Pectin and xylan are generally considered as separate cell wall glycan networks distinct from cell wall proteins. This work describes a cell wall proteoglycan with pectin and arabinoxylan covalently attached to an arabinogalactan protein, identifying a cross-linked matrix polysaccharide wall protein architecture with implications for wall structure, function, and synthesis.
Abstract
Plant cell walls are comprised largely of the polysaccharides cellulose, hemicellulose, and pectin, along with ∼10% protein and up to 40% lignin. These wall polymers interact covalently and noncovalently to form the functional cell wall. Characterized cross-links in the wall include covalent linkages between wall glycoprotein extensins between rhamnogalacturonan II monomer domains and between polysaccharides and lignin phenolic residues. Here, we show that two isoforms of a purified Arabidopsis thaliana arabinogalactan protein (AGP) encoded by hydroxyproline-rich glycoprotein family protein gene At3g45230 are covalently attached to wall matrix hemicellulosic and pectic polysaccharides, with rhamnogalacturonan I (RG I)/homogalacturonan linked to the rhamnosyl residue in the arabinogalactan (AG) of the AGP and with arabinoxylan attached to either a rhamnosyl residue in the RG I domain or directly to an arabinosyl residue in the AG glycan domain. The existence of this wall structure, named ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1 (APAP1), is contrary to prevailing cell wall models that depict separate protein, pectin, and hemicellulose polysaccharide networks. The modified sugar composition and increased extractability of pectin and xylan immunoreactive epitopes in apap1 mutant aerial biomass support a role for the APAP1 proteoglycan in plant wall architecture and function.
INTRODUCTION
The plant primary cell wall is a matrix of the polysaccharides cellulose, hemicellulose, and pectin with ∼10% of its mass comprised of enzymes and structural proteins, such as the Hyp-rich glycoproteins (HRGPs), including extensins and arabinogalactan proteins (AGPs) (Burton et al., 2010). The prevailing tethered network model of the cell wall depicts the type I primary cell wall as a xyloglucan tethered cellulose network embedded in an independent pectin gel (reviewed in Albersheim et al., 2011), while Type II primary wall models consist of a xylan-tethered cellulose network and a reduced pectin component. Cell wall components are proposed to interact both covalently and noncovalently to form the functional wall. Noncovalent interactions include ionic bonds between pectic homogalacturonan (HG) domains (Caffall and Mohnen, 2009) and hydrogen bonds between cellulose chains and between cellulose and regions of xyloglucan or xylan (Pauly et al., 1999). In addition to noncovalent interactions, covalent bonds appear to cross-link at least some of the wall polymers into a durable and rigid structure that can withstand extreme turgor pressure. The full extent of these covalent cross-links is not known, but they include Tyr-based linkages between the family of wall HRGPs known as extensins (Fry, 1982; Held et al., 2004); borate esters between pectin rhamnogalacturonan II monomers (Ishii et al., 1999); and ester linkages between polysaccharides and phenolic moieties of lignin (Ishii, 1997; Ishii and Hiroi, 1990a, 1990b; Harris and Trethewey, 2010). Here, we report the identification of a type of covalent linkage between cell wall polymers in which the matrix polysaccharides pectin and xylan are linked to AGP.
Current models of xylan and pectin structure portray independent wall polysaccharides. Xylan is a major hemicellulosic polysaccharide in secondary walls and in grass primary walls and is also present in reduced amounts (∼5%) in dicot primary walls (Darvill et al., 1980). Xylan is composed of a backbone of 1→4-linked β-d-xylopyranosyl residues that may be partially glycosylated at O-2 or O-3 with arabinofuranosyl residues and/or at O-2 with 4-O-methyl glucuronosyl residues to form arabinoxylan and/or glucuronoarabinoxylan (reviewed in Doering et al., 2012; Kulkarni et al., 2012). Arabinoxylan in grasses is commonly arabinosylated at Xyl O-3 and may, to a lesser degree, include O-2 and O-3 di-arabinosyl substitution. Dicot xylan is less frequently arabinosylated, with reported arabinosylation generally at the O-2 of Xyl (reviewed in Scheller and Ulvskov, 2010). Xylan may also be acetylated at O-3.
Pectin is a family of galacturonic acid (GalA)-rich polysaccharides that accounts for 30 to 35% (w/w) of primary cell walls in dicots and nongraminaceous monocots but is also present in secondary walls and in grasses. The most abundant pectic polysaccharide, HG, is a linear homopolymer of 1→4 linked α-d-GalA residues that accounts for ∼65% of pectin. HG may reach lengths of 100 residues or more (Thibault et al., 1993). The other major pectin, rhamnogalacturonan I (RG-I), comprises 20 to 35% of pectin and consists of a repetitive [-2-α-l-Rhap-1→4-α-d-GalAp-1-] disaccharide backbone with 20 to 80% of the rhamnosyl residues decorated by side chains of 1,5-arabinans, 1,4-galactans, and type I and type II arabinogalactans (Mohnen, 2008). The GalA residues in HG may also be substituted with four complex side chains to form rhamnogalacturonan II, representing ∼10% of wall pectin, or to a lesser degree with terminal-xylosyl or apiosyl residues (Mohnen, 2008; Harholt et al., 2010). Acetylation and methylation of pectin in vivo may change the charge and hydrophobicity of the polysaccharides and, hence, its roles in the plant.
The AGPs are highly glycosylated HRGPs that consist of up to 95% carbohydrate. Clustered noncontiguous Hyp residues in the AGP protein backbone usually have covalently attached type II arabinogalactan (AG) polysaccharides. Individual AG glycans in an AGP consist of up to 150 sugar residues and are rich in Ara and Gal (Kieliszewski, 2001; Showalter, 2001). An individual AG glycan consists of a β-1,3-galactan backbone with β-1,6-galactosyl branches that are decorated with arabinosyl residues and occasionally with minor sugar residues, such as glucuronic acid (GlcA), rhamnose (Rha), and Fuc (Carpita and Gibeaut, 1993; Ellis et al., 2010; Tan et al., 2010). However, as noted above, type II AGs are also found as side chains of the pectin RG-I (Caffall and Mohnen, 2009) and as free polysaccharides (Ponder and Richards, 1997).
Although AGPs account for <10% of the wall matrix, they have many diverse biological roles that span plant embryogenesis through multiple stages of plant development (Ellis et al., 2010). The molecular basis for these biological activities is not known. Here, we describe the identification and structural characterization of an Arabidopsis thaliana AGP found in two glycoforms, YS1 and YS2, both of which contain pectin and arabinoxylan glycan domains. We present direct evidence for the covalent attachment of these wall matrix polysaccharides to the AGP. The identified structure demonstrates that an AGP can serve as a type of proteoglycan cross-link between cell wall matrix polysaccharides and a wall glycoprotein and provides a possible mechanism for some of the biological activities of AGPs.
RESULTS
Identification of a Xyl-Rich and GalA-Containing AGP
During an extensive purification and analysis of AGPs from Arabidopsis suspension culture medium, seven AGP-containing protein fractions were collected (Figure 1A). Neutral sugar and uronic acid analyses showed that the major glycosyl residues in each fraction were those expected for AGPs (see Supplemental Table 1 online). These analyses also revealed that fraction 3 (peak 3 in Figure 1A) contained 45% (molar percentage) Xyl (see Supplemental Table 1 online), an uncharacteristically high amount of Xyl for AGPs. Attempts to purify the Xyl-rich material from AGPs in fraction 3 by precipitation with β-Gal Yariv reagent, a method that precipitates most AGPs, resulted in the recovery of the Xyl-rich material in the Yariv-soluble fraction, and this fraction was named YS. The Yariv solubility of YS suggested either that YS did not contain an AGP or that one or more AGPs in YS had unusual glycosylation or glycan modifications and thus was not accessible for precipitation with Yariv reagent. The YS fraction was further purified by size-exclusion and reverse-phase chromatography into two YS populations: a major UV-220 nm absorbing protein fraction (abbreviated YS1) and a minor fraction (YS2) (Figure 1B). Both YS1 and YS2 eluted during Superose-12 gel chromatography with a molecular size of between 75 and 100 kD.
Figure 1.
Reverse-Phase HPLC Chromatography of APAP1 and Separation of Hyp-O-Glycosides from APAP1 by Size Exclusion Chromatography.
(A) Reverse-phase PRP-1 chromatography profile of AGP-enriched material from Arabidopsis suspension culture medium (modified from Xu et al., 2008). AGPs from the medium of 10-d-old Arabidopsis suspension-cultured cells were purified by sequential DEAE anion exchange and Superose-12 SEC and separated into seven 220 nm-absorbing peaks by reverse-phase PRP-1 chromatography (Xu et al., 2008). The yield of peak 3 (5 mg/5 liters media) was <1% (w/w) of the total AGPs recovered.
(B) The material in peak 3 from (A) was treated with Yariv reagent to yield a Yariv precipitant and a Yariv-soluble fraction. The Yariv-soluble fraction was separated by PRP-1 reverse-phase chromatography into two subfractions, major peak YS1 (1 mg/5 liters media) and shoulder peak YS2 (0.6 to 0.7 mg/5 liters media).
(C) YS1 was hydrolyzed with 0.44 n NaOH at 105°C for 18 h, each hydrolysate was neutralized with 1 n HCl on ice, and freeze-dried residues were dissolved and separated over an analytical Superdex-75 column as described (see Methods). An aliquot (10% volume) of each collected fraction was assayed for Hyp and pentose. Red squares represent absorbance at 560 nm to detect Hyp derivatives; blue diamonds represent absorbance at 665 nm to detect pentose derivatives. The asterisks show the fractions used for more extensive sugar analyses (red asterisks, fractions 22 and 50) and NMR analyses (blue asterisks, fractions 26 and 27).
(D) YS2 hydrolyzed and analyzed as described in (C).
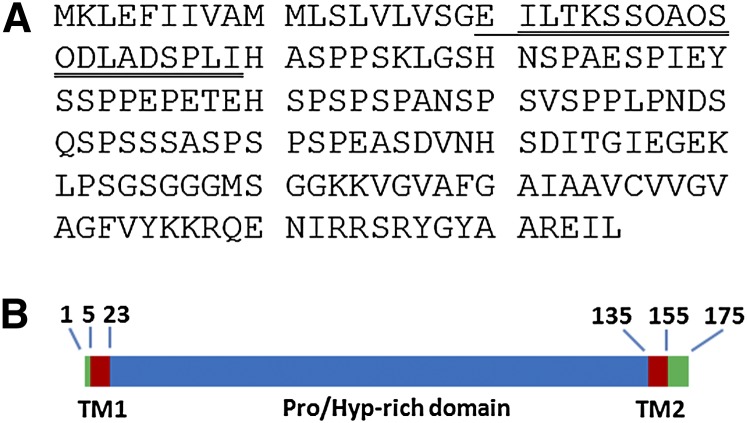
To identify any protein component in YS1 and YS2, native YS1 and anhydrous hydrogen fluoride (HF)-deglycosylated YS2 were subjected to N-terminal amino acid sequencing, yielding an N-terminal amino acid sequence of EILTKSSOAOSODLADSPLI (O as Hyp) for YS1 and an almost identical N-terminal sequence of ILTKSSOAOSODLADSPLI for YS2. A comparison of these sequences with the translated Arabidopsis genome database revealed a perfect match with Arabidopsis hydroxyproline-rich glycoprotein family protein At3g45230, which is annotated as an HRGP family protein and as AGP AGP57C (Showalter et al., 2010) (Figure 2). The identified N-terminal sequences of YS1 and YS2 represent cleavage at amino acids 19 (Gly) and 20 (Glu), respectively, of the predicted 175 amino acid protein encoded by gene At3g45230 (Figure 2). In agreement with this observation, the protein encoded by At3g45230 is predicted by PSORT (http://psort.hgc.jp/) to have a potential signal sequence cleavage site at amino acid 19. The PSORT and TmHMM_V2 (http://www.cbs.dtu.dk/services/TMHMM/) programs also predict an α-helix membrane-spanning domain between C-terminal amino acids 135 and 154, suggesting a possible signal sequence for glycosylphosphatidylinositol (GPI) anchoring. Taken together, these results show that YS1 and YS2 contain the same AGP protein core.
Figure 2.
Protein Sequences Deduced from YS1 and YS2 Exactly Match a Portion of the Arabidopsis Gene At3g45230.
(A) The gene encodes a Ser/Pro-rich AGP, At-AGP57C. N-terminal sequencing of YS1 yielded a 20–amino acid sequence that matched the underlined sequence of the mature Ser/Pro-rich protein. O stands for Hyp, as confirmed by amino acid sequencing. The double underlined sequence matched the N-terminal sequence of anhydrous HF deglycosylated YS2.
(B) Domain structure diagram of At-AGP57C. Red indicates two predicted transmembrane domains (TM) of APAP1 calculated by the transmembrane prediction programs TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). The TM1 regions include the predicted N-terminal TM from amino acids 5 to 22 and the predicted C-terminal TM, TM2, from amino acids 135 to 154. Green indicates non-Pro/Hyp-rich regions, both predicted to have the opposite topology of the Pro/Hyp-rich domain (marked in blue). The N-terminal TM was predicted to be a signal peptide by SignalP (http://www.cbs.dtu.dk/services/SignalP). The C-terminal TM indicated a possible signal sequence for GPI anchor modification (http://mendel.imp.ac.at/gpi/plant_server.html).
Sugar composition analyses revealed that YS1 and YS2 contained 63 and 97% sugar residues (w/w), respectively, including GalA and Xyl (Table 1). This high sugar content explained why these relatively small polypeptides (Figure 2) eluted as much larger proteins (75 to 100 kD) upon size-exclusion chromatography (SEC). The presence of high levels of Xyl and GalA in YS1 and YS2 (Table 1) was intriguing because these sugars are commonly present in xylan and pectin, respectively, but are not generally found in these amounts in typical AGPs. Sugar linkage analyses confirmed the presence of type II AG, based on the existence of terminal Galp and 3-, 6- and 3,6-Galp in YS1 and YS2 (Table 1) and also confirmed the identity of these proteoglycans as AGPs. However, the glycosyl linkage analyses also indicated the existence of 2- and 2,4-Rhap, 4-GalAp, and 4- and 2,4-Xylp, indicating the presence of the pectins RG-I and HG and of substituted xylan in the YS1 and YS2 preparations. This led to the question of whether the pectin and xylan-like material was covalently attached to the AGP in YS1 and YS2 or, alternatively, whether free pectin and xylan polysaccharides had cofractionated with the AGPs.
