Light inhibits hypocotyl growth and the expression of genes that stimulate it. This study reveals that the CHD3 chromatin remodeling factor PKL/EPP1 is recruited by HY5, a master transcription factor of the light signaling pathway and represses H3K27me3 modification of cell elongation–related loci. PKL/EPP1 and HY5 work together to fine-tune hypocotyl cell elongation in response to light.
Abstract
Photomorphogenesis is a critical plant developmental process that involves light-mediated transcriptome changes, histone modifications, and inhibition of hypocotyl growth. However, the chromatin-based regulatory mechanism underlying this process remains largely unknown. Here, we identify ENHANCED PHOTOMORPHOGENIC1 (EPP1), previously known as PICKLE (PKL), an ATP-dependent chromatin remodeling factor of the chromodomain/helicase/DNA binding family, as a repressor of photomorphogenesis in Arabidopsis thaliana. We show that PKL/EPP1 expression is repressed by light in the hypocotyls in a photoreceptor-dependent manner. Furthermore, we reveal that the transcription factor ELONGATED HYPOCOTYL5 (HY5) binds to the promoters of cell elongation–related genes and recruits PKL/EPP1 through their physical interaction. PKL/EPP1 in turn negatively regulates HY5 by repressing trimethylation of histone H3 Lys 27 at the target loci, thereby regulating the expression of these genes and, thus, hypocotyl elongation. We also show that HY5 possesses transcriptional repression activity. Our study reveals a crucial role for a chromatin remodeling factor in repressing photomorphogenesis and demonstrates that transcription factor–mediated recruitment of chromatin-remodeling machinery is important for plant development in response to changing light environments.
INTRODUCTION
Unlike animals, plants respond to the surrounding environments by shaping their growth and development. A developmental transition from heterotrophic growth in subterranean darkness to autotrophic growth upon reaching the light is a critical event in the life cycle of plants. Dark-grown seedlings undergoing skotomorphogenesis (etiolation) are characterized by elongated hypocotyls, apical hooks with unexpanded cotyledons, undifferentiated chloroplasts, and repression of light-regulated genes. Light triggers photomorphogenesis, resulting in seedlings with deetiolated morphologies (i.e., short hypocotyls and open cotyledons) that contain differentiated chloroplasts and undergo chlorophyll and anthocyanin biosynthesis (Von Arnim and Deng, 1996). Plants have evolved an array of photoreceptors, including the red and far-red light-absorbing phytochromes (phyA to phyE) and the blue/UV-A light-absorbing cryptochromes (cry1 and cry2), to perceive and transduce light signals that ultimately modulate the transcriptome and trigger photomorphogenic growth and development (Chen et al., 2004).
Numerous studies demonstrate that phytochromes and cryptochromes suppress two main branches of light signaling during photomorphogenesis (Lau and Deng, 2010). A group of CONSTITUTIVE PHOTOMORPHOGENIC (COP)/DEETIOLATED (DET)/FUSCA proteins acts as central repressors downstream of the photoreceptors (Wei and Deng, 1996). Among them, COP1 functions as a RING finger E3 ubiquitin ligase that targets a number of positive factors, such as ELONGATED HYPOCOTYL5 (HY5), for degradation, and thus desensitizes light signaling (reviewed in Henriques et al., 2009; Lau and Deng, 2012). HY5 is a basic domain/Leucine zipper transcription factor that plays a key role in promoting photomorphogenesis in all light conditions by regulating the transcription of a wide range of genes, largely by binding to consensus sequences, such as the G-box (Oyama et al., 1997; Lee et al., 2007). HY5 is stabilized by light, and its abundance correlates inversely with the extent of hypocotyl growth (Osterlund et al., 2000). HY5 and its close homolog HYH can act as heterodimers and homodimers, thus mediating light-regulated expression of overlapping as well as distinct target genes (Holm et al., 2002). Furthermore, a group of basic helix-loop-helix transcription factors, named PHYTOCHROME-INTERACTING FACTORs (PIFs), is active in darkness and thus regulates gene expression to promote the skotomorphogenic response (Leivar et al., 2008). Under light, PIFs interact with photoactivated phytochromes and result in their phosphorylation and subsequent degradation (Leivar and Quail, 2011). Extensive studies have also identified dozens of intermediates in the light signaling pathway and revealed the importance of transcriptional regulatory networks in controlling photomorphogenesis (Jiao et al., 2007; Chory, 2010).
Chromatin remodeling plays a central role in establishing specific gene expression patterns and maintaining transcriptional states in eukaryotes (Jarillo et al., 2009; Ho and Crabtree, 2010). Accumulating evidence suggests that light-mediated gene expression and responses also require chromatin reorganization (Fisher and Franklin, 2011). The first piece of evidence came from the observation that increased acetylation of histone H3 and H4 of a pea (Pisum sativum) plastocyanin promoter correlates with light-induced transcription (Chua et al., 2003). It was shown that DET1, a repressor of photomorphogenesis, binds to unacetylated histone tails, consistent with its role in transcriptional repression (Benvenuto et al., 2002). Furthermore, histone acetylation, which is mediated by histone acetyltransferase and deacetylase, is required for regulating light-responsive gene expression and seedling growth (Bertrand et al., 2005; Benhamed et al., 2006). This notion is further supported by a study that showed that rapid light-mediated changes in phyA transcript levels are correlated with alterations of specific histone modifications (Jang et al., 2011). Two recent genome-wide analyses suggest the existence of a chromatin-based program that regulates photomorphogenesis (Guo et al., 2008; Charron et al., 2009). However, the mechanism by which chromatin configuration controls light-regulated gene expression and photomorphogenic responses is largely unknown.
ATP-dependent chromatin remodeling enzymes use the energy released by ATP hydrolysis to alter histone-DNA contacts and, thus, the accessibility of genomic regions to the transcriptional machinery or transcription factors. Although ATP-dependent chromatin remodeling enzymes are well characterized in animals, only a few have been functionally studied in plants (Clapier and Cairns, 2009). The SWITCH/SUCROSE NONFERMENTING (SWI/SNF) family of ATP-dependent chromatin remodelers has been studied extensively. Two members of this family, BRAHMA (BRM) and SPLAYED (SYD), are involved in regulating diverse developmental programs, such as seed maturation, root development, floral development, and biotic stress (Farrona et al., 2004, 2011; Hurtado et al., 2006; Tang et al., 2008; Walley et al., 2008). The CHD (for chromodomain, helicase/ATPase, and DNA binding domain) proteins belong to the SNF2-like family of ATPases and have been identified in a variety of organisms (Hall and Georgel, 2007; Hargreaves and Crabtree, 2011). Arabidopsis thaliana PICKLE (PKL; also named GYMNOS/Suppressor of slr2 [SSL2]/CYTOKININ-HYPERSENSITIVE2) is a member of the CHD3 subfamily that shares similarity with Mi-2 in fly and human genomes (Clapier and Cairns, 2009) and possesses nucleosome-stimulated ATPase activity (Ho et al., 2012). PKL regulates multiple plant developmental processes, including embryonic development, seed germination, and root meristem activity (Ogas et al., 1999; Fukaki et al., 2006; Perruc et al., 2007; Aichinger et al., 2011).
Through forward genetic screening, we identify the enhanced photomorphogenic1 (epp1) mutant and reveal that EPP1 encodes PKL and acts as an important negative regulator of photomorphogenesis. We demonstrate that PKL/EPP1 physically interacts with HY5 to directly regulate hypocotyl cell elongation by repressing trimethylation of histone H3 Lys 27 (H3K27me3) on the regulatory regions of several cell elongation–related genes in response to changing light conditions.
RESULTS
EPP1 Is a Repressor of Photomorphogenesis
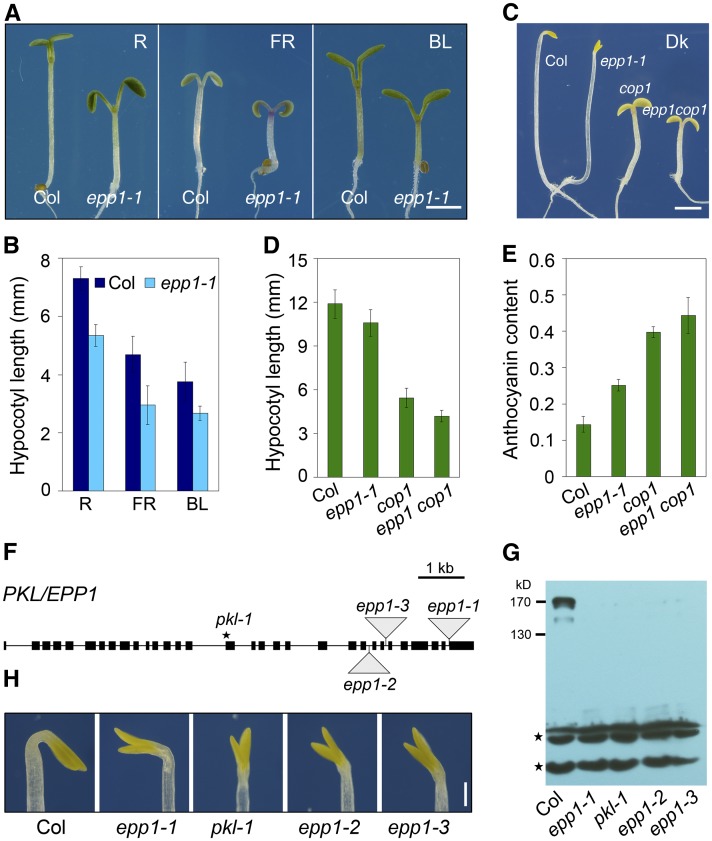
To identify novel components of the light signaling pathway, we performed a genetic screen for Arabidopsis mutants with an epp phenotype in a T-DNA mutant pool (Weigel et al., 2000). Mutants exhibiting short hypocotyls and/or large cotyledons under red light were chosen and the phenotypes were further confirmed by analyzing the next generation of mutants under red, far-red, and blue light. One of the mutants, referred to as epp1-1, exhibited shorter hypocotyls and more expanded cotyledons in far-red, red, and blue light conditions than the Columbia (Col) wild type (Figures 1A and 1B; see Supplemental Figure 1A online). Remarkably, the epp1-1 seedlings had hypocotyls that were slightly reduced in length, opened cotyledons, and unfolded apical hooks in darkness (Figures 1C and 1D). In addition, epp1-1 accumulated increased levels of chlorophyll and anthocyanin compared with the wild type (see Supplemental Figures 1B and 1C online). Furthermore, the expression of two light-inducible genes, CHS (encoding chalcone synthase) and CAB2 (encoding chlorophyll a/b binding protein), was increased in epp1-1 in both light and darkness (see Supplemental Figure 1D online). Together, these results suggest that epp1-1 is a weak cop-like mutant.
