HDA18 has deacetylase activity and affects epidermal cell patterning of Arabidopsis roots by regulating the transcription of a set of kinase genes. This set of kinase genes functions in a positional information relay system. Both down- and upregulation of HDA18 expression result in the same phenotype and increase the expression the kinase genes.
Abstract
The differentiation of hair (H) and non-hair (N) cells in the Arabidopsis thaliana root epidermis is dependent on positional relationships with underlying cortical cells. We previously found that histone acetylation relays positional information and that a mutant altered in the histone deacetylase gene family member HISTONE DEACETYLASE 18 (HDA18) exhibits altered H and N epidermal cell patterning. Here, we report that HDA18 has in vitro histone deacetylase activity and that both mutation and overexpression of HDA18 led to cells at the N position having H fate. The HDA18 protein physically interacted with histones related to a specific group of kinase genes, which are demonstrated in this study to be components of a positional information relay system. Both down- and upregulation of HDA18 increased transcription of the targeted kinase genes. Interestingly, the acetylation levels of histone 3 lysine 9 (H3K9), histone 3 lysine 14 (H3K14) and histone 3 lysine 18 (H3K18) at the kinase genes were differentially affected by down- or upregulation of HDA18, which explains why the transcription levels of the four HDA18-target kinase genes increased in all lines with altered HDA18 expression. Our results reveal the surprisingly complex mechanism by which HDA18 affects cellular patterning in Arabidopsis root epidermis.
INTRODUCTION
How cells are arranged in a particular spatial order is fundamental to the development of multicellular organisms. The single-layered Arabidopsis thaliana root epidermis consists of two types of cells, hair (H) and non-hair (N) cells, in an arrangement that depends on their spatial relationships with underlying cortical cells. Cells residing over the intercellular spaces between cortical cells (the H position) differentiate into H cells, while those directly over cortical cells (the N position) differentiate into N cells (Dolan et al., 1994). This well-characterized position-dependent cellular differentiation serves as a model system for understanding pattern formation in plants (Schiefelbein et al., 2009). Position-dependent cellular differentiation is known to be regulated by a hierarchical regulatory system consisting of at least three levels. The first level is a GL2-centered transcription factor network (TFN) that includes transcription factor genes such as GLABRA2 (GL2), WEREWOLF (WER), CAPRICE/TRIPTYCHON/ENHANCER OF TRY AND CPC1 (CPC/TRY/ETC1), TRANSPARENT TESTAGLABRA (TTG), and GLABRA3/ENHANCER OF GLABRA3 (GL3/EGL3). Referred to as pattern genes, these factors determine the fates of cells located at the N and H positions in root epidermis. Additional players in this fate specification include MYB23 (Tominaga et al., 2007; Kang et al., 2009; Schiefelbein et al., 2009), GL2 EXPRESSION MODULATOR (GEM) (Caro et al., 2007), and ADENOSINE DIMETHYL TRANSFERASE 1A (DIM1A) (Wieckowski and Schiefelbein, 2012). The second level of regulation comprises the plasma membrane–localized receptor-like kinases, such as SCRAMBLED (SCM) and BRASSINOSTEROID INSENSITIVE 1 (BRI1), that sense the N and H positions (Kwak et al., 2005; Kwak and Schiefelbein, 2007, 2008; Kuppusamy et al., 2009). The third level concerns the proper differentiation of cortical cells, where positional information presumably originates, in which JACKDAW (JKD) is involved (Hassan et al., 2010). However, some key questions remain unanswered, including how positional information sensed in the plasma membrane is transduced to the nucleus where pattern genes function.
Previously, we found that the application of trichostatin A (TSA), a specific inhibitor of histone deacetylase (HDAC), alters cellular patterning in the Arabidopsis root epidermis (i.e., confers H fate to cells at the N position) by affecting the expression of pattern genes (Xu et al., 2005). This finding suggests that histone acetylation is involved in relaying positional information during cellular patterning. Further supporting this, cells at the N position are also converted to H fate in a mutant containing defective HDA18 (Xu et al., 2005). Arabidopsis HDA18 contains an HDAC domain and was therefore annotated as a member of the HDAC family (Figure 1A; see Supplemental Figure 1 online; Pandey et al., 2002). Although HDA18 is closely grouped with HDA5 and HDA15 in Class II of the RPD3/HDA1 subfamily (Pandey et al., 2002), it differs in function from HDA5 and HDA15, as no abnormal cellular patterning in the root epidermis was observed in mutants for the latter two genes (Xu et al., 2005). Alinsug et al. (2009) reported the presence of multiple conserved domains in the HDA18 protein with various predicted biological functions, based on similar domains in yeast and mammalian cells (Yang and Seto, 2008). However, none of these functions have been experimentally verified in HDA18.
Figure 1.
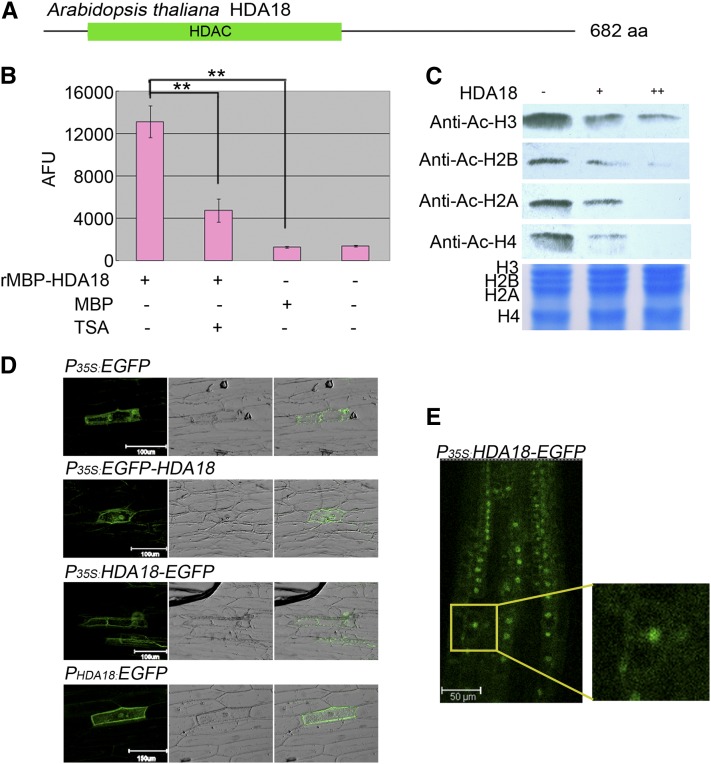
HDA18 Protein Has HDAC Activity and Localizes to Both the Nucleus and Cytoplasm.
(A) Schematic of HDA18 protein consisting of 682 amino acids with a conserved HDAC domain (green bar). aa, amino acids.
(B) In vitro HDAC activity assay with fluorescent substrate using a recombinant protein consisting of the HDAC domain of HDA18 fused to a maltose binding protein (MBP) tag. The HDAC activity of the recombinant protein was inhibited by TSA, an HDAC-specific inhibitor. Data show mean ± sd from four independent experiments (**P < 0.01; Student’s t test). AFU, arbitrary fluorescence units.
(C) In-gel assay revealing that the HDAC domain of HDA18 has similar deacetylation activity on the four core histones.
(D) Transient expression of HDA18 protein in onion epidermis revealed that HDA18 protein localizes to both the nucleus and cytoplasm (left column, dark field with florescence of GFP; middle column, bright field; right column, merged).