Table 1. Glycosyl Composition and Linkage Analysis of Sugar Residues in YS1, YS2, and Ara101P.
| Glycosyl Residuea | YS1a,b | YS2a,c | Ara101Pa,d | Glycosyl Linkage | YS1b,e | YS2c,e | Ara101Pd,e |
|---|---|---|---|---|---|---|---|
| Ara | 37.7 | 39.4 | 64.8 | t-Araf | 3.0 | 22.0 | 15.7 |
| Xyl | 23.4 | 39.2 | Trace | 3-Araf | 0.8 | 1.9 | 0.4 |
| GalA | 11.0 | 6.6 | 11.4 | 5-Araf | 10.4 | 8.2 | 8.4 |
| Rha | 5.7 | 5.6 | 10.4 | 3,5-Araf | 5.5 | 2.3 | 1.9 |
| Gal | 19.9 | 6.8 | 13.4 | 2,5-Araf | – | 3.6 | 4.2 |
| GlcA | – | 1.1 | – | t-Arap | – | 5.1 | 0.4 |
| Fuc | – | 0.3 | – | 2,3,4-Arap | 4.8 | 3.3 | 4.3 |
| Glc | – | 1.0 | – | 2,4-Arap | 5.0 | – | – |
| O-methyl sugar | 1.7 | – | – | t-Xylp | – | 4.2 | – |
| Unknown sugar | 0.6 | – | – | 4-Xylp | 5.7 | 16.9 | – |
| 2,4-Xylp | 4.2 | 7.5 | – | ||||
| 4-GalAp | 13.4 | 6.3 | 8.2f | ||||
| 4,6-GalAp | – | – | 10.7 | ||||
| t-Rhap | – | – | 0.2 | ||||
| 2-Rhap | 6.6 | 3.1 | 15.3 | ||||
| 4-Rhap | – | – | 0.3 | ||||
| 2,3-Rhap | 1.2 | – | 0.3 | ||||
| 2,4-Rhap | 4.3 | 1.2 | 12.3 | ||||
| 2,3,4-Rhap | 0.8 | – | – | ||||
| t-Galp | 2.2 | 2.3 | 9.4 | ||||
| 3-Galp | 4.9 | 3.9 | 2.0 | ||||
| 4-Galp | 3.3 | 1.9 | f | ||||
| 6-Galp | 10.0 | 1.6 | 2.6 | ||||
| 3,6-Galp | 13.9 | 3.2 | 2.2 | ||||
| 3,4,6-Galp | – | – | 0.2 | ||||
| t-GlcAp | – | – | 0.3 | ||||
| 4-GlcAp | – | 1.6 | 0.2 | ||||
| 4-Manp | – | – | 0.3 | ||||
| 3,4-Glcp | – | – | 0.4 |
All numbers are mole percentages. The high percentage of t-Ara in YS2 resulted from the 17% of Hyp residues in APAP1 branched with Ara2 or Ara residues, as well as the t-Ara residues that serve as side chains of arabinoxylan and AG. A dash designates that the sugar level was below detection limits.
Glycosyl residue composition by GC-MS analysis of trimethylsilyl derivatives. Data are averages of two separate analyses by CCRC Analytical Services.
Total average weight percentage of carbohydrate of YS1 was 63.2%.
Total average weight percentage of carbohydrate of YS2 was 97.7%.
Total average weight percentage of carbohydrate of Ara101P was 78.0%.
Sugars were converted to partially methylated alditol acetates and analyzed by GC-MS.
Total molar percentage of 4-GalAp and 4-Galp was 8.2%.
Evidence for Covalent Attachment of Xylan and Pectin to the AGP in YS
Two methods were used to determine whether the xylan- and pectin-like glycans in YS were free polysaccharides or covalently attached to the AGPs. If free pectin and/or xylan polysaccharides were present in YS, they would be detectable by reducing end assays or reducing end labeling due to the presence of terminal anomeric carbons not bound in a glycosidic bond. The reducing end of polysaccharides can be labeled with the hydrophobic fluor 2-amino-benzoamide (2-AB) and detected by a gain in absorbance at UV-254 nm (Ishii et al., 2002). Following incubation of YS2 with 2-AB and purification of the treated YS2 by SEC and reverse-phase chromatography, the resulting 2-AB–treated YS2 showed no gain in absorbance at 254 nm and had the same retention time as nontreated YS2. These results indicated that there was no covalent attachment of the hydrophobic 2-AB to any component in YS2. Conversely, 2-AB labeling of the control polysaccharide, polygalacturonic acid (PGA), resulted in a gain of UV absorbance at 254 nm. In addition, 2-AB labeling caused the PGA to bind to the reverse-phase column, while nonlabeled PGA did not, indicating increased hydrophobicity upon the attachment of the hydrophobic 2-AB to the control PGA. Finally, sugar composition analyses of 2-AB–treated and nontreated YS2 after sequential SEC and reverse-phase chromatography indicated that 2-AB labeling resulted in no change in sugar composition of 2-AB–treated YS2 compared with YS2. The results indicated that there were no free reducing ends present in YS2 and did not support the presence of any free pectin or xylan polysaccharides in the YS2 preparation.
Another method used to test if the pectin and xylan were covalently attached to the YS AGP involved selective attachment of the protein component of the AGP to a resin and the washing away of any nonbound material. The AGP protein component in YS2 was covalently bound to glutaraldehyde-activated amine magnetic beads via a Schiff base reaction with the primary amine groups in the N-terminal and intrapeptide Lys residues present in the protein. The YS2 beads were extensively washed with wash buffer (10 mM EDTA, 1 M NaCl, and 0.1% BSA) to dissociate noncovalently bound material, and the washed YS2-beads were hydrolyzed in 2 n trifluoroacetic acid (TFA) to release monosaccharides that had been covalently attached to the protein. Glycosyl residue composition analysis revealed that the sugar component on the YS2 beads was identical to that of the starting YS2 material (see Supplemental Table 2 online). The lack of any loss of sugar upon washing of the bound beads indicated that there were no free polysaccharides present in YS2. Control reactions in which a monosaccharide mixture (Ara, Gal, Xyl, GalA, and Rha) and a yeast 1,3;1,6-glucan were incubated with activated beads, as well as incubation of YS2 with nonactivated beads, resulted in no sugar residues being bound to the beads. These data confirmed that there were no free polysaccharides present in the YS2 preparation and that the glycans in YS2 were covalently attached to the protein. Taken together, these results support the conclusion that the xylan and pectin-like glycans, AG, and polypeptide moieties in YS2 are covalently linked together.
Xylan in YS Is Arabinoxylan That Is Attached in Two Different Ways to the AGP with Approximately Half of the Xylan Attached to the RG-I Domain
To probe the identity and structure of the glycans in relation to the typical AGP core, YS2 was hydrolyzed using glycan-specific enzymes. Rhamnogalacturonan hydrolase (RGH) is an endohydrolytic enzyme that cleaves the repeating [-2-α-l-Rha-1→4-α-d-GalA-1→] linkages present in the backbone of RG-I, yielding fragments with Rha at the nonreducing end. Because RG-I may contain 1,5-arabinan, 1,4-galactan, and/or type II AG side chains, the RGH treatment was also predicted to release arabinosyl- and galactosyl-containing RG-I fragments from YS2. Indeed, treatment of YS2 with RGH released more than 99% of the Rha, 99% of the GalA, 61% of the Ara, and 35% of the Gal residues from YS2 (see Supplemental Table 3 online). These results provided initial evidence that the Xyl-rich YS2 contained a glycan structure typical of RG-I. There was also a concomitant release of 44% of the Xyl residues upon cleavage of YS2 with RGH. Because almost half of the xylan in YS2 was released by the RGH treatment, while about half remained associated with the remaining YS2, we postulated that some xylan-like structure was linked to RG-I and some xylan-like structure was linked to the remaining part of YS2 in a different manner (i.e., either to the AG portion of the AGP or to the protein backbone of the AGP).
Glycosyl residue linkage analyses indicated that the GalA residues in YS2 were 4-linked and that the Rha residues were either 2-linked or 2,4-linked. Thus, one possible attachment site of xylan to RG-I was at the 4-position of Rha in the RG-I backbone. The attachment of short β-1,4-linked Xyl oligosaccharides to the 4-position of the RG-I backbone has previously been reported in soybean (Glycine max; Nakamura et al., 2002), but there have been no prior reports of this structure associated with AGPs.
After RGH treatment and reverse-phase column purification of RGH-treated YS2 (see Supplemental Table 3 online), the treated YS2 was able to be precipitated with β-Gal Yariv reagent. These results indicated that the Yariv-precipitable AG glycan portion of the AGP was made accessible following removal of the pectin and xylan-like glycans upon RGH treatment, thereby resulting in precipitation of the treated YS2 (Xyl-containing AGP) by Yariv reagent. The fully glycosylated form of YS2 prior to RGH treatment was resistant to precipitation. These results provided additional support for the covalent attachment of xylan and RG-I glycans to the AGP.
To investigate the YS2 structure further, untreated YS2 was incubated with three endo-β-d-xylanases. Approximately one-third of the Xyl was released from YS2 upon treatment with either of two different endo-β-d-xylanases: CAZy (Cantarel at al., 2009) family GH10 xylanase Cj-Xyn10A (Biely et al., 1997) and GH11 xylanase Np-Xyn11A (Paës et al., 2012) (see Supplemental Table 3 online). These enzymes hydrolyze low-substituted xylan and require at least two or three, respectively, unsubstituted Xyl residues for cleavage. The results suggested that 33% of the xylan in YS2 was low to moderately substituted, while the remaining ∼67% was more highly substituted, which was consistent with the 22% of terminal-Araf in the linkage analysis of YS2 (Table 1). However, YS2 was not cleaved by a GH5 family endo-β-d-xylanase (Clostridium thermocellum Xyl5A) (Correia et al., 2011). Ct-Xyl5A specifically cleaves highly arabinosylated xylan, generating oligosaccharide products with Ara-1→3-Xyl at the reducing end, a structure that has been proposed as a specificity determinant for the enzyme. We interpret the lack of cleavage of YS2 by Ct-Xyl5A as indicating that YS2 does not contain appreciable amounts of O-3-linked Xyl. This conclusion is in agreement with the YS2 sugar linkage data showing that 26% of Xyl in YS2 is O-2 substituted (Table 1). Taken together, these results indicate that the xylan in YS2 is partially O-2 arabinosylated.
To further study the nature of the pectin and arabinoxylan regions in the AGP structure, YS2 was probed for cross-reactivity with 47 antibodies raised against RG-I, xylan, and AG (Pattathil et al., 2010). Reactivity of YS2 against antibody classes AG-2, AG-4, RG-I/AG, RG-I backbone, linseed (Linum usitatissimum) mucilage RG-I, and xylan 7 (Pattathil et al., 2010) confirmed the presence of RG-I, xylan, and AG glycan epitopes in YS2 (see Supplemental Figure 1 and Supplemental Table 4 online). Among the four selected antixylan antibodies, only CCRC-M149 showed a strong reaction with YS2. We propose that reaction with the other xylan-reactive antibodies may have been limited due to the high level of arabinosylation and acetylation (see below) of xylan in YS2.
These results led us to propose that YS was an AGP proteoglycan with two arabinoxylan-containing regions, one attached to a pectin moiety and one not. To test this, we first asked how the pectin-arabinoxylan domain and the separate arabinoxylan domain (henceforth referred to as the arabinoxylan1 domain) were linked to the AGP. We postulated that the pectin-arabinoxylan domain and the arabinoxylan1 domain were linked either to the AG domain or to the polypeptide portion of the AGP.
Hyp Residues Are the Attachment Sites for the Glycan to the Protein Core in YS, and YS Is Named ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1
To determine if the glycan in YS1 and YS2 was O-glycosylated to Hyp, to Ser/Thr, or to both types of amino acids in the AGP protein core, a β-elimination reaction known to remove sugars O-linked to Ser/Thr was used (Kieliszewski et al., 1992). AGPs are known to contain long AG glycans and short oligoarabinosides attached to Hyp residues (Kieliszewski, 2001), and these Hyp-O-linked glycans/oligosaccharides are stable to β-elimination (Lamport and Miller, 1971; Tan et al., 2004). β-Elimination of YS1 resulted in only a small decrease in the mole percentage of Gal (see Supplemental Table 5 online) and a total sugar weight decrease from 53.3% (intact) to 50.9%. These results agree with prior reports showing that Ser/Thr residues of AGPs are usually glycosylated with single Gal residues (Kieliszewski, 2001). The results further suggested that the glycan in YS1 was attached via O-glycosylation to Hyp in the AGP57C polypeptide.
To confirm that the AG, RG-I, and arabinoxylan glycan domains were indeed directly or indirectly O-linked to Hyp, the glycosides attached to Hyp (Hyp-O-glycosides) were released from YS1 and YS2 by treatment with mild base, separated, and analyzed (Tan et al., 2004, 2010). The Hyp-O-glycoside profile revealed that ∼70% of Hyp residues in both YS1 and YS2 are attachment sites for large polysaccharides. The remaining Hyp residues are in part glycosylated with oligoarabinosides (∼11 to 13%), with single Ara residues (5 to 7%), or are present as free Hyp (nonglycosylated) (∼11%) (see Supplemental Table 6 online). These results confirmed that YS1 and YS2 are AGPs with both long glycan and oligoarabinose substitutions at Hyp residues.