Figure 1.
Loss of EPP1, Encoding PKL, Triggers a Constitutive Photomorphogenic-Like Response.
(A) The epp1-1 mutant is hyposensitive to red (R), far-red (FR), and blue (BL) light compared with the Col wild type. Bar = 2 mm.
(B) Hypocotyl length of epp1-1 and wild-type seedlings under various light conditions, as shown in (A). Data are mean ± sd of 30 seedlings.
(C) Phenotypes of Col, epp1-1, cop1, and epp1 cop1 seedlings in darkness (Dk). Bar = 2 mm.
(D) Hypocotyl length of wild-type and mutant seedlings in darkness. Data represent the mean ± sd of 30 seedlings.
(E) Anthocyanin content [(A530 − 0.25 × A657)/100 seedlings] of wild-type and mutant seedlings in far-red light. Data represent the mean ± sd of triplicate assays.
(F) Diagram of PKL/EPP1 and positions of the mutations. Black boxes represent exons, and lines between the boxes indicate introns. Triangles denote T-DNA insertions, and the star indicates a point mutation.
(G) Immunoblot analysis showing the presence of PKL in the wild type but not in the epp1 mutants. Stars indicate cross-reacting bands used for a loading control.
(H) Multiple epp1/pkl mutant alleles displaying open cotyledons without an apical hook under dark conditions. Bar = 0.5 mm.
All seedlings in (A) to (E), (G), and (H) were grown in the light or dark conditions for 5 d.
[See online article for color version of this figure.]
Next, we generated double mutants and tested for genetic interaction between epp1-1 and the photoreceptor mutants. The epp1 phyA, epp1 phyB, and epp1 cry1 double mutants had slightly shorter hypocotyls than the photoreceptor parent mutants under far-red, red, and blue light conditions, respectively (see Supplemental Figures 2A and 2B online), indicating that EPP1 is a positive regulator of hypocotyl growth that acts downstream of both phytochromes and cryptochromes. Because COP1 acts as a central repressor in the light signaling pathway, we then examined the relationship between epp1 and cop1. The epp1 cop1 double mutant had much shorter hypocotyls and more expanded cotyledons and accumulated more anthocyanin than either of its parent mutants (Figures 1C to 1E; see Supplemental Figure 2C online), suggesting that EPP1 functions in parallel with COP1.
EPP1 Encodes PKL, an ATP-Dependent Chromatin Remodeling Factor
Using inverse PCR, we identified a single T-DNA insertion in the last exon of At2g25170 in the epp1-1 mutant (Figure 1F). At2g25170 encodes PKL, an ATP-dependent chromatin remodeling factor exhibiting a high level of similarity to human Mi2/CHD4 and CHD3 (Ogas et al., 1999). A 4.3-kb region of genomic DNA containing the entire open reading frame of PKL/SSL2 driven by its own promoter (Fukaki et al., 2006) was able to complement the short hypocotyl phenotype of epp1-1 (see Supplemental Figure 1E online). Next, we examined other epp1 alleles, including pkl-1, epp1-2, and epp1-3 (Figure 1F). These mutants did not accumulate PKL; an immunoblotting analysis using antibodies that detected the C-terminal region of PKL (residues 1178 to 1384 amino acids) recognized a specific endogenous band of around 170 kD only in the wild type, although a weak band of 150 kD was also present (Figure 1G). pkl-1, epp1-2, and epp1-3 phenocopied epp1-1 in terms of hypocotyl length and cotyledon angle (Figure 1H; see Supplemental Figures 1F and 1G online). Therefore, these data confirm that EPP1 is indeed PKL.
Light Represses PKL Expression in the Hypocotyls in a Photoreceptor-Dependent Manner
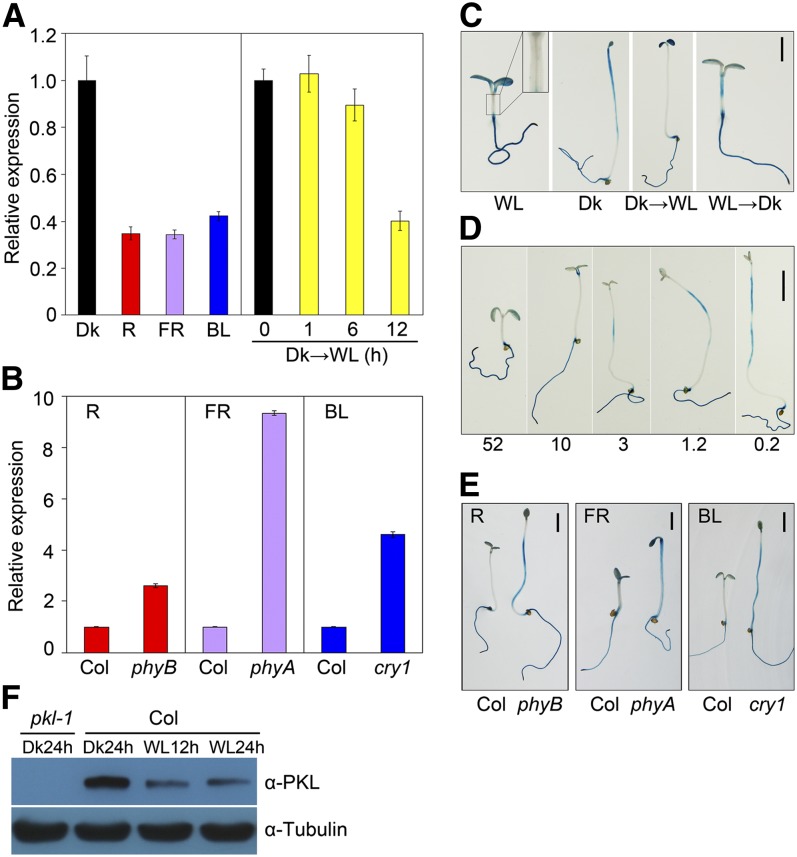
To further reveal how light influences PKL expression, we first compared the mRNA levels of PKL in 5-d-old Col seedlings grown under dark, far-red, red, and blue light by quantitative real-time PCR with reverse transcription (qRT-PCR). As shown in Figure 2A, PKL expression levels were dramatically lower in the light conditions tested than in darkness. Furthermore, PKL transcript levels were decreased within 12 h of transfer from dark to light, although the difference was not evident during the first six hours (Figure 2A). In addition, compared with those in the wild type, the PKL transcripts were greatly upregulated in the phyB, phyA, and cry1 mutants in red, far-red, and blue light, respectively (Figure 2B), suggesting that the light-mediated repression of PKL is facilitated by phytochrome and cryptochrome photoreceptors.
Figure 2.
PKL Expression in the Hypocotyls Is Repressed by Light.
(A) A qRT-PCR assay showing PKL transcript levels in wild-type seedlings subjected to red (R), far-red (FR), or blue (BL) light or darkness (Dk) for 5 d or in 5-d-old dark-grown seedlings transferred to white light (WL) for the indicated periods of time (0 to 12 h). Data represent the mean ± sd of three biological replicates.
(B) qRT-PCR assay showing increased PKL expression, relative to the Col wild type, in phyB, phyA, and cry1 photoreceptor mutants grown in red, far-red, and blue light conditions, respectively, for 5 d. For (A) and (B), relative expression was normalized to that of UBQ1, and data represent the mean ± sd of three biological replicates.
(C) GUS staining of ProPKL:GUS transgenic seedlings. Seedlings were grown in white light or darkness for 5 d, 4-d-old dark-grown seedlings were exposed to white light for an additional 1 d (Dk→WL), or 4-d-old light-grown seedlings were transferred to darkness for an additional 1 d (WL→Dk). The inset in the WL panel is an enlargement of the hypocotyl. Bar = 2 mm.
(D) GUS staining gradually increased in the hypocotyls of seedlings grown in a decreasing series of light intensities (μmol m−2 s−1; indicated below) for 5 d. Bar = 2 mm.
(E) GUS staining in the hypocotyls was significantly enhanced in the photoreceptor mutants compared with the Col wild type. Seedlings harboring the ProPKL:GUS reporter were grown under the indicated light conditions for 5 d. Bar = 2 mm.
(F) Immunoblotting of PKL protein. Seedlings were grown in long-day conditions (16 h day/8 h night) for 3 d and were then transferred to darkness or white light for the indicated period of time at the end of the day. Hypocotyls and cotyledons were detached and harvested for protein isolation. Immunoblotting against the tubulin antibody served as a loading control.
PKL was previously shown to be ubiquitously expressed in different tissues of the adult plant (Ogas et al., 1999). We decided to examine whether PKL undergoes tissue-specific regulation in response to light. To this end, transgenic plants expressing ProPKL:GUS (a β-glucuronidase reporter gene driven by a 2.0-kb promoter fragment of PKL) were generated. The GUS reporter was expressed in the cotyledons and roots regardless of the type of light treatments tested (Figure 2C). Interestingly, GUS was strongly expressed in the uppermost regions of hypocotyls in darkness and disappeared upon exposure to light. However, GUS expression was weak in light but became stronger when transferred to darkness (Figure 2C). In addition, GUS activity in the hypocotyls increased with the light intensity decreased (Figure 2D), indicating that PKL expression correlates positively with the extent of hypocotyl elongation. Next, we introduced ProPKL:GUS into the phyB, phyA, and cry1 mutants and tested the expression of the transgene in red, far-red, and blue light conditions, respectively. As shown in Figure 2E, the photoreceptor mutants displayed significantly enhanced GUS expression compared with the wild-type control in the region of the hypocotyls but not in the cotyledons and roots. Together, these observations indicate that, through phytochromes and cryptochromes, light represses PKL expression specifically in the hypocotyls, reinforcing a role for PKL in promoting hypocotyl growth.