(E) Observation of EGFP localization in a T2 transgenic Arabidopsis line. The small image at the right is an enlargement of the squared area in the left image.
Here, we report that HDA18 encodes a protein with HDAC activity. Either reducing or increasing the expression of HDA18 resulted in conversion of cells at the N position to H fate. Unexpectedly, we found that the HDA18 protein did not directly bind to and regulate the transcription of pattern genes, but instead it directly bound to and affected the transcriptional regulation of other types of genes, including several kinase genes. These data demonstrate that HDA18 affects cellular patterning by regulating the transcription of a group of kinase genes through histone acetylation.
RESULTS
HDA18 Encodes a Protein with HDAC Activity
HDA18 is located on chromosome 5. The genomic sequence contains 3202 bp, and the coding sequence contains 2049 bp, including eight exons. Analysis of data at AREX (Birnbaum et al., 2003; Brady et al., 2007; http://www.arexdb.org) indicated that HDA18 is expressed at low levels, with no tissue preference. These results are consistent with previous in situ hybridization observations (Xu et al., 2005). Because of the similarity between the hda18 mutant phenotype and the effects of the HDAC inhibitor TSA on cellular patterning of the root epidermis, we tested recombinant HDA18 protein for HDAC activity. The recombinant protein had significant HDAC activity, which was concentration dependent and inhibited by TSA (Figures 1B and 1C; for detailed data, see Supplemental Figure 1 online). An in-gel assay, using hyperacetylated histones extracted from HeLa cells as substrates and commercially available histone-specific (but site nonspecific) antibodies as indicators, revealed that the recombinant protein deacetylated all four core histones (Figure 1C), suggesting that the HDAC domain of HDA18 does not have preferential activity on any of the core histones.
Transient expression of enhanced green fluorescent protein (EGFP)–labeled HDA18 (see Supplemental Figure 2 online) in onion (Allium cepa) epidermis revealed that the HDA18 protein was localized to both the nucleus and cytoplasm (Figure 1D), which is consistent with bioinformatic predictions (Alinsug et al., 2009). This result was further verified by observation of EGFP localization in stable transgenic Arabidopsis lines containing the same construct for EGFP-labeled HDA18 (Figure 1E; see Supplemental Figure 2 online).
Both Down- and Upregulation of HDA18 Convert Some N Cells to H Fate
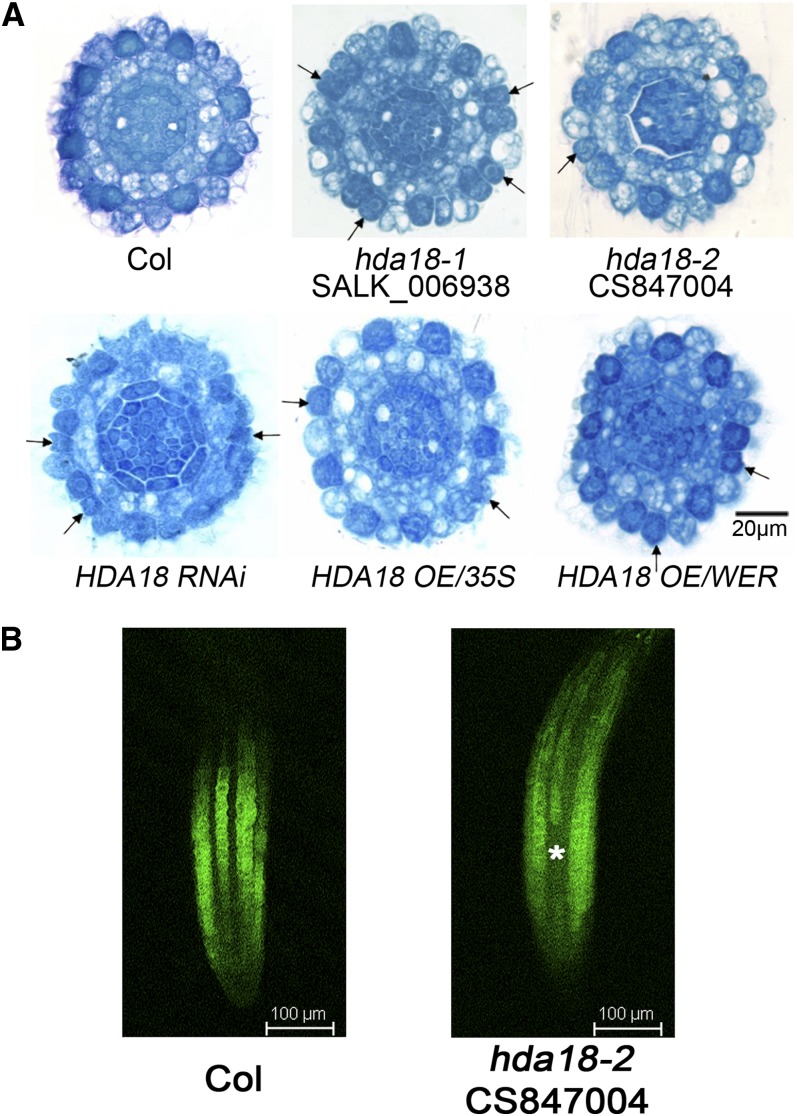
To understand further the role of HDA18 in cellular patterning, we analyzed the phenotypes of a T-DNA insertion mutant (hda18-2), an RNA interference (RNAi) line (HDA18 RNAi), an overexpression (OE) line driven by the 35S promoter P35S:HDA18 (HDA18 OE/35S), and an OE line driven by the promoter of the WEREWOLF gene PWER:HDA18-GUS (HDA18 OE/WER; see Supplemental Figure 2 online). The hda18-2 mutant and HDA18 RNAi line exhibited conversion of some cells at N positions to H fate, similar to the previously described hda18-1 mutant line (Salk_006938) (Xu et al., 2005). Unexpectedly, however, the two HDA18 OE lines also displayed this phenotype (Figure 2A). The number of cells at N positions that were converted to H fate significantly increased in both mutants and overexpressing lines based on statistics analysis compared with wild-type plants (see Supplemental Table 1 online). Introduction of an N cell–specific reporter, the GL2 promoter driving green fluorescent protein (GFP) expression in the hda18-2 mutant, enabled us to identify cell fate patterns (Figure 2B, asterisk; see Supplemental Figure 3 online), further indicating the conversion of cells at the N position to H fate.
Figure 2.
Altered HDA18 Expression Confers H Fate to Some Cells at the N Position.
(A) Cross sections of root tips of the wild type (Columbia [Col], top row, left), mutants (top row, middle and right), RNAi (bottom row, left), and OE lines (bottom row, middle and right) at the ends of the root caps, showing cells with H fate at N positions (arrows). The regular pattern of epidermal cells in the wild type (Col) is that cells reside over the intercellular space between cortical cells (H position) differentiating to H cells were darkly stained, while those directly over cortical cells (N position) differentiating to N cells were less stained.
(B) PGL2:GFP expression in roots of mutant (hda18-2) and the wild type (Col). Since GL2 is specifically expressed in N cells, and root cells form continuous files, the florescence of GFP generated by PGL2:GFP presents as continuous files longitudinally (Col). An asterisk indicates the cells at N positions converted to H fate (no GFP signal detected).