SEC of the Hyp-O-polysaccharides released from YS1 and YS2 by base hydrolysis, combined with colorimetric analyses, revealed that pentose residues and Hyp residues cochromatographed (Figures 1C and 1D), as expected for glycans covalently attached to Hyp (Tan et al., 2004). Furthermore, sugar composition analyses of YS1 and YS2 Hyp-O-polysaccharide fractions representing large and small Hyp-O-polysaccharides, fractions 22 and 50, respectively (Figures 1C and 1D; see Supplemental Table 7 online), showed that the large Hyp-O-polysaccharides still contained the characteristic Xyl, uronic acid, and Rha as well as Gal and Ara. Importantly, based on 2-AB reducing end labeling tests, no reducing end sugar residues were detected in representative YS2 Hyp-O-polysaccharide fraction 24 (Figure 1D). Specifically, 2-AB–treated Hyp-O-polysaccharides had a maximum UV absorbance of only ∼220 nm, while 2-AB–labeled control PGA or RG-I oligomers displayed maximum UV absorbances at 254 nm. These results confirmed the direct or indirect covalent attachment of the arabinoxylan, RG-I, and AG glycan domains to Hyp residues in AGP57C.
The combined results provided compelling evidence that AGP57C, encoded by Arabidopsis gene At3g45230, is the core of a proteoglycan with covalently attached AG, arabinoxylan, and pectin domains. To distinguish the AGP core protein from the highly glycosylated proteoglycan structure identified here, from this point forward, we refer to the AGP protein core as AGP57C and to the full proteoglycan structure as ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1 (APAP1).
The AG Glycans in YS1 and YS2 Are Attached Directly to Hyp in the Protein Core
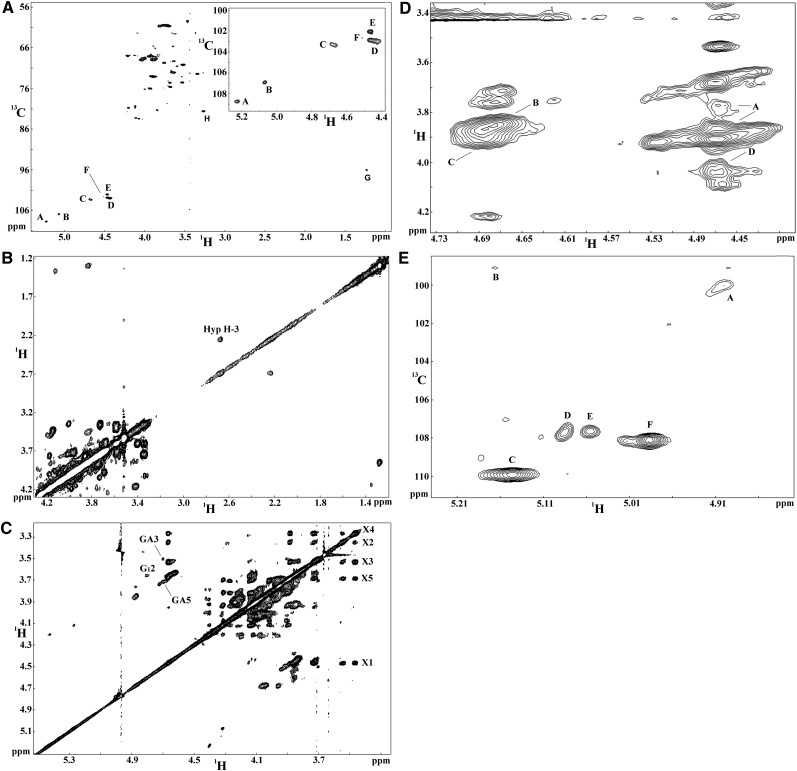
We next sought to determine which of the three glycan domain(s) in YS (i.e., the AG, pectin, and/or arabinoxylan domains) were directly attached to Hyp residues in the protein core of APAP1. To achieve this, YS1 Hyp-O-polysaccharide fractions 26 and 27 (named YS1-HP2627; see Figure 1C) were pooled for NMR analysis because of their relative abundance. Sugar analyses of YS1-HP2627 revealed a Gal:Ara:Xyl:GalA:Rha molar ratio of 14:10.2:3.1:1.5:1. The mild base treatment used to generate YS1-HP2627 almost completely β-eliminated Rha and GalA residues, thus simplifying its structure and the linkages between Hyp, AG, and Xyl residues. The heteronuclear single quantum coherence (HSQC) spectrum of YS1-HP2627 (Figure 3A) resembled those reported for Hyp-O-arabinogalactan (Hyp-O-AG) polysaccharides (Tan et al., 2004, 2010), except for the additional signals attributed to 1,4-β-d-Xylp, suggesting the presence of a Hyp-O-AG structure in this Hyp-O-polysaccharide fraction. Furthermore, the identification of Hyp protons from the 1H homonuclear correlation spectroscopy (COSY) spectrum (Figure 3B) and the unique Gal residue O-linked to Hyp from the total correlation spectroscopy (TOCSY) spectrum (Figure 3C) established the Hyp-O-Gal linkage in YS1-HP2627 (Tan et al., 2004, 2010). These results indicated that it was the AG domain that was directly linked to the protein core in YS1. The Hyp-O-Gal linkage was also identified in Hyp-O-polysaccharide fraction YS2-HP3132 from YS2 (fractions 31 and 32 in Figure 1D). YS2-HP3132 had a Gal:Ara:Xyl:uronic acid:Rha molar ratio of 3:2:7:1.4:1, indicating that this particular YS2 Hyp-O-polysaccharide fraction was more highly xylosylated than that from YS1 (see above).
Figure 3.
NMR Spectra of YS1-HP2627 Collected at 25°C on a Varian VNMRS 800 Instrument.
(A) HSQC spectrum. The inset is the enlarged anomeric region. Signals A and B were identified as the anomeric C/H of α-l-Araf residues; C, as the anomeric C/H of the 1,3-galactan backbone Gal (Galbb); D, as the anomeric C/H of the AG side chain Gal (Galsc); E, as the anomeric C/H of β-d-Xylp residues; F, as the anomeric C/H of β-d-GlcAp; G, as the C/H-6 of α-l-Rhap residues (the signal of G is folded in this HSQC spectrum due to the small 13C spectral width used; as such, 80 ppm should be subtracted from the respective 13C chemical shifts); H, as the C/H-4 of 4-Xylp.
(B) COSY spectrum (partial) collected at 25°C. The correlation between Hyp H-3ax and Hyp H-3ex is labeled.
(C) TOCSY spectrum identified the protons of Xyl residues, GlcA, and the Gal residue attached to Hyp. The labels X1 to X5 represent H-1 to H-5 of 4-Xyl; GA3 and GA5 represent H-3 and H-5 of GlcA; and G12 represents H-2 of Gal attached to Hyp. A similar pattern was also observed in TOCSY recorded on YS2-HP2627.
(D) NOESY spectrum (partial) (mixing time 100 ms). Cross-peak A shows the NOE between Xyl H-1 to Ara H-5, which established the β-d-Xylp-1→5-α-l-Araf linkage; cross-peak B shows NOEs between Galbb H1 and Galbb H3, suggesting the Galbb-1→3-Galbb linkage; cross-peak C (NOE between Galbb H1 and Galbb H6) supports the Galbb-1→6-Galbb linkage; NOEs in D suggest the Galsc-1→6-Galbb linkage.
(E) HMQC spectrum (partial) collected at 30°C. Signal A represents the anomeric 1H/13C signals of 2-α-l-Rha, B the anomeric 1H/13C signals of α-d-GalA, and C to F the anomeric 1H/13C signals of α-l-Araf. Hyp-glycoside YS1-HP2627 produced by SEC chromatography of base hydrolyzed YS1 as depicted in Figure 1C.
The type II AG linkages in YS1-HP2627 were further supported by nuclear Overhauser effect (NOE) assignments in the nuclear Overhauser effect spectroscopy (NOESY) spectrum (Figure 3D), including assignments for Gal residues (Galbb) in the galactan backbone (β-d-Galbb-1→3-β-d-Galbb and β-d-Galbb-1→6-β-d-Galbb) and side chain Gal (Galsc) residues (β-d-Galsc-1→6-β-d-Galbb). These data showed that the AG domain in YS1 and YS2 was linked to the protein core in the same way as in common AGPs (Kieliszewski, 2001). Taken together, the above results indicated that the AG domain in APAP1 was linked to Hyp in the polypeptide core and that the pectin arabinoxylan and arabinoxylan1 domains were linked in some manner to the AG domain. To identify the precise covalent linkages of the pectin arabinoxylan and arabinoxylan1 domains to the AG domain, we performed a series of NMR and mass spectrometry–based analyses.
Pectin Is Attached to the Rhamnosylglucuronosyl Side Chain of AG
AGPs isolated from many plant materials have been shown to have t-Rha-1→4-β-d-GlcA-1→6-β-d-Galsc as one of the bifurcate side chains of the AG glycan domain (Defaye and Wong, 1986; Gane et al., 1995; Tan et al., 2004). A GlcA residue was detected in YS2 by the trimethylsilyl composition method but was not detected in YS1 due to its lower abundance. However, signals characteristic for 4-β-d-GlcAp were obtained in HSQC and TOCSY spectra of YS1-HP2627 (Figures 3A and 3C), which yielded 1H/13C signals of ∼4.51/∼102.1 (H/C-1), 3.713/75.317 (H/C-5), and 3.501 (H-3) ppm (Tan et al., 2004). Furthermore, NOEs between GlcA H-1 and the AG side chain Gal (Galsc) H-6 (3.92 ppm) in YS1 also supported the GlcA-1→6-Galsc linkage (Figure 3D) in YS1.
A heteronuclear multiple quantum coherence (HMQC) spectrum of YS1-HP2627 (Figure 3E) revealed anomeric 1H/13C signals at 4.903/100.077 ppm, typical 2-α-l-Rha signals for RG-I (Zheng and Mort, 2008), suggesting the residual Rha on the typical t-Rha-1→4-β-d-GlcA-1→6-β-d-Galsc AG side chain was O-2-substituted in YS1-HP2627. In addition, anomeric signals (1H/13C 5.167/99.115 ppm) for α-d-GalA residues were also found in the HMQC spectrum of YS1 (see Supplemental Table 8 online). Taken together, the identification of the AG side chain 4-β-d-GlcA and characteristic 2-α-l-Rha and α-d-GalA of pectin, as well as the establishment of the Xyl to Ara linkage in YS1-HP2627 as shown below, are consistent with the existence in YS1-HP2627 of the following structural unit as a side chain of the AG: α-d-GalA-1→2-α-l-Rha-1→4-β-d-GlcA-1→6-β-d-Galsc. It also indicates that the nonreducing end Rha of the AG side chain may serve as the attachment site of pectin.
The base treatment that generated YS2 Hyp-O-polysaccharide fractions 26 and 27 (YS2-HP2627) did not completely β-eliminate the pectic 2(4)-α-Rha and 4-α-GalA residues and even some acetyl groups remained intact after base treatment, as evidenced in the HSQC spectrum (see Supplemental Figure 2A and Supplemental Table 9 online). 4-β-GlcA 1H/13C signals were also identified in the HSQC spectrum. Because the AG was the glycan domain attached to the polypeptide as demonstrated above and there was no t-Rha in YS2-HP2627, these results provided further evidence for the occurrence of -4-α-d-GalA-1→2-α-l-Rha-1→4-β-d-GlcA-1→6-β-d-Galsc structures in YS. Furthermore, the 1:1 ratio of anomeric protons between α-l-Rha and α-d-GalA in YS2-HP2627 indicated that the remaining pectic fragment on YS2-HP2627 was RG-I. These results support the conclusion that the rhamnosyl residue in the rhamnosylglucuronsyl side chain of AG serves as an attachment site for RG-I.
The higher percentages of GalA than Rha in YS suggest the existence of HG in the pectin domain of APAP1 (Table 1). Oligosaccharides released from YS2 by treatment with RGH and analyzed by collision-induced dissociation (CID)–tandem mass spectrometry (MS/MS) yielded fragments that indicated the presence of RG-I and HG domains in YS2. The MS/MS fragmentation of parent ions at a mass-to-charge ratio (m/z) of 1608 and 1389 yielded a series of B, C, Y, and Z ions (see Supplemental Figures 3A and 3B online). Together with the sugar linkage analysis of YS2 and the hydrolytic features of RGH, these MS/MS fragments are consistent with the oligomer structures Rha-GalA-Rha-GalA-GalA4-GalA-[-O-COCH3] ([M+H+Na]+, m/z 1608) and Rha-GalA-Rha-GalA-(GalA)3-GalA ([M+H+Na]+, m/z 1389), respectively. The results indicate that at least short HG regions are part of the pectin domain in APAP1 and that they form a continuous pectin backbone with RG-I in APAP1.
Arabinoxylan1 Is 1→5 O-Linked to an Arabinosyl Residue in the AG Domain of APAP1
Four types of β-d-Xylp residues were identified in the NMR spectra of YS2 (see Supplemental Figure 4 online), including 3-OAc-(2,4)-β-Xylp, 3-OAc-(4)-β-Xylp, 2,4-β-d-Xylp, and 4-β-d-Xylp (see Supplemental Table 10 online). The -β-Xylp-1→4-β-Xylp- linkages in the arabinoxylan domain of YS2 were confirmed by the heteronuclear multiple-bond correlation spectroscopy (HMBC) connections. Furthermore, the HMBC connection of 2,4-β-Xylp H-1 to Araf C-5 showed that some of the 2,4-β-Xylp residues were 1→5 attached to α-l-Araf in YS2. These results indicated that arabinoxylan was either attached to Ara residues present in the AG domain or to 1,5-arabinan in a side chain of RG-I.
Further evidence for the attachment of arabinoxylan to the AG domain was provided by NOEs between β-d-Xylp H-1 and unique α-l-Araf H-5 of YS1-HP2627 in the NOESY spectrum (Figure 3D) (Tan et al., 2004). The 13C/1H signals of Xylp were assigned based on the TOCSY and the HSQC spectra (Figure 3; see Supplemental Table 8 online). From the 13C signals, we conclude that the Xyl residues in YS1-HP2627 are 4-linked because there were no 2,4-bisubstituted Xylp residues with two characteristic 13C chemical shifts at 80 and 78 ppm (Mazumder and York, 2010). Because only AG and xylan domains were predominant in YS1-HP2627, the Xylp-1→5-α-l-Araf linkage indicates that the xylan oligomers were attached to the arabinosyl residues in the AG domain. Linkage of xylan to Ara in the AG domain in APAP1 explains why substantial amounts of Xyl remained in YS2 after RGH treatment, while RG-I–attached Xyl residues were removed as described above.