To investigate whether light regulates PKL abundance, Col wild-type seedlings were grown in long-day conditions for 3 d. At the end of the light period, seedlings were kept in darkness or exposed to white light for an additional 12 or 24 h. We then performed an immunoblotting assay with PKL antibody and found that PKL protein level was drastically decreased in seedlings exposed to light compared with those in darkness (Figure 2F), suggesting that light also represses PKL at the protein level.
PKL Directly Activates Cell Elongation–Related Genes
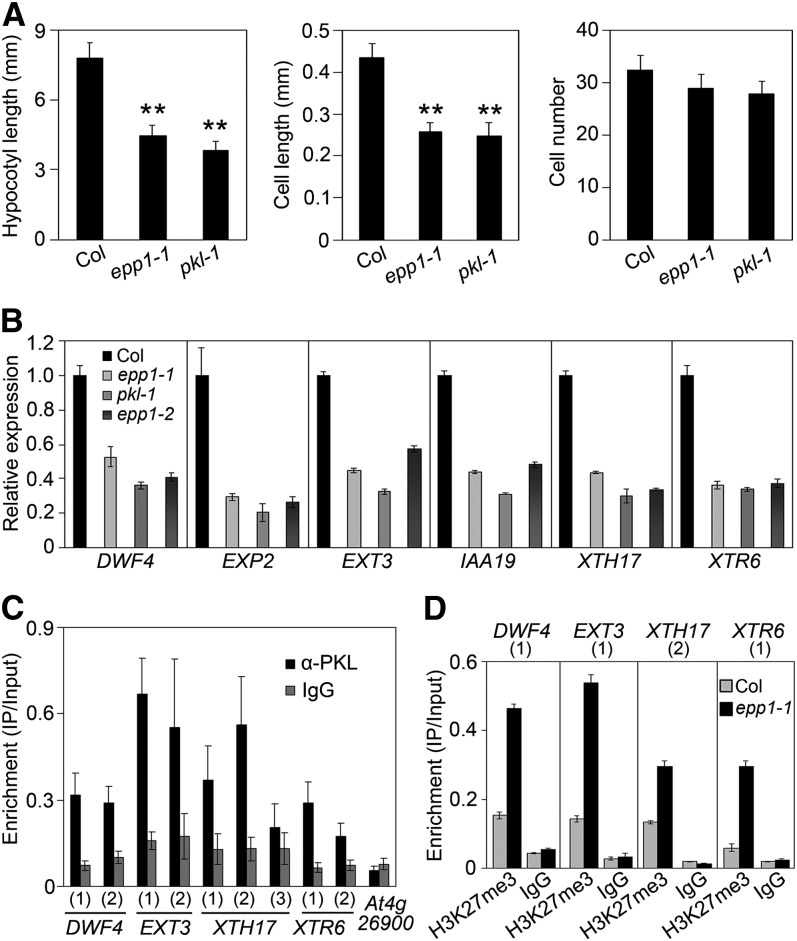
Plant growth involves both cell elongation and proliferation. To assess the contribution of defects in cell elongation and/or proliferation to the short hypocotyls of epp1/pkl mutants, we examined cell length and cell number along the vertical axes of the hypocotyls. The average cell length of both epp1-1 and pkl-1 hypocotyls was significantly decreased to 60% of the wild type, correlating with the decrease in overall hypocotyl length, whereas the number of cells did not differ significantly between epp1/pkl and the wild type (Figure 3A). This finding indicates that PKL predominantly regulates cell elongation, but not cell proliferation, during hypocotyl growth.
Figure 3.
PKL Directly Promotes the Expression of Cell Elongation Genes.
(A) The short hypocotyls of the epp1 mutants correlate with a reduction in cell elongation but not in cell number. Seedlings were grown in white light for 5 d. Data represent the mean ± sd of 30 seedlings. Asterisks indicate significant difference from the wild type at P < 0.01 using Student’s t test.
(B) qRT-PCR assay showing reduced expression of various genes involved in cell elongation in 5-d-old white light–grown epp1 mutants relative to the Col wild type. Relative expression was normalized to that of UBQ1. Data represent the mean ± sd of biological triplicates.
(C) ChIP-qPCR assay showing enrichment of various cell elongation genes in DNA samples pulled down by PKL antibody or IgG control. Numbers in parentheses indicate regions for amplification, as shown in Supplemental Figure 3A online. The Col wild-type seedlings were grown under white light for 5 d. At4g26900 served as a negative control. Data represent the mean ± sd of triplicates.
(D) ChIP-qPCR assay using the H3K27me3 antibody, showing relatively high enrichment of DWF4, EXT3, XTH17, and XTR6 in the epp1-1 mutant compared with the Col wild type. Numbers in parentheses indicate regions for amplification, as shown in Supplemental Figure 3A online. Precipitation by IgG preimmune serum served as a control. IP, immunoprecipitation. Data represent the mean ± sd of triplicates.
To further explore the molecular mechanism by which PKL regulates hypocotyl cell elongation, we attempted to identify its putative direct target genes from the public data available. Candidate genes were selected based on the following criteria: (1) they should encode cell wall loosening or hydrolytic enzymes that are implicated directly in regulating cell elongation (Cosgrove, 2005); (2) they should encode components involved in the biosynthesis or signaling of brassinosteroids or auxin (Sun et al., 2010; Chapman et al., 2012); and (3) if the genes satisfy criteria (1) and (2), expression of the genes should be repressed by light in a manner similar to EPP1/PKL (Ma et al., 2005). Accordingly, six genes, including EXTENSIN3 (EXT3), EXPANSIN2 (EXP2), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE17 (XTH17), XYLOGLUCAN ENDOTRANSGLYCOSYLASE6 (XTR6), DWARF4 (DWF4), and INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19) were selected for further analysis. We performed qRT-PCR analysis to determine whether these genes are regulated by PKL. As shown in Figure 3B, the transcript levels of these genes were drastically decreased in the epp1-1, pkl-1, and epp1-2 mutants compared with the wild type, suggesting that PKL does indeed contribute to the activation of these genes.
To test whether PKL activates the expression of EXT3, EXP2, XTH17, XTR6, DWF4, and IAA19 directly, we performed chromatin immunoprecipitation with quantitative PCR (ChIP-qPCR) using DNA isolated from Col wild type and the PKL antibody. Indeed, the promoter and coding regions of DWF4, EXT3, XTH17, and XTR6 was greatly enriched in DNA precipitated using the PKL antibody, but not in DNA pulled down with the IgG serum control (Figure 3C; see Supplemental Figure 3A online; EXP2 and IAA19 were analyzed below in detail). As a negative control, the enrichment of the At4g26900 promoter (Lee et al., 2007) was very low and indistinguishable between the PKL antibody and IgG. These data indicate that PKL directly associates with the genomic sequences of these cell elongation–related genes. In support of our finding that these genes are downstream targets of PKL, loss-of-function mutants of EXP2 and DWF4 also displayed short hypocotyls when grown in light (see Supplemental Figure 4 online). Furthermore, IAA19 expression in the hypocotyls was repressed by light in a manner similar to PKL (Tatematsu et al., 2004) and is also involved in hypocotyl cell elongation (Oh et al., 2012).
Previous studies documented that H3K27me3 plays an important role in PKL-mediated transcriptional regulation (Zhang et al., 2008, 2012; Aichinger et al., 2009). A ChIP assay using the H3K27me3 antibody followed by quantitative PCR revealed that EXT3, XTH17, XTR6, DWF4, EXP2, and IAA19 were all marked by H3K27me3; however, H3K27me3 levels were substantially increased in the epp1-1 mutant (Figure 3D; EXP2 and IAA19 were analyzed below). Taken together, these results demonstrate that PKL promotes cell elongation likely by inhibiting H3K27me3 of regions of chromatin that are specifically associated with the target genes involved in cell elongation.