We attempted to rescue the mutant phenotype by complementation. However, possibly because of the low native expression level of HDA18, all transgenic lines were found to overexpress the gene and thus exhibited the corresponding phenotype (see Supplemental Figure 4 and Supplemental Table 1 online).
To test whether the effect of HDA18 could be enhanced by HDA5 and HDA15, single mutants that do not have altered cellular patterning in the root epidermis (Xu et al., 2005), we introduced artificial microRNA constructs targeting HDA5 and HDA15 into the hda18-2 mutant. We also introduced artificial microRNA constructs targeting HDA5 and HDA18 into the hda15 mutant (Salk_004027). No enhanced phenotypes were observed in transgenic lines in which all three targeted genes were downregulated (see Supplemental Figure 4 online). These results suggest that the role of HDA18 in the production of the mutant phenotype is independent of HDA5 and HDA15, its two most closely related members of the Class II RPD3/HDA1 subfamily (Alinsug et al., 2009).
HDA18 Does Not Affect Cellular Patterning through Direct Regulation of Pattern Genes
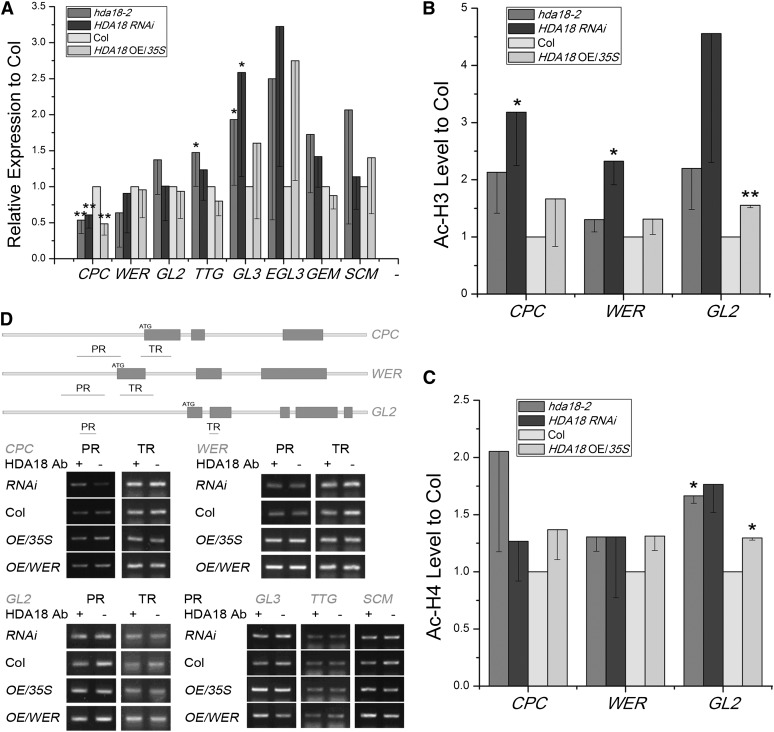
To test whether altered HDA18 expression affected the GL2-centered TFN, we examined the expression of five representative pattern genes (CPC, WER, GL2, TTG, and GL3) and related genes (EGL3, GEM, and SCM) in hda18-2, HDA18 RNAi, and HDA18 OE/35S. We did not further analyze hda18-1 or HDA18 OE/WER lines at the molecular level because the former is a weak allele and the latter has a similar phenotype to the HDA18 OE/35S line, with no obvious differential expression of HDA18 in the H and N position (see Supplemental Figure 2 online). We found that the expression of CPC was significantly downregulated in all three lines (Figure 3A). Expressions of TTG and GL3 were upregulated in the mutant line, while those of WER GL2, EGL3, GEM, and SCM were not changed significantly (Figure 3A). These results reveal that the pattern of gene expression was affected differentially by altered levels of HDA18 expression.
Figure 3.
Representative Pattern Gene Expression Levels and Histone Acetylation Status of CPC, WER, and GL2 in Lines with Altered HDA18 Expression.
(A) Expression levels of pattern genes were altered in root tips of hda18-2 mutant, RNAi, and OE lines. Data show mean ± sd from four to seven independent real-time PCR experiments, each with three replicates (*P < 0.05 and **P < 0.01; Student’s t test). Col, Columbia.
(B) and (C) Histone acetylation status of H3 and H4, examined by ChIP-PCR with AcH3 and AcH4 antibodies, at CPC, WER, and GL2 sequences in root tips of hda18-2 mutant, RNAi, and OE lines. Data show mean ± sd from three independent experiments, each with three replicates (*P < 0.05 and **P < 0.01; Student’s t test).
(D) HDA18 protein does not bind pattern genes based on ChIP-PCR analysis of regions shown in orange. PR, promoter region; TR, transcribed region.
We previously determined that the histone acetylation status of pattern genes was altered upon treatment with the HDAC inhibitor TSA (Xu et al., 2005), and we confirmed here that HDA18 has in vitro HDAC activity. We therefore examined whether HDA18 expression affects the histone acetylation status of pattern genes. Using the previously identified DNA fragments that displayed altered levels of histone acetylation in response to TSA treatment (Xu et al., 2005), we found that upon down- or upregulation of HDA18 expression, the acetylation levels of H3 and H4 increased to some extent in three representative pattern genes, CPC, WER, and GL2. The increases in acetylation of H3 at CPC and WER were statistically significant (P < 0.05) in the HDA18 RNAi line, while those of H3 and H4 at GL2 were statistically significant (P < 0.05) in hda18-2 and HDA18 OE/35S lines (Figures 3B and 3C). These data reveal that although the downregulation of HDA18 indeed increased histone acetylation, consistent with the previously observed TSA effects, the increased histone acetylation status of the HDA18 OE/35S line could not be directly explained by HDA18 functioning as an HDAC.
To clarify further if HDA18 is involved in the transcriptional regulation of pattern genes, we examined whether the HDA18 protein directly binds to chromatin, thus altering pattern gene–related histone acetylation, using an antibody against HDA18 (see Supplemental Figure 5 online). Surprisingly, none of the DNA fragments tested were enriched in this chromatin immunoprecipitation (ChIP) experiment (Figure 3D). Because the primers used in the ChIP-PCR assay covered only limited areas of chromatin, we performed a ChIP-chip screen to search for possible binding sites at the genome level (see Supplemental Figure 5 online). Among the 286 enriched DNA fragments (representing putative HDA18 protein binding sites), none was related to any of the pattern genes (see Supplemental Data Set 1 online).
Taken together, we conclude that although altered HDA18 expression affects the cellular patterning of the root epidermis, this effect does not appear to be due to the direct transcriptional regulation of pattern genes.
Identification of HDA18 Target Genes
As HDA18 did not directly regulate the expression of pattern genes, we explored how altered expression of HDA18 was linked to cellular patterning. We searched among the 286 DNA fragments identified in the HDA18 ChIP-chip experiment for genes with annotated functions potentially related to cellular patterning of the root epidermis and found that 13 of these fragments encoded kinases, in addition to nine encoding transcription factors, eight encoding chromatin modification factors, four functioning in root development, and one encoding a phosphatase (see Supplemental Data Set 1 online). These data suggest that HDA18 can directly bind to genes, as is common for HDAC proteins, and may affect cellular patterning via these target genes.