The Pectin Arabinoxylan Moiety in APAP1 Consists of Arabinoxylan 2 Attached to Rha in the RG-I Domain
The NOESY spectrum (see Supplemental Figure 2B online) of YS2-HP2627 clearly showed unique NOEs between some Xyl H-1 (4.597, 4.621, and 4.638 ppm) and Rha/GalA H-1 (5.402 ppm), indicating a Xyl-O-Rha/or GalA linkage. Because only 2,4-Rha residues but not 2,4-GalA residues were detected in YS2, these NOEs suggested that some Xyl residues were attached to the Rha in the RG-I domain in YS2 through -4-β-d-Xylp-1→4-[-α-d-GalA-1→2-]-α-l-Rhap- linkage. The Xyl to Rha linkages were further supported by CID-MS/MS analysis identifying fragment ions of oligosaccharides released from YS2 with RGH. The MS/MS fragmentation of parent ions at m/z 1348 and 1216 showed a range of Y and Z and B and C ions. These fragment ions were consistent with the following structures: Xylp-(Xylp)3-Xylp-(AcO)Rha-GalA-Rha-GalA ([M-H2O+2H]+, m/z 1348) and Xylp-(Xylp)2-Xylp-(AcO)Rha-GalA-Rha-GalA ([M-H2O+2H]+, m/z 1216) (see Supplemental Figures 3C to 3E online). These data establish the arabinoxylan (named arabinoxylan 2) to RG-I covalent attachment through the Xyl-1→4-Rha linkage and explain why RGH released 44% of the Xyl from YS2 (see Supplemental Table 3 online).
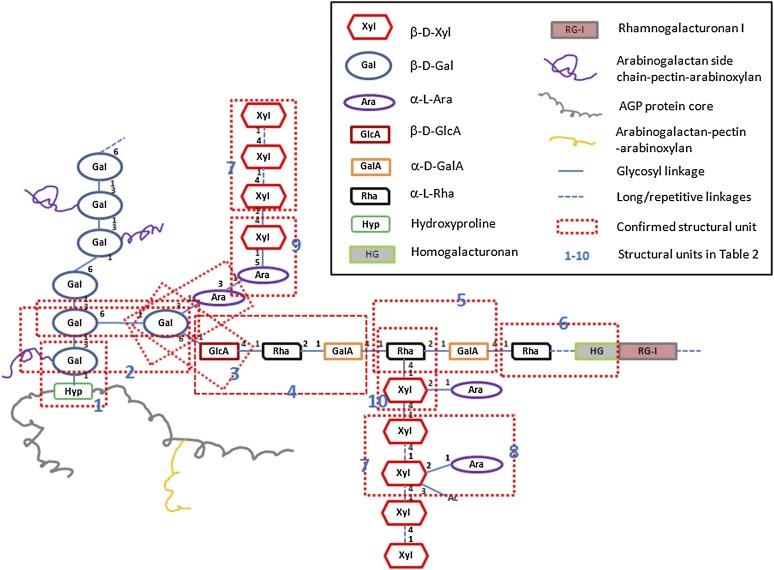
Proposed Structure of APAP1
Based on the above results as summarized in Table 2, we propose the following structure for APAP1 (Figure 4). APAP1 is built on an AGP core (encoded by At3g45230) to which are attached the classic type II AG polysaccharides. The pectin domain of APAP1 is composed of both RG-I and HG, with relatively small HG oligosaccharides flanked by RG-I oligomers on the pectin backbone. The range of lengths of the RG and HG domains is not known. The reducing end GalA residue of an RG-I/HG polysaccharide is linked to a nonreducing end Rha residue in a Rha-1→4-GlcA AG side chain. The data strongly indicate that a typical [→2-α-l-Rha-1→4-α-GalA-1→] disaccharide repeat RG-I backbone is extended from this. Arabinoxylan1 is directly attached to a side chain Ara residue of the AG domain through a 1→5 linkage. Arabinoxylan2 is linked to the O-4 of Rha in the RG-I domain of APAP1 (Figure 4).
Table 2. Summary of NMR and Mass Spectrometry Evidence for Structural Units in APAP1, All of Which Is Consistent with the Proposed APAP1 Structure Shown in Figure 4.
| Structure Unit | Identified Structures | Techniques Used |
|---|---|---|
| 1 AG-to-Hyp linkage | Hyp-O-Galp | TOCSY and HSQC spectra of YS1-HP2627 and YS2-HP3132 |
| Hyp-O-AG | TOCSY and HSQC spectra of YS1-HP2627 and YS2-HP3132 | |
| 2 Type II AG | -β-d-Galpbb-1→3-β-d-Galpbb-1- | HMBC spectrum of YS2 and NOESY spectrum of YS1 HP-2627 |
| -β-d-Galpbb-1→6-β-d-Galpbb-1- | NOESY spectrum of YS1-HP2627 | |
| -β-d-Galpsc-1→6-β-d-Galpbb-1- | NOESY spectrum of YS1-HP2627 | |
| 3 GlcA side chain of AG | -β-d-GlcAp-1→6-β-d-Galpsc-1- | NOESY spectra of YS1-HP2627 and YS2-HP2627 |
| 4 Pectin to AG GlcA linkage | α-d-GalAp-1→-2-α-L-Rhap-1→4-β-d-GlcAp | HMQC spectrum of YS1-HP2627 |
| 5 RG-I linkage | −2-α-l-Rhap-1→4-α-d-GalAp-1- | HMBC spectrum of YS2 |
| 6 HG-RG-I attachment | Rha-GalA-Rha-GalA-(GalA)4-GalA-(OAc)-OH | CID-MS/MS of RGH released oligomers from YS2 |
| Rha-GalA-Rha-GalA-(GalA)3-GalAOH | CID-MS/MS of RGH released oligomers from YS2 | |
| 7 Different linkages of Xyls in arabinoxylan | −4[α-l-Araf-1→2,AcO-3]-β-d-Xylp-1→4-β-d-Xylp-1- | HMBC spectrum of YS2 |
| −4[AcO-3]-β-d-Xylp-1→4-β-d-Xylp-1- | HMBC spectrum of YS2 | |
| −4-β-d-Xylp-1→4-β-d-Xylp-1- | HMBC spectrum of YS2 | |
| Xylp6, Xylp5, Xylp4 | CID-MS/MS of RGH released oligomers from YS2 | |
| 8 Arabinose to Xyl linkage in arabinoxylan | α-l-Araf-1→2-β-d-Xylp- | HMBC spectrum of YS2 and NOESY spectrum of YS2-HP2627 |
| 9 Arabinoxylan to AG arabinosyl side chain linkage | β-Xylp-1→5-α-l-Araf | HMBC spectrum of YS2 and NOESY spectrum of YS1-HP2627 |
| 10 Arabinoxylan to RG-I rhamnosyl residue linkage | β-Xylp-1→4-α-l-Rhap | NOESY spectrum of YS2-HP2627 |
| Xylp-(Xylp)3-Xylp-(AcO)Rha-GalA-Rha-GalA | CID-MS/MS of RGH released oligomers from YS2 | |
| Xylp-(Xylp)2-Xylp-(AcO)Rha-GalA-Rha-GalA | CID-MS/MS of RGH released oligomers from YS2 | |
| 11 β-1,4-Galactan side chain of RG-I | →4-β-d-Galp-1→4-α-(2-)-Rhap | NOESY spectrum of YS2-HP2627 |
Galbb represents Gal residues in the β-1→3-galactan backbone of the AG domain. Galsc represents side chain β-Gal residues connected 1→6 to the β-1→3-galactan backbone. Structural unit numbers listed in this table are the same as those marked in Figure 4. YS1-HP2627, YS2-HP2627, and YS2-HP3132 represent the Hyp-O-polysaccharide fractions 26 plus 27, and 31 plus 32, generated from YS1 and YS2, respectively. HMBC is heteronuclear multiple-bond correlation spectroscopy.
Figure 4.
Proposed Structural Model for APAP1.
RG-I is attached to type II AG through an α-d-GalA-1→2-α-l-Rha-1→4-β-d-GlcA-1→6-Gal structural unit (box 4). HG, with at least four to five GalA residues is embedded within RG-I (box 6). The length of flanking RG-I and HG domains is unknown. Arabinoxylan1 is attached to type II AG through a β-d-Xylp-1→5-α-l-Araf- linkage (box 9). However, only one out of three possible types of Ara residues (based on the common sizes of the arabinosyl side chains: Ara1, Ara2, and Ara3) for this attachment is shown in this model. Furthermore, some of the Xyl residues in arabinoxylan1 are arabinosylated at position 2. Arabinoxylan2 is attached to RG-I through a β-d-Xylp-1→4-α-l-Rha linkage (box 10). The AGP protein core in APAP1 is AGP57C encoded by At3g45230. Bolded blue numbers (1 to 10) represent the identified structural units as listed in Table 2. Based on the amount of Hyp and monosaccharides in YS1 and YS2, a given Hyp residue in At-AGP57C in YS1 is attached with, on average, ∼24 Gal, 45 Ara, 28 Xyl, seven Rha, and 13 GalA/GlcA, while in YS2, there are 11 Gal, 65 Ara, 65 Xyl, nine Rha, and 13 GalA/GlcA residues. Due to the heterogeneity of glycosylation, this proposed model reflects representative, and not exact, numbers and length of the AG-pectin-(arabino)xylan chains in APAP1.
Identification of APAP1-Like Proteoglycan in Cell Walls of Arabidopsis Suspension Culture Cells
APAP1 was originally isolated from the medium surrounding Arabidopsis suspension cultured cells. To determine if APAP1 is present in plant cell walls, we analyzed an RG-I–enriched fraction prepared from an independent Arabidopsis suspension culture line (Guillaumie et al., 2003). Cell walls were isolated, treated with mild base to remove methyl and acetyl esters, incubated with endopolygalacturonase to obtain a pectin-enriched fraction, and then fractionated to yield an RG-I–enriched cell wall preparation as previously described (York et al., 1985). Further purification of this material using a method similar to the one used to isolate APAP1 (see Methods) yielded a UV-220 nm absorbing protein fraction that was named Ara101P (see Supplemental Figure 5 online). Glycosyl residue composition and glycosyl linkage analyses showed that Ara101P consisted of sugar residues consistent with type II AG and RG-I structures (Table 1), suggesting that Ara101P was an APAP1-like proteoglycan. Furthermore, attachment of Ara101P to glutaraldehyde-activated magnetic amine beads, in a manner similar to that done for APAP1, followed by monosaccharide analysis of the Ara101P beads (see Supplemental Table 2 online), demonstrated that the AG and RG-I glycans were covalently attached to the polypeptide in Ara101P.
Liquid chromatography–MS/MS analysis of tryptic peptides from deglycosylated Ara101P identified a 22–amino acid sequence, SSOAOSODLADSOLIHASOOSK (O as Hyp). This peptide is in the N-terminal region of At-AGP57C, the same protein core of YS1 and YS2. These results provided independent evidence that the core protein of APAP1, At-AGP57C, was attached to pectin in an RG-I–enriched preparation released from cell walls of a different Arabidopsis suspension cell line than that used to identify APAP1. Importantly, these results demonstrate that At-AGP57C exists in cell walls in an APAP1-like form (i.e., RG-I/HG-AGP proteoglycan). Without diagnostic tools to probe intact APAP1 and APAP1-like molecules in intact cell walls, a difficult goal due to the highly glycosylated nature of APAP1, it is not possible at this time to quantify the amount of such proteoglycans among all wall polymers or to determine the proportion of APAP1-like proteoglycan that exists linked to pectin and xylan in the wall. The method used to isolate Ara101P selectively enriched for pectin-containing fractions, which are relatively easily released from the wall without harsh chemical treatments. Based on the amount of APAP1-like proteoglycan in the total RG-I–enriched pectin preparation, we calculate that more than 95% of the RG-I released from cell walls of Arabidopsis suspension cultured cells is covalently attached to AGPs.
Identification of Two Arabidopsis apap1 Homozygous Mutant Lines
The above results indicated that APAP1 is a proteoglycan that connects an AGP to matrix polysaccharides in the wall. To determine the effect of reduced expression of the APAP1 core protein on cell wall structure in the plant, homozygous lines for two Arabidopsis apap1 homozygous SALK T-DNA insertion mutants (SALK_070113c/apap1-3 and SALK_002144/apap1-4) (see Supplemental Figure 6A online) were identified. RT-PCR of RNA prepared from leaves of the wild type and two mutants showed reduced APAP1/At-AGP57C transcript in apap1-3 and complete absence of APAP1/At-AGP57C transcript in apap1-4 (see Supplemental Figure 6B online). Further quantitative PCR (qPCR) analysis of the RNA confirmed apap1-4 as a knockout line and apap1-3 as a knockdown line with a significant (∼31%) reduction in APAP1/At-AGP57C transcript (see Supplemental Figure 6C online). The overall morphology of the apap1 mutants was comparable to that of the wild type. However, growth measurements of 10 individual sets of the wild type versus apap1-3 and apap1-4 mutants revealed that in 50% of the experiments there was a 5 to 25% significant increase in inflorescence stem height in the mutants compared with the wild type and in the remaining experiments there was a nonsignificant trend for increased stem growth in the mutants.