HY5 Physically and Genetically Interacts with PKL
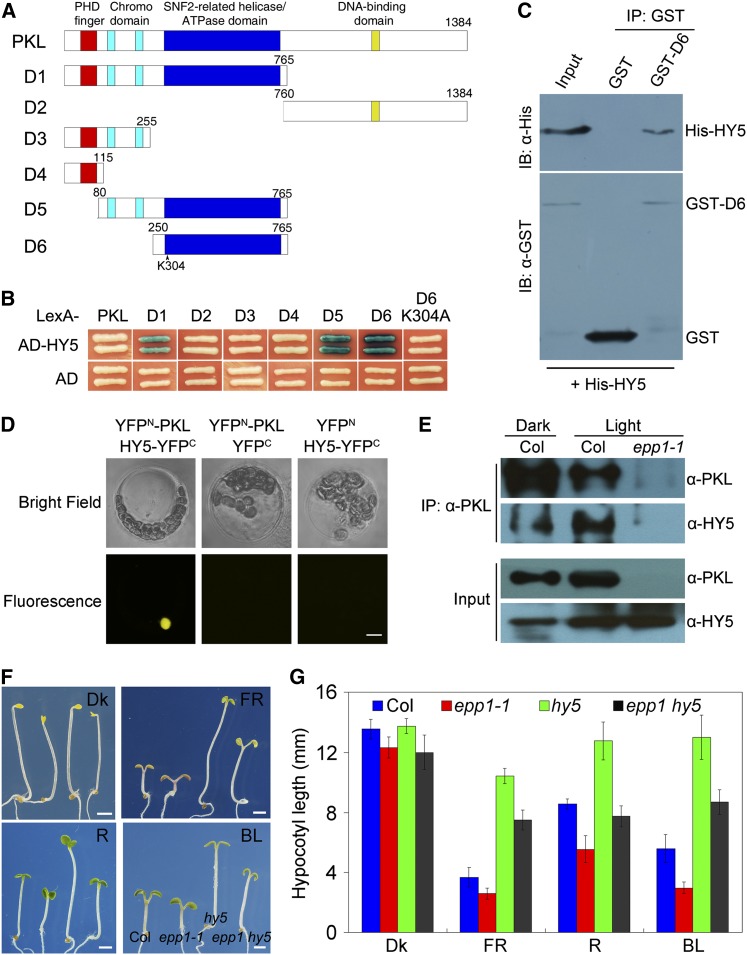
In mammals, the PKL homolog Mi-2b (CHD4) acts as an integral subunit of different complexes and interacts with transcription factors to modulate gene expression (Denslow and Wade, 2007). We reasoned that PKL might also have interacting partners involved in the regulation of light signaling. Given that PKL represses photomorphogenesis under all light conditions and that HY5 is a central transcription factor in the light signaling pathway, HY5 was a good candidate partner. To investigate whether PKL and HY5 interact with each other, we generated LexA fusions of full-length PKL and of the N-terminal (D1) and C-terminal (D2) fragments of PKL for yeast two-hybrid analysis with B42 activation domain–tagged HY5 (AD-HY5) (Figure 4A). However, neither full-length PKL (LexA-PKL) nor the C-terminal domain (LexA-D2) showed a positive interaction with AD-HY5. Notably, coexpression of LexA-D1 (including the PHD domain, chromodomains, and ATPase domain) and AD-HY5 activated the LacZ reporter gene (Figure 4B). To further determine which domain(s) of PKL is required for binding to HY5, a series of deletion constructs was generated (Figure 4A). As shown in Figure 4B, deletion of the ATPase domain (D3) or of both the ATPase domain and the chromodomain (D4) abolished the PKL-HY5 interaction. Intriguingly, the ATPase domain (D6) was necessary and sufficient for the interaction with HY5 and also with HYH, the close homolog of HY5 (see Supplemental Figure 5 online). However, a point mutation (Lys changed to Ala) of an evolutionarily conserved amino acid, Lys-304, which is predicted to bind ATP within the ATPase domain, abolished the interaction (see Supplemental Figure 6 online; Figure 4B). Next, we performed a pull-down assay and found that glutathione S-transferase (GST)-D6 successfully precipitated His-tagged HY5 (Figure 4C). Together, these results demonstrate that PKL physically interacts with HY5 through the ATPase domain.
Figure 4.
PKL and HY5 Interact with Each Other.
(A) Diagram of the domain structures of PKL and various PKL deletions (D1-6).
(B) Yeast two-hybrid assay between various fragments or mutant forms of PKL shown in (A) fused to the LexA DNA binding domain and AD-tagged HY5 (AD-HY5) or AD alone.
(C) Pull-down assay showing direct interaction between GST-PKL-D5 and His-HY5 fusion proteins in vitro. IB, immunoblotting; IP, immunoprecipitation.
(D) BiFC assay showing that YFPN-PKL and HY5-YFPC interact to form a functional YFP in the nucleus. Bar = 5 μm.
(E) Coimmunoprecipitation assay showing that the PKL antibody could precipitate HY5 in 5-d-old Col wild-type seedlings grown in both white light (1.5 μmol m−2 s−1) and darkness.
(F) Seedling phenotypes of hy5, epp1-1, and epp1 hy5 mutants and the Col wild type after exposure to red (R), far-red (FR), blue light (BL), or darkness (Dk) for 5 d. The seedlings are arranged in identical order in each panel. Bars = 2 mm.
(G) Quantification of hypocotyl length of the wild-type and mutant seedlings shown in (F). Data represent the mean ± sd of 30 seedlings.
To confirm the PKL-HY5 interaction in vivo, we performed a bimolecular fluorescence complementation (BiFC) assay in Arabidopsis protoplasts and found that coexpression of PKL-YFPN (fused with the N-terminal half of yellow fluorescent protein) and HY5-YFPC (fused with C-terminal half of YFP) reconstituted a functional YFP in the nucleus, whereas the controls did not (Figure 4D). Furthermore, coimmunoprecipitation analysis showed that PKL was able to precipitate HY5 in Col seedlings grown in either light or dark conditions but not in the epp1-1 mutant (Figure 4E). Collectively, these results indicate that PKL constitutively interacts with HY5 in the nucleus, regardless of the type of light treatment.
To study the functional relationship between PKL and HY5, we constructed an epp1 hy5 double mutant by crossing epp1-1 with hy5-215 (Oyama et al., 1997). The epp1 hy5 seedlings displayed intermediate hypocotyl elongation in all light conditions and were indistinguishable from the Col wild type in red light (Figures 4F and 4G), suggesting that PKL and HY5 antagonistically regulate hypocotyl growth.
PKL Negatively Regulates HY5 by Repressing H3K27me3 on Target Loci
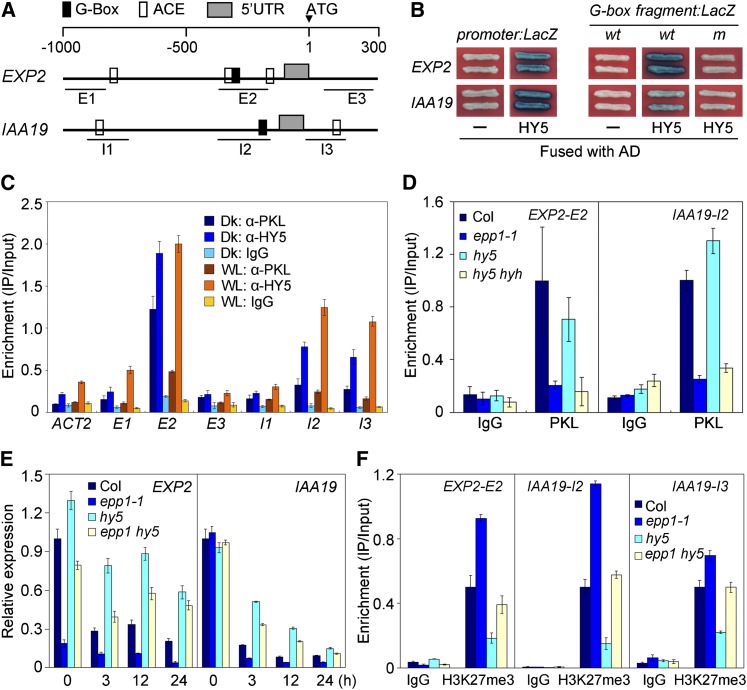
A previous study reported that HY5 directly binds target genes with ACGT-containing elements or a G-box (CACGTG) in their promoter sequences (Lee et al., 2007). Promoter analysis revealed that EXT3, EXP2, XTH17, XTR6, DWF4, and IAA19 contain a putative G-box or ACGT-containing elements within their regulatory regions and coding regions close to translational start site (Figure 5A; see Supplemental Figure 3A online). ChIP-qPCR assay showed that these genes were highly enriched in DNA samples precipitated with HY5 antibody (see Supplemental Figure 3B online; Figure 5B), suggesting that, similar to PKL, HY5 associates with the DNA sequences of these cell elongation–related genes. In this study, we selected EXP2 and IAA19 as examples for further detailed molecular analysis.
Figure 5.
PKL Negatively Regulates HY5 by Inhibiting H3K27me3 of Target Loci.
(A) Diagram of promoter structures of IAA19 and EXP2. ACE, ACGT-containing cis-element; UTR, untranslated region. E1 to E3 and I1 to I3 indicate fragments for ChIP-qPCR amplification.
(B) Yeast one-hybrid assay showing that AD-HY5 binds to the promoter regions of IAA19 and EXP2 via the G-box. wt and m indicate wild-type and mutant forms of the G-box–containing fragments, respectively. “–” means empty AD fusion.
(C) ChIP assay showing the relative enrichment of IAA19 (I1 to I3) and EXP2 (E1 to E3) genomic fragments upon precipitation with PKL or HY5 antibodies in light- or dark-grown seedlings. Dk, dark; WL, white light.
(D) ChIP assay showing that binding of PKL to the regulatory regions of IAA19 and EXP2 was compromised in the hy5 hyh double mutant, as in the epp1-1 mutant. DNA samples were pulled down with PKL antibody or the IgG sera control.
(E) Relative gene expression of EXP2 and IAA19 in hy5, epp1, and epp1 hy5 mutants and the Col wild type following transition from darkness to light for the indicated number of hours.
(F) ChIP assay with DNA isolated from Col, epp1-1, hy5, and epp1 hy5 seedlings using anti-H3K27me3 antibody or the IgG control. In (C) to (F), data represent the mean ± sd of triplicates.
Yeast one-hybrid assays showed that AD-HY5 fusion proteins, but not AD alone, bound to the promoters (1.0 and 1.7 kb upstream from the ATG start site, respectively) or to a 39-bp fragment containing the G-box motif of EXP2 and IAA19 (wild type) and strongly activated the expression of the LacZ reporter gene (Figure 5B). However, point mutations in the G-box (m, CACGTG changed to TTTTTG) of both genes abolished LacZ activation (Figure 5B). These observations indicate that HY5 specifically binds to the regulatory regions of IAA19 and EXP2 via the G-box motif.
ChIP-qPCR was performed to investigate the in vivo binding of IAA19 and EXP2 to HY5 and PKL in light- and dark-grown Col seedlings. Relative to the IgG serum, HY5 antibody markedly pulled down the G-box–containing fragments of EXP2 (E2) and IAA19 (I2) in both darkness and light and even bound to the coding sequence of IAA19 (I3) but did not pull down the actin2 negative control (Figure 5C). Intriguingly, these pulled-down fragments were also drastically enriched in samples precipitated by antibodies against PKL. In addition, there was a slight increase in samples pulled down by the HY5 antibody in seedlings grown in light versus darkness, and the opposite was true for PKL (Figure 5C). We further found that the ability of PKL to bind to target loci was compromised in the hy5 hyh double mutant, as it was in epp1-1, but not in the hy5 single mutant (Figure 5D). These results together suggest that PKL and HY5 (and HYH) likely associate with the cell elongation–related gene loci at the same time during light-mediated seedling development. The recruitment of PKL requires the presence of both HY5 and HYH, consistent with their redundant function in regulating hypocotyl growth (Holm et al., 2002).
