seedlings were grown for 7 d from germination in the presence or absence of DEX ([E] to [L]).
alp, abnormal leaf primordia; mlp, malformed leaf primordia; P1-P5, plastochrons 1 to 5; WT, the wild type. Bars = 50 µm in (A), (B), (D), (F), and (H) to (L), 1 mm in (C), and 1 cm in (E) and (G).
To elucidate further the function of FCP1 and WOX4 (see below), we developed an inducible expression system in rice using a modified pINDEX vector that induces the gene of interest in trans by application of dexamethasone (DEX) (see Supplemental Figure 1 online). In the DEX induction experiment, we first selected transformants (T0 plants) in which each transgene was effectively induced by application of DEX and then examined effect of DEX induction by comparing T1 siblings derived from the same T0 plant throughout in this study (see Methods).
T1 seedlings with pACT1-GVG>FCP1 were germinated and grown for 7 d in the presence or absence of DEX. The shoot phenotype and the SAM morphology in seedlings without DEX treatment were indistinguishable from those in seedlings under normal growth conditions (Figures 1A, 1B, 1E, and 1F). By contrast, abnormal shoots were produced in DEX-treated seedlings: The leaves were small and pale green, and crown roots were not formed (Figure 1G). Observation of a longitudinal section of the shoot apex revealed that the SAM was morphologically abnormal and exhibited a reduced number of cells. The leaf primordia were enlarged and had large vacuolated cells (Figure 1H).
Next, we examined two marker genes of the meristem, OSH1 and FON2. OSH1 was expressed in the meristem except for the L1 layer and leaf initiation sites in both seedlings grown in soil and those without DEX treatment (Figures 1B and 1I). Expression of OSH1 was downregulated, and its spatial expression pattern was disturbed in the SAM in seedlings grown in the presence of DEX (Figure 1J). FON2 is expressed in a small number of cells in the apical region of the meristem, probably corresponding to the stem cell region (Suzaki et al., 2006). This expression pattern of FON2 was confirmed in mock-treated seedlings (Figure 1K). By contrast, FON2 expression disappeared completely from the SAM in DEX-treated seedlings (Figure 1L). These results suggest that stem cell maintenance is compromised by induced overexpression of FCP1.
RNA Silencing of FCP1 and FCP2
We tried to examine the effect of loss of function of FCP1. Despite extensive screening, however, we failed to obtain any knockout lines of FCP1. We therefore inhibited expression of the endogenous FCP1 gene by inducible RNA silencing (see Supplemental Figure 2 online; pACT1-GVG>FCP1:RNAi) using a pAID3 vector, which was made in this study (see Supplemental Figure 1 online). Although slight abnormalities, such as twisted leaves, were observed in the seedlings with induced silencing of FCP1, the SAM was largely normal in morphology and size, and leaf primordia were initiated normally (Figures 2A and 2B). We hypothesized that this finding was due to the existence of another CLE gene, FCP2, the CLE domain of which is highly similar to that of FCP1 with only one amino acid difference (Suzaki et al., 2006).
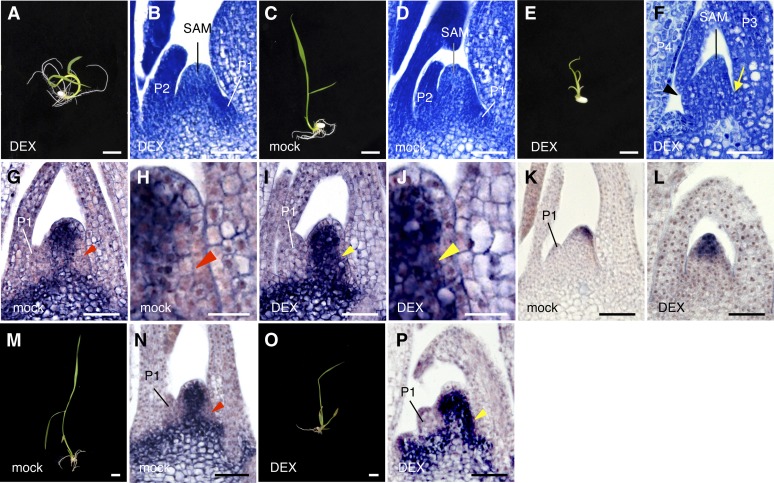
Figure 2.
Effects of Inducible Silencing of FCP1 and Cosilencing of FCP1-FCP2.
(A) and (B) Transgenic shoot with pACT1-GVG>FCP1:RNAi (25 µM DEX).
(C) and (D) Transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (mock).
(E) and (F) Transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (25 µM DEX). Arrow and arrowhead indicate the positions for the formation of P1 and P2 primordia, respectively. Note that no leaf primordium is observed in these positions.
(G) and (H) Spatial expression pattern of OSH1 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (mock). (H) is a magnified view of part of (G). OSH1 is downregulated in the P0 site (red arrowheads).
(I) and (J) Spatial expression pattern of OSH1 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (50 µM DEX). (J) is a magnified view of part of (I). Note that OSH1 is expressed in the P0 site (yellow arrowheads).
(K) Spatial expression pattern of FON2 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (mock).
(L) Spatial expression pattern of FON2 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (50 µM DEX).
(M) and (N) Shoot (M) and spatial expression pattern of OSH1 (N) in a transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (mock). OSH1 is downregulated in the P0 site (red arrowhead in [N]).
(O) and (P) Shoot (O) and spatial expression pattern of OSH1 (P) in a transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi (25 µM DEX). Yellow arrowhead in (P) indicates the P0 site, where OSH1 is ectopically expressed.
Seedlings were grown for 7 d from germination in the presence or absence of DEX ([A] to [L]) or for 5 d from germination in MS medium and treated without or with DEX for 5 d ([M] to [P]). Bars = 1 cm in (A), (C), (E), (M), and (O), 50 µm in (B), (D), (F), (G), (I), (K), (L), (N), and (P), and 20 µm in (H) and (J).
We therefore made a construct to silence FCP1 and FCP2 simultaneously using pAID3 (pACT1-GVG>FCP1-FCP2:RNAi) (see Supplemental Figure 2 online) and examined transgenic plants (T1) with pACT1-GVG>FCP1-FCP2:RNAi (Figures 2C to 2P). A longitudinal section revealed that leaf initiation was inhibited: P1 and P2 leaf primordia were not detected in the DEX-treated seedlings (Figure 2F). The P3 and P4 leaves that were observed would have been formed before a significant effect of cosilencing of FCP1 and FCP2. Similar morphological abnormalities in the SAM and leaf primordia were observed in other DEX-treated seedlings (see Supplemental Figures 3A to 3G online). In rice, three true leaves are formed in embryogenesis (Itoh et al., 2005). Therefore, the phenotype of small yellow seedlings suggests that growth of these leaves is inhibited by cosilencing of FCP1 and FCP2 (Figure 2E).