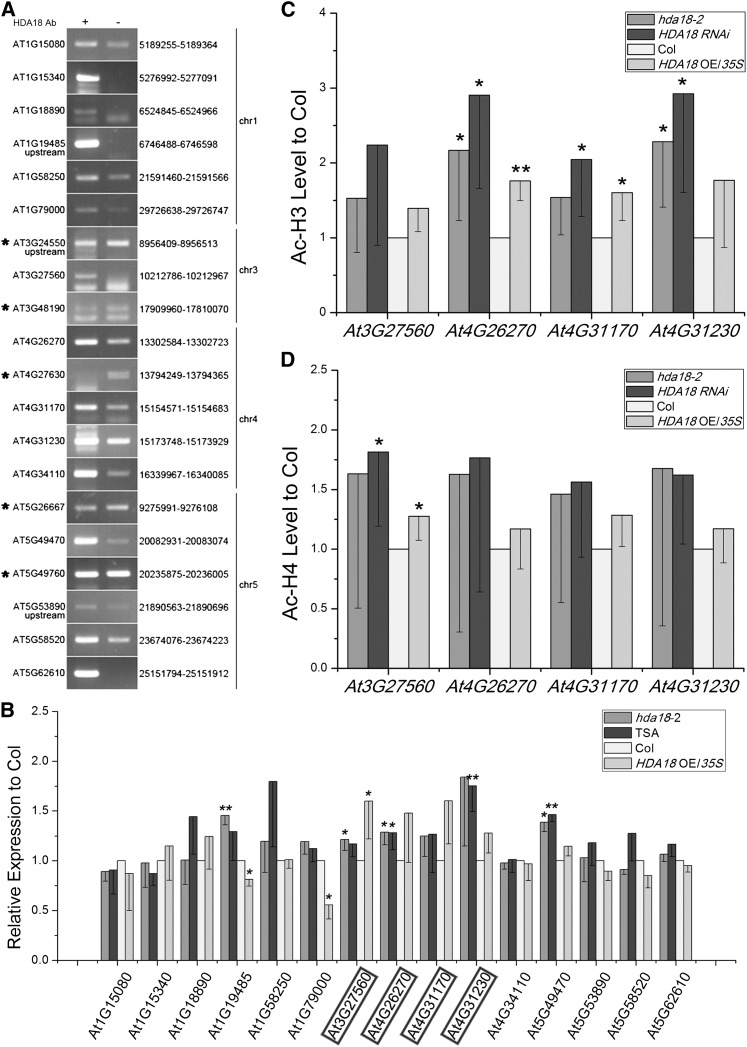
Because both SCM and BRI1 are plasma membrane–localized receptor-like kinases, we reasoned that there must be some downstream components that transduce the positional information sensed by the receptors. The preferential enrichment of kinase genes in the HDA18 ChIP-chip data suggested that HDA18 might function in the relay of positional information by regulating the transcription of these kinase genes. To test this possibility, we first verified that HDA18 binds to the 20 candidate DNA fragments, including all 13 putative kinase genes, by performing ChIP-PCR. We found that 15 candidates were enriched, whereas five were not (Figure 4A).
Figure 4.
HDA18 Protein Binds and Affects Transcription of Certain Kinase Genes by Changing Their Histone Acetylation Status.
(A) ChIP-PCR verification of binding of HDA18 protein to putative targets identified by ChIP-chip. The genes marked with asterisks were false positives. The numbers listed to the right indicate the sites identified in the ChIP-chip analysis (for detailed information, see Supplemental Data Set 1 online).
(B) Expression levels of HDA18-targeted genes in root tips with altered HDA18 expression, showing upregulation of four kinase genes (highlighted with frames) regardless of increased or decreased expression of HDA18. Data show mean ± sd from three to four independent experiments, each with three replicates (*P < 0.05 and **P < 0.01; Student’s t test). Col, Columbia.
(C) and (D) Histone acetylation status of H3 and H4 correspond to altered HDA18 expression, examined by ChIP-PCR with AcH3 and AcH4 antibodies at the identified binding sites of selected kinase genes. Data show mean ± sd from five independent experiments, each with three replicates (*P < 0.05 and **P < 0.01; Student’s t test).
Both Down- and Upregulation of HDA18 Increase Expression of Some Targeted Kinase Genes
To examine whether HDA18 is involved in the transcriptional regulation of the targeted genes, we examined the expression of the verified HDA18-bound genes in hda18-2, HDA18 OE/35S, and TSA-treated wild-type plants. Among the 15 genes that were tested, two showed changes in expression that were correlated with HDA18 expression level (At1G19485 and At1G79000), five showed upregulation to various extents regardless of HDA18 expression levels in all lines (At3G27560, At4G26270, At4G31170, At4G31230, and At5G49470), and the remaining eight showed no significant changes in their expression levels that could be correlated with HDA18 expression (Figure 4B).
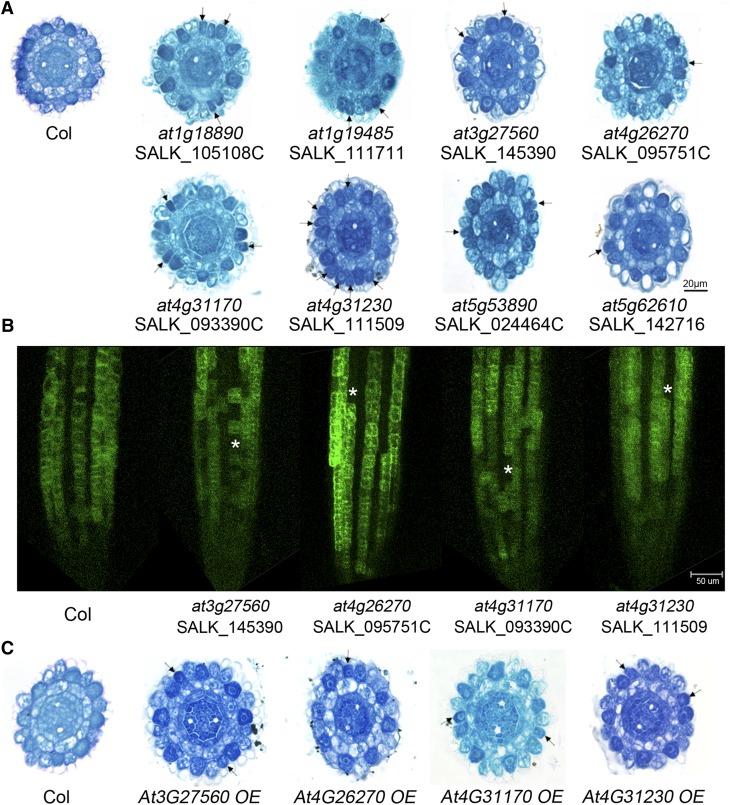
We then analyzed the mutant phenotypes of all 15 HDA18-targeted genes (see Supplemental Figure 6 online). We found that mutations in eight of the 15 genes led to the conversion of some cells at the N position to H fate (Figure 5A; see Supplemental Tables 2 and 3 online). Among these eight genes, six were kinases, one was a WD-40 gene, and one was a basic HELIX-LOOP-HELIX (bHLH) gene (Figure 5A; see Supplemental Data Set 1 online). Mutations in At5G49470 and At5G58520, both bound by HDA18 and annotated as kinase genes, did not induce cell fate conversion phenotypes in the root (see Supplemental Data Set 1 and Supplemental Table 3 online), indicating that these HDA18-targeted genes may either not be involved in the patterning process or may be involved in a redundant manner.
Figure 5.
Identification of Kinase Genes as Components Involved in Cellular Patterning of the Root Epidermis.