Mutation of APAP1 Affects Cell Wall Properties
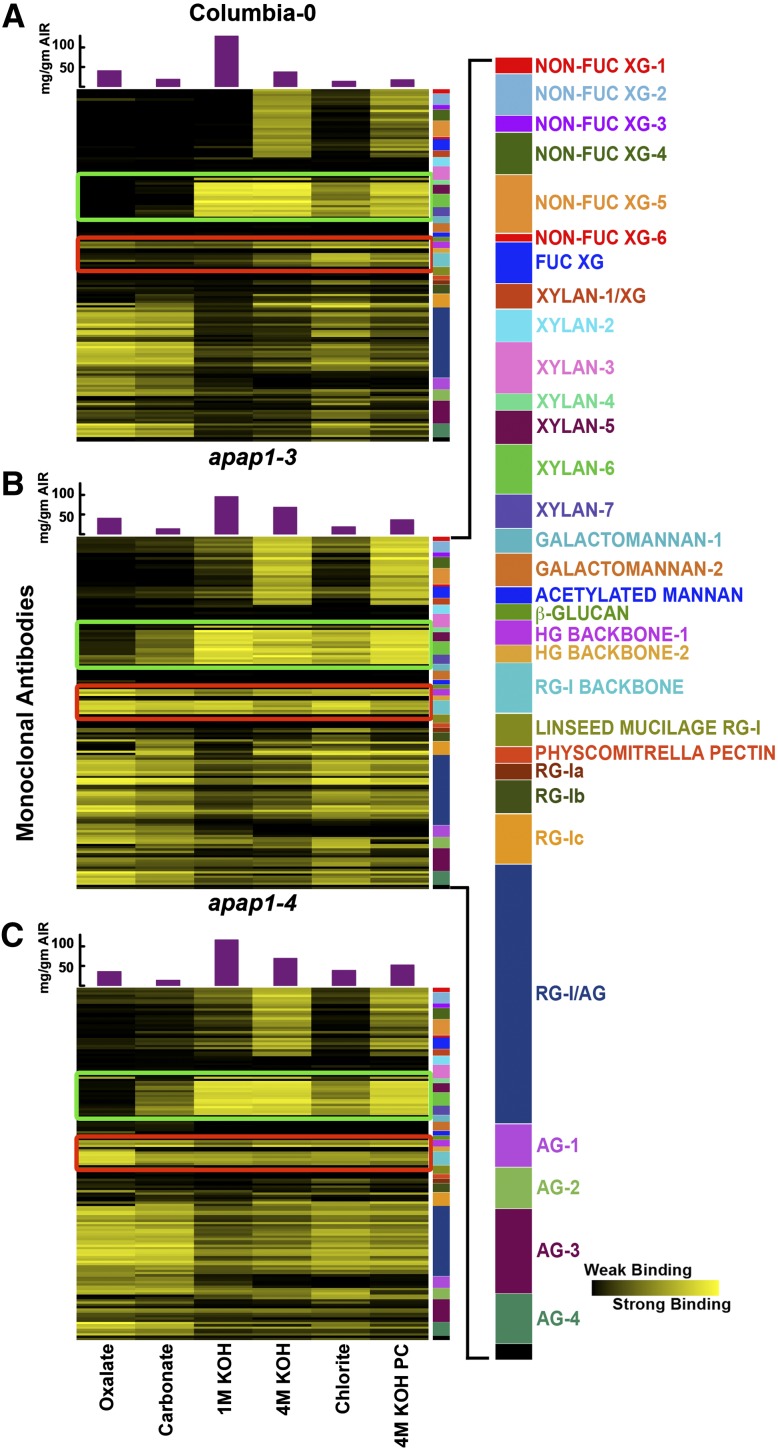
To determine if the apap1 mutation affected wall structure, glycome-profiling (Zhu et al., 2010) was used to compare extractable polysaccharides from Arabidopsis apap1 mutant versus wild-type walls. We reasoned that if the apap1 mutation reduced covalent linkages between cell wall proteins, pectins, and xylans, one or more components of the wall should be more easily extracted from the apap1 mutant walls. Because APAP1/At-AGP57C transcript is expressed in multiple Arabidopsis tissues, including rosette leaves, siliques, seeds, flowers, and the shoot apex region of the inflorescence stem (eFP browser; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), cell walls from both whole aerial tissues and from stems were analyzed. Cell walls from aerial tissues of 8-week-old apap1-3 and apap1-4 mutant plants and from the wild type were sequentially extracted with increasingly harsh solvents to release wall polymers (Zhu et al., 2010), and the wall extracts were analyzed using a comprehensive array of 155 monoclonal antibodies (Pattathil et al., 2010) reactive against diverse epitopes present on most major plant cell wall glycans (Figure 5; see Supplemental Figure 7 and Supplemental Table 4 online). There was a 5- to 83-fold (for different antibodies) increase of RG-I backbone and HG epitopes in the milder solvent (oxalate, carbonate, and 1 M KOH) fractions from apap1-3 and apap1-4 aerial tissue walls compared with the wild type (see red box, Figure 5). Likewise, a 9- to 49-fold increase in xylan-4 through xylan-7 epitopes (see green box, Figure 5) was obtained in the milder solvent (oxalate and carbonate) fractions. The two mutant lines exhibited similar changes in their glycome profiles.
Figure 5.
Glycome Profiling of Sequential Cell Wall Extracts from 8-Week-Old Arabidopsis apap1-3 and apap1-4 Mutant and Wild-Type Plants.
(A) Glycome profiling of the wild type. Panels show analyses of representative examples from multiple biological replicates (see Supplemental Figure 7 online for full set of results). Sequential cell wall extracts were prepared from the aerial portion (above the rosette leaves) of 8-week-old Arabidopsis wild-type (Columbia-0) (A), apap1-3 mutant (B), and apap1-4 mutant (C) plants. Labels at the bottom show reagents used for the different extraction steps. The amounts of material extracted in each extraction step are indicated in the bar graphs above the heat maps. Extracts were ELISA screened using 155 plant cell wall glycan-directed monoclonal antibodies (see Supplemental Table 4 online; Pattathil et al., 2010). Data are represented as heat maps. The panel on the right of the heat maps shows the antibodies used, color-coded as groups based on the principal cell wall glycans recognized by each antibody group (see Supplemental Table 4 online; Pattathil et al., 2010). Major changes in binding of specific antibodies to different mutant versus wild-type wall extracts are outlined in green (xylan groups 3 to 7) and red (HG backbone-1 and RG-1 backbone). The strength of the ELISA signal is indicated by a yellow-black scale, with bright yellow depicting strongest binding and black indicating no binding.
(B) Glycome profiling of the apap1-3 mutant as described in (A).
(C) Glycome profiling of the apap1-4 mutant as described in (A).
Monosaccharide composition analyses of the wall extracts confirmed the presence of significantly more Rha, GalA, and Xyl in the mild oxalate extract from apap1-3 mutant walls compared with the wild type (Table 3), indicating more pectin and xylan in these fractions and supporting the glycome profiling results. Because there was no significant difference in the total amount of material extracted from the walls of the wild type (269 ± 14.1 μg) versus the apap1 mutant (248.7 ± 22.4 μg), the glycome profiling and sugar composition analyses indicate that specific HG, RG-I, and xylan fractions are more easily extracted from apap1 walls. This result supports the hypothesis that some of the pectin and xylan in the apap1 mutants is held less tightly in the wall because the mutants lack the core protein of the APAP1 structure (Figure 4). Similar glycome profiling analyses of 8-week-old apap1-3 mutant and wild-type stems indicated more subtle differences in the distribution of wall glycan epitopes in cell wall extracts from mutant versus wild-type stems (see Supplemental Figure 8 online). There was a twofold to 13-fold increase in HG and RG-I backbone epitopes in the most easily extracted (oxalate) cell wall fractions from the mutant, consistent with the conclusion that a loss of the Arabidopsis APAP1 core protein AGP57C results in reduced covalent linkages in at least a fraction of pectin in Arabidopsis stem cell walls. Taken together, the results indicate that APAP1 is a structural proteoglycan in plant cell walls.
Table 3. Sugar Composition Analysis of Sequential Solvent Extracts of AIRs (Cell Walls) from apap1-3 Aerial Tissues versus Wild-Type Aerial Tissues.
| Sample |
Ara | Xyl | Fuc | Rha | Glc | Gal | Man | GlcA | GalA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| μg/mg of Each Sample | |||||||||||||||||
| Oxalate | apap1-3 | 61.6 ± 12.6 | 10.3 ± 0.3* | 5.6 ± 0.3 | 67.3 ± 5.6* | 12.0 ± 5.6 | 60.4 ± 14.2 | 6.8 ± 2.5 | 59.4 ± 14.3 | 450.8 ± 79.4* | |||||||
| Wild type | 33.7 ± 20.0 | 7.6 ± 0.8* | 4.8 ± 1.8 | 28.9 ± 4.8* | 6.2 ± 0.1 | 36.1 ± 17.7 | 2.5 ± 0.2 | 94.8 ± 19.2 | 145.1 ± 29.4* | ||||||||
| Carbonate | apap1-3 | 94.0 ± 21.6 | 49.1 ± 21.4 | 5.4 ± 2.3 | 62.9 ± 6.5 | 55.6 ± 39.2 | 62.6 ± 12.4 | 8.0 ± 2.3 | 30.0 ± 11.3 | 181.2 ± 12.9* | |||||||
| Wild type | 43.1 ± 22.4 | 12.6 ± 7.8 | 2.5 ± 0.4 | 39.3 ± 13.3 | 11.7 ± 0.7 | 45.2 ± 17.3 | 5.5 ± 3.7 | 34.5 ± 14.7 | 57.9 ± 36.3* | ||||||||
| 1 M KOH | apap1-3 | 23.9 ± 11.1 | 218.0 ± 16.5 | 0.7 ± 0.4 | 15.3 ± 3.0 | 61.6 ± 24.2 | 13.4 ± 3.3 | 3.8 ± 1.3 | 11.8 ± 5.5 | 23.4 ± 6.1 | |||||||
| Wild type | 22.6 ± 11.9 | 239.2 ± 72.9 | 1.2 ± 1.2 | 7.7 ± 1.1 | 110.3 ± 69.3 | 19.2 ± 7.6 | 2.0 ± 0.1 | 19.0 ± 0.4 | 23.0 ± 8.9 | ||||||||
| 4 M KOH | apap1-3 | 29.8 ± 6.0 | 364.4 ± 99.6 | 10.4 ± 3.2 | 20.3 ± 4.4 | 85.0 ± 22.7 | 42.2 ± 11.5 | 17.4 ± 4.3 | 25.2 ± 6.1 | 41.4 ± 14.0 | |||||||
| Wild type | 17.5 ± 11.3 | 201.7 ± 48.5 | 6.0 ± 2.8 | 12.7 ± 5.5 | 60.3 ± 17.3 | 36.5 ± 18.5 | 22.8 ± 20.3 | 16.0 ± 5.5 | 36.5 ± 14.9 | ||||||||
| Chlorite | apap1-3 | 92.4 ± 21.5 | 22.0 ± 6.5 | 5.0 ± 3.4 | 43.0 ± 14.7 | 164.1 ± 48.7 | 39.1 ± 4.9 | 3.7 ± 0.8 | 22.8 ± 13.9 | 74.0 ± 23.1 | |||||||
| Wild type | 53.5 | 5.2 | 5.2 | 25.6 | 35.0 | 45.3 | 1.3 | 7.7 | 62.6 | ||||||||
| PC 4 M KOH | apap1-3 | 68.0 ± 20.0 | 388.9 ± 82.0 | 9.9 ± 2.1 | 49.4 ± 17.4 | 83.1 ± 20.0 | 40.5 ± 5.8 | 12.8 ± 2.4 | 19.5 ± 11.9 | 96.7 ± 11.4 | |||||||
| Wild type | 18.1 ± 15.0 | 91.5 ± 46.1 | 2.5 ± 2.5 | 14.9 ± 9.5 | 25.0 ± 12.2 | 17.4 ± 11.7 | 10.8 ± 8.9 | 14.2 ± 0.2 | 37.2 ± 18.9 | ||||||||
Three biological samples (plants) of mutant and two wild-type biological samples (plants) were analyzed and the data were averaged, respectively. Data are μg/mg of each extract. The same extracts were also analyzed in the glycome profiling analyses shown in Figure 5 and Supplemental Figure 7 online. The differences between bold mutant and wild type numbers for each solvent set are statistically significant (two-tailed t test, *P value < 0.05). The numbers are presented as mean ± se. PC, postchlorite.
DISCUSSION
The core protein of Arabidopsis APAP1 has recently been classified as one of 22 classical AGPs, named AGP57C, in a bioinformatics analysis (Showalter et al., 2010). The mature AGP described here, after posttranslational modification, is a large proteoglycan that harbors three different glycan domains: type II AG, pectin, and arabinoxylan. Among the 23 Pro residues in AGP57C, 12 to 14 are predicted to be hydroxylated from which eight Hyp residues are predicted to be attachment sites for AG addition (Kieliszewski, 2001). This matches the Hyp-O-glycoside profile of APAP1 reported in this article in which 70% of the Hyp residues were shown to contain attached polysaccharides.
The two different glycosylation forms of APAP1, YS1 and YS2, contain similar amounts of acidic sugar residues (8.72 μg/100 μg YS1 and 9.57 μg/100 μg YS2) and thus have similar overall charges. However, they have significantly different glycan content, suggesting heterogeneity in their physical and chemical properties. A comparison of the Hyp:Gal:Ara:Xyl:Rha:GalA/GlcA ratios of YS1 and YS2 indicate that on average, a given Hyp residue in At-AGP57C of YS1 has ∼24 Gal, 45 Ara, 28 Xyl, seven Rha, and 13 GalA/GlcA, while in YS2 there are on average 11 Gal, 65 Ara, 65 Xyl, nine Rha, and 13 GalA/GlcA residues. Because linkage analyses indicated a terminal Xyl to internal 4-Xyl/2,4-Xyl ratio of above 10 for YS1 and approximately six for YS2, the results suggest that YS1 has two to three arabinoxylan oligomer chains of an average degree of polymerization of 11 on each Hyp-O-polysaccharide, while YS2 has six to seven arabinoxylan/xylan oligomer chains of an average degree of polymerization of 7 per Hyp-O-polysaccharide.
Because APAP1 was isolated from suspension culture media, a question that arises is whether APAP1 is a glycoconjugate generated by enzymes in the culture medium or rather is a glycoconjugate that exists in the walls of tissues in the plant. Numerous prior reports have suggested the possible existence of such complex structures in plant tissues, as summarized in Supplemental Table 11 online. Among these complex structures, pectic AGP fractions have been broadly reported from different plant tissues, including seeds of Coix lacryma-jobivar (Yamada et al., 1987), maize (Zea mays) shoots (Kato and Nevins, 1992), grape (Vitis vinifera; Pellerin et al., 1995), roots of Angelica acutiloba (Zhang et al., 1996), Humulus lupulus spent hops (Oosterveld et al., 2002), roots of Vernonia kotschyana (Nergard et al., 2005), Beta vulgaris (McKenna et al., 2006), and carrot (Daucus carota) taproot (Immerzeel et al., 2006). Also, the Xyl-to-pectin linkage was identified in a pectic AG polysaccharide isolated from leaves of Diospyros kaki (Duan et al., 2004) and might also exist in the Xyl-rich pectins purified from pea (Pisum sativum) hulls (Renard et al., 1997) and flax seed mucilage (Naran et al., 2008). Furthermore, Selvendran and colleagues (Stevens and Selvendran, 1984; Ryden and Selvendran, 1990) systematically studied wall polysaccharides extracted from diverse tissues of different plants, including runner bean (Phaseolus coccineus), mung bean (Vigna radiata), cabbage (Brassica oleracea), and tomato (Solanum lycopersicum). Based on these studies, the authors suggested the presence of wall polysaccharide-protein and wall polysaccharide-polyphenol-Hyp–rich glycoprotein complexes in those plant tissues.
The challenges associated with the definitive identification of covalent linkages between the polysaccharides and proteins in the earlier studies prevented firm conclusions to be made regarding the existence of polysaccharide-protein proteoglycans in plants. However, the early studies from multiple independent labs using diverse plant tissues and plant types support the existence of APAP1-like complexes in the walls of plant tissues and, hence, their function and biosynthesis in plants (Tan et al., 2012). Multiple and diverse methods and numerous controls were used in this study to show that the pectin and arabinoxylan domains in APAP1 are covalently attached to the AGP. These results suggest that some AGPs may serve as a type of cross-linker in cell walls, connecting at least some of the pectin and hemicellulose polysaccharides with at least one AGP, thereby providing the possibility of forming a continuous network between wall polysaccharides and wall structural proteins. This type of plant cell wall model lies between that proposed by Keegstra et al. (1973) and the more current tethered network model (Park and Cosgrove, 2012).
Why can APAP1 be secreted into the culture medium and why is APAP1 present at such a low concentration in the culture medium compared with normal AGPs? As we calculated above, a Hyp residue in APAP1 that bears a polysaccharide chain has an average of 13 GalA/GlcA residues and multiple xylan chains with an average length of seven to 11 xylosyl residues. This suggests that both the pectin and xylan domains of APAP1 are much shorter than those of native pectins (minimum of 70 to 100 GalA residues for HG regions) (Thibault et al., 1993) and xylans (up to 500 kD) (Porchia and Scheller, 2000). We hypothesize that APAP1 with longer pectin and arabinoxylan chains may exist in the cell walls and that such macromolecules are highly cross-linked and interwoven in the walls and thus cannot be released into the culture media. However, further research on such structures is needed to test this.
The location of APAP1 synthesis is yet to be determined. Pectin and xylan synthesis occurs in the Golgi (Mohnen, 2008; Brown et al., 2011), and AGP synthesis initiates in the endoplasmic reticulum and continues in the Golgi (Oka et al., 2010; Wu et al., 2010). Thus, one possible mechanism for APAP1 biosynthesis is the addition of glycans onto AGP57C by either the consecutive addition of one residue at a time or by block addition, possibly occurring within the Golgi apparatus (Atmodjo et al., 2011, 2013). This mechanism would require that the AGP, pectin, and hemicellulose biosynthetic enzymes work in a spatially and temporally coordinated fashion to synthesize such proteoglycans. Alternatively, pectin and arabinoxylan glycans may become attached to AGP in the extracellular matrix by endotransglycosylases, in a manner comparable to xyloglucan restructuring in the wall (Fry et al., 1992; Rose et al., 2002).
It has recently been proposed that some noncellulosic polysaccharides, like (1,3;1,4)-β-glucans, may be synthesized via a two-phase mechanism where oligosaccharides are synthesized in the Golgi apparatus as oligosaccharides attached to a recyclable lipid or protein, transported via vesicles to the wall, and then transferred to a growing polymer chain via a second stage of synthesis at the plasma membrane (Burton et al., 2010). Evidence to support this model is wanting. However, the transfer of pectin and arabinoxylan glycans from glycolipid intermediates onto AGP in the Golgi or to GPI-anchored AGP on the plasma membrane, followed by subsequent cleavage of the GPI anchor and deposition into the cell wall, are possible mechanisms for the synthesis of APAP1-like molecules. The results presented here do not indicate how many diverse AGPs act as core(s)/acceptor(s) for the synthesis of diverse proteoglycan structures nor do they reveal how much structural variation there is within diverse APAP1-like structures.
AGPs, pectin, and to a lesser extent arabinoxylans exist in a spectrum of structural forms in cell walls, culture media, and plant extracellular exudates, such as gums. Their multiple functions include providing structural support to the wall and serving as carbohydrate reserves, signaling molecules, cell–cell adhesion factors, and water retention polymers (Ellis et al., 2010). The identification of APAP1 suggests that at least one AGP serves as a cross-linker for at least a subfraction of pectin and arabinoxylan. The existence of APAP1 in plant walls has significant consequences for our understanding of wall architecture and function and potentially for engineering plant cell walls for improved agronomic and renewable biomaterial uses.
METHODS
Isolation of YS1 and YS2 from Culture Media
Arabidopsis thaliana (cv Columbia-0) cells were cultured as described earlier (Xu et al., 2008). AGP fractions from Arabidopsis culture media were purified and separated into seven subfractions by diethylaminoethyl anion exchange (16 × 700 mm; Amersham-Pharmacia Biotech), Superose 12 size exclusion (16 × 500 mm; Amersham-Pharmacia Biotech), and PRP-1 reverse phase (10 µm, C-18, 7 × 305 mm; Hamilton) chromatography as previously reported (Xu et al., 2008). The seven fractions were identified as containing AGPs based on precipitate formation upon addition of β-Gal Yariv reagent to aliquots from each fraction. Fraction 3 from the PRP-1 column (Figure 1A) was shown to contain Xyl residues by glycosyl residue composition analysis of alditol acetates. The lyophilized fraction 3 (70 mg) was dissolved in 70 mL of water and mixed with an equal volume of 1 mg/mL β-galactosyl Yariv reagent. After incubation (25°C, 250 rpm) for 1 h, the mixture was centrifuged (10,000g, 10 min). The supernatant (termed YS) was removed from the pellet (termed Yariv precipitate) and dialyzed (10,000 molecular weight cut-off; Spectrum) extensively against double-distilled water until the solution became colorless. The YS dialysate was lyophilized, dissolved in 3 mL Superose buffer (200 mM sodium phosphate, pH 7), and separated on a preparative Superose-12 column (16 × 500 mm; Amersham-Pharmacia Biotech; equilibrated with the same buffer at 1 mL/min). The column effluent was monitored by UV detection, and two fractions absorbing at 220 nm were detected and collected. The fraction representing the first peak was collected (YS-S12). The second peak was residual Yariv reagent.
The YS-S12 fraction was further purified on an analytical PRP-1 reverse phase column (5 µm, C-18, 4.1 × 150 mm; Hamilton), with a gradient from solvent A (0.1% [v/v] TFA aqueous) to 50% solvent B (80% [v/v] acetonitrile in 0.1% [v/v] TFA aqueous) over 100 min at 0.5 mL/min. Two fractions (peak 1, designated YS1; and shoulder peak 2, designated YS2 [Yariv soluble 1 and 2, respectively]) with UV absorbance at 220 nm were collected and freeze-dried. The material at the cross region between YS1 and YS2 was not collected.
Protein Deglycosylation via Anhydrous HF Treatment
Freeze-dried YS2 (9 mg) was solubilized in 500 μL of anhydrous HF containing 10% (v/v) anhydrous methanol and the peptide deglycosylated on ice as previously described (Tan et al., 2003).
Automated Edman Degradation
N-terminal amino acid sequencing of the intact glycoprotein and the HF-deglycosylated protein was done on a 477A Applied Biosystems gas phase sequencer at the Macromolecular Facility, Michigan State University (East Lansing, MI).
Hyp-O-Glycoside Profiling
Fourteen milligrams of YS1 and 13.6 mg of YS2 were each hydrolyzed in 2 mL 0.44 n NaOH (105°C, 18 h) and then chilled at 4°C for 10 min. The chilled hydrolysates were neutralized with 1 M HCl, freeze-dried, redissolved in 500 μL of double-distilled water, and loaded onto a Chromo-Beads C2 cation exchange chromatography column (75 × 0.6 cm; Technicon) previously equilibrated with double-distilled water at 0.5 mL/min. Hyp-O-glycosides were separated and detected with an automated Hyp analyzer as previously described (Lamport and Miller, 1971).
Glycosyl Residue Composition and Linkage Analysis
For neutral sugar analysis, 60 μg of each sample was hydrolyzed in 200 μL of 2 n TFA at 121°C for 1 h. The hydrolysate was dried at 50°C under a stream of nitrogen gas, and the free sugars were converted to alditol acetate derivatives and analyzed by gas chromatography–flame ionization detection as described (Tan et al., 2003). Total uronic acids were measured colorimetrically using 100 μg of each sample with m-hydroxydiphenyl and using GlcUA as the standard as described earlier (Tan et al., 2003).
A Dionex ICS-3000 HPLC system was used to determine the monosaccharide composition of some samples. The TFA hydrolysates were separated on a PA20 column (Dionex) at high pH and detected using an electrochemical detector in the carbohydrate mode. The buffers were buffer A, nanopure water; buffer B, 200 mM NaOH; and buffer C, 1 M NaOAc. The gradient was as follows: 0 min, 1% buffer B; 0.1 min, 10% buffer B; 2 min, 10% buffer B; 4 min, 1% buffer B; 15 min, 0% buffer B; 25 min, 5% buffer B and 10% buffer C; 30 min, 5% buffer B and 50% buffer C; and 35 min, 1% buffer B. The flow rate was at 0.5 mL/min.
Total sugar analysis for quantification of both neutral and acidic sugars by gas chromatography–mass spectrometry (GC-MS) analysis of trimethylsilyl derivatives and linkage analysis were conducted at the Complex Carbohydrate Research Center Analytical Services, University of Georgia. Glycosyl residue composition analysis was performed by combined GC-MS of per-O-trimethylsilyl derivatives of monosaccharide methyl glycosides produced from the sample by acidic methanolysis. For glycosyl linkage analysis, the samples were permethylated, depolymerized, reduced, and acetylated, and the resulting partially methylated alditol acetate residues were analyzed by GC-MS (York et al., 1985).
Fractionation of Hyp-O-Glycosides on Superdex-75 Column
Hyp-O-polysaccharides (750 µg) from YS1 or YS2 isolated following Chromo-Beads C2 cation exchange chromatography were dissolved in 1 mL of double-distilled water and further fractionated on an analytical Superdex-75 size exclusion column (10-mm i.d., 300 mm in length; Amersham-Pharmacia). The column eluent was acetonitrile/water (20/80, v/v) at 0.3 mL/min, and fractions (0.3 mL/fraction) were collected. All fractions were freeze-dried and those with abundant materials were analyzed for neutral sugar and uronic acid compositions (Tan et al., 2004).
β-Elimination of YS1 and YS2
One milligram each of YS1 and YS2 was dissolved in 1 mL of 0.2 M NaOH in 1 M Na2SO3 solution. The mixtures were incubated at 50°C for 5 h, cooled on ice, and neutralized with 0.1 M HCl (Kieliszewski et al., 1992). The glycoproteins after β-elimination were repurified on an analytical PRP-1 column as described above.
Preparation of RG-I from Cell Walls of Arabidopsis Suspension-Cultured Cells
The RG-I preparation Ara101 was isolated from the walls of suspension-cultured Arabidopsis wild-type cells (Guillaumie et al., 2003) basically as described (York et al., 1985) with the following modifications. In brief, the cells were harvested, and alcohol-insoluble residue (AIR) of the cell walls was prepared. The dried AIR was deesterified overnight at pH 12.0 at 4°C. The reaction was stopped by lowering the pH to 5.2 with 1 M HCl and the mixture dialyzed and lyophilized. The pectic polysaccharides were solubilized from the AIR by treatment with a purified Aspergillus niger endo-α-1,4-polygalacturonase (gift of Carl Berfmann, Complex Carbohydrate Research Center), and the solubilized material was dialyzed and lyophilized. The resulting pectic mixture was fractionated in 50 mM sodium acetate, pH 5.2, by SEC on a Sephadex G-75 column (GE Healthcare Biosciences), yielding three populations based on uronic acid colorimetric assays. The material in the largest population size (named Ara101, RG-I) was pooled, dialyzed, and freeze-dried.
Ten milligrams of Ara101 was dissolved in double-distilled water and purified on a reverse-phase column using the same conditions as described above for YS1 and YS2. The resulting pectic proteoglycan was named Ara101P.
Proteomic Analysis of Ara101P
One milligram of Ara101P was deglycosylated with anhydrous HF as described above. The HF-treated material was dried under nitrogen and dissolved in 400 μL of 2% NH4HCO3 in 5 mM CaCl2. A 30-μL aliquot of 0.1 µg/μL of sequence-grade trypsin was added to the solution, followed by incubation at room temperature for 24 h. The tryptic peptides of HF-treated Ara101P were analyzed on an Agilent 1100 capillary LC interfaced directly to an LTQ linear ion trap mass spectrometer (Thermo Electron). Buffers A and B were 0.1% formic acid in water and 0.1% formic acid in acetonitrile, respectively. Peptides were eluted from a C18 column into the mass spectrometer during a 75-min linear gradient from 5 to 60% buffer B at a flow rate of 4 μL/min. The instrument was set to acquire MS/MS spectra on the nine most abundant precursor ions from each mass spectrometry scan with a repeat count of 3 and repeat duration of 10 s. Dynamic exclusion was enabled for 200 s. Generated raw MS/MS were converted into mzXML format and then into PKL format using ReAdW followed by mzMXL2Other (Pedrioli, et al., 2004). The peak lists were searched using Mascot 2.2 software (Matrix Science).
Database searches were performed against the annotated proteins from Arabidopsis genes obtained from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) using the following parameters: full tryptic enzymatic cleavage with two possible missed cleavages, peptide tolerance of 1000 ppm, and fragment ion tolerance of 0.6 D. Variable modification was set as carbamidomethyl due to carboxyamidomethylation of Cys residues (+57 D), oxidation of Met residues (+16 D), and hydroxylation of Pro (+16 D).