OSH1 was expressed more strongly and widely in DEX-treated seedlings with pACT1-GVG>FCP1-FCP2:RNAi than in mock-treated seedlings (Figures 2G and 2I). In mock-treated seedlings, OSH1 expression was downregulated at the future leaf initiation site (P0; Figures 2G, 2H, and 2N), as in normal seedlings (Figure 1B). This downregulation of OSH1 was invariably detected in mock-treated seedling (see Supplemental Figures 4A to 4D online). By contrast, OSH1 was ectopically expressed at this P0 site in DEX-treated pACT1-GVG>FCP1-FCP2:RNAi seedlings (Figures 2I and 2J). We often observed this ectopic OSH1 expression in other DEX-treated seedlings with this construct (see Supplemental Figures 4E to 4G online). These observations suggest that the SAM fails to recruit leaf founder cells and that the undifferentiated state was maintained throughout the meristem due to cosilencing of FCP1 and FCP2. In addition, expression of FON2 was elevated and the expression domain was expanded in the DEX-treated seedlings (Figure 4L), compared with the mock-treated ones (Figure 4K), suggesting that the stem cell population was likely to be increased.
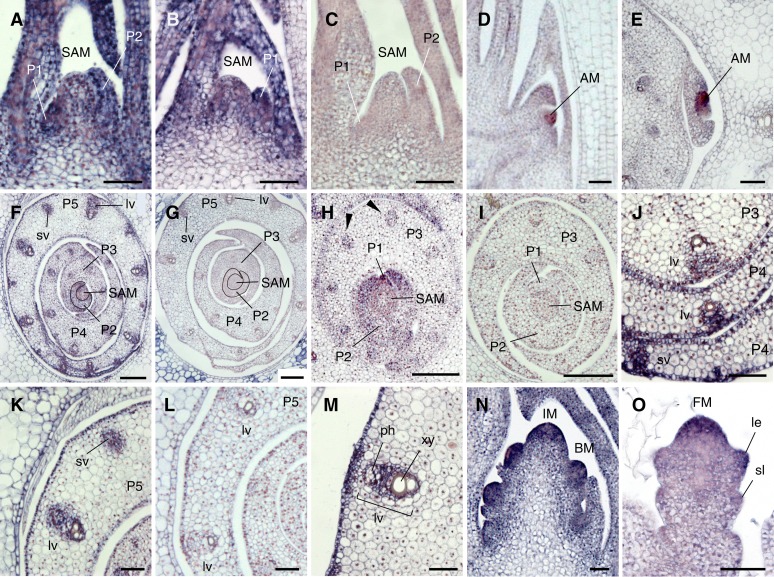
Figure 4.
Temporal and Spatial Expression Pattern of WOX4 in the Shoot Apex and Developing Leaf.
(A) and (C) Longitudinal section of the shoot apex of a seedling grown in soil for 14 d.
(B) Longitudinal section of the shoot apex of a control seedling grown in MS medium without DEX 7 d after germination.
(D) and (E) Longitudinal (D) and transverse (E) section of an axillary bud.
(F) to (I) Transverse section of the shoot apex with leaf primordia and developing young leaves. Arrowheads in (H) indicate putative procambium. P2 primordium is outlined in (F) and (G).
(J) to (M) Close-up view of vascular bundles in developing young leaves.
(N) Longitudinal section of a young inflorescence.
(O) Longitudinal section of a developing flower.
(A), (B), (D) to (F), (H), (J), (K), and (M) to (O) Antisense probe of WOX4.
(C), (G), (I), and (L) Sense probe of WOX4.
AM, axillary meristem; BM, branch meristem; FM, floral meristem; IM, Inflorescence meristem; lv, large vascular bundle; le, lemma; ph, phloem; P1 to P5, plastochrons 1 to 5; sl, sterile lemma; sv, small vascular bundle; xy, xylem. Bars = 50 µm in (A) to (E) and (J) to (O) and 100 µm in (F) to (I).
We then checked the effect of DEX treatment on the SAM in a mild condition. Seedlings were grown for 5 d in the absence of DEX, transferred to a new solution containing 25 μM DEX, and then grown for a further 5 d. The apparent shoot phenotype was largely normal, although the leaves were a bit smaller (cf. Figure 2O with Figure 2M). However, in these seedlings, OSH1 was also strongly expressed at the P0 site (Figure 2P), confirming the above result.
Taking into account the results of induced FCP1 overexpression, FCP1 seems to be a negative regulator of meristem maintenance.
Identification of WOX4 and Its Spatial Expression Patterns
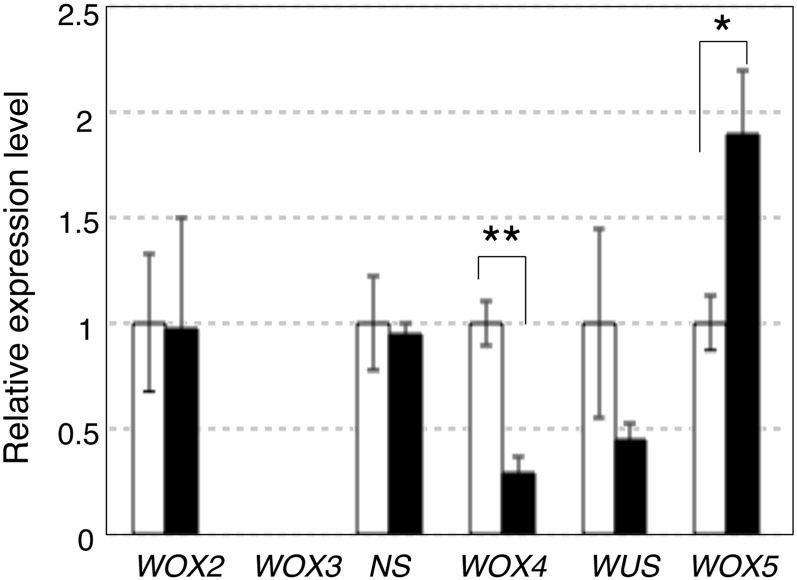
Because FCP1 negatively regulates stem cell maintenance, it is possible that genes responsible for the positive regulation of meristem maintenance would be downregulated in the regenerating shoots with pACT1-FCP1 (Figure 1C). Here, we focused on the rice WOX genes, especially those encoding proteins containing a WUS box (Haecker et al., 2004). We examined expression levels of WOX genes in pACT1-FCP1 regenerating shoots by quantitative real-time PCR (qRT-PCR) analysis. We found that WOX4, an ortholog of Arabidopsis WOX4 (Nardmann and Werr, 2006), was significantly downregulated in the regenerating shoots with pACT1-FCP1, compared with those with empty vector (Figure 3). An increase in WOX5 expression was observed in pACT1-FCP1 shoots. This upregulation probably results from the ectopic expression of WOX5 in the adventitious root primordia, which were included in the regenerating shoot samples, because the WOX5 (QHB) expression domain was highly expanded by the application of FCP1 CLE peptide in our previous study (Suzaki et al., 2008). The change in the levels of WUS expression was not statistically significant (Figure 3).