(A) Cross sections of root tips of mutants of verified HDA18 target genes at the ends of root caps, showing cells with H fate at N positions (arrows). Col, Columbia.
(B) PGL2:GFP expression in roots of mutants (at3g27560, at4g26270, at4g31170, and at4g31230) and the wild type (Col). Asterisks represent the cells at N positions converted to H fate (no GFP signal detected).
(C) Cross sections of root tips of OE lines of selected HDA18 target kinase genes at the ends of root caps, showing cells with H fate at N positions (arrows).
Although mutations of At1G18890 and At5G53890 produced the cellular patterning phenotype, the transcription levels of these genes were not significantly affected by that of HDA18. We therefore focused our further investigations on the remaining kinase genes (i.e., At3G27560, At4G26270, At4G31170, and At4G31230). First, we verified the phenotype by introducing the cell type–specific reporter PGL2:GFP into the corresponding mutants. We found that GFP expression patterns were altered in the kinase mutants (Figure 5B; see Supplemental Figure 6 online), consistent with the phenotypes revealed by paraffin sectioning (Figure 5A).
We next examined the histone acetylation status of the four kinase genes in hda18-2, HDA18 RNAi, and HDA18 OE/35S at the verified HDA18 binding sites. The levels of H3 acetylation were significantly increased at At4G26270 in all three lines, at At4G31170 in HDA18 RNAi and HDA18 OE/35S, and at At4G31230 in hda18-2 and HDA18 RNAi (Figure 4C). H4 acetylation was significantly increased at At3G27560 in HDA18 RNAi and HDA18 OE/35S (Figure 4D).
Taken together, these data indicate that HDA18 binds to and is involved in the transcriptional regulation of some genes, including At3G27560, At4G26270, At4G31170, and At4G31230.
HDA18-Targeted Kinase Genes Mediate the Conversion of Cells at the N Position to H Fate by either Down- or Upregulating mRNA Levels
We next explored how the cell conversion phenotype could result from both the upregulation of target kinase genes (caused by altered HDA18 expression) and the downregulation of these same genes (caused by the loss-of-function mutations). We chose the straightforward approach of constructing OE transgenic lines for these kinase genes to determine whether increased expression of these genes might produce a similar phenotype as loss-of-function mutations. OE lines for each of the four HDA18-targeted kinase genes, At3G27560, At4G26270, At4G31170, and At4G31230 (see Supplemental Figure 6 and Supplemental Table 3 online), indeed caused some cells at the N position to convert to H fate (Figure 5). These results demonstrate that both down- and upregulation of kinase gene expression could result in this phenotype, indicating that homeostasis of expression of the kinase genes is required for cellular patterning in the Arabidopsis root epidermis and implicating these kinase genes as components relaying positional information during this process.
Down- and Upregulation of HDA18 Increase Kinase Gene Transcription by Altering H3 Acetylation at Lysine-9 (Lys-9), Lys-14, and Lys-18 in Distinct Patterns
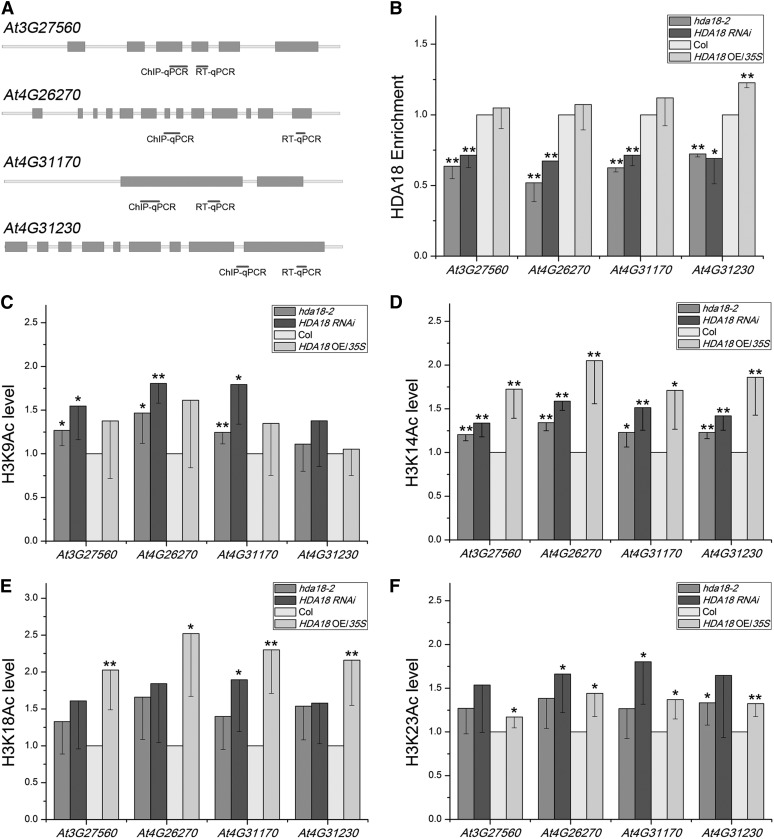
It is intriguing that both the down- and upregulation of HDA18 resulted in the cell conversion phenotype (Figure 2A). To address the underlying mechanisms of this, we examined the efficiency of enrichment of the HDA18 protein on the kinase genes. Figure 6B shows that as expected, the enrichment of HDA18 protein on the DNA of all four kinase genes was significantly reduced in the lines with reduced expression of the HDA18 gene. However, unexpectedly, the HDA18 enrichment level was similar to that of the wild type in the lines with increased HDA18 expression. These observations suggest that the phenotype of the upregulated HDA18 expression lines cannot simply be explained by decreased target binding by the HDA18 protein.
Figure 6.
Effects of HDA18 Levels on the Acetylation of H3K9, H3K14, H3K18, and H3K23 at the Four HDA18-Target Kinase Genes.
(A) Regions selected for ChIP-PCR analyses are indicated with short black bars. qPCR, quantitative PCR.
(B) The enrichment of DNA related to the four kinase genes in ChIP assays using HDA18 protein in the wild type and altered HDA18 expression lines. Data show mean ± sd from three independent experiments, each with three replicates (*P < 0.05 and **P < 0.01; Student’s t test). Col, Columbia.
(C) to (F) The changes in acetylation of the H3 Lys residues at the four kinase genes under down- and upregulated HDA18 expression examined using antibodies specific to H3K9, H3K14, H3K18, and H3K23. Data show mean ± sd from four independent experiments, each with three replicates (*P < 0.05 and **P < 0.01; Student’s t test).
Next, we analyzed the histone acetylation status of specific amino acid residues in the lines with altered HDA18 expression. As indicated in Figure 4, the overall histone acetylation at the kinase genes increased to various levels. As more significantly increased acetylation levels were observed in H3 than in H4, we chose H3 for further analysis. Among the seven known Lys residues available for acetylation in the H3 N-terminal tail, acetylated Lys-9, Lys-14, Lys-18, and Lys-23 are responsible for promoting transcription (Berger, 2007; Chen and Tian, 2007; Choi and Howe, 2009). Therefore, we examined the acetylation levels at Lys-9, Lys-14, Lys-18, and Lys-23 of H3 at the four kinase genes in the hda18-2 mutant, the RNAi line (HDA18 RNAi), and the OE line (HDA18 OE/35S).