Separation of PGA by Reverse Phase Chromatography
One milligram of crude Sigma-Aldrich PGA (P-1879; >66 kD) was dissolved in 400 μL of double-distilled water and separated on an analytical PRP-1 reverse-phase column as described above. The PGA portion that voided on the column was collected and lyophilized.
Attachment of YS2 to Magnetic Amine Beads
The covalent attachment of amino-containing material in YS2 onto BioMag Plus Amine beads (Bangs Laboratories) was done following the manufacturer’s manual. Briefly, the beads in their free amino group form were incubated with rotation with 5% (v/v) glutaraldehyde for 6 h at room temperature on a rotary shaker. The supernatant was discarded and the beads were washed six times with pyridine wash buffer (10 mM pyridine adjusted to pH 6.0 with 6 n HCl). YS2 (100 μg) was dissolved in 1 mL pyridine wash buffer, mixed with 1 mg of the above washed beads, and incubated overnight at room temperature on a rotary shaker. Following incubation, the unbound material was removed and 0.5 mL of quenching solution (1 M Gly) was added. The reaction was rotated for 1 h at room temperature to block any unreacted glutaraldehyde residues on the bead surface. The quenching solution was discarded, the beads were washed 10 times with 1.5 mL wash buffer (10 mM Tris-HCl, pH 7.4, 0.1% [w/v] sodium azide, 0.1% [w/v] BSA, 1 M NaCl, and 10 mM M EDTA), and the sample was stored in wash buffer at 4°C until use.
2-AB Labeling
The reductive alkylation labeling of free reducing ends of YS2, YS2 Hyp-O-polysaccharide fraction 24, purified PGA (Sigma-Aldrich; P-1879), and an RG-I–enriched fraction (gift of C. Deng, University of Georgia) was performed by reaction with 2-AB as described (Ishii et al., 2002). About 100 μg of each sample was used in the labeling reactions, and after 1 h at 80°C, the products from each reaction were purified on a Sephadex LH-20 column (1 ×30 cm; GE Healthcare) to separate free 2-AB from labeled product. The labeled product was eluted by gravity using 20% (v/v) aqueous ethanol. The lyophilized 2-AB–treated product was further purified on the analytical PRP-1 column as described above.
Enzymatic Digestion of APAP1
One hundred micrograms of YS2 was dissolved in 200 μL of 50 mM NaOAc, pH 5.5. One microliter of the corresponding enzyme (RGH [Novozyme, 1 μg/μL; Kofod et al., 1994], endo-β-d-xylanase GH10 [Cj-Xyn10A, 10 mM], endo-β-d-xylanase GH11 [Np-Xyn11A, 15 mM], and endo-β-d-xylanase GH5 [Ct-Xyl5A, 1 mM] [the latter three gifts of Harry Gilbert]) was added individually to tubes and the tubes incubated at 37°C for 24 h on a rotary shaker. The released oligosaccharides voided on a PRP-1 analytical reverse-phase column and separated from the remaining bound proteoglycan using the conditions described above.
CID-MS/MS Analysis of YS2 Oligomers Released by RGH
The glycan fragments released from 100 μg of YS2 pretreated with RGH (see above) were analyzed on a linear ion trap mass spectrometer (LTQ; ThermoFisher). The glycans were dissolved in a total of 50 μL of solvent (15 μL of 100% methanol and 35 μL of 1 mM NaOH in 50% methanol) and infused directly via a syringe pump (0.4 μL/min) into the mass spectrometer using a nanospray ion source with a fused-silica emitter (360 × 75 × 30 μm; SilicaTip; New Objective) at 2.2 kV capillary voltage and 200°C capillary temperature. Full ion trap mass spectrometry spectra performed in positive mode and profile mode were collected between 400 and 2000 m/z for 30 s with five microscans and 150-ms maximum injection time. The centroid MS/MS spectra following CID were obtained from 400 to 2000 m/z at 40% normalized collision energy, 0.25 activation Q, and 30.0-ms activation time by total ion mapping. Parent mass step size and isolation width were set at 2.0 and 2.8 m/z, respectively, for automated MS/MS spectra with total ion mapping scans.
NMR Analysis
Five milligrams of YS2 were dissolved in 500 μL of 99.996% deuterium oxide. Three hundred micrograms of Hyp-O-polysaccharide fractions 26 and 27 (YS1-HP2627), generated from YS1 by base treatment and fractionated on the Superdex 75 column as described above, was dissolved in 150 μL of 99.996% deuterium oxide, as was 400 μg of YS2 Hyp-O-polysaccharide fractions 26 and 27 (YS2-HP2627). Samples were analyzed either on a Bruker-DRX800 equipped with a cryoprobe at the Campus Chemical and Instrument Center, Ohio State University, or on a Varian VNMRS 600 or VNMRS 800 equipped with a 3-mm cold probe at the Complex Carbohydrate Research Center. NMR experiments were performed either at 25°C or higher (30 to 55°C). The one-dimensional 1H NMR, two-dimensional 1H homonuclear COSY, TOCSY (DIPSI2 mixing time 60 ms), NOESY (NOE mixing time 200 or 100 ms), two-dimensional heteronuclear 1H-13C HSQC, HMQC, and HMBC experiments were performed as described previously (Tan et al., 2010).
Plant Genotyping and Transcript Analysis by qPCR
Seeds from SALK T-DNA insertion lines SALK_070113c/apap1-3 and SALK_002144/apap1-4 were obtained from the ABRC. Seeds from wild-type Arabidopsis (Columbia-0) and the two insertion lines were sterilized and germinated on half-strength Murashige and Skoog plates as described (Persson et al., 2007). Two-week-old seedlings were transferred to soil and maintained in a 14/10 light/dark cycle (14 h at 19°C; 150 µEi m−2 s−1/10 h at 15°C).
Fresh leaf samples (1 cm2) from individual wild-type and mutant plants were collected, and DNA was isolated and genotyped using a REDExtract-N-Amp plant PCR kit (Sigma-Aldrich). The following primers were used for PCR reactions: primers for SALK_002144 left primer (LP), 5′-CAAGTGTTTCGCACCTTTCTC-3′; right primer (RP), 5′-TTCATATCAAACAATATTTTTGAACTC-3′, for SALK_070113c (LP, 5′-GGTTCGGATATCTTTCGGTTC-3′; RP, 5′-GCAACTCCAACTTTCTTCCC-3′), and the T-DNA primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′).
Total RNA was isolated from plant leaves using the RNeasy plant mini kit (Qiagen Sciences). First-strand cDNA was synthesized from 850 ng of RNA using the SuperScript III first-strand synthesis super mix (Invitrogen) in reactions incubated for 50 min at 50°C. One microliter of cDNA from each sample was amplified using primer 1, 5′-GATGCTAAGTCTCGTACTCGTC-3′, and primer 2, 5′-CTCTTGCCGTTTCTTGTACAC-3′, with an annealing temperature of 55°C and 28 cycles of 95°C, 30 s; 55°C, 30 s; and 72°C, 1 min.
SYBR Green–based qPCR was performed with q-primer-F, 5′-TCGCCGGATTTGTGTACAAG-3′, and q-primer-R, 5′-AATCTCTCTGGCGGCGTAAC-3′ (generating a 74-bp amplicon) on a CFX96TM real-time PCR detection system (Bio-Rad) following the manufacturer’s instructions. ACTIN2 was used as the reference gene. The qPCR primers for ACTIN2 included the sense primer 5′-GGTAACATTGTGCTCAGTGGTGG-3′ and the antisense primer 5′-AACGACCTTAATCTTCATGCTGC-3′. Each reaction was repeated three times using cDNA generated from independently isolated RNA preparations. The PCR cycles included initial polymerase activation at 95°C for 3 min, then 40 cycles of 10 s at 95°C and 30 s at 60°C. Melt curve analyses from 65 to 95°C were performed after each run to ensure single size amplicon production. Data are mean ± sd of two biological samples. The data were analyzed as described by Livak and Schmittgen (2001).
Cell Wall Fractionation for Glycome Profiling
Fresh samples (all plant materials above rosette leaves) from each plant were collected and ground immediately under liquid nitrogen to a fine powder. Each powder was sequentially washed with 80% (v/v) ethanol, 95% (v/v) ethanol, CHCl3/methanol (1:1, v/v), and acetone with agitation. The insoluble residues (AIRs) were air-dried in a hood. Sequential extraction of cell walls (AIR) was done with increasingly harsh reagents in the following order: 50 mM ammonium oxalate (oxalate), 50 mM sodium carbonate with 0.5% (w/v) of sodium borohydride (carbonate), 1 M KOH with 1% (w/v) of sodium borohydride, 4 M KOH with 1% (w/v) of sodium borohydride, 100 mM sodium chlorite (Chlorite), and postchlorite 4 M KOH with 1% (w/v) of sodium borohydride treatment (4 M KOH PC) to isolate fractions enriched in cell wall components as previously described (Zhu et al., 2010). The 1 M KOH, 4 M KOH, and 4 M KOH PC fractions were neutralized using glacial acetic acid. All extracts were dialyzed with four changes of de-ionized water (sample:water ∼1:60) at room temperature for a total of 48 h and then lyophilized.
Total sugars in the cell wall fractions were quantified using the phenol sulphuric acid method (Masuko et al., 2005), and ELISA analyses were done as previously described (Pattathil et al., 2010; Zhu et al., 2010).
Monoclonal Antibodies
Monoclonal antibodies against different cell wall glycans were obtained as hybridoma cell culture supernatants from laboratory stocks at the Complex Carbohydrate Research Center (CCRC). The CCRC, JIM, and MAC series of antibodies are available from CarboSource Services (www.carbosource.net). LAMP and BG-1 antibodies are available from Biosupplies Australia, Parkville, Victoria, Australia, and were used as per the manufacturer’s instructions. ELISA analyses of YS2 with different antibodies were done as previously described (Pattathil et al., 2010; Zhu et al., 2010).
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under accession number At3g45230.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ELISA Analysis of YS2 Using 47 Monoclonal Antibodies Reactive against Plant Cell Wall Glycans.
Supplemental Figure 2. NMR Spectra of YS2-HP2627 Collected at 25°C on a Varian VNMRS 800 Instrument.
Supplemental Figure 3. CID-MS/MS Analysis of Oligosaccharides Released from YS2 by Treatment with RGH Indicates the Presence of RG-I and HG Domains in YS2 and the Existence of a Covalent Xylan to RG-I Linkage in YS2.
Supplemental Figure 4. NMR Spectra of YS2 Collected at 55°C on a Bruker DRX800 Instrument.
Supplemental Figure 5. Purification of Arabidopsis Cell Wall RG-I–Enriched Fraction.
Supplemental Figure 6. Analysis of APAP1 Transcript Level in apap1 Mutants.
Supplemental Figure 7. Glycome Profiling of Sequential Cell Wall Extracts from 8-Week-Old Arabidopsis thaliana apap1-3 and apap1-4 Mutant and Wild-Type Plants.
Supplemental Figure 8. Difference Heat Map of the Glycome Profiling Data from 8-Week-Old Arabidopsis thaliana apap1 and Wild-Type Stems.
Supplemental Table 1. Glycosyl Residue Composition (Mole Percentage) of Neutral and Acidic Sugars in Arabidopsis AGPs Isolated by a Combination of Anion Exchange, Gel Filtration, and Reverse-Phase Liquid Chromatography.
Supplemental Table 2. Sugar Composition of YS2 and Ara101P after Covalent Attachment onto Magnetic Amine Beads.
Supplemental Table 3. Sugar Composition of YS2 before and after Treatment with Rhamnogalacturonan Hydrolase and Endo-β-d-Xylanase GH10.
Supplemental Table 4. List of Plant Cell Wall Glycan-Directed Monoclonal Antibodies Used for ELISA (Supplemental Figure 1) and Glycome Profiling Analyses (Figure 5; Supplemental Figures 7 and 8).
Supplemental Table 5. Sugar Composition (Mole Percentage) of APAP1 YS1 and β-Eliminated YS1.
Supplemental Table 6. Hyp-Glycoside Profile of YS1 and YS2.
Supplemental Table 7. Sugar Composition of YS1 and YS2 Hyp-O-Polysaccharides.
Supplemental Table 8. Chemical Shift Assignments of YS1-HP2627.
Supplemental Table 9. Chemical Shift Assignment of YS2-HP2627.
Supplemental Table 10. Chemical Shift Assignments of YS2.
Supplemental Table 11. Prior Published Evidence Suggesting the Existence of APAP1-Like Wall Polymer Complex Structure in Plants.
Acknowledgments
We thank C. Bergman for use of HPLC equipment and H. Gilbert for the enzymes Cj-Xyn10A, Np-Xyn11A, and Ct-Xyl5A. We thank Daniel Cosgrove, Maor Bar-Peled, and William York for critical reading of the article. This research was supported by a National Science Foundation award (NSF-MCB 0646109 to D.M.) and partially by the Department of Energy Center Grant DOE DE-FG02-09ER20097 and BioEnergy Science Center Grant DE-AC05-00OR22725. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science. The generation of the CCRC series of glycan-directed monoclonal antibodies used in this work was supported by the National Science Foundation Plant Genome Program (Award DBI-0421683 to M.G.H.).
AUTHOR CONTRIBUTIONS
All the authors designed the research. L.T., S.E., S.P., C.W., J.G., C.Y., Z.H., X.Z., U.A., J.S.M., D.B., and C.P. performed the experiments. L.T., S.P., J.G., C.Y., Z.H., X.Z., M.G.H., M.J.K, and D.M. analyzed the data and contributed to data interpretation and the article preparation. L.T. and D.M. supervised the study and wrote the article.