Figure 3.
Real-Time PCR Analysis of Rice WOX Genes.
RNA was isolated from regenerating shoots with empty vector (white) or those with pACT1-FCP1 (black) (see Figure 1C), and the relative expression of each WOX gene was measured by real-time PCR (n = 4; four RNA samples independently isolated). Student’s t test; **P < 0.01 and *P < 0.05. The difference in WUS expression is not statistically significant.
Thus, we focused on WOX4 for further work. In rice and Arabidopsis, WOX4 proteins are well conserved with respect to not only homeoboxes, but also 5′WOX1/4 and WUS boxes (see Supplemental Figure 5 online) (Haecker et al., 2004; Vandenbussche et al., 2009). We analyzed the spatial expression patterns of WOX4 by in situ hybridization. In the shoot apex, WOX4 was expressed in the meristem and the leaf primordia in the vegetative shoot apex (Figures 4A and 4B). No signal was detected when sense probe was used (Figure 4C). Expression of WOX4 was also detected in the axillary meristems (Figures 4D and 4E). Although WOX4 was expressed in the whole region of the P1 and P2 leaf primordia, its expression was restricted to narrow regions in subsequent leaf development (Figures 4F and 4H). Rice develops two types of vasculature, large and small vascular bundles, in the leaf (Nishimura et al., 2002). In the P3 primordia, WOX4 expression was detected in the putative procambium of the large vascular bundle (Figure 4H). WOX4 was subsequently expressed in developing large vascular bundles (Figures 4J and 4K), and the expression became restricted to the cells between the xylem and the phloem in a later stage (Figure 4M). WOX4 expression was also detected in putative procambium of the small vascular bundle in P4 and P5 (Figures 4F and 4K). These WOX4 signals were not detected when sense probe was used (Figures 4G, 4I, and 4L). These observations suggest that WOX4 is also involved in vascular development in rice, as in other plants such as Arabidopsis.
In the reproductive phase, WOX4 was expressed predominantly in the inflorescence and branch meristems (Figure 4N). During flower development, strong expression was detected in the floral meristem, whereas weaker expression was observed in the primordia of the lateral floral organs such as the lemma and sterile lemma (Figure 4O).
Downregulation of WOX4 Fails to Maintain the SAM
To elucidate the function of WOX4, we first downregulated WOX4 expression constitutively. Although shoot-like structures were regenerated from calli with pACT1-WOX4:RNAi, the morphology of these structures was abnormal (Figure 5A). A cross section of the shoot apex revealed that the SAM was reduced in size or flattened, suggesting premature termination of the meristem (Figures 5B and 5C). These shoot phenotypes and the SAM morphology in pACT1-WOX4:RNAi plants was similar to those in pACT1-FCP1 plants (Figures 1C and 1D), suggesting that FCP1 and WOX4 play opposite functions in maintaining the SAM of regenerated shoots.
Figure 5.
Effects of Constitutive and Inducible RNA Silencing of WOX4.
(A) to (C) Transgenic shoot with pACT1-WOX4:RNAi.
(D) to (F) Transgenic shoot with pACT1-GVG>WOX4:RNAi (mock).
(G) to (I) Transgenic shoot with pACT1-GVG>WOX4:RNAi (25 µM DEX).
(J) to (L) Spatial expression pattern of OSH1 in the shoot apex of a transgenic shoot with pACT1-GVG>WOX4:RNAi. Mock (J); 25 µM DEX ([K] and [L]).
(M) Shoot phenotypes of a transgenic shoot with pACT1-GVG>WOX4:RNAi.
(N) and (O) Spatial expression pattern of OSH1 in the shoot apex of a transgenic shoot with pACT1-GVG>WOX4:RNAi. Mock (N); 25 µM DEX (O).
(P) and (Q) Spatial expression pattern of FON2 in the shoot apex of a transgenic shoot with pACT1-GVG>WOX4:RNAi. Mock (P); 25 µM DEX (Q).
(R) Spatial expression pattern of FON2 in the axillary bud of a transgenic shoot with pACT1-GVG>WOX4:RNAi (25 µM DEX).
The seedlings were grown for 7 d from germination in the presence or absence of DEX ([D] to [L] and [P] to [R]) or for 5 d from germination in MS medium and treated without or with DEX for 5 d ([M] to [O]). alp, abnormal leaf primordia; AM, axillary meristem; aSAM, abnormal SAM; mlp, malformed leaf primordia. Bars = 1 mm in (A), 1 cm in (D), (G), and (M), 50 µm in (B), (C), (E), (F), (H) to (L), and (N) to (R).
We then examined the effect of silencing WOX4 using an inducible RNA interference (RNAi) system (see Supplemental Figure 2 online; pACT1-GVG>WOX4:RNAi). qRT-PCR analysis indicated that WOX4 was clearly downregulated by the application of DEX in pACT1-GVG>WOX4:RNAi lines (see Supplemental Figure 6A online). When pACT1-GVG>WOX4:RNAi transgenic plants were grown in the presence of DEX for 7 d from germination, the shoots were very small (Figure 5G), compared with mock-treated plants (Figure 5D). In addition, the SAM became smaller and morphologically abnormal, and the leaf primordium did not initiate properly (Figures 5H and 5I; compare with mock-treated shoot apex in Figures 5E and 5F). In addition, gaps were observed among the cells (Figure 5H). These gaps were observed in other pACT1-GVG>WOX4:RNAi transgenic seedlings treated with DEX (see Supplemental Figure 7 online) but not observed in the mock-treated seedlings (Figure 5E). The formation of gaps may reflect uncoordinated growth of cells and tissues in the plants with a reduced level of WOX4.