Figures 6C to 6F show that although there were some variations, the acetylation levels at the H3 Lys residues changed in similar patterns: Except for At4G31230, the acetylation levels of H3K9 at the kinase genes were significantly enhanced under reduced levels of HDA18 but were not significantly changed with increased HDA18 expression (Figure 6C). The acetylation levels of H3K14 in all four kinase genes were significantly enhanced under both reduced and increased HDA18 levels (Figure 6D). In contrast with H3K9, the acetylation levels of H3K18 in all four kinase genes were significantly enhanced under increased HDA18 levels but not under decreased HDA18 levels, except in At4G31170 (Figure 6E). The H3K23 acetylation levels were all enhanced to some extent whether HDA18 was down- or upregulated (Figure 6F). The above data suggest that the down- and upregulation of HDA18 expression (i.e., the reduced or increased levels of HDA18 protein) affect the acetylation of the four detected H3 Lys residues differentially.
DISCUSSION
HDA18 Protein Has HDAC Activity and Is Involved in Transcriptional Regulation
HDA18 was previously identified as a member of the HDAC gene family based on sequence annotation. Here, we demonstrated that this protein does indeed have HDAC activity. Based on our ChIP-chip, ChIP-PCR, transcription, and mutant phenotype analysis of HDA18-targeted genes, we demonstrated that HDA18 plays a regulatory role in the transcription of its target kinase genes. The role of HDA18 in the regulation of other targeted genes remains to be examined.
In our previous work, we reported the involvement of HDA18 in cellular patterning of the root epidermis (Xu et al., 2005). Here we not only confirmed this observation, but also found that down- and upregulation of HDA18 expression resulted in similar phenotypes. With the observation that HDA18 affects cellular patterning through the function of target kinase genes, not known pattern genes, the results of this study reveal the complexity of the mechanisms for relaying positional information between plasma membrane–located receptor-like kinases, such as SCM and BRI1, and the GL2-centered TFN, consisting of nucleus-localized transcriptional factors. In addition, our findings present opportunities to decipher this complicated signaling network.
Four Kinase Genes Are Involved in Positional Information Signaling during Cellular Patterning of Arabidopsis Root Epidermis
We found that four kinase genes regulated by HDA18 (i.e., At4G26270, At3G27560, At4G31170, and At4G31230) are involved in the relay of positional information for cellular patterning in the root epidermis. It is likely that these kinases are downstream components in transmitting the positional information received by SCM or BRI1. Detailed root gene expression data provided by AREX LITE (Birnbaum et al., 2003; Brady et al., 2007; http://www.arexdb.org) revealed further expression information of the four genes in H and N cells. While the expression level of At4G31230 was quite low, those of the other three genes were at a comparable range to that of SCM (see Supplemental Table 4 online). These data are consistent with the roles of the four HDA18 targeted genes, which affect epidermal cell fate determination.
Information is limited regarding to the functions of these kinase genes. Whereas At4G26270 is annotated as phosphofructokinase 3, and the cytoplasmic localization and tissue preferential expression patterns of this gene have been reported (Mustroph et al., 2007), the other three genes have only annotation information. At3G27560 is annotated as an ATN1-like kinase (Tregear et al., 1996; Heazlewood et al., 2004; Rudrabhatla et al., 2006), At4G31170 has STY-like kinase characteristics (Rudrabhatla et al., 2006), and At4G31230 belongs to the 1.4.1 (receptor-like cytoplasmic kinase IX) family (Kerk et al., 2003).
To understand the relationship between these kinase genes and pattern genes, we examined pattern gene expression in various transgenic lines, with both down- and upregulated kinase genes. The results were complex, and it was difficult to establish a simple causal relationship fitting the current model of the GL2-centered TFN for the expression of kinase genes and pattern genes (see Supplemental Figure 7 online). According to the current model of the GL2-centered TFN, which is well established based on mutant analysis, changes in the expression levels of pattern genes are considered to be key criteria for evaluating the potential involvement of other genes in cellular patterning of the root epidermis (Wieckowski and Schiefelbein, 2012). However, changes in the expression of pattern genes are not absolutely related to cellular patterning phenotypes. For example, pattern gene expression was significantly altered in the scr mutant, but no cellular patterning phenotype was observed (see Supplemental Figure 8 online). To explain the phenotypic changes in cellular patterning that resulted from altered kinase gene expression, three possibilities should be considered: The first is the homeostasis of the GL2-centered TFN, including the differential expression of the genes in H and N cells; the second is the complexity of a multifactor positional information signaling network, in which the kinase genes are integrated in various ways, with varying levels of expression of each gene affecting the operation of the network; and the third is that the kinase genes may influence genes involved in H cell differentiation downstream of the GL2-centered TFN. Clearly, our findings present tremendous challenges as well as opportunities for future investigations.
Complicated Mechanisms Underlying the Regulation of Transcription of the Kinase Genes by HDA18
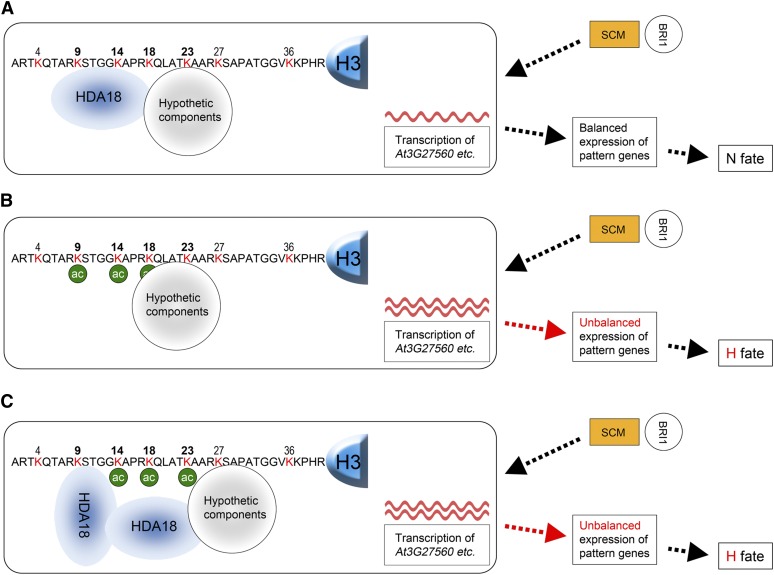
Our findings that the four kinase genes are involved in positional information signaling explain why altered HDA18 expression converts cells at the N position to H fate. However, it is intriguing that down- and upregulated HDA18 expression affect acetylation related to the four kinase genes at H3K9, Lys-14, and Lys-18 differentially. As shown in the proposed model in Figure 7, these site-specific differential changes of acetylation levels can explain why both down- and upregulation of HDA18 result in similar changes of target gene transcription levels, thereby leading to a similar cellular patterning phenotype.
Figure 7.
Model of Possible Mechanism for HDA18 Regulation of Transcription of the Four Kinase Genes.
The HDA18 target kinase genes function between the membrane-localized receptor-like kinases, such as SCM (orange rectangles) and BRI1 (open circles), and the pattern genes in the GL2-centered TFN (open rectangles).
(A) In the wild type, the HDA18 protein, perhaps with other unknown components, binds to the region containing H3K9 through Lys-23 of the H3 related to the four target kinase genes and maintains the acetylation of these residues at the proper levels for balanced transcription of the kinase genes.
(B) In the hda18 mutant, the quantity of HDA18 protein is reduced and the region containing H3K9 through K23 of the H3 related to the four kinase genes cannot be properly covered. The acetylation levels are therefore increased (green circles), which results in increased transcription of the four kinase genes (red waves). The latter disturbs the previously balanced expression of the pattern genes and results in the cell conversion phenotype.
(C) In the HDA18 OE line, surplus HDA18 protein may interfere with the interaction of HDA18 with the unknown components and lead to an abnormal structure, leaving H3K14 and H3K18 unbound by HDA18. This would result in increased acetylation at those sites, which results in increased transcription of the four kinase genes. The latter disturbs the previously balanced expression of pattern genes and results in the cell conversion phenotype similar to that seen in the hda18 mutants.
We hypothesize that HDA18 interacts with H3 associated with the four kinase genes at the region centered at H3K14, along with one or more hypothetical component(s), as a complex. This structure could maintain relatively low acetylation levels of H3K9, Lys-14, and Lys-18 because of the HDAC activity of HDA18 (Figure 7A). In downregulated HDA18 lines, the quantity of HDA18 protein is reduced, as shown in Figure 7B. Without HDA18, H3K9 and Lys-14 would not be deacetylated, which would result in increased acetylation as found in Figures 6C and 6D. The lack of significant enhancement of H3K18 acetylation levels suggests that other components may be involved in H3K18 deacetylation. Alternatively the HDA18-deficient structure may interfere with the efficiency of putative acetyltransferase functions. In upregulated HDA18 lines, the quantity of HDA18 protein is increased, possibly leading to the formation of an abnormal structure (Figure 7C). While HDA18 covers H3K9 in this structure, although perhaps not as efficiently as in the wild type, the additional HDA18 may make the hypothetical complex crowded and leave H3K14 and K18 untouched by HDA18 protein. This scenario is consistent with the enhanced acetylation levels of H3K14 and K18 detected in the upregulated HDA18 line (Figures 6D and 6E). This working model can successfully explain why both down- and upregulation of HDA18 resulted in upregulation of the kinase genes and, therefore, the cell conversion phenotype.
In conclusion, this study revealed the mechanism of HDA18’s role as an HDAC that affects cellular patterning in Arabidopsis root epidermis. We demonstrated the HDAC activity of HDA18, identified its target genes, among which four kinase genes were demonstrated to be components of a positional information relay system, and demonstrated that HDA18 affects cellular patterning by regulating transcription of its target kinase genes through histone acetylation. Furthermore, we solved the puzzle of how both down- and upregulation of HDA18 expression converted N cells to H fate by discovering that under both situations, altered levels of HDA18 led to increased kinase gene transcription by altering H3 acetylation at Lys-9, Lys-14, and Lys-18 in distinct patterns. These findings not only present an opportunity for understanding how positional information is relayed during cellular patterning but also provide a case study for elucidating the underlying mechanism of the phenomenon that up- and downregulation of same gene cause the same phenotype.
METHODS
Plant Materials
All Arabidopsis thaliana kinase mutant seeds were obtained from the ABRC and are listed in Supplemental Table 2 online. SALK mutant line CS847004 is designated as hda18-2 herein, whereas hda18-1 represents the previously described SALK_006938 mutant line (Xu et al., 2005). The PGL2:GFP marker line was described previously (Lin and Schiefelbein, 2001). Plants were grown under the same conditions described previously (Xu et al., 2005).
Generation of Recombinant HDA18 and HDAC Activity Assay
A recombinant HDA18 protein was constructed with the HDAC domain of the HDA18 sequence at the C terminus and a maltose binding protein tag at the N terminus (see Supplemental Figures 5 and 9 online). The in vitro HDAC activity assays were conducted according to Xu et al. (2005) and Rundlett et al. (1996). TSA was purchased from Sigma-Aldrich (catalog number T8552).
Construction of Transgenic Plants and Characterization of Phenotypes
HDA18 OE construct was produced by cloning HDA18 coding sequence into pCAMBIA binary vector, driven by the cauliflower mosaic virus 35S promoter (CaMV35S) (HDA18 OE/35S) or 4-kb WER promoter fragment (Lee and Schiefelbein, 1999; Kwak and Schiefelbein, 2007) (HDA18 OE/WER). HDA18 RNAi silencing was performed by cloning HDA18 coding region 1294 to 2026 into pCAMBIA vector in antisense/sense directions, respectively, to generate small hairpin RNA under the CaMV35S promoter. For HDA18 localization study, the HDA18 coding sequence was placed under the CaMV35S promoter and fused to 5′ and 3′ end of EGFP, respectively, to generate P35S:HDA18-EGFP and P35S:EGFP-HDA18 construct. In addition, the HDA18 promoter region, containing 529 bp 5′ upstream, 1st exon, and 1st intron, was cloned and placed before the EGFP coding region. To rescue HDA18 depletion, 3.8-kb genomic fragment from HDA18 was cloned into pDR vector and then transformed into hda18 (SALK_006938). For kinase OE analysis, kinase coding sequence was cloned into pDR binary vector, driven by the CaMV35S promoter or kinase genomic fragment from At3G27560 was cloned into pDR vector. All the primer pairs for plasmids construction are listed in Supplemental Data Set 2 online.
Binary plasmids were introduced into Agrobacterium tumefaciens stain GV3101 and transformed into plants using the floral dip method (Duan et al., 2008). T1 seeds were harvested and screened by germinating on Murashige and Skoog solid medium with antibiotic or herbicide selection. The resistant seedlings were replanted on wet soil and verified by PCR to detect T-DNA insertion and gene expression. Thermal asymmetric interlaced–PCR was performed to identify T-DNA insertion location in selfed transgenic plants (Liu et al., 1995). To ensure that the phenotypes of the transgenic plants did not result from the interruption of other genes due to T-DNA insertion, we examined the insertion sites of our transgenic plants (see Supplemental Table 5 online). After verification, seeds collected from T1 plants were cultivated and processed to paraffin section phenotype inspection and other experiments.
Transient expression of HDA18 for subcellular localization in onion (Allium cepa) epidermal cells was conducted according to Das et al. (2009) with a Bio-Rad PDS-1000/He biolistic particle delivery system and observed with a Leica TCS-SP confocal microsystem. Introduction of the PGL2:GFP marker to the hda18-2 and mutants of kinase genes was achieved by crossing (see Supplemental Figures 3 and 6 online). Design of artificial microRNAs was according to WMD3-Web MicroRNA Designer (http://wmd3.weigelworld.org) (Schwab et al., 2006). Epidermal cellular patterns were observed according to Xu et al. (2005) for paraffin sectioning and to Kang et al. (2009) for confocal microscopy.
Real-Time PCR Analysis and ChIP
Examination of expression levels in root tips of plants with various genetic backgrounds was conducted with real-time PCR according to Xu et al. (2005). ChIP was also conducted according to Xu et al. (2005). The HDA18 antibody was raised in mouse (for detailed information, see Supplemental Figure 5 and Supplemental Table 6 online). Antibodies used for examining histone acetylation status by ChIP are listed in Supplemental Table 7 online. Primers used are listed in Supplemental Data Set 2 online.
ChIP-on-Chip
DNA fragments enriched by the HDA18 antibody were prepared as above. Hybridization of enriched DNA fragments with the Affymetrix GeneChip Arabidopsis Tiling 1.0R array (catalog number AFFY-900594) was conducted by Beijing Capital Bio Corporation. Microarray data were analyzed by tiling analysis software supplied by Affymetrix. The final processed data are listed in Supplemental Data Set 1 online. All data comply with MIAME and were submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE41991.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.Arabidopsis.org/) or GenBank/EMBL data libraries under the following accession numbers: HDA18 (At5G61070), HDA5 (At5G61060), HDA15 (At3G18520), CPC (At2G46410), WER (At5G14750), GL2 (At1G79840), TTG (At5G24520), GL3 (At5G41315), EGL3 (At1G63650), GEM (At2G22475), SCM (At1G11130), and SCR (At3G54220); accession numbers of HDA18 target genes are list in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. HDA18 Has an HDAC Domain and Therefore Has Concentration-Dependent and TSA-Inhibited Histone Deacetylase Activity.
Supplemental Figure 2. Construction and Analysis of Different HDA18 Transgenic Lines.
Supplemental Figure 3. Introducing the PGL2:GFP Transcriptional Reporters into hda18 (CS847004) Mutant by Genetic Crosses.
Supplemental Figure 4. The Phenotype of the Transgenic Lines in Which the hda18 Mutant Is Rescued Is Similar to HDA18 OE, and the Phenotype of Transgenic Lines Containing Knocked Down HDA5/HDA15/HDA18 Is Similar to That of hda18.
Supplemental Figure 5. ChIP-Chip Assays to Search for DNA Fragments Bound by HDA18.
Supplemental Figure 6. Expression of Kinase Genes Is Reduced in Mutants and Elevated in Overexpression Lines.
Supplemental Figure 7. Kinase Genes Affect Expression of Pattern Genes in Arabidopsis Root Epidermis.
Supplemental Figure 8. SCR Affects Expression of Pattern Genes but Not Cellular Patterning.
Supplemental Figure 9. Purification of rMBP-HDA18 to Perform in Vitro HDAC Activity Assays.
Supplemental Table 1. Root-Hair and Non-Hair Cell Specification in the Root Epidermis of Wild-Type, hda18, RNAi, HDA18 OE, and Rescue Lines.
Supplemental Table 2. Mutant Screening to Test Participation of HDA18 and Its Target Genes in Cellular Patterning of Arabidopsis Root Epidermis.
Supplemental Table 3. Root-Hair and Non-Hair Cell Specification in the Root Epidermis of the Wild Type, Mutants, and Overexpression Lines of Some HDA18 Targeted Genes.
Supplemental Table 4. Gene Expression Information about the HDA18 Targeted Genes Retrieved from AREX LITE (http://www.arexdb.org/).
Supplemental Table 5. Genome Positions of T-DNA Inserts in Transgenic Plants.
Supplemental Table 6. Procedure for Generation of the HDA18 Antibody.
Supplemental Table 7. Antibodies Used in the Experiments.
Supplemental Data Set 1. HDA18 Binding DNA Fragments by ChIP-Chip and ChIP-PCR–Verified DNA Fragments.
Supplemental Data Set 2. Primers Used in the Experiments.
Acknowledgments
We thank John Schiefelbein (University of Michigan) and Philip Benfey (Duke University) for their advice on this project and discussions of the article. We also thank them for kindly providing the marker line used in this study. We thank Martin Hulskamp (University of Koln) for discussions of the article. We thank Xiao-Dong Su, Jian-Guo Ji, and their students Cong Liu and Hai Pu (College of Life Sciences, PKU) for their help in generating and analyzing HDA18 recombinant protein and Li-Jia Qu, Yu-Xian Zhu, and Jin-Dong Zhao of the College of Life Sciences for kindly sharing their equipment. We thank Min-Ping Qian and Ming-Hua Deng and their students Lin Wan and Quan Wang (School of Mathematical Sciences, PKU) for their help in analyzing ChIP-chip data. This work was supported by grants from the National Natural Science Foundation (30393114 and 30570901) and the Ministry of Science and Technology of People's Republic of China (2003CB715906; to S.-N.B.).
AUTHOR CONTRIBUTIONS
C.L., L.-C.L., and S.-N.B. designed the experiments. C.L. and L.-C.L. performed most of the experiments, analyzed the data, and prepared the figures. W.-Q.C. performed some experiments. S.-N.B., X.C., and Z.-H.X. wrote the article. All authors contributed to analyzing the article.
Glossary
- H
hair
- N
non-hair
- TFN
transcription factor network
- TSA
trichostatin A
- HDAC
histone deacetylase
- EGFP
enhanced green fluorescent protein
- RNAi
RNA interference
- OE
overexpression
- GFP
green fluorescent protein
- ChIP
chromatin immunoprecipitation
- CaMV35S
cauliflower mosaic virus 35S promoter
References
- Alinsug M.V., Yu C.W., Wu K. (2009). Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol. 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Caro E., Castellano M.M., Gutierrez C. (2007). A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447: 213–217 [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Tian L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.K., Howe L.J. (2009). Histone acetylation: Truth of consequences? Biochem. Cell Biol. 87: 139–150 [DOI] [PubMed] [Google Scholar]
- Das P., Ito T., Wellmer F., Vernoux T., Dedieu A., Traas J., Meyerowitz E.M. (2009). Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 136: 1605–1611 [DOI] [PubMed] [Google Scholar]
- Dolan L., Duckett C.M., Grierson C., Linstead P., Schneider K., Lawson E., Dean C., Poethig S., Roberts K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Duan Q.H., Wang D.H., Xu Z.H., Bai S.N. (2008). Stamen development in Arabidopsis is arrested by organ-specific overexpression of a cucumber ethylene synthesis gene CsACO2. Planta 228: 537–543 [DOI] [PubMed] [Google Scholar]
- Hassan H., Scheres B., Blilou I. (2010). JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137: 1523–1529 [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J.S., Gout A.M., Day D.A., Whelan J., Millar A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J. (2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D., Bulgrien J., Smith D.W., Gribskov M. (2003). Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol. 131: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K.T., Chen A.Y., Nemhauser J.L. (2009). Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 106: 8073–8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2007). The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev. Biol. 302: 118–131 [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2008). A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr. Biol. 18: 1949–1954 [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Shen R., Schiefelbein J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Lin Y., Schiefelbein J. (2001). Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128: 3697–3705 [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Mustroph A., Sonnewald U., Biemelt S. (2007). Characterisation of the ATP-dependent phosphofructokinase gene family from Arabidopsis thaliana. FEBS Lett. 581: 2401–2410 [DOI] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrabhatla P., Reddy M.M., Rajasekharan R. (2006). Genome-wide analysis and experimentation of plant serine/threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 60: 293–319 [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen A.A., Kobayashi R., Bavykin S., Turner B.M., Grunstein M. (1996). HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93: 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Kwak S.H., Wieckowski Y., Barron C., Bruex A. (2009). The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 60: 1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Okada K., Wada T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19: 2264–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregear J.W., Jouannic S., Schwebel-Dugu N., Kreis M. (1996). An unusual protein kinase displaying characteristics of both the serine/threonine and tyrosine families is encoded by the Arabidopsis thaliana gene ATN1. Plant Sci. 117: 107–119 [Google Scholar]
- Wieckowski Y., Schiefelbein J. (2012). Nuclear ribosome biogenesis mediated by the DIM1A rRNA dimethylase is required for organized root growth and epidermal patterning in Arabidopsis. Plant Cell 24: 2839–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.R., Liu C., Wang Y.L., Li L.C., Chen W.Q., Xu Z.H., Bai S.N. (2005). Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc. Natl. Acad. Sci. USA 102: 14469–14474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Seto E. (2008). The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9: 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]