Abstract
Intrinsically disordered proteins (IDPs) are highly abundant in eukaryotic proteomes. Plant IDPs play critical roles in plant biology and often act as integrators of signals from multiple plant regulatory and environmental inputs. Binding promiscuity and plasticity allow IDPs to interact with multiple partners in protein interaction networks and provide important functional advantages in molecular recognition through transient protein–protein interactions. Short interaction-prone segments within IDPs, termed molecular recognition features, represent potential binding sites that can undergo disorder-to-order transition upon binding to their partners. In this review, we summarize the evidence for the importance of IDPs in plant biology and evaluate the functions associated with intrinsic disorder in five different types of plant protein families experimentally confirmed as IDPs. Functional studies of these proteins illustrate the broad impact of disorder on many areas of plant biology, including abiotic stress, transcriptional regulation, light perception, and development. Based on the roles of disorder in the protein–protein interactions, we propose various modes of action for plant IDPs that may provide insight for future experimental approaches aimed at understanding the molecular basis of protein function within important plant pathways.
INTRODUCTION
Traditionally, protein function has been closely associated with well-defined three-dimensional structure. For those proteins with well-defined globular structure, the paradigm that sequence leads to a unique three-dimensional structure and, therefore, a specific function has become predominant. Contrary to this paradigm, some protein regions (or sometimes entire proteins) lack stable secondary and/or tertiary structure under physiological conditions, yet possess crucial biological functions. It is increasingly clear that these intrinsically disordered regions (IDRs) or proteins (IDPs) are vital to many biological processes, particularly in stress responses, transcriptional regulation, signaling, and disease. The existence of IDRs or IDPs raises intriguing questions about the roles of protein intrinsic disorder in biological processes and the potential for correlation between the structure of these unusual regions and function. In the late 1990s, studies of disordered yet functional proteins emerged as a new research field, extending the traditional paradigm to include a more comprehensive view of protein structure-function relationships (Wright and Dyson, 1999; Uversky et al., 2000; Dunker et al., 2001; Tompa, 2002). Examples of functional proteins with regions of disorder are shown in Figure 1. The discovery and characterization of IDRs and IDPs is a rapidly growing area of protein science, and the potential impact of this new area is becoming more widely recognized (Dunker et al., 2008; Uversky, 2010; Chouard, 2011).
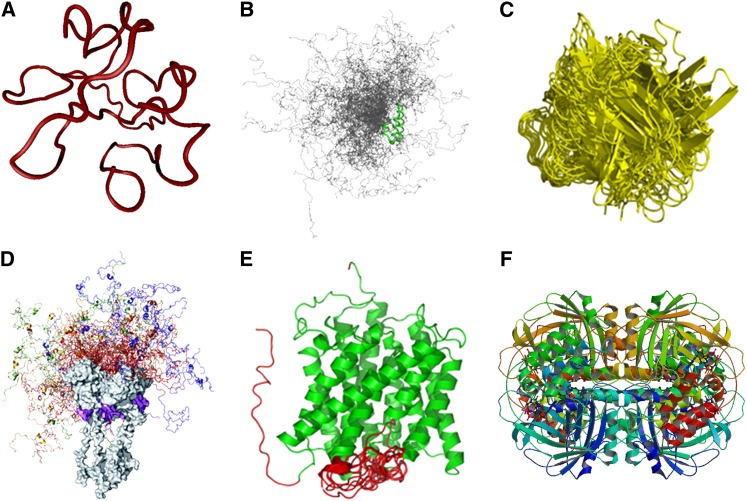
Figure 1.
Functional Proteins with Different Proportions of Intrinsic Disorder.
(A) Random coil. The structure shown is one of the 10 conformers in an ensemble of solution NMR structure of HIV-1 Tat protein (Protein Data Bank code: 1TIV).
(B) Premolten globule, which is more compact than random coil but still mostly disordered with some local residual secondary structures. The example shown is the conformational ensemble containing 200 conformers of the natively disordered region of PX, the nucleocapsid binding domain of Sendai virus phosphoprotein (Bernadó et al., 2005); the regions in green indicate the local ordered structures along the flexible chain of PX.