In situ experiments showed that OSH1 expression was downregulated in the SAM of these DEX-treated shoots (Figures 5J to 5L). This OSH1 downregulation was also observed similarly in the shoot apex of seedlings that were mildly treated with DEX (Figures 5M to 5O). Expression of FON2 was also downregulated by DEX treatment (Figures 5P to 5R). These results suggest that WOX4 is required for maintenance of the undifferentiated cell state and stem cell identity in rice.
In addition to the abnormalities in the SAM, cells in the leaf primordia were enlarged and vacuolated, implying that expression of WOX4 in the leaf primordia is necessary for normal development of the leaf (Figures 5K, 5L, 5Q, and 5R). These results resembled those of the experiments using pACT1-GVG>FCP1 plants.
Taking these results altogether, it is likely that WOX4 is a positive regulator of meristem maintenance and probably antagonizes FCP1 function.
FCP1 Negatively Regulates WOX4 Expression
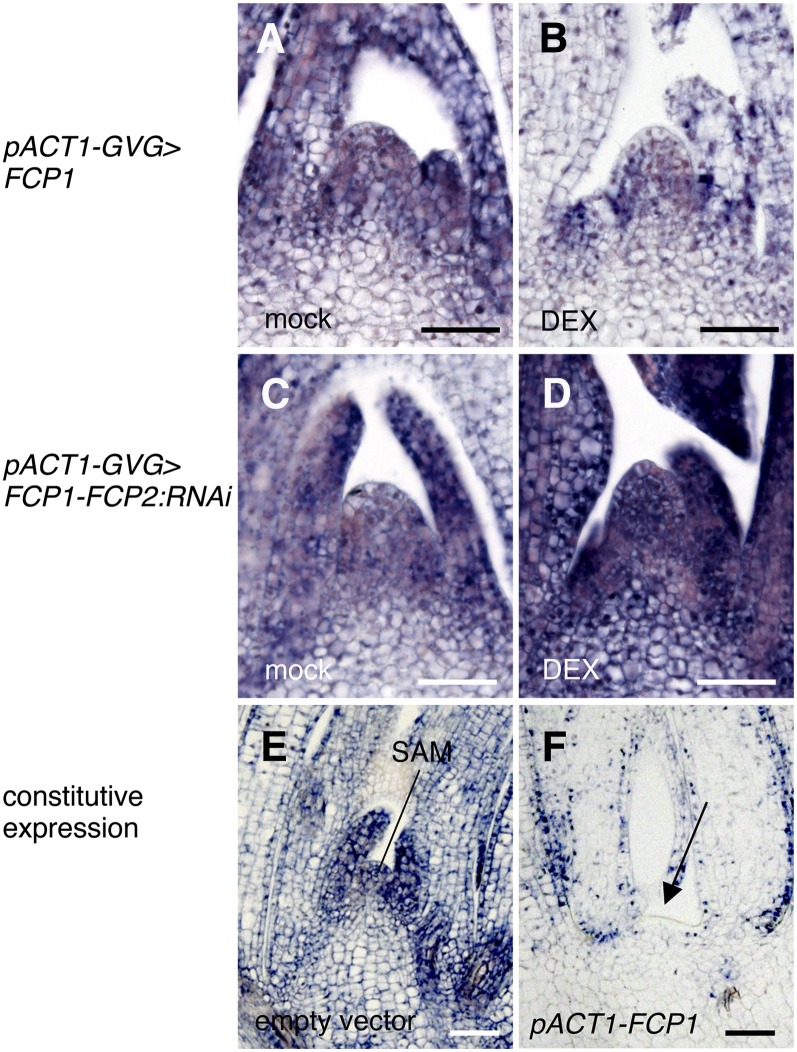
To examine the genetic relationship between WOX4 and FCP1, we examined the expression pattern of WOX4 after induced overexpression or silencing of FCP1. In the presence of DEX, expression of WOX4 was downregulated in both the meristem and the leaf primordia of transgenic seedlings carrying pACT1-GVG>FCP1 (Figures 6A and 6B). By contrast, WOX4 expression was upregulated in transgenic seedlings with pACT1-GVG>FCP1-FCP2:RNAi (Figures 6C and 6D). Strong downregulation of WOX4 was observed in abnormal shoot-like structures formed in calli constitutively expressing FCP1 (Figures 6E and 6F). These results suggest that expression of WOX4 is negatively regulated by FCP1.
Figure 6.
Analysis of the Effect of the FCP1 and FCP2 Genes on the Expression of WOX4.
(A) and (B) Expression of WOX4 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1. Mock (A); 10 µM DEX (B).
(C) and (D) Expression of WOX4 in the shoot apex of a transgenic shoot with pACT1-GVG>FCP1-FCP2:RNAi. Mock (C); 10 µM DEX (D).
(E) and (F) Expression of WOX4 in the shoot apex of regenerating shoot with empty vector (E) or pACT1-FCP1 (F). Arrow indicates a terminated meristem (F).
Bars = 50 µm.
To confirm negative regulation of WOX4 by FCP1, we performed qRT-PCR analysis. The results indicated that induction of FCP1 by the application of DEX caused downregulation of WOX4 expression (see Supplemental Figure 6B online), confirming the results of in situ analysis. By contrast, WOX4 was not changed by DEX application in the pACT1-GVG>FCP1-FCP2:RNAi seedlings (see Supplemental Figure 6C online). This inconsistency seems to result from a difference in temporal expression pattern of WOX4 and FCP1 in leaf development: FCP1 expression is restricted to only the P1 and P2 primordia (Suzaki et al., 2008), whereas WOX4 expression was maintained in the later stages of leaf development (Figure 4). The young leaves, in which WOX4 expression should not be regulated by FCP1, occupied a large portion of the shoot apices used for the qRT-PCR experiments.
Constitutive Expression of WOX4 Mimics Cytokinin Action in Calli
Next, we examined the effects of constitutive expression of WOX4. Calli with pACT1-WOX4 proliferated both in the callus induction medium and in the selection medium, as did ordinary calli, such as nontransformed calli or calli transformed with genes that do not inhibit shoot regeneration. After transferring the calli into regeneration medium, the ordinary calli produced green spots, from which shoots were regenerated. By contrast, although the calli with pACT1-WOX4 produced green spots, they either remained green without regeneration or regenerated morphologically abnormal shoot-like structures (Figure 7A).