The interaction between PKL and HY5 as well as their association with the target chromatin prompted us to analyze how EXP2 and IAA19 expression was coregulated by these proteins. As shown in Figure 5E, the expression of EXP2 and IAA19 was dramatically decreased after the transition from darkness to light. Remarkably, EXP2 and IAA19 expression was stronger in hy5 but weaker in epp1 compared with the Col wild type. Loss of HY5 activity caused a substantial induction of EXP2 and IAA19 expression in the epp1 mutant background (Figure 5E). Thus, PKL and HY5 act antagonistically to regulate downstream gene expression.
This antagonistic regulation led us to further investigate the levels of H3K27me3 on IAA19 and EXP2 loci. ChIP-qPCR assays using the H3K27me3 antibody were performed in hy5 and/or epp1 mutants and also in the Col wild type. Mutation of PKL caused a remarkable increase in H3K27me3 levels at critical IAA19 and EXP2 regulatory regions, as was observed for other cell elongation–related genes (Figure 3D). However, H3K27me3 levels were reduced by more than 50% in the hy5-215 mutant compared with the wild type (Figure 5F). Strikingly, in the epp1 hy5 double mutant, similar amounts of DNA were pulled down by H3K27me3 antibody as in the wild type (Figure 5F). These data suggest that PKL antagonizes with HY5 in regulating the level of H3K27me3 at the cell elongation gene loci, whereas HY5 promoters H3K27me3 at these loci, correlating with their antagonistic role in modulating gene expression. Consistent with the expression pattern of EXP2 and IAA19, the H3K27me3 repressive mark was markedly recruited to these loci in light conditions but was low in the dark (see Supplemental Figure 7A online). It is noteworthy that the transcript level of EXP2, but not of IAA19, was drastically inhibited in epp1-1 compared with the wild type in darkness (Figure 5E). Accordingly, a ChIP assay showed that relatively more H3K27me3 marks were deposited on EXP2 locus, but not on IAA19, in dark-grown epp1-1 than those were in the wild type (see Supplemental Figure 7B online).
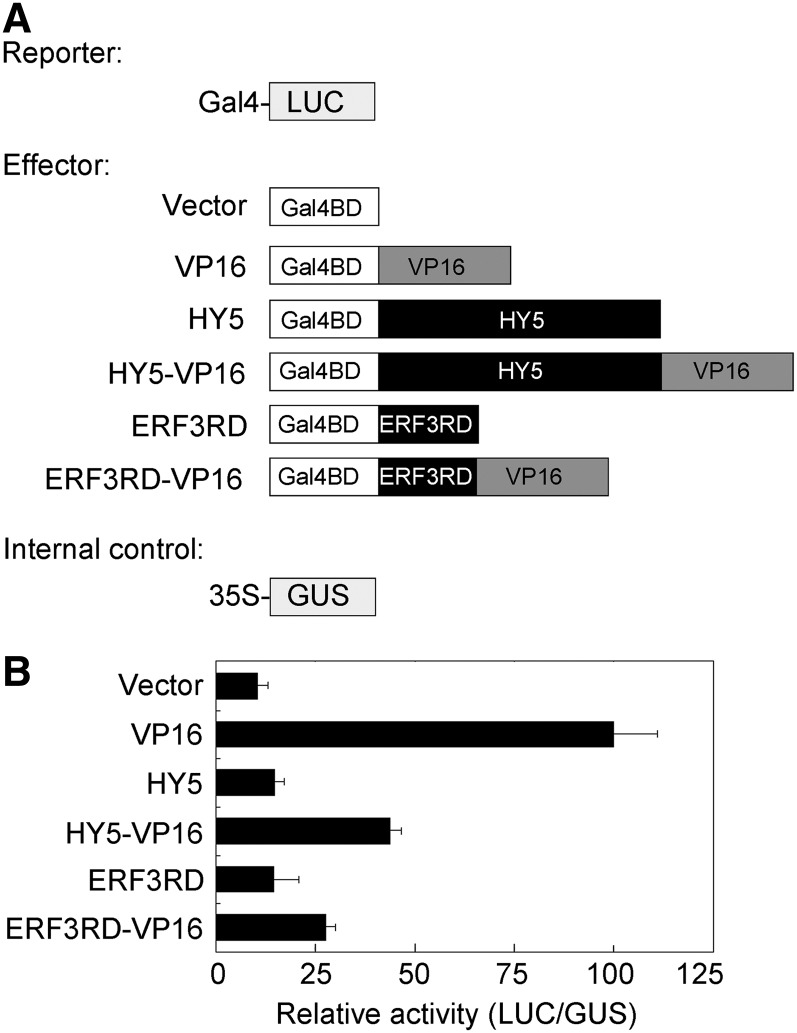
HY5 Possesses Transcriptional Repression Activity
The fact that HY5 directly binds to the promoters of EXP2 and IAA19 and that it represses their expression prompted us to investigate whether HY5 has transcriptional repression activity. Li et al. (2010) failed to detect any transcriptional repression activity of HY5 in yeast and plant cells. We thus fused HY5 with the VP16 activation domain and tested the activity in a transient luciferase (LUC) expression system (Figure 6A). As shown in Figure 6B, BD-HY5 (fused with GAL4 DNA binding domain) did not affect transcription of the ProGAL4:LUC reporter gene. BD-VP16 strongly activated the LUC reporter gene; however, this activity was drastically inhibited by HY5 (BD-HY5-VP16), in a manner similar to the function of the repression domain of ERF3 (Ohta et al., 2001). Therefore, this result suggests that HY5 indeed has intrinsic transcriptional repression activity in vivo.
Figure 6.
HY5 Has Transcriptional Repression Activity.
(A) Diagram of various constructs used in this assay.
(B) Plasmid combinations of LUC reporter, GUS internal control, and effectors were cotransformed into Arabidopsis protoplasts. The protoplasts were incubated in weak light for 16 h, and relative activity was expressed as the ratio of LUC to GUS activity. Data represent the mean ± sd of three biological replicates.
DISCUSSION
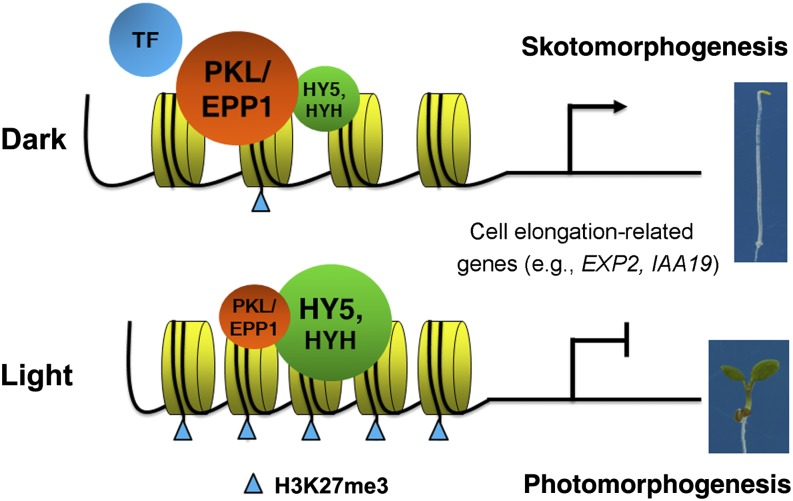
Extensive studies have established a light signaling framework composed of photoreceptors, key repressors (e.g., COP1), and downstream transcription regulators (Lau and Deng, 2010). Our study identifies the CHD3 chromatin remodeling factor PKL/EPP1 as an important repressor of light signaling. Although both COP1 and PKL/EPP1 interact with HY5 and regulate HY5 activity to repress photomorphogenesis, their underlying molecular mechanisms are distinct. COP1 is known to be an E3 ubiquitin ligase that targets positive regulators of photomorphogenesis, such as HY5, HYH, LONG AFTER FAR-RED LIGHT1, and LONG HYPOCOTYL IN FAR RED1, for degradation at the posttranslational level (Lau and Deng, 2012). However, PKL/EPP1 acts as an ATP-dependent chromatin remodeling factor that alters the pattern of histone marks on HY5 target genes involved in cell elongation. The additive phenotype of the epp1 cop1 double mutant supports the notion that PKL/EPP1 and COP1 have additive effects. Using the molecular and biochemical evidence presented in this study, we therefore propose a model in which HY5 (and HYH) constitutively binds to the proximal regulatory regions of cell elongation–related genes and recruits PKL/EPP1 through direct interaction. PKL/EPP1 in turn represses the H3K27me3 repressive mark, resulting in the activation of target genes and the promotion of hypocotyl elongation in the dark. Light stabilizes HY5 and decreases the transcript and protein levels of PKL/EPP1, leading to the recruitment of more H3K27me3 mark at cell elongation loci and the reduction of hypocotyl elongation (Figure 7).
Figure 7.
A Proposed Model of the Action of PKL/EPP1 and HY5 in Regulating Arabidopsis Hypocotyl Growth.
In darkness, HY5 (and HYH) is largely degraded by the 26S proteasome–mediated pathway, whereas PKL/EPP1 is strongly expressed in the hypocotyl. A small pool of HY5 binds to the proximal promoter of cell elongation–related genes, including IAA19 and EXP2, which enables the recruitment of the chromatin remodeling factor PKL/EPP1 to these target loci through physical interaction. This largely prevents H3K27me3 formation and thereby activates cell elongation genes, leading to the promotion of hypocotyl growth. Other transcription activator(s) (indicated as TF) might also be involved in the activation of gene expression. Light triggers the stabilization of HY5 but reduces the level of PKL/EPP1. This allows the recruitment of more H3K27me3 marks on histones at cell elongation–related loci, leading to their repression and the inhibition of hypocotyl elongation.
The biochemical functions of HY5 and PKL/EPP1 are opposite; whereas HY5 promotes H3K27me3 deposition on cell elongation target loci and represses their gene expression, PKL represses this mark on cell elongation target loci and activates gene expression. Surprisingly, full-length PKL exhibited transcriptional repression activity in a protoplast transient expression system (see Supplemental Figure 8 online). The other transcription activator(s) might be required to induce the expression of target genes. Since HY5 is a strong, positive master regulator of the light signaling pathway, the recruitment of PKL/EPP1 might dampen HY5 activity and thereby prevent inadvertent entry into photomorphogenesis, providing an additional regulatory mechanism to fine-tune the pathway at the epigenetic level in response to environmental light intensity. Therefore, compared with the strong and pleiotropic effects of the cop1 mutation, the phenotype of epp1/pkl plants is mild under dark condition. In support of this model, the H3K27me3 repressive mark is strongly associated with EXP2 and IAA19 in light conditions, but only weakly in the dark (see Supplemental Figure 7A online). Furthermore, more than 30% of genes that exhibited significantly decreased transcript levels in pkl plants were enriched for H3K27me3 (Zhang et al., 2012). In addition, the protein-coding genes exhibited higher levels of H3K27me3 after the dark-to-light transition than in darkness (Charron et al., 2009). Our study provides strong evidence that chromatin remodeling factor–mediated histone methylation plays a crucial role in determining cell elongation and plant growth.
Phylogenetic analysis revealed that PICKLE RELATED1 (PKR1), PKR2, and CHROMATIN REMODELING5 (CHR5) are close homologs of PKL (Shaked et al., 2006). The mild phenotype of epp1/pkl mutants may also be caused by redundancy with PKL homologs. Mutants lacking PKR1, PKR2, or CHR5 were indistinguishable from the wild type in terms of hypocotyl elongation and cotyledon opening. Strikingly, the pkl pkr1, pkl pkr2, and pkl chr5 double mutants exhibited slightly but repeatable shorter hypocotyls than the pkl single mutant in both light and darkness, suggesting that PKR1, PKR2, and CHR5 may play redundant roles with PKL in regulating hypocotyl growth (see Supplemental Figure 9 online). However, the cotyledon phenotype of pkl was not enhanced in these double mutants. Previous studies also documented that PKL and PKR2 redundantly regulate root cell identity and root growth (Aichinger et al., 2009, 2011). Further studies using higher order mutants are needed to elucidate the redundant functions of PKL and its homologs.
Although HY5 functions as a master transcription factor that acts downstream in the photomorphogenic pathway and the hy5 null mutant displays elongated hypocotyls under light conditions, the cellular mechanism by which HY5 mediates hypocotyl growth remains unknown (Oyama et al., 1997; Lau and Deng, 2010). This study uncovers a direct link between HY5-mediated repression of cell elongation genes and hypocotyl growth in the light. This notion is further supported by the observation that HY5 possesses intrinsic transcriptional repression activity and that loss of HY5 reduces H3K27me3 at the cell elongation genes whose expression is inhibited by light. Conversely, numerous photosynthesis-related light-induced HY5 targets are marked by histone acetylation, the active mark, during seedling deetiolation (Guo et al., 2008; Charron et al., 2009). Thus, histone modifications are essential for differentially determining the transcriptional activity of HY5 targets.
Chromatin remodeling plays an important role in specifying gene expression states and patterns. Compared with extensive research in animals, only a small body of evidence has documented the role of chromatin-remodeling factors in regulating plant growth and development (Clapier and Cairns, 2009; Jarillo et al., 2009). For instance, the SWI2/SNF2 ATP-dependent chromatin-remodeling enzyme BRM plays crucial roles in vegetative, embryonic, and reproductive plant development by repressing specific target genes (Farrona et al., 2004, 2011; Kwon et al., 2006; Tang et al., 2008). SYD is required for shoot apical meristem maintenance, flower patterning, and biotic stress signaling (Kwon et al., 2005, 2006; Walley et al., 2008; Wu et al., 2012). Previous studies on PKL mostly focused on its involvement in regulating embryonic identity and root meristem activity (Ogas et al., 1999; Henderson et al., 2004; Li et al., 2005; Fukaki et al., 2006; Perruc et al., 2007; Aichinger et al., 2011). However, its role in the hypocotyl/shoot remains obscure. Here, we demonstrate that PKL/EPP1 participates in the repression of photomorphogenesis by directly promoting hypocotyl cell elongation, thus elucidating an important function for CHD chromatin-remodeling enzymes in regulating cell elongation that does not involve cell differentiation and proliferation (Hall and Georgel, 2007; Ho and Crabtree, 2010). Consistent with this, a genome-wide analysis revealed that genes exhibiting hypocotyl-specific expression are overrepresented among the genes that display decreased levels in the pkl mutant (Zhang et al., 2012). Although PKL targets H3K27me3-enriched loci, such as LEAFY COTYLEDON1 (LEC1), LEC2, and PHERES1, and acts as a transcriptional corepressor to inhibit their gene expression during germination (Zhang et al., 2008, 2012), our accumulated data strongly suggest that this protein represses H3K27me3 modification and activates the expression of target genes involved in cell elongation.
Consistently, studies also showed that PKL reduces the level of H3K27me3 on root stem cell and meristem marker genes and promotes their expression (Aichinger et al., 2009, 2011). Thus, PKL either promotes or represses H3K27me3 and acts as a transcriptional corepressor or coactivator, respectively. These opposing roles of PKL might be determined by sequence-specific transcription factors that it associates with and their targets and/or by the recruitment of different histone modification enzymes that catalyze methylation or demethylation. PKL function is probably also affected by endogenous and exogenous cues associated with particular tissues and developmental processes. It would be of great interest to uncover the underlying mechanisms for chromatin remodeling factors that mediate a specific biological function. The PHD superfamily of proteins is proposed to function as readers of histone methylation (Liu et al., 2010). In agreement with this, a rice (Oryza sativa) CHD3 protein, CHR729, can interact with H3K27me3 and H3K4me2 via its PHD finger and chromodomains, respectively (Hu et al., 2012). PKL likely acts as a reader that recognizes histone marks.
Our study points to an example of a member of plant CHD-type chromatin remodeling factors, PKL/EPP1, directly interacting with a key transcription factor, HY5, to control cell growth. Although HY5 is largely degraded in darkness, a small pool of less active phosphorylated HY5 might be responsible for interacting with PKL/EPP1 (Hardtke et al., 2000). The SNF2-related helicase/ATPase domain of PKL/EPP1 is necessary and sufficient for the interaction with HY5, suggesting that this domain mediates protein–protein interactions besides its function in ATP hydrolysis. Interestingly, full-length PKL/EPP1 did not interact with HY5, and the N-terminal fraction (D1) interacted less strongly with HY5 than did the D5 and D6 fragments in the yeast two-hybrid assay (Figure 4B). This is likely because the yeast two-hybrid assay is a transcription-based method, and full-length PKL may repress the transcriptional activity in yeast cells. Similar interactions have been observed for animal CHD3 chromatin remodelers. For instance, Drosophila melanogaster Mi-2 physically interacts with the transcription factor dDREF to regulate cell development (Hirose et al., 2002). A recent study also documented that SYD and BRM interact with LEAFY and SEPALLATA3 transcription factors to regulate floral organ identity (Wu et al., 2012). It is thus speculated that chromatin remodeling factors might determine the target and regulatory specificity by interacting with specific transcription factors in a spatial and temporal manner, underlining the fundamental importance of chromatin configuration during development. Consistent with this, PKL/EPP1 expression is repressed by light specifically in the hypocotyl regions in a photoreceptor-dependent manner and negatively correlates with the extent of photomorphogenesis. Similarly, HY5 abundance peaks in early seedling development and is positively correlated with photomorphogenesis (Hardtke et al., 2000; Osterlund et al., 2000). Both components act together for the fine regulation of downstream genes in response to light. Future studies should investigate how photoreceptors inhibit PKL/EPP1 expression in the hypocotyls. It is possible that PKL modulates transcriptional activities in a range of processes (e.g., embryo and root meristem development) by interacting with other transcription factors as well.
The Polycomb-repressive Complex2 (PRC2) catalyzes the trimethylation of H3K27 and acts as a global repressor of transcription (Schubert et al., 2006). Since the level of H3K27me3 on the regulatory regions of several cell elongation–related genes is enhanced by the epp1 mutation, it is tempting to speculate that PKL antagonizes the function of PRC2 components to repress H3K27me3 and mediate light signaling. This possibility is supported by the antagonistic regulation of root meristem activity and floral organ identity by CURLY LEAF (a PRC2 component) and PKL or SYD, respectively (Aichinger et al., 2011; Wu et al., 2012). Besides preventing methylation, PKL could also promote the demethylation of genes involved in cell elongation and thereby activate them. Future studies are required to distinguish between these possibilities. Additionally, PKL homologs, Mi-2b (CHD4) and CHD3, belong to the integral subunits of the Mi-2/NuRD (for nucleosome-remodeling and histone deacetylase) complex that functions in transcription regulation (Denslow and Wade, 2007). Arabidopsis histone deacetylase HD1 is closely related to HDAC1 and HDAC2, two NuRD components in human. A previous study provided evidence that HD1 is required for histone H3 and H4 Lys deacetylation in several light-responsive genes and that the hd1 mutant displayed hyposensitivity to all light treatments, just like the epp1 mutant (Benhamed et al., 2006). These observations argue for a possible relationship between HD1 and PKL/EPP1 in the regulation of histone modification and light signaling, although PKL is unlikely to be a member of the plant equivalent NuRD complex (Ho et al., 2012). Nevertheless, the identification of more factors involved in chromatin modification will enable us to better understand the chromatin remodeling–mediated epigenetic control of plant plasticity in response to ambient light conditions.
METHODS
Plant Materials and Growth Conditions
The epp1-1 mutant was originally isolated from activation-tagging T-DNA insertion stocks (Weigel et al., 2000). pkl-1 (Ogas et al., 1999), epp1-2 (SAIL_73_H08), epp1-3 (Salk_033554C), exp2-1 (Salk_137972C), dwf4-11 (SAIL_713_F05), phyA-211 (Reed et al., 1994), phyB-9 (Reed et al., 1993), cry1-304 (Mockler et al., 1999), hy5-215 (Oyama et al., 1997), hyh (CS849765), cop1-4 (McNellis et al., 1994), pkr1-1, pkr2-2 (Aichinger et al., 2009), chr5-1 (Salk_087282), and ProPKL:PKL-4Myc (Fukaki et al., 2006) are of the Col ecotype. The T-DNA mutants from the Arabidopsis Biological Resource Center were confirmed by PCR genotyping and sequencing. Double mutants were generated by genetic crossing and were verified by phenotype inspection, antibiotic selection, PCR genotyping, and/or sequencing. After sterilization, seeds were sown onto Murashige and Skoog (MS) medium containing 1% Suc and 0.8% agar and were incubated at 4°C in darkness for 3 d, followed by various light treatments. Far-red (12 μmol m−2 s−1), red (10 μmol m−2 s−1), and blue (14 μmol m−2 s−1) light were supplied by light-emitting diode light sources, or the light intensities were otherwise indicated. White light (60 μmol m−2 s−1) was supplied by cool white fluorescent lamps.
Phenotypic Analysis
After growth under appropriate light conditions, the seedlings were transferred to MS plates and were photographed with a digital camera (Olympus). Cell length and number were determined by inspection under a microscope. The hypocotyl length, cotyledon angle, and cell length were measured using NIH Image J software (http://rsbweb.nih.gov/ij/).
Anthocyanin and Chlorophyll Measurements
Anthocyanin and chlorophyll measurements were conducted as previously described (Lin et al., 2007). Anthocyanin content was expressed as (A530 − 0.25 × A657) per 100 seedlings. Chlorophyll content was expressed as micrograms per 100 seedlings. All experiments were performed in triplicate.
Inverse PCR Cloning
The T-DNA insertion site in epp1-1 was identified by inverse PCR analysis. Genomic DNA was extracted from the epp1-1 seedlings and digested with BamHI or SpeI. After self-ligation with T4 DNA ligase, PCR was performed to amplify the T-DNA flanking sequence using T3 and pSKI-RB primers. The resulting PCR product was cloned into the pEASY vector (TransGen) and subjected to DNA sequencing with the T3 primer.
Plasmid Construction
To obtain the open reading frame (ORF), first-strand cDNA was reverse transcribed using oligo(dT)18 primer from total RNA extracted from Col wild-type seedlings. The ORFs of PKL and HY5 were amplified using high fidelity Pfu DNA polymerase (Invitrogen) and cloned into the pEASY vector, resulting in pEASY-PKL and pEASY-HY5, respectively. To facilitate the follow-up cloning, two restriction sites within the PKL ORF, MfeI and XhoI, were mutagenized without changing the amino acid sequence by site-directed mutagenesis (Takara). The resulting plasmid was designated pEASY-PKLm. To generate a series of PKL deletions, D1, D2, D3, D4, D5, and D6 fragments, as shown in Figure 4A, were amplified using the pEASY-PKLm plasmid as template and the corresponding primer pairs and cloned into the pEASY vector, resulting in pEASY-D1/D2/D3/D4/D5/D6, respectively. To generate a point mutation derivative within the D6 fragment, D6K304A, site-directed mutagenesis was performed using pEASY-D6 as template. The resulting vector was named pEASY-D6K304A.
To construct vectors for the yeast two-hybrid assay, the pEASY-PKLm/D1/D2/D3/D4/D5/D6/D6K304A plasmids were cut with MfeI and XhoI, and the corresponding fragments were ligated into EcoRI/XhoI-digested pEG202 vector (Clontech) to give rise to pLexA-PKL/D1/D2/D3/D4/D5/D6/D6K304A, respectively. The HY5 fragment was released from EcoRI/XhoI-cut pEASY-HY5 and ligated into the pJG4-5 vector digested with the same enzymes to generate pAD-HY5.
To prepare constructs for the BiFC assay, the fragment liberated from MfeI/XhoI-cut pEASY-PKLm was cloned into the pUC-SPYNE vector (Walter et al., 2004) digested with EcoRI and XhoI to generate pYFPN-PKL. pEASY-HY5 plasmid was cut with EcoRI and XhoI, and the HY5 fragment was cloned into the pUC-SPYCE vector digested with EcoRI and XhoI to give rise to pYFPC-HY5.
To generate bacterial expression vectors, the pEASY-D6 plasmid was digested with MfeI and XhoI, and the D6 fragment was inserted into the pGEX-5X-1 vector (GE Healthcare) digested with EcoRI and XhoI, resulting in pGEX-D6. pEASY-HY5 plasmids were cut with EcoRI and XhoI, and the HY5 ORF was ligated into the pET-28a vector (Novagen) digested with the same enzymes to generate pHis-HY5. To make the construct for generating PKL antibody, a DNA fragment corresponding to amino acids 1178 to 1384 was amplified and cloned into pEASY, resulting in pEASY-PKL-CT. pEASY-PKL-CT was then digested with BamHI and XhoI, and the PKL-CT fragment was inserted into pET-28a cut with BamHI and XhoI to give rise to pHis-PKL-CT.
To prepare constructs for the yeast one-hybrid assay, 1.0- and 1.7-kb fragments of the EXP2 and IAA19 promoters, respectively, were PCR amplified from Col genomic DNA and ligated into the pEASY vector to give rise to pEASY-EXP2p and pEASY-IAA19p, respectively. The EXP2 and IAA19 promoter fragments were released from pEASY-EXP2p (cut with EcoRI/XhoI) and pEASY-IAA19p (cut with MfeI/XhoI) and inserted into pLacZi2μ (Lin et al., 2007) to generate ProEXP2:LacZ and ProIAA19:LacZ, respectively. To construct LacZ reporter genes driven by the subfragments of the EXP2 and IAA19 promoters (including the wild-type and mutant form of the G-box), oligonucleotides were synthesized as two complementary oligo primers with an EcoRI site overhang at the 5′-end and an XhoI site overhang at the 3′-end. The oligo primers were annealed and the double-stranded oligonucleotides were ligated into the EcoRI-XhoI sites of pLacZi2μ to produce EXP2wt:LacZ, EXP2m:LacZ, IAA19wt:LacZ, and IAA19m:LacZ, respectively.
To generate constructs for the transient expression assay, a fragment encoding the ERF3 repression domain (ERF3RD) was amplified from Col DNA, and a fragment encoding the VP16 activation domain was amplified from pGEM-VP16A (Lin et al., 2007). These genes were cloned into pEASY to generate pEASY-ERF3RD and pEASY-VP16, respectively. The GAL4 DNA binding domain was amplified from the pGBKT7 vector (Clontech). The fragment was then digested with NcoI and EcoRI and ligated into the pSAT vector (Tzfira et al., 2005) cut with the same enzymes, resulting in pSAT-GAL4BD. The HY5 and ERF3RD fragments were released from pEASY-HY5 and pEASY-ERF3RD digested with EcoRI and XhoI and inserted into the EcoRI-SalI sites of pSAT-GAL4BD to generate pGAL4DB-HY5 and pGAL4DB-ERF3RD, respectively. pEASY-VP16 was cut with BamHI and SalI to release VP16, which was then inserted into the BamHI-SalI sites of pSAT-GAL4BD, resulting in pGAD4BD-VP16. pEASY-HY5 and pEASY-ERF3RD plasmids were cut with EcoRI and XhoI, and the fragments were then inserted into EcoRI-SalI–cut pGAD4BD-VP16 to generate pGAD4BD-HY5-VP16 and pGAD4BD-ERF3RD-VP16, respectively. VP16 was amplified and digested with KpnI and BamHI, and the fragment was inserted into pSAT-GAL4BD cut with the same enzymes to generate pGAL4BD-VP16-2. pEASY-PKLm was cut with MfeI and XhoI and the PKL fragment was cloned into EcoRI-SalI–cut pSAT-GAL4BD and pGAL4BD-VP16-2, resulting in pGAL4DB-PKL and pGAL4DB-PKL-VP16, respectively.
To construct ProPKL:GUS, a fragment spanning the region 2 kb upstream of the ATG start site of the PKL coding sequence was amplified by PCR and cloned into pEASY, resulting in pEASY-PKLp. pEASY-PKLp plasmid was digested with SalI and BamHI to release the PKL promoter, which was then ligated into the pRI101-AN vector (Takara) digested with the same enzymes to generate ProPKL:GUS. The primers used for cloning are listed in Supplemental Table 1 online. All amplified fragments were validated by sequencing.
The binary constructs were electroporated into Agrobacterium tumefaciens strain GV3101 and then introduced into destination plants via the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS plates in the presence of 50 mg/L kanamycin.
Gene Expression Analysis
Seedling growth conditions are described in the text. Plant total RNA was isolated by RNA extraction kit (Tiangen), and the first-strand cDNA was synthesized by reverse transcriptase (Invitrogen). Real-time PCR was performed with the SYBR Premix ExTaq kit (Takara) following the manufacturer’s instructions. Primers are listed in Supplemental Table 1 online. Three biological replicates were performed for each sample and the expression level was normalized to that of a UBQ control.
GUS Histochemical Assay
Seedlings of the ProPKL:GUS transgenic line were harvested and incubated overnight in 0.1 M sodium phosphate buffer containing 50 mM K3Fe(CN)6, 50 mM K4Fe(CN)6, and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide at 37°C. GUS expression was examined under a dissecting microscope (Olympus), and images were captured by a digital camera (Olympus).
LUC Activity Assay
The LUC activity assay was performed as previously reported (Tang et al., 2012). LUC reporter activity was detected with a luminescence kit using the LUC assay substrate (Promega). Relative reporter gene expression levels are expressed as the ratio of LUC to GUS.
Purification of Recombinant Protein and Antibody Preparation
GST, GST-D6, His-PKL-CT, and His-HY5 recombinant fusion proteins were induced by isopropyl β-d-1-thiogalactopyranoside and expressed in the Escherichia coli BL21 (DE3) strain. The proteins were then purified using Glutathione Sepharose 4B beads (GE Healthcare; for GST fusions) or Ni-NTA agarose (Qiagen; for His fusions) following the manufacturer’s instructions. His-PKL-CT and His-HY5 were then used to raise PKL and HY5 polyclonal antibodies in rabbits, respectively. For tubulin antibody, a synthetic peptide (EEVGAEGGDDEDDEGEEY) derived from amino acids 433 to 450 of tubulin 4 was conjugated with KLH (Cali-Bio) and used for injecting rabbit.
ChIP Assays
ChIP assays were performed as described previously (Tang et al., 2012). Chromatin complexes were incubated with anti-PKL, anti-HY5, anti-H3 (Millipore 07-690), or anti-H3K27me3 (Millipore 07-449) polyclonal antibodies or IgG serum. The precipitated DNA fragments were recovered and quantified by quantitative PCR with the primers shown in Supplemental Table 1 online. Relative enrichment is expressed as the ratio of DNA amount after immunoprecipitation to that in input.
Pull-Down and Coimmunoprecipitation
The procedures used for pull-down, coimmunoprecipitation, and immunoblot assays were described previously (Tang et al., 2012). For the pull-down assay, ∼2 µg of purified recombinant bait protein (GST-D6 or GST) and 2 µg of prey protein (His-HY5) were incubated in binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.6% Triton X-100) for 2 h at 4°C. Glutathione Sepharose 4B beads (Roche) were added and reactions were incubated for 1 h. After washing with binding buffer, precipitated proteins were eluted in 2× SDS loading buffer. The proteins were then size fractionated on a 10% SDS-PAGE gel and immunoblotted with anti-His or anti-GST antibodies (Abcam ab19256). For the coimmunoprecipitation assay, seedlings were grown in the light for 4 d. Total proteins were extracted with extraction buffer and incubated with 2 μg anti-PKL antibody for 2 to 3 h at 4°C. Fifty microliters of protein G-Sepharose beads (Roche) was added and the reaction was incubated for a further 2 to 3 h. The beads were washed three times with coimmunoprecipitation buffer, and the precipitated proteins were eluted in 2× SDS loading buffer by boiling for 10 min. The proteins were separated on a 6 or 8% SDS-PAGE gel and detected by immunoblotting with anti-PKL and anti-HY5 antibodies.
BiFC Assays
For BiFC assays, plasmids of N- and C-terminal fusions of YFP were cotransformed into Arabidopsis thaliana protoplasts as previously described (Walter et al., 2004). The protoplasts were incubated under weak light for 12 to 16 h before observation. The YFP fluorescence was determined using a confocal microscope (Olympus).
Yeast Interaction Assays
Yeast one-hybrid and two-hybrid assays were performed according to the Yeast Protocols Handbook (Clontech). Briefly, for the yeast one-hybrid assay, the AD fusion constructs were cotransformed with various LacZ reporter plasmids into yeast strain EGY48. Transformants were grown on SD/-Trp-Ura dropout plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) for blue color development. For yeast two-hybrid analysis, the AD fusion constructs were transformed into the strain Ym4271, and LexA-fusion plasmids were cotransformed with a LexAop:LacZ (Clontech) reporter into the strain EGY48. After mating, transformants were grown on SD/-Trp-Ura-His dropout plates with X-gal for blue color development.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: PKL/EPP1 (At2g25170), HY5 (At5g11260), HYH (At3g17609), EXP2 (At5g05290), IAA19 (At3g15540), DWF4 (At3g50660), EXT3 (At1g21310), XTH17 (At1g65310), XTR6 (At4g25810), CHS (At5g13930), CAB2 (At1g29920), PKR1 (At5g44800), PKR2 (At4g31900), CHR5 (At2g13370), ERF3 (AT1G50640), UBQ1 (At3g52590), ACT2 (At3g18780), and TUA4 (At1g04820). Mutants investigated in this study are listed in Methods.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of epp1 Mutants.
Supplemental Figure 2. Genetic Relationship between PKL/EPP1 and the Photoreceptors and COP1.
Supplemental Figure 3. ChIP-qPCR Assay Using HY5 Antibody.
Supplemental Figure 4. Photomorphogenic Phenotype of the exp2 and dwf4 Loss-of-Function Mutants.
Supplemental Figure 5. HYH Interacts with PKL in Yeast Cells.
Supplemental Figure 6. Amino Acid Alignment of Truncated ATP Binding/Helicase Domains of CHD3/CHD4.
Supplemental Figure 7. ChIP Assay Using H3K27me3 Antibody of Samples Grown under Light and Dark Conditions.
Supplemental Figure 8. Transient Transcriptional Activity Assay of PKL.
Supplemental Figure 9. Phenotype of pkr1, pkr2, and chr5 Single Mutants and Their Double Mutants with pkl.
Supplemental Table 1. List of Primers Used in This Study.
Acknowledgments
We thank Claudia Köhler for providing the pkl, pkl pkr1, and pkl pkr2 mutants, Hidehiro Fukaki for providing the ProPKL:PKL-4Myc plasmid, and the Arabidopsis Biological Resource Center for providing the T-DNA mutants. This work was supported by grants from the National Science Foundation of China (31000530) to Y.J. and the State Basic Research Development Program (2009CB118500), the National Science Foundation of China (31170221), and the Chinese Academy of Sciences to R.L.
AUTHOR CONTRIBUTIONS
Y.J. and R.L. designed the research. Y.J., D.Z., X.W., W.T., W.W., J.H., G.X., D.C., and Y.L. performed the research. Y.J. and R.L. analyzed the data and wrote the article.
Glossary
- H3K27me3
H3 Lys 27
- Col
Columbia
- qRT-PCR
quantitative real-time PCR with reverse transcription
- GUS
β-glucuronidase
- ChIP-qPCR
chromatin immunoprecipitation with quantitative PCR
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- MS
Murashige and Skoog
- ORF
open reading frame
- GST
glutathione S-transferase
References
- Aichinger E., Villar C.B.R., Di Mambro R., Sabatini S., Köhler C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E., Villar C.B.R., Farrona S., Reyes J.C., Hennig L., Köhler C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto G., Formiggini F., Laflamme P., Malakhov M., Bowler C. (2002). The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 12: 1529–1534 [DOI] [PubMed] [Google Scholar]
- Bertrand C., Benhamed M., Li Y.F., Ayadi M., Lemonnier G., Renou J.P., Delarue M., Zhou D.X. (2005). Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 280: 1465–1473 [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Greenham K., Castillejo C., Sartor R., Bialy A., Sun T.-P., Estelle M. (2012). Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J.-B., He H., Elling A.A., Deng X.W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chory J. (2010). Light signal transduction: An infinite spectrum of possibilities. Plant J. 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua Y.L., Watson L.A., Gray J.C. (2003). The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Denslow S.A., Wade P.A. (2007). The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438 [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J.L., Reyes J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975 [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., March-Díaz R., Schmitz R.J., Florencio F.J., Turck F., Amasino R.M., Reyes J.C. (2011). Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS ONE 6: e17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A.J., Franklin K.A. (2011). Chromatin remodelling in plant light signalling. Physiol. Plant. 142: 305–313 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Taniguchi N., Tasaka M. (2006). PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 48: 380–389 [DOI] [PubMed] [Google Scholar]
- Guo L., Zhou J., Elling A.A., Charron J.B., Deng X.W. (2008). Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 147: 2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.A., Georgel P.T. (2007). CHD proteins: A diverse family with strong ties. Biochem. Cell Biol. 85: 463–476 [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D.C., Crabtree G.R. (2011). ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J.T., Li H.-C., Rider S.D., Mordhorst A.P., Romero-Severson J., Cheng J.-C., Robey J., Sung Z.R., de Vries S.C., Ogas J. (2004). PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134: 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Jang I.-C., Chua N.-H. (2009). Regulated proteolysis in light-related signaling pathways. Curr. Opin. Plant Biol. 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Hirose F., Ohshima N., Kwon E.-J., Yoshida H., Yamaguchi M. (2002). Drosophila Mi-2 negatively regulates dDREF by inhibiting its DNA-binding activity. Mol. Cell. Biol. 22: 5182–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho K.K., Zhang H., Golden B.L., Ogas J. (November 2, 2012). PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim. Biophys. Acta http://dx.doi.org/. 10.1016/j.bbagrm.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Crabtree G.R. (2010). Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Ma L.-G., Qu L.-J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Liu D., Zhong X., Zhang C., Zhang Q., Zhou D.-X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. USA 109: 5773–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L., Farrona S., Reyes J.C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62: 291–304 [DOI] [PubMed] [Google Scholar]
- Jang I.-C., Chung P.J., Hemmes H., Jung C., Chua N.-H. (2011). Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell 23: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo J.A., Piñeiro M., Cubas P., Martínez-Zapater J.M. (2009). Chromatin remodeling in plant development. Int. J. Dev. Biol. 53: 1581–1596 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Chen C., Wagner D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.S., Hibara K., Pfluger J., Bezhani S., Metha H., Aida M., Tasaka M., Wagner D. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.-C., Chuang K., Henderson J.T., Rider S.D., Jr, Bai Y., Zhang H., Fountain M., Gerber J., Ogas J. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.-H., Zhang H., Shen Y., Wang H., Deng X.W. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Ma L., Sun N., Liu X., Jiao Y., Zhao H., Deng X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T.C., Guo H., Yang H., Duong H., Lin C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y., Wang Z.-Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E., Kinoshita N., Lopez-Molina L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52: 927–936 [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H., Avivi-Ragolsky N., Levy A.A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173: 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., Lin R. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Hou A., Babu M., Nguyen V., Hurtado L., Lu Q., Reyes J.C., Wang A., Keller W.A., Harada J.J., Tsang E.W., Cui Y. (2008). The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 147: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Kumagai S., Muto H., Sato A., Watahiki M.K., Harper R.M., Liscum E., Yamamoto K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and forma-tion of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Tian G.-W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005). pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Von Arnim A., Deng X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 215–243 [DOI] [PubMed] [Google Scholar]
- Walley J.W., Rowe H.C., Xiao Y., Chehab E.W., Kliebenstein D.J., Wagner D., Dehesh K. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4: e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wei N., Deng X.W. (1996). The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112: 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.-F., Sang Y., Bezhani S., Yamaguchi N., Han S.-K., Li Z., Su Y., Slewinski T.L., Wagner D. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bishop B., Ringenberg W., Muir W.M., Ogas J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159: 418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rider S.D., Jr, Henderson J.T., Fountain M., Chuang K., Kandachar V., Simons A., Edenberg H.J., Romero-Severson J., Muir W.M., Ogas J. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283: 22637–22648 [DOI] [PMC free article] [PubMed] [Google Scholar]