Glossary
- HRGP
Hyp-rich glycoprotein
- AGP
arabinogalactan protein
- HG
homogalacturonan
- GalA
galacturonic acid
- RG-I
rhamnogalacturonan I
- AG
arabinogalactan
- Rha
rhamnose
- GlcA
glucuronic acid
- GPI
glycosylphosphatidylinositol
- 2-AB
2-amino-benzoamide
- PGA
polygalacturonic acid
- RGH
rhamnogalacturonan hydrolase
- HSQC
heteronuclear single quantum coherence
- Hyp-O-AG
Hyp-O-arabinogalactan
- COSY
1H homonuclear correlation spectroscopy
- TOCSY
total correlation spectroscopy
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- HMQC
heteronuclear multiple quantum coherence
- CID
collision-induced dissociation
- MS2
mass spectrometry
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
- qPCR
quantitative PCR
- HMBC
heteronuclear multiple-bond correlation
- TFA
trifluoroacetic acid
- GC-MS
gas chromatography–mass spectrometry
- AIR
alcohol-insoluble residue
- SEC
size-exclusion chromatography
- HF
hydrogen flouride
References
- Albersheim, P., Darvill, A., Roberts, K., Sederoff, R., and Staehelin, A. (2011). Plant Cell Walls. (New York: Garland Science, Taylor & Francis Group). [Google Scholar]
- Atmodjo, M.A., Hao, Z., and Mohnen, D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64: in press. [DOI] [PubMed]
- Atmodjo M.A., Sakuragi Y., Zhu X., Burrell A.J., Mohanty S.S., Atwood J.A., III, Orlando R., Scheller H.V., Mohnen D. (2011). Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:Galacturonosyltransferase complex. Proc. Natl. Acad. Sci. USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biely P., Vrsanská M., Tenkanen M., Kluepfel D. (1997). Endo-β-1,4-xylanase families: Differences in catalytic properties. J. Biotechnol. 57: 151–166 [DOI] [PubMed] [Google Scholar]
- Brown D., Wightman R., Zhang Z., Gomez L.D., Atanassov I., Bukowski J.P., Tryfona T., McQueen-Mason S.J., Dupree P., Turner S. (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 66: 401–413 [DOI] [PubMed] [Google Scholar]
- Burton R.A., Gidley M.J., Fincher G.B. (2010). Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 6: 724–732 [DOI] [PubMed] [Google Scholar]
- Caffall K.H., Mohnen D. (2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 37 (Database issue): D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N.C., Gibeaut D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Correia M.A.S., Mazumder K., Brás J.L.A., Firbank S.J., Zhu Y., Lewis R.J., York W.S., Fontes C.M.G.A., Gilbert H.J. (2011). Structure and function of an arabinoxylan-specific xylanase. J. Biol. Chem. 286: 22510–22520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvill J.E., McNeil M., Darvill A.G., Albersheim P. (1980). Structure of plant cell walls XI. Glucuronoarabinoxylan, a second hemicellulose in primary cell walls of suspension-cultured sycamore cells. Plant Physiol. 66: 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defaye J., Wong E. (1986). Structural studies of gum arabic, the exudate polysaccharide from Acacia senegal. Carbohydr. Res. 150: 221–231 [Google Scholar]
- Doering A., Lathe R., Persson S. (2012). An update on xylan synthesis. Mol. Plant 5: 769–771 [DOI] [PubMed] [Google Scholar]
- Duan J., Zheng Y., Dong Q., Fang J.-N. (2004). Structural analysis of a pectic polysaccharide from the leaves of Diospyros kaki. Phytochemistry 65: 609–615 [DOI] [PubMed] [Google Scholar]
- Ellis M., Egelund J., Schultz C.J., Bacic A. (2010). Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S.C. (1982). Isodityrosine, a new cross-linking amino acid from plant cell-wall glycoprotein. Biochem. J. 204: 449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S.C., Smith R.C., Renwick K.F., Martin D.J., Hodge S.K., Matthews K.J. (1992). Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 282: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gane A.M., Craik D., Munro S.L.A., Howlett G.J., Clarke A.E., Bacic A. (1995). Structural analysis of the carbohydrate moiety of arabinogalactan-proteins from stigmas and styles of Nicotiana alata. Carbohydr. Res. 277: 67–85 [DOI] [PubMed] [Google Scholar]
- Guillaumie F., Sterling J.D., Jensen K.J., Thomas O.R.T., Mohnen D. (2003). Solid-supported enzymatic synthesis of pectic oligogalacturonides and their analysis by MALDI-TOF mass spectrometry. Carbohydr. Res. 338: 1951–1960 [DOI] [PubMed] [Google Scholar]
- Harholt J., Suttangkakul A., Vibe Scheller H. (2010). Biosynthesis of pectin. Plant Physiol. 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P.J., Trethewey J.A.K. (2010). The distribution of ester-linked ferulic acid in the cell walls of angiosperms. Phytochem. Rev. 9: 19–33 [Google Scholar]
- Held M.A., Tan L., Kamyab A., Hare M., Shpak E., Kieliszewski M.J. (2004). Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J. Biol. Chem. 279: 55474–55482 [DOI] [PubMed] [Google Scholar]
- Immerzeel P., Eppink M.M., de Vries S.C., Schols H.A., Voragen A.G.J. (2006). Carrot arabinogalactan proteins are interlinked with pectins. Physiol. Plant. 128: 18–28 [Google Scholar]
- Ishii T. (1997). Structure and function of feruloylated polysaccharide. Plant Sci. 127: 111–127 [Google Scholar]
- Ishii T., Hiroi T. (1990a). Linkage of phenolic acids to cell-wall polysaccharides of bamboo shoot. Carbohydr. Res. 206: 297–310 [DOI] [PubMed] [Google Scholar]
- Ishii T., Hiroi T. (1990b). Isolation and characterization of feruloylated arabinoxylan oligosaccharides from bamboo shoot cell-walls. Carbohydr. Res. 196: 175–183 [DOI] [PubMed] [Google Scholar]
- Ishii T., Ichita J., Matsue H., Ono H., Maeda I. (2002). Fluorescent labeling of pectic oligosaccharides with 2-aminobenzamide and enzyme assay for pectin. Carbohydr. Res. 337: 1023–1032 [DOI] [PubMed] [Google Scholar]
- Ishii T., Matsunaga T., Pellerin P., O’Neill M.A., Darvill A., Albersheim P. (1999). The plant cell wall polysaccharide rhamnogalacturonan II self-assembles into a covalently cross-linked dimer. J. Biol. Chem. 274: 13098–13104 [DOI] [PubMed] [Google Scholar]
- Kato Y., Nevins D.J. (1992). Structural characterization of an arabinoxylan-rhamnogalacturonan complex from cell walls of Zea shoots. Carbohydr. Res. 227: 315–329 [DOI] [PubMed] [Google Scholar]
- Keegstra K., Talmadge K.W., Bauer W.D., Albersheim P. (1973). The structure of plant cell walls III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 51: 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski M.J. (2001). The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57: 319–323 [DOI] [PubMed] [Google Scholar]
- Kieliszewski M.J., Kamyab A., Leykam J.F., Lamport D.T.A. (1992). A histidine-rich extensin from Zea mays is an arabinogalactan protein. Plant Physiol. 99: 538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofod L.V., Kauppinen S., Christgau S., Andersen L.N., Heldt-Hansen H.P., Dorreich K., Dalboge H. (1994). Cloning and characterization of two structurally and functionally divergent rhamnogalacturonases from Aspergillus aculeatus. J. Biol. Chem. 269: 29182–29189. [PubMed] [Google Scholar]
- Kulkarni A.R., et al. (2012). The ability of land plants to synthesize glucuronoxylans predates the evolution of tracheophytes. Glycobiology 22: 439–451 [DOI] [PubMed] [Google Scholar]
- Lamport, D.T.A., and Miller, D.H. (1971). Hydroxyproline arabinosides in the plant kindom. Plant Physiol. 48: 454–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Masuko T., Minami A., Iwasaki N., Majima T., Nishimura S.I., Lee Y.C. (2005). Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 339: 69–72 [DOI] [PubMed] [Google Scholar]
- Mazumder K., York W.S. (2010). Structural analysis of arabinoxylans isolated from ball-milled switchgrass biomass. Carbohydr. Res. 345: 2183–2193 [DOI] [PubMed] [Google Scholar]
- McKenna C., Al-Assaf S., Phillips G.O., Funami T. (2006). The protein component in pectin: Is it an AGP? FFI J. 211: 264–274 [Google Scholar]
- Mohnen D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Furuta H., Maeda H., Takao T., Nagamatsu Y. (2002). Analysis of the molecular construction of xylogalacturonan isolated from soluble soybean polysaccharides. Biosci. Biotechnol. Biochem. 66: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Naran R., Chen G., Carpita N.C. (2008). Novel rhamnogalacturonan I and arabinoxylan polysaccharides of flax seed mucilage. Plant Physiol. 148: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergard C.S., Matsumoto T., Inngjerdingen M., Inngjerdingen K., Hokputsa S., Harding S.E., Michaelsen T.E., Diallo D., Kiyohara H., Paulsen B.S., Yamada H. (2005). Structural and immunological studies of a pectin and a pectic arabinogalactan from Vernonia kotschyana Sch. Bip. ex Walp. (Asteraceae). Carbohydr. Res. 340: 115–130 [DOI] [PubMed] [Google Scholar]
- Oka T., Saito F., Shimma Y.I., Yoko-o T., Nomura Y., Matsuoka K., Jigami Y. (2010). Characterization of endoplasmic reticulum-localized UDP-D-galactose:hydroxyproline O-galactosyltransferase using synthetic peptide substrates in Arabidopsis. Plant Physiol. 152: 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveld A., Voragen A.G.J., Schols H.A. (2002). Characterization of hop pectins shows the presence of an arabinogalactan-protein. Carbohydr. Polym. 49: 407–413 [Google Scholar]
- Pattathil S., et al. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paës G., Berrin J.G., Beaugrand J. (2012). GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol. Adv. 30: 564–592 [DOI] [PubMed] [Google Scholar]
- Park Y.B., Cosgrove D.J. (2012). Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 158: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M., Albersheim P., Darvill A., York W.S. (1999). Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J. 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Pedrioli P.G.A., et al. (2004). A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22: 1459–1466 [DOI] [PubMed] [Google Scholar]
- Pellerin P., Vidal S., Williams P., Brillouet J.-M. (1995). Characterization of five type II arabinogalactan-protein fractions from red wine of increasing uronic acid content. Carbohydr. Res. 277: 135–143 [DOI] [PubMed] [Google Scholar]
- Persson S., Caffall K.H., Freshour G., Hilley M.T., Bauer S., Poindexter P., Hahn M.G., Mohnen D., Somerville C. (2007). The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder G.R., Richards G.N. (1997). Arabinogalactan from western larch, Part III: Alkaline degradation revisited, with novel conclusions on molecular structures. Carbohydr. Polym. 34: 251–261 [Google Scholar]
- Porchia A.C., Scheller H.V. (2000). Arabinoxylan biosynthesis: identification and partial characterization of a β-1,4-xylosyltransferase from wheat. Physiol. Plant. 110: 350–356 [Google Scholar]
- Renard C.M.G.C., Weightman R.M., Thibault J.-F. (1997). The xylose-rich pectins from pea hulls. Int. J. Biol. Macromol. 21: 155–162 [DOI] [PubMed] [Google Scholar]
- Rose J.K.C., Braam J., Fry S.C., Nishitani K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Ryden P., Selvendran R.R. (1990). Cell-wall polysaccharides and glycoproteins of parenchymatous tissues of runner bean (Phaseolus coccineus). Biochem. J. 269: 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller H.V., Ulvskov P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Showalter A.M. (2001). Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 58: 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A.M., Keppler B., Lichtenberg J., Gu D., Welch L.R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153: 485–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B.J.H., Selvendran R.R. (1984). Hemicellulosic polymers of cabbage leaves. Phytochemistry 23: 339–347 [Google Scholar]
- Tan L., Leykam J.F., Kieliszewski M.J. (2003). Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol. 132: 1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Qiu F., Lamport D.T.A., Kieliszewski M.J. (2004). Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279: 13156–13165 [DOI] [PubMed] [Google Scholar]
- Tan, L., Showalter, A.M., Egelund, J., Hernandez-Sanchez, A., Doblin M.S., and Bacic, A. (2012). Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 3: 140. [DOI] [PMC free article] [PubMed]
- Tan L., Varnai P., Lamport D.T.A., Yuan C., Xu J., Qiu F., Kieliszewski M.J. (2010). Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285: 24575–24583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault J.-F., Renard C.M.G.C., Axelos M.A.V., Roger P., Crepeau M.-J. (1993). Studies of the length of homogalacturonic regions in pectins by acid hydrolysis. Carbohydr. Res. 238: 271–286 [Google Scholar]
- Wu Y.Y., Williams M., Bernard S., Driouich A., Showalter A.M., Faik A. (2010). Functional identification of two nonredundant Arabidopsis α(1,2)fucosyltransferases specific to arabinogalactan proteins. J. Biol. Chem. 285: 13638–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Tan L., Lamport D.T.A., Showalter A.M., Kieliszewski M.J. (2008). The O-Hyp glycosylation code in tobacco and Arabidopsis and a proposed role of Hyp-glycans in secretion. Phytochemistry 69: 1631–1640 [DOI] [PubMed] [Google Scholar]
- Yamada H., Yanahira S., Kiyohara H., Cyong J.-C., Otsuka Y. (1987). Characterization of anti-complementary acidic heteroglycans from the seeds of Coix lacryma-jobivar Ma-yuen. Phytochemistry 26: 3269–3275 [Google Scholar]
- York W.S., Darvill A.G., McNeil M., Stevenson T.T., Albersheim P. (1985). Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118: 3–40 [Google Scholar]
- Zhang Y., Kiyohara H., Sakurai M.H., Yamada H. (1996). Complement activating galactan chains in a pectic arabinogalactan (AGIIb-1) from the roots of Angelica acutiloba Kitagawa. Carbohydr. Polym. 31: 149–156 [Google Scholar]
- Zheng Y., Mort A. (2008). Isolation and structural characterization of a novel oligosaccharide from the rhamnogalacturonan of Gossypium hirsutum L. Carbohydr. Res. 343: 1041–1049 [DOI] [PubMed] [Google Scholar]
- Zhu X., Pattathil S., Mazumder K., Brehm A., Hahn M.G., Dinesh-Kumar S.P., Joshi C.P. (2010). Virus-induced gene silencing offers a functional genomics platform for studying plant cell wall formation. Mol. Plant 3: 818–833 [DOI] [PubMed] [Google Scholar]