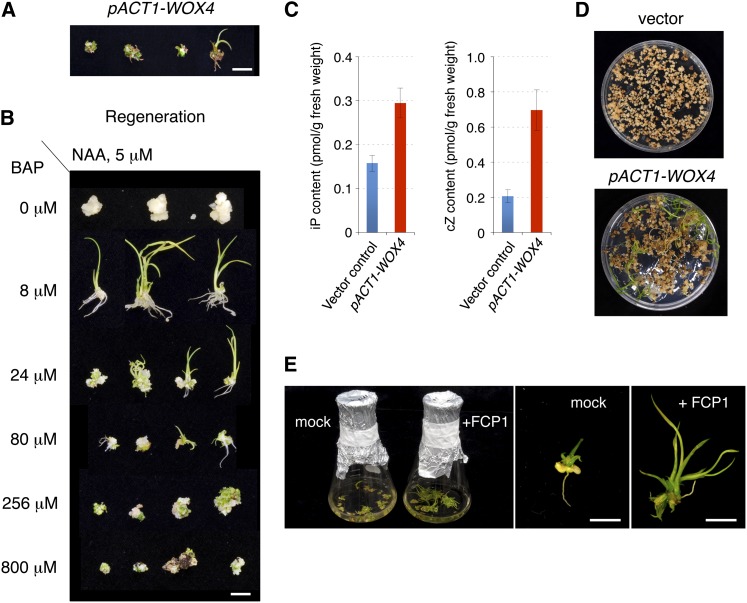
Figure 7.
WOX4 Overexpression and Cytokinin Action.
(A) Phenotypes of calli constitutively overexpressing WOX4.
(B) Phenotypes of calli and regenerating shoots grown on different concentrations of BAP. NAA, naphthalene acetic acid.
(C) Content of iP and cZ in calli with empty vector (blue) or with pACT1-WOX4 (red). tZ was undetectable in either type of calli. Error bars indicate se; n = 6.
(D) Shoot formation from calli overexpressing WOX4 in a hormone-free medium.
(E) Effect of FCP1 peptide on calli overexpressing WOX4. Calli with abnormal shoot-like structures (as in [A]) were grown in regeneration liquid medium in the absence or presence of FCP1 peptide (20 µM) for 7 d with shaking (left). Shoot regeneration (shown on the right) was observed in the presence of FCP1 peptide, whereas no change was observed in the absence of FCP1.
Bars = 5 mm in (A), (B), and (E).
The phenotypes of WOX4-expressing calli were similar to those of calli grown on culture medium containing a high concentration of cytokinin (Figure 7B). Under an appropriate concentration of cytokinin (8 μM), shoots were regenerated. At higher concentrations of cytokinin, by contrast, normal shoot formation was inhibited or only green calli proliferated. Thus, this finding raised the possibility that green calli or abnormal shoot-like structures overexpressing WOX4 contained a higher content of cytokinin.
We therefore determined the content of cytokinin in the calli. Although the content varied depending on the callus sample (probably due to the state of individual calli), overall, cytokinins, including active forms such as isopentenyl adenine (iP) and cis-zeatin (cZ), accumulated at higher levels in WOX4-expressing calli than in calli with control vector (Figure 7C; see Supplemental Table 1 online). Whereas iP and trans-zeatin (tZ)-type cytokinins are the major forms in Arabidopsis, cZ is also an active cytokinin in rice and maize (Veach et al., 2003; Kudo et al., 2012). The level of tZ was undetectable in calli with pACT1-WOX4 and in those with empty vector (see Supplemental Table 1 online). Consistent with the higher cytokinin levels, shoots were regenerated from WOX4-expressing calli that were placed on hormone-free media, whereas the ordinary calli did not show any shoot regeneration (Figure 7D).
We tested the effect of FCP1 peptide on calli expressing WOX4. Calli with abnormal shoot-like structures that had been grown on the regeneration agar medium for 1 month were transferred into regeneration liquid medium and grown in the presence or absence of FCP1 peptide (20 μM) for 7 d with shaking (Figure 7E). As a result, normal shoots with expanded leaves were grown from the abnormal shoot-like structures in the presence of FCP1 peptide, but such shoots were not observed in the absence of FCP1 peptide. Thus, it seems likely that FCP1 peptide suppresses the effect of WOX4 overexpression in the abnormal shoot-like structures.
DISCUSSION
FCP1/FCP2 Is Likely to Be a Negative Regulator of Meristem Maintenance in Rice
We have shown that, similar to CLV3 in Arabidopsis, FON2 negatively regulates stem cell identity in the floral meristem of rice (Chu et al., 2006; Suzaki et al., 2006). Although the floral meristem is strongly affected in null fon2 mutants or FON2-overexpressing transgenic plants, no defects are observed in the meristem of their vegetative phases, implying that there are other factors that regulate maintenance of the vegetative SAM. In a previous study (Suzaki et al., 2008), we suggested that FCP1 is a candidate to be such a factor, based on the observation that regenerated shoots overexpressing FCP1 fail to maintain the SAM. In this article, using an inducible gene expression system, we demonstrated that FCP1 is a negative regulator in maintenance of the vegetative SAM. First, induction of FCP1 in seedlings germinated from seeds inhibited maintenance of the SAM that had formed normally during embryogenesis. Second, cosilencing of FCP1 and FCP2 resulted in promotion of meristem activity. The shoot phenotypes and the SAM morphologies in both FCP1-overexpresssing and FCP1/2-silenced plants are consistent with changes in the expression of meristem markers, such as OSH1 and FON2.
OSH1 promotes indeterminacy of the meristem and maintains the undifferentiated state of the cells (Sato et al., 1996; Tsuda et al., 2011). A loss-of-function mutant of OSH1 fails to maintain the SAM after germination (Tsuda et al., 2011). A reduction or enhancement in the expression levels of OSH1 was observed in the vegetative SAM of FCP1-overexpressing or FCP1/2-silenced plants, respectively. It is therefore likely that OSH1 may be involved in the action of FCP1/FCP2 in the meristem to maintain the undifferentiated state of cells. In addition, failed downregulation of OSH1 in the leaf founder cells (P0 site) was observed in FCP1/2-silenced plants. This suggests that FCP1/2 probably acts not only on the central region of the meristem, but also on the peripheral region where leaves initiate. Thus, FCP1/2 may be required to function in the downregulation of OSH1 in the P0 site to establish founder cells for leaf differentiation in wild-type rice.
FON2 is expressed in the tip of the meristem, and its expression domains are expanded in the floral meristem of the fon1 mutant (Suzaki et al., 2006). This FON2 behavior, similar to that of CLV3 in Arabidopsis, suggests that the region expressing FON2 corresponds to stem cells in rice. Expression of FON2 was strongly reduced by the induced overexpression of FCP1, whereas the expression domain of FON2 was expanded by the induced cosilencing of FCP1 and FCP2. Therefore, it is likely that FCP1 (and probably FCP2) negatively regulates stem cell identity.
Both plants overexpressing FCP1 and those with FCP1/2 silenced by DEX induction showed similar phenotypes of small shoots with small pale green leaves; however, these phenotypic similarities seem to result from different consequences of the loss or gain of FCP1/2 function. In FCP1-overexpressing plants, leaf initiation would be inhibited because of a reduction in meristematic activity, as shown by reduced OSH1 expression. In FCP1/2-silenced plants, by contrast, the SAM may fail to recruit leaf founder cells because of the ectopic expression of OSH1, which would invade the P0 site. As a result, FCP1/2 may promote leaf initiation by repressing OSH1 expression properly at the flank of the meristem.
WOX4 Seems to Be a Positive Regulator of Maintenance of the SAM
The induced RNA silencing of WOX4 led to small aberrant shoots with yellow leaves, a reduction in SAM size, and a decrease in the expression level of both OSH1 and FON2, suggesting that the function of SAM was compromised. Therefore, WOX4 is likely to be involved in the positive regulation of meristem maintenance in rice. The upregulation and downregulation of WOX4 in FCP1-silenced and FCP1/2-overexpressing seedlings, respectively, demonstrates that WOX4 is likely to be negatively regulated by FCP1. This idea is supported by the observation that exogenous application of FCP1 peptide suppressed the effect of WOX4 overexpression in calli. The induced overexpression of FCP1 and RNA silencing of WOX4 resulted in the downregulation of OSH1 and FON2, suggesting that FCP1 and WOX4 have opposite roles in SAM maintenance. Taking these observations together, it is probable that one of the roles of FCP1 in meristem maintenance is regulation of WOX4, which promotes meristem activity (see Supplemental Figure 8 online).
The function of WOX4 may be associated with cytokinin action. Constitutive expression of WOX4 resulted in increased levels of active cytokinins, such as isopentenyl adenine and cis-zeatin. It has been shown that OSH1 promotes the expression of genes involved in cytokinin biosynthesis (Sakamoto et al., 2006). Therefore, an increase in cytokinin may result from the upregulation of OSH1 expression in calli that overexpress WOX4 (see Supplemental Figure 8 online). Consistent with increased levels of cytokinin, calli overexpressing WOX4 showed phenotypes similar to those of calli grown in a higher concentration of cytokinin and were able to regenerate shoots without the addition of cytokinin to the culture medium.
The combined action of FCP1 and WOX4 in rice is similar to that of CLV3 and WUS that regulates stem cell maintenance in Arabidopsis. Although the FCP1-WOX4 pathway is likely to be involved in meristem maintenance as discussed above, there seem to be a few differences between FCP1-WOX4 and CLV-WUS. First, no enlargement of the SAM was observed in FCP1/2-silenced plants. This contrasts with the enlargement of the SAM caused by overproliferation of stem cells in a loss-of-function mutant of CLV3 in Arabidopsis (Clark et al., 1995; Reddy and Meyerowitz, 2005). Downregulation of FCP1/2 may not be directly linked to overproliferation of stem cells, although upregulation of FON2 expression was observed in FCP1/2-silenced plants. Second, leaf initiation was inhibited in FCP1/2-silenced plants, whereas no such inhibition is observed in the Arabidopsis clv3 mutant where, instead, an increased number of floral organs is formed from the floral meristem. As discussed above, inhibition of leaf initiation in FCP1/2-silenced plants may result from the ectopic expression of OSH1 in the P0 site. Third, expression of WOX4 was seen throughout the SAM, and this expression overlaps with that of FCP1 (Suzaki et al., 2008). This contrasts with the expression patterns of CLV3 and WUS, which are involved in communication between the different domains of the meristem. Thus, the FCP1-WOX4 pathway must act on the meristem in rice in a manner that differs from that of the CLV3-WUS pathway in Arabidopsis.
In rice, loss of function of FON2 causes an enlargement of the floral meristem and an increase in the number of floral organs. Therefore, FON2 seems to act on the floral meristem in rice, as CLV3 does in Arabidopsis. It would be of great interest to identify the gene that is involved in positive regulation of the floral meristem, similar to Arabidopsis WUS, and to elucidate the regulatory genetic pathway that maintains stem cells.
Roles of WOX4 in Leaf Development
WOX4 is likely to be involved in leaf development. WOX4 was strongly expressed in the P1 and P2 primordia. Downregulation of WOX4 by induced RNA silencing of WOX4 or by overexpression of FCP1 caused malformed leaf primordia with enlarged and vacuolated cells. Therefore, WOX4 is likely to be required to maintain the leaf primordia during early developmental stages, when cells divide and proliferate properly, probably by inhibiting premature differentiation of the cells.
In the P3 primordia, expression of WOX4 was restricted to the putative procambium and subsequently to the region between the differentiating xylem and phloem. In Arabidopsis, WOX4 promotes stem cell proliferation in the procambium and cambium (Hirakawa et al., 2010). The temporal and spatial expression patterns of rice WOX4 are highly similar to those of Arabidopsis WOX4, suggesting that these orthologous genes have similar roles in the regulation of vascular development. Although, like other monocots, rice has no distinct layer corresponding to the cambium, the expression pattern of rice WOX4 implies that the cells between the xylem and the phloem might have characteristics similar to those of the procambium or cambium in eudicots. Further studies would be required to clarify whether WOX4 is involved in vascular development in rice.
The similar expression patterns of WOX4 in developing leaves of both rice and Arabidopsis imply that the ancestral function of WOX4 might be associated with the regulation of vascular development. Unlike in rice, WOX4 is not expressed in the SAM in Arabidopsis, and no abnormalities in the SAM have been reported in the null wox4 mutant (Hirakawa et al., 2010). It is therefore possible that WOX4 might have been recruited to regulate meristem maintenance during the evolution of rice. Alternatively, meristem maintenance might be an ancestral function of WOX4 that has been lost in the lineage of Arabidopsis. It would be of great interest to know how the WOX4 function associated with meristem maintenance is conserved among Angiosperms.
METHODS
Plasmid Construction
pINDEX4 was a kind gift from P. Ouwerkerk (Leiden University) (Ouwerkerk et al., 2001). The rice (Oryza sativa) ACTIN1 promoter (Zhang et al., 1991), which induces strong expression in rice, was inserted in the ApaI site of pINDEX4, and the resulting plasmid was named pAID1 (see Supplemental Figure 1 online). To make constructs easily, the Gateway rfC cassette was inserted in the SpeI site of pAID1 (named pAID2; see Supplemental Figure 1 online). The cassette for RNAi silencing, which was derived from the pANDA vector (Miki and Shimamoto, 2004), was inserted in the same site of pAID1 (pAID3; see Supplemental Figure 1 online). Full or a partial cDNA of WOX4 was amplified using the primers listed in Supplemental Table 2 online and inserted in the Gateway entry vector pENTR-D/TOPO (Invitrogen). Concerning FCP1 and FCP2, plasmids constructed in a previous study were used for further construction (Suzaki et al., 2006). FCP1 cDNA was inserted in pAID2 (pACT1-GVG>FCP1; see Supplemental Figure 2 online) by the LR reaction. For inducible RNA silencing, a partial FCP1 cDNA, a ligated fragment of partial FCP1 and FCP2 cDNA, and a partial cDNA fragment specific to WOX4 (341 bp; from the initiation codon, 20 to 361) were inserted in pAID3 (pACT1-GVG>FCP1:RNAi, pACT1-GVG>FCP1-FCP2:RNAi, and pACT1-GVG>WOX4:RNAi; see Supplemental Figure 2 online) by the LR reaction. A partial WOX4 cDNA was similarly inserted in pANDA-EG1 (pACT1-WOX4:RNAi; see Supplemental Figure 2 online) (Suzaki et al., 2008).
Transformation and Tissue Culture
The rice cultivar Taichung65 (T65) was used as a host for the transformation. The constructs were introduced into Agrobacterium tumefaciens strain EHA101 or EHA103 and transformed into scutellum-derived calli of T65, according to the method described by Hiei et al. and/or Ozawa (Hiei et al., 1994; Ozawa, 2012). To make the transgenic plants, shoots were regenerated from hygromycin-resistant calli in regeneration medium containing 8 μM 6-benzylaminopurine (BAP) and 5 µM naphthalene acetic acid.
To examine the effect of cytokinin on shoot regeneration, different concentrations of BAP were used instead of 8 μM BAP in regeneration medium (Hiei et al., 1994). FCP1 peptides (REVPTGDPIHH) were synthesized and purified to at least 95% using HPLC by TaKaRa and added to the medium as described previously (Suzaki et al., 2006). WOX4-overexpressing calli with abnormal shoot-like structures (grown in regeneration agar medium for 30 days) were transferred into liquid regeneration medium (20 mL) either containing or not containing 20 μM FCP1 peptide in a 100-mL conical flask. The flask was incubated at 28°C under 16 h light/8 h dark cycles with shaking (100 rotations/minute).
Selection of Transgenic Plants Used for DEX Induction
For inducible overexpression or RNAi silencing, transformants that showed efficient DEX induction were selected. Leaf sections (∼1-cm length, four to five sections for each sample) cut off from regenerated shoots (T0 generation) were immersed in sterile water in the presence (50 μM) or absence of DEX overnight. RNA was isolated, and the levels of RNA transcript from the cDNA or the DNA fragment (for RNA silencing) under the control of 4UAS in each construct were analyzed by RT-PCR. For each construct, five T0 lines showing high induction of transgene expression by DEX application were selected from ∼25 to 30 independent transformants.
The effect of DEX induction was examined using T1 seedlings germinated from seeds, which were harvested from the same T0 plants. The concentration of DEX and length of treatment are indicated in each figure legend.
In Situ Hybridization
OSH1 and FON2 probes were prepared using the plasmid described in the original studies (Sato et al., 1996; Suzaki et al., 2006). To generate probes for the WOX4 transcript, partial cDNA fragments (341 bp; from the initiation codon, 20 to 361) were amplified with the primers listed in Supplemental Table 2 online and cloned into pENTR-D/TOPO (Invitrogen). This fragment contained the WOX4-specific region (∼250 bp) and a part of the homeodomain region in which similarity is not high among rice WOX members. After linearization of the plasmids with NotI, RNA was transcribed by T7 RNA polymerase and labeled with digoxigenin (Roche). Preparation of sections and hybridization experiments were performed by the methods described by Suzaki et al. (2004).
qRT-PCR Analysis
To determine the expression levels of WOX genes in regenerating shoots, whole shoots including adventitious root primordia, which carried pACT1-FCP1 and empty vector, were used. For DEX induction experiments, the shoot apices (<3 mm diameter × 7- to 8-mm height), which include leaf primordia and a few young leaves, were excised from the DEX- or mock-treated seedlings and used for RNA isolation. Total RNA was isolated using Treasure (Bioline), and the RNA extract was treated with DNase I (Invitrogen) to remove genomic DNA. The first strand of cDNA was synthesized from 1 µg of total RNA using a PrimScript RT reagent kit (TaKaRa). The primers used for RT-PCR are listed in Supplemental Table 2 online. qRT-PCR was performed using a 7300 Real-Time PCR system (Applied Biosystems) with Power SYBR Green RT-PCR Master Mix (Applied Biosystems). Four RNA samples, which were independently isolated from three to five regenerated shoots, were used for qRT-PCR analysis, and the PCR reaction was performed twice for each sample. The ACTIN1 gene was used as a control.
Cytokinin Quantification in the Calli Overexpressing WOX4
Calli carrying pACT1-WOX4 and empty vector were grow on selection medium containing auxin (2 mg/L 2,4-D) for 30 d (Hiei et al., 1994) and then on hormone-free Murashige and Skoog (MS) medium containing 3% Suc for 30 d. A few calli (corresponding to 80 to 100 mg) were harvested and used for cytokinin quantification. The quantification was repeated six times for each construct. Extraction and determination of cytokinins from calli were performed as described previously using ultraperformance liquid chromatography–tandem mass spectrometry (AQITY UPLC system/Xevo-TQS; Waters) with an ODS column (Aquity UPLC BEH C18, 1.7 µm, 2.1 × 100 mm; Waters) (Kojima et al., 2009).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: AB354584 (FCP1), AB354585 (FCP2), AJ556181 (Os WOX4), NM_103605 (At WOX4), AB218894 (Os WUS), AM234749 (Os WOX2), AM490244 (Os WOX3), AM490243 (Os NS), AK102203 (QHB), AB245090 (FON2), and AK107637 (OSH1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Vectors Used for Inducible Overexpression or Downregulation.
Supplemental Figure 2. Plasmid Constructs Used in This Study.
Supplemental Figure 3. Morphologies of the Shoot Apex in Seedlings Carrying pACT1-GVG>FCP1-FCP2:RNAi after DEX Treatment.
Supplemental Figure 4. OSH1 Expression Patterns in the Shoot Apex of Seedlings Carrying pACT1-GVG>FCP1-FCP2:RNAi.
Supplemental Figure 5. Comparison of Rice WOX4 and Arabidopsis WOX4.
Supplemental Figure 6. qRT-PCR Analysis of the WOX4 and WUS Genes of Rice.
Supplemental Figure 7. The Shoot Apices of pACT1-GVG>WOX4:RNAi Transgenic Plants after DEX Treatment.
Supplemental Figure 8. Model of Genetic Interactions Involved in Maintenance of the Vegetative Meristem in Rice.
Supplemental Table 1. Cytokinin Content in Calli with pACT1-WOX4 or Empty Vector.
Supplemental Table 2. Primers Used in This Study.
Acknowledgments
We thank Pieter Ouwerkerk for kindly providing the pINDEX vectors, Kenjiro Ozawa for providing the protocol for rice transformation before publication, and Hiroo Fukuda for encouragement and critical reading of the article. We thank Takuya Suzaki for technical support and Keiko Ohsawa-Yamamoto and Eiko Oki for technical assistance. Hormone analysis was supported by the Japan Advanced Plant Science Network.
AUTHOR CONTRIBUTIONS
Y.O. and H.-Y.H. designed the research. Y.O., W.T., M.K., H.S., and H.-Y.H. performed the research. Y.O. and H.-Y.H. wrote the article.
Glossary
- SAM
shoot apical meristem
- OC
organizing center
- DEX
dexamethasone
- qRT-PCR
quantitative real-time PCR
- RNAi
RNA interference
- BAP
6-benzylaminopurine
- MS
Murashige and Skoog
References
- Aichinger E., Kornet N., Friedrich T., Laux T. (2012). Plant stem cell niches. Annu. Rev. Plant Biol. 63: 615–636 [DOI] [PubMed] [Google Scholar]
- Barton M.K. (2010). Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Betsuyaku S., Takahashi F., Kinoshita A., Miwa H., Shinozaki K., Fukuda H., Sawa S. (2011). Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 52: 14–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P., Lunde C., Nardmann J., Vollbrecht E., Running M., Jackson D., Hake S., Werr W. (2005). thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Chickarmane V.S., Gordon S.P., Tarr P.T., Heisler M.G., Meyerowitz E.M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Qian Q., Liang W., Yin C., Tan H., Yao X., Yuan Z., Yang J., Huang H., Luo D., Ma H., Zhang D. (2006). The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142: 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067 [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Dai M., Hu Y., Zhao Y., Liu H., Zhou D.-X. (2007). A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells J.P., Turner S.R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Chickarmane V.S., Ohno C., Meyerowitz E.M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Fletcher J.C. (2010). Shoot apical meristem form and function. Curr. Top. Dev. Biol. 91: 103–140 [DOI] [PubMed] [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Itoh J.-I., Nonomura K.-I., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Ji J., Strable J., Shimizu R., Koenig D., Sinha N., Scanlon M.J. (2010). WOX4 promotes procambial development. Plant Physiol. 152: 1346–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Nagasaki H., Morikami A., Sato Y., Matsuoka M. (2003). Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 35: 429–441 [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kojima M., Kamada-Nobusada T., Komatsu H., Takei K., Kuroha T., Mizutani M., Ashikari M., Ueguchi-Tanaka M., Matsuoka M., Suzuki K., Sakakibara H. (2009). Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kudo T., Makita N., Kojima M., Tokunaga H., Sakakibara H. (2012). Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-Zeatin-O-glucosyltransferase in rice. Plant Physiol. 160: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J.P.C., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mayer K.F.X., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J., Werr W. (2006). The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 23: 2492–2504 [DOI] [PubMed] [Google Scholar]
- Nishimura A., Ito M., Kamiya N., Sato Y., Matsuoka M. (2002). OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. 30: 189–201 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Ouwerkerk P.B., de Kam R.J., Hoge J.H., Meijer A.H. (2001). Glucocorticoid-inducible gene expression in rice. Planta 213: 370–378 [DOI] [PubMed] [Google Scholar]
- Ozawa K. (2012). A high-efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Methods Mol. Biol. 847: 51–57 [DOI] [PubMed] [Google Scholar]
- Reddy G.V., Meyerowitz E.M. (2005). Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310: 663–667 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Sakakibara H., Kojima M., Yamamoto Y., Nagasaki H., Inukai Y., Sato Y., Matsuoka M. (2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Sato Y., Hong S.K., Tagiri A., Kitano H., Yamamoto N., Nagato Y., Matsuoka M. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. USA 93: 8117–8122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Song S.-K., Lee M.M., Clark S.E. (2006). POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133: 4691–4698 [DOI] [PubMed] [Google Scholar]
- Suer S., Agusti J., Sanchez P., Schwarz M., Greb T. (2011). WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23: 3247–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Ohneda M., Toriba T., Yoshida A., Hirano H.-Y. (2009). FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice. PLoS Genet. 5: e1000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Sato M., Ashikari M., Miyoshi M., Nagato Y., Hirano H.-Y. (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131: 5649–5657 [DOI] [PubMed] [Google Scholar]
- Suzaki T., Toriba T., Fujimoto M., Tsutsumi N., Kitano H., Hirano H.-Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Suzaki T., Yoshida A., Hirano H.-Y. (2008). Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell 20: 2049–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F., Yuan Z., Hake S., Jackson D. (2001). The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Ito Y., Sato Y., Kurata N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Horstman A., Zethof J., Koes R., Rijpkema A.S., Gerats T. (2009). Differential recruitment of WOX transcription factors for lateral development and organ fusion in petunia and Arabidopsis. Plant Cell 21: 2269–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach Y.K., Martin R.C., Mok D.W.S., Malbeck J., Vankova R., Mok M.C. (2003). O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 131: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Williams R.W., Wilson J.M., Meyerowitz E.M. (1997). A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 94: 10467–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dabi T., Weigel D. (2005). Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 15: 436–440 [DOI] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Tavakkoli M., Reddy G.V. (2010). WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137: 3581–3589 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Zhang W., McElroy D., Wu R. (1991). Analysis of rice Act1 5′ region activity in transgenic rice plants. Plant Cell 3: 1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L., Zhou D.-X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]