(C) Molten globular state, which retains all secondary structures and compact shape with the side chains changing from rigid to nonrigid packing. This state is responsible for biological functions such as translocation of proteins across membranes. The example shown is the unstable conformers of Allergen PHL P2 (Protein Data Bank code: 1WHO) generated from molecular dynamic simulations during interaction with receptor (Liang et al., 2009).
(D) Protein with ordered domain and premolten globule-like domain. The example shown is full-length human tumor repressor p53 tetramer-DNA complex; the C-terminal ordered domains (gray) and DNA (magenta) are shown in space-fill mode. The conformational ensemble of the disordered N-terminal domains from the four different monomers is shown in different colors, and 20 conformers are shown for each monomer (Wells et al., 2008).
(E) Protein with ordered domain and random coil. The example shown is an integral membrane protein involved in forming an ammonia channel (Protein Data Bank code: 2NMR); the helical bundle is shown in green and the disordered regions in red (Xue et al., 2009).
(F) Ordered protein. The example shown is the crystal structure of beef liver catalase with an NADPH binding site (Protein Data Bank code: 8CAT).
In this review, we focus on evidence for the importance of IDPs in several areas of plant biology and evaluate the functions of intrinsic disorder in five different families of plant proteins that have been experimentally confirmed to be IDPs (or proteins containing IDRs) and have been subjected to substantial functional study. We emphasize the role of disorder in protein–protein interactions within plant stress response, signaling, and molecular recognition pathways. We also propose various modes of action for plant IDPs as a guide for future research to build a better understanding of the molecular basis of protein action within important plant pathways.
IDPs AND THEIR FUNCTIONAL ADVANTAGES
IDPs exist in all kingdoms of life and are particularly widespread in eukaryotic proteomes (Xue et al., 2012). Bioinformatics studies have predicted that 23 to 28% of eukaryotic proteins are mostly disordered (Oldfield et al., 2005) and >50% of eukaryotic proteins and 70% of signaling proteins contain long IDRs (Iakoucheva et al., 2002). Some gene families are known to be particularly dominated by IDPs (e.g., 82 to 94% of transcription factors (TFs) from three TF data sets possess extended IDRs) (Liu et al., 2006). The propensity for intrinsic disorder is encoded in the peculiarities of amino acid sequences. Statistical studies reveal that hydrophobic residues (order-promoting residues [i.e., Trp, Cys, Phe, Ile, Tyr, Val, Leu, His, Thr, and Asn]) are substantially underrepresented in most IDPs, whereas polar and charged residues plus Pro (disorder-promoting residues [i.e., Lys, Glu, Pro, Ser, Gln, Arg, Asp, and Met]) are enriched and amino acids Ala and Gly are neutral with regard to order and disorder (Radivojac et al., 2007). In addition, all IDPs exhibit lower sequence complexity (Romero et al., 2001). These special sequence features of IDPs have been used together with experimental databases and algorithms to develop various bioinformatics tools for evaluating the intrinsic disorder of a given sequence on a per-residue basis (He et al., 2009). A partial list of predictors can be found in the DisProt database (http://www.disprot.org/predictors.php). For brevity, we will refer to proteins either completely disordered or containing IDRs as IDPs unless specified otherwise.
As short interaction-prone segments located within IDRs, molecular recognition features (MoRFs) often contain a conformational preference for the structure it will take upon binding to its specific partners (i.e., α-helix [α-MoRFs], β-strand [β-MoRFs], or an irregular structure [ι-MoRFs]) (Fuxreiter et al., 2004; Vacic et al., 2007). Specialized tools for identifying MoRFs as potential binding sites in IDRs have been developed (e.g., MoRF-I and II for prediction of α-MoRFs [Cheng et al., 2007] and the general MoRF predictor MoRFpred [Disfani et al., 2012]). Another disorder-based binding site predictor is ANCHOR (Dosztányi et al., 2009). In addition, motif-based tools are used for finding possible binding sites involving short linear regions of proteins, either eukaryotic linear motifs (ELMs) (Gould et al., 2010) or short linear motifs (SLiMs) (Davey et al., 2010). Like MoRFs, ELMs and SLiMs occur mostly in disordered regions. There is therefore considerable overlap between the binding sites found by MoRF predictors and those found by either of the motif-based methods (Fuxreiter et al., 2007), suggesting that ELMs, SLiMs, and MoRFs are all similar.
Order from Disorder: Binding Promiscuity and Plasticity
IDPs possess both binding promiscuity (ability to interact with multiple partners) and binding plasticity (ability to undergo binding-induced folding to accommodate diverse binding sites for different partners), two key factors conferring IDPs functional advantages in stress responses, signaling, and regulation (Oldfield et al., 2008; Sun et al., 2012b). Hub proteins play a central role in cellular biological processes by interacting with multiple partners to connect various biological molecules in protein interaction networks. This high level of connectivity is reflected in their protein structures, and intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes (Haynes et al., 2006). Disorder confers such hub proteins with the binding promiscuity necessary to interact with multiple structurally diverse partners (Patil and Nakamura, 2006).
On the other hand, binding plasticity enables IDPs to play crucial roles in molecular recognition during binding to their partners. MoRFs initiate recognition by undergoing disorder-to-order transitions upon binding to their specific partners. Such structural elements allow IDPs to interact with many distinct partners by folding into different ordered conformations. This in turn leads to specific signals induced by the formation of complexes between IDPs and their various binding partners (Wright and Dyson, 2009). A large decrease in conformational entropy due to folding of the disordered regions can uncouple binding specificity from binding affinity, resulting in a high specificity but low affinity interaction (Schulz, 1979) between an IDP and its partner. Such high specificity, low affinity binding is required for transient protein–protein interactions during signal transduction steps within regulation events (Uversky et al., 2005). Highly specific and easily dispersed binding is essential in signaling networks since activating and terminating a signal are equally important for signaling cascades (Dunker et al., 2002). Thus, both binding promiscuity and binding plasticity facilitate recognition by IDPs of their biological targets, allowing the formation of a flexible interaction network (Oldfield et al., 2008).
Intrinsic Disorder Facilitates Phosphorylation
Reversible phosphorylation is an important posttranslational modification in eukaryotic organisms and provides a regulatory mechanism for controlling the activity, function, and translocation of many proteins. Bioinformatic analysis of the GRAS (for GIBBERELLIC ACID INSENSITIVE, REPRESSOR of GAI, and the SCARECROW) proteins (Sun et al., 2011) and human proteome data mining (Fukuchi et al., 2011) indicate that phosphorylation occurs with ∼2 to 3 times higher frequency within IDRs than in ordered regions. It has been observed that intrinsic disorder is an important element facilitating the phosphorylation in IDRs; both Ser and Thr clusters that are specifically enriched in IDRs and the open structure caused by disorder in local regions around the potential phosphorylation sites contribute to IDR phosphorylation (Iakoucheva et al., 2004). Disorder-assisted phosphorylation (or dephosphorylation) can mediate interaction and recognition between IDPs and their partners as well as lead to different charge distributions that may influence protein folding and/or interaction. For example, disorder-assisted phosphorylation has been shown in a model of dynamic disordered protein complexes to be efficiently used as a means to fine-tune the electrostatic interactions of disordered protein regions for signal transduction (Mittag et al., 2010).
Phosphorylation is a functional characteristic of a number of IDPs, including LATE EMBRYOGENESIS ABUNDANT (LEA) proteins, basic domain/leucine zippers (bZIPs), and GRAS family members, all discussed in more detail below. Among LEA proteins, phosphorylation-regulated ion binding is exhibited by the acidic group 2b LEA dehydrins (Alsheikh et al., 2005); phosphorylation modulates the binding-induced folding of Thellungiella salsuginea DEHYDRIN-1 (DHN-1) and DHN-2 associated with membranes (Rahman et al., 2011a) and the membrane binding of Arabidopsis thaliana dehydrin Lti30 (Eriksson et al., 2011); and phosphorylation of the maize (Zea mays) LEA protein Rab17 regulates its cellular localization in that unphosphorylated Rab17 is retained in the nucleolus, whereas phosphorylated Rab17 is mainly cytoplasmic (Riera et al., 2004). Phosphorylation in the disordered N-terminal domain of bZIP protein HY5 (for LONG HYPOCOTYL 5) by a light-regulated kinase activity affects its stability and activity in Arabidopsis (Hardtke et al., 2000). In parallel, unphosphorylated HY5, the preferred substrate for degradation, shows stronger interaction with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), has higher affinity to target promoters, and is physiologically more active than the phosphorylated version. Therefore, phosphorylation provides an added level of light-mediated regulation of HY5 stability and activity on top of nuclear COP1 levels (Hardtke et al., 2000). Another plant bZIP, Opaque-2 from maize, is likely phosphorylated by casein kinase II. Diurnal oscillation between hyper- and hypophosphorylated isoforms of Opaque-2 results in accumulation of the phosphorylated isoform during the night, and the DNA binding potential of the phosphorylated form is diminished (Ciceri et al., 1997). With regard to GRAS proteins, phosphorylation and dephosphorylation states of the DELLA subfamily are correlated with their stability, plant growth repressive activity, and gibberellic acid (GA)-induced degradation (Hussain et al., 2005; Itoh et al., 2005); reversible phosphorylation is required for the plant stress–induced response of tobacco (Nicotiana tabacum) GRAS1 (Czikkel and Maxwell, 2007); rice (Oryza sativa) CIGR1 and CIGR2 (for Chitin-inducible gibberellin-responsive protein) are induced by GA signals, depending on both phosphorylation and dephosphorylation events (Day et al., 2004). The light-dependent phosphorylation of the disordered C-terminal domains of cryptochromes (CRYs) plays a key role in their regulatory mechanisms (Li and Yang, 2007). A link between disorder and phosphorylation has also been postulated recently for the Remorin family, members of which are likely to perform key roles in membrane lipid rafts and signaling roles in both symbiotic and pathogenic plant–microbe interactions (Jarsch and Ott, 2011). For these proteins, most of the experimentally detected phosphorylation sites overlap with the predicted disordered N-terminal region of the group 1b (and other) Remorins (Marín and Ott, 2012).
As one of the primary determinants for phosphorylation, intrinsic disorder has been integrated with position-specific amino acid frequency and local sequence similarity to known phosphorylation sites to enable more comprehensive prediction of phosphorylation sites (Iakoucheva et al., 2004). Disorder-assisted prediction of phosphorylation sites, such as by Musite (Yao et al., 2012), may provide a useful alternative approach with increased accuracy for investigating the roles of phosphorylation in plant protein interactions or annotation of whole plant proteomes.
LEA PROTEIN FUNCTION IN RESPONSE TO ABIOTIC STRESS
Plants have evolved effective mechanisms that help them adapt to abiotic stresses such as drought, low temperature, and high salinity. One of these mechanisms revolves around the LEA proteins (Figure 2A), a plant protein family with the largest number of known IDPs. Under normal growth conditions, LEA proteins are highly abundant during the late stages of plant seed development when the embryo becomes desiccation tolerant. They are also expressed constitutively in some actively dividing tissues, such as the tips of roots and leaves (Nylander et al., 2001). Most LEA proteins are classified into three loosely defined major groups (groups 1, 2, and 3) by the presence of particular sequence motifs (Tunnacliffe and Wise, 2007). In addition, some minor groups, such as group 5, are composed of atypical LEA proteins (Haaning et al., 2008; Boucher et al., 2010). Group 2 LEA proteins, also called DHNs, are further divided into two subsets (groups 2a and 2b) based on distinct expression patterns and sequence motifs, including the K-segment (Lys-rich motif), the Y-segment (an N-terminal motif), the S-segment (a stretch of Ser residues), and the φ-segment (rich in polar residues and Gly or a combination of Pro and Ala residues) (Close, 1997). The members of group 2a appear in the late stages of embryogenesis, are neutral or basic overall, and are likely to have a Y-segment. By contrast, members of group 2b in fact are not embryogenic but are associated with cold and/or drought tolerance, are acidic, and are unlikely to have a Y-segment. A new classification has recently been developed to place LEA proteins into 12 nonoverlapping classes with distinct physico-chemical properties (Jaspard et al., 2012).
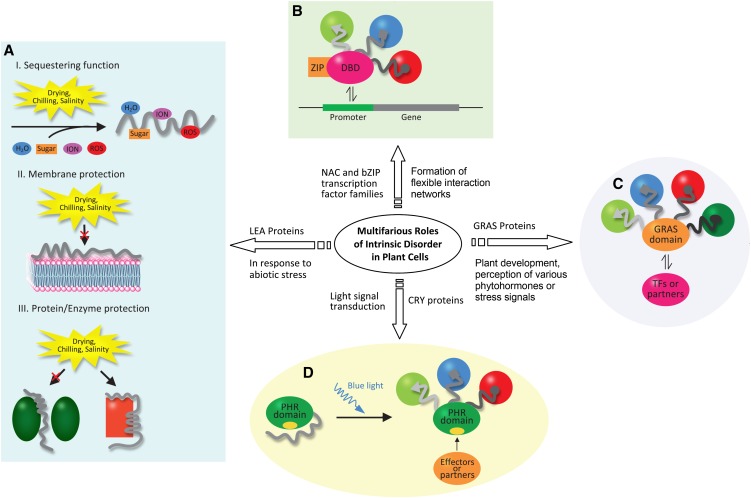
Figure 2.
Multifarious Roles of Intrinsic Disorder in Five Plant Protein Families.
(A) Intrinsically disordered LEA proteins (gray coil) protect plant cells against abiotic stress through sequestering functions (I), binding membranes (II), and safeguarding active enzymes (green ovals) by molecular shielding, as well as refolding misfolded proteins (red rectangle with entropy transfer action shown on the top) by acting as a disordered chaperone (III). ROS, reactive oxygen species.
(B) The disordered TRD of a TF (gray coil) helps form flexible transcriptional regulatory networks by interacting with various partners (spheres in different colors) under different conditions.
(C) Disordered N-domains of GRAS proteins control plant development by interacting with various partners (spheres in different colors) for perception of phytohormones and environmental signals, which coordinate interactions between the GRAS domain and TFs or protein partners.
(D) Disordered C-terminal domains of CRYs act as light-dependent switches that transduce the signal to a specific protein–protein interaction between the C-terminal domain of a CRY and partner (spheres in different colors) or between the PHR of a CRY and an effector/partner, which initiates the photomorphogenic program. The yellow oval in the PHR represents an active site masked by a disordered C-terminal domain when in darkness.
In (B) to (D), the gray circle, oval, square, and triangle represent various binding motifs or MoRFs in disordered domains for a specific interaction.
A wide range of evidence from both experimental and bioinformatic analyses has shown that LEA proteins are either entirely or partly disordered, sharing properties of a flexible conformation in solution, a high proportion of disorder-promoting residues, and a lack of stable structure or only some residual secondary structure scattered within long IDRs (Battaglia et al., 2008; Hundertmark and Hincha, 2008) (see Table 1 for details). The functions of LEA proteins have been characterized mainly by in vitro experiments. These include binding of metal ions (Heyen et al., 2002; Hara et al., 2005; Rahman et al., 2011b) or lipid vesicles (Koag et al., 2003; Kovacs et al., 2008; Rahman et al., 2010), hydration or ion sequestration (McCubbin et al., 1985; Tompa et al., 2006), and, remarkably, stabilization of proteins and membranes in adaptation to abiotic stress (Figure 2A, I to III). The latter function includes protection of various proteins against heat-induced inactivation or aggregation under stress conditions (Haaning et al., 2008; Boucher et al., 2010) and protection of membranes against drying and chilling (Tolleter et al., 2007; Rahman et al., 2010; Tolleter et al., 2010). LEA proteins also aid in the formation and stability of an intracellular glassy state that is indispensable for survival of the dry state in plant propagules (Tunnacliffe et al., 2010).
Table 1. Experimentally Characterized IDPs from the LEA Family.
| PFAMa | Groupb | Classc | Protein | Species | Secondary Structures and Transitions | Methodsd | Reference |
|---|---|---|---|---|---|---|---|
| PF00477 | 1 | 5 | Em | Wheat (Triticum aestivum) | 17% β-Sheet, 13% α-helix; 29% α-helix with TFE | CD | McCubbin et al. (1985) |
| p11 | Pea (Pisum sativum) | Largely unstructured in solution; conformation resistant to heat | CD | Russouw et al. (1997) | |||
| rGmD-19 | Soybean (Glycine max) | Highly disordered, 6 to 14% poly (l-Pro)-type structure (PII); 30% α-helix with TFE | DSC, CD | Soulages et al. (2002) | |||
| Mt-Em6 | M. truncatula | 37% α-helix, 10% β-sheet; increase to 60% α-helix upon drying | FTIR | Boudet et al. (2006) | |||
| PF00257 | 2 | 1 | Dsp16 | Craterostigma plantagineum | Largely unstructured with some local residual structural elements; TFE promotes its folding | NMR | Lisse et al. (1996) |
| 4 | Cowpea dehydrin | Vigna unguiculata | Largely unstructured; folded into α-helix with SDS | CD | Ismail et al. (1999) | ||
| 3 | Cu-COR19 | C. unshiu | Largely unstructured; α-helix induced by SDS | CD | Hara et al. (2001) | ||
| 2(DHNs) | Cor47 | Arabidopsis thaliana | 5% α-Helix, 15% PII; 50% α-helix with TFE | CD | Mouillon et al. (2006) | ||
| 4 | Lti30 | 0.3% α-Helix, 14% PII; 30% α-helix with TFE | |||||
| 2 | Lti29 | 0.8% α-Helix, 12% PII; 30% α-helix with TFE | |||||
| 1 | Rab18 | 12% PII; no α-helix induced with TFE | |||||
| 2 1 | ERD10 ERD14 | Largely unstructured; α-helix induced by TFE, no structural changes binding to lipid vesicles | NMR and CD | Kovacs et al. (2008) | |||
| 1 | K2 | Vitis riparia | Highly disordered in the middle Φ-region, transient α-helices in K-regions at both ends | NMR | Hughes and Graether (2011) | ||
| 1 | DHN1 | Maize | Largely unstructured; 9.5% α-helix induced by lipid vesicles or SDS | CD | Koag et al. (2003) | ||
| 4 | Gm-DHN1 | Soybean | 27% PII at 12°C, 15% PII at 80°C; TFE and SDS induce only moderate α-helix | DSC and CD | Soulages et al. (2003) | ||
| 2 | Ts-DHN-1 Ts-DHN-2 | T. salsuginea | Both largely unstructured; 31% β-sheet, 18% α-helix for TsDHN-1, and 24% β-sheet, 28% α-helix for TsDHN-2 with lipid vesicles | FTIR and CD | Rahman et al. (2010) | ||
| PF02987 | 3 | 6 | D-7 | Typha latifolia | Largely unstructured; 24% β-sheet, 51% α-helix upon fast drying, 45% β-sheet, 40% α-helix upon slow drying | FTIR | Wolkers et al. (2001) |
| 6 | COR15A COR15B | Arabidopsis | Both highly disordered; 65% α-helix (COR15A) and 57% α-helix (COR15B) in the dry state | CD | Thalhammet al. (2010) | ||
| 6 | LEA7 | Arabidopsis | Largely unstructured in solution; 15% β-sheet, 27% α-helix in dry state, α-helix is promoted with lipid vesicles | FTIR and CD | Popova et al. (2011) | ||
| 6 | LEAM | Pea | Largely unstructured in solution; 50% α-helix with SDS, 70% α-helix with TFE or in dry state | FTIR and CD | Tolleter et al. (2007) | ||
| PF03760 | 4 | 10 | Gm-PM16 | Soybean | 25% α-helix; 90% α-helix with SDS, TFE or upon drying | FTIR and CD | Shih et al. (2004) |
| 10 | LEA18 | Arabidopsis | Largely unstructured; 35% β-sheet, 20% α-helix with negatively charged lipid vesicles | CD | Hundertmark et al. (2011) | ||
| PF04927 | 5 | 11 | Mt-PM25 | M. truncatula | Mostly unstructured in solution; increase to 56% α-helix and 25% β-sheet upon drying | FTIR and CD | Boudet et al. (2006); Boucher et al. (2010) |
| PF03242 | 5 | 9 | Lj-IDP1 | Lotus japonicas | Largely unstructured in solution; 40% α-helix, 15% β-sheet induced by TFE or upon drying | CD, NMR, and FTIR | Haaning et al. (2008) |
This is a major but not exhaustive list for the experimentally characterized IDPs of LEA family.
Protein family (Pfam) ID.
Former LEA protein groups.
New classification according to Jaspard et al. (2012).
Circular dichroism (CD), differential scanning calorimetry (DSC), Fourier transform infrared spectroscopy (FTIR), and nuclear magnetic resonance (NMR).
Structurally, intrinsic disorder and high hydrophilicity facilitate the protective functions of LEA proteins by promoting association with membrane surfaces or protein partners. For example, dehydrin Lti30 (for LOW TEMPERATURE-INDUCED 30) functions in cold tolerance by lowering the temperature of the lipid phase transition, and the membrane–Lti30 interaction is regulated by the pH-dependent His switch and phosphorylation of the disordered dehydrin (Eriksson et al., 2011). Disordered structures of LEA proteins impart the ability to sequester water and sugars in a tight hydrogen-bonded network to form stable hydrated gels. As a result, LEA proteins become resistant to structural collapse under the conditions of abiotic stress (Wolkers et al., 2001; Tompa et al., 2006; Mouillon et al., 2008).
The importance of intrinsic disorder in LEA proteins is reflected in their striking functional versatility, a common characteristic of IDPs. A dehydrin from citrus not only plays a role in scavenging hydroxyl and peroxyl radicals to protect membrane lipids against peroxidation (Hara et al., 2003, 2004) but also binds metal ions (Hara et al., 2005), which can catalyze the generation of reactive oxygen species under dehydration conditions. Unlike other LEA proteins, LEA18 from Arabidopsis specifically destabilizes model membranes (Hundertmark et al., 2011), suggesting that it may play a role in vesicle-mediated transport rather than membrane stabilization. Different dehydrins have also been found to have multiple modes of ion binding: The acidic group 2b LEA dehydrins show phosphorylation-dependent metal-ion binding (Alsheikh et al., 2005), whereas Cu-COR15 (for Cold-Regulated) from Citrus unshiu shows metal-ion binding without phosphorylation (Hara et al., 2005). Some LEA proteins acquire secondary structure through binding-induced folding upon binding to partners, metal ions, and model membranes (Koag et al., 2009; Rahman et al., 2010, 2011b) or upon drying (Boudet et al., 2006; Thalhammer et al., 2010; Popova et al., 2011), whereas for other dehydrins, neither temperature, metal ions, nor stabilizing agent promoted structural folding (Mouillon et al., 2006). It seems the role of the conserved segments within these nonfoldable dehydrins is to act as beads on a string for specific recognition or interaction with membranes rather than promoting tertiary structure formation.
In addition to ensuring functional versatility of LEA proteins, intrinsic disorder and high hydrophilicity facilitate a diversity of mechanisms for the protective function of LEA proteins. Two models have been proposed for LEA proteins to protect cellular proteins or enzymes against drying, freezing, or heat-induced aggregation: the disordered chaperone (Tompa and Kovacs, 2010) and molecular shield (Tunnacliffe and Wise, 2007) models, both based on the high water binding capacity of intrinsically disordered polypeptide chains (Figure 2A, III). The disordered chaperone activities of some dehydrins have been observed to have rather wide substrate specificity. This distinguishes these LEA proteins from traditional molecular chaperones, which generally have specific interactions with a limited number of client proteins. (Kovacs et al., 2008). In the disordered chaperone model, LEA proteins may directly interact with the hydrophilic surface of a substrate in the native state for stabilization or with exposed hydrophobic patches to recognize the partially unfolded state of a substrate. Furthermore, the disordered chaperone may also have transient interactions with a misfolded substrate to accomplish entropy transfer in which the disordered segment of the chaperone undergoes a disorder-to-order transition and the misfolded substrate becomes partially unfolded, enabling a structural search within the available conformational space (Tompa and Kovacs, 2010). Whereas the entropy transfer mechanism awaits experimental confirmation, it is consistent with the recognition function of IDRs.
The molecular shield model suggests that some LEA proteins may entropically fill the space and prevent aggregation caused by collision between substrate molecules, especially unfolded proteins, similar to the steric stabilization of colloidal dispersion by polymer (Tunnacliffe and Wise, 2007; Hughes and Graether, 2011). In fact, the antiaggregation functions of LEA protein Mt-PM25 from Medicago truncatula satisfies both models (Boucher et al., 2010). According to the molecular shielding model, intrinsically disordered Mt-PM25 prevents aggregation of substrate through unspecific coating during freezing, heating, and drying, thereby exerting a steric stabilization effect. On the other hand, Mt-PM25 might efficiently dissolve aggregates after stress since in the hydrated state, the protein has high water binding capacity, attributed to its intrinsically disordered nature. Therefore, it might destabilize large aggregates by interacting with interfacial water around the hydrophilic and hydrophobic moieties of the substrate (i.e., acting as a disordered chaperone).
In our view, these two mechanisms, like the dual role of the citrus dehydrin in antioxidant effects by sequestering metal ions and scavenging radicals, can be unified in a broad landscape of modes of action originating from the intrinsically disordered nature of LEA proteins. In other words, the multifunctional capacity of LEA proteins arises from their intrinsically disordered nature in solution. Given that LEA proteins have been observed to interact with other proteins, membranes, sugars, nucleic acids, and metal ions, it is envisaged that disordered LEA proteins, like other IDPs, may act as hub proteins to coordinate crosstalk with other signals and pathways that are needed to respond appropriately to stress conditions.
TF FUNCTION
TFs are modular proteins that typically contain a DNA binding domain (DBD), a transcription regulatory domain (TRD), and sometimes a third domain for dimerization. TFs are usually grouped into different families on the basis of sequence similarity in the DBD, the defining feature of TFs. TFs from the same family use their similar DBDs to bind gene promoters containing a consensus DNA sequence. TFs are also characterized by their TRDs that, unlike the DBDs, do not have well-defined structures and often are classified according to their amino acid profiles, such as acidic, Gln-, Pro-, or Ser/Thr-rich regions (Triezenberg, 1995). Eukaryotic transcriptional regulation studies have shown the structural and functional modularity of TFs, since DBDs can be separated from TRDs without loss of function of either module (Johnston et al., 1986). A bioinformatic study of selected TF data sets revealed that most TFs possess extended regions of intrinsic disorder within the TRDs, where numerous α-MoRFs were also found (Liu et al., 2006).
Plant-Specific NAC Family
NAC (for NO APICAL MERISTEM, ATAF, CUP-SHAPED COTYLEDON) proteins constitute one of the largest families of plant-specific TFs, found in a wide range of land plants. The plant-specific NAC TFs are key regulators of developmental processes (Olsen et al., 2005), abiotic stress responses (Jensen et al., 2010), plant defense (Seo et al., 2010), senescence processes (Kjaersgaard et al., 2011), and xylem formation (Ko et al., 2007). Consistent with the structural modularity of TFs, most NAC proteins have a conserved N-terminal DBD domain, known as the NAC domain, that consists of a twisted β-sheet packed against an α-helix on both sides and binds a consensus core sequence CGT(GA) (Ernst et al., 2004). Unlike the structurally folded and conserved NAC domain, the C-terminal domains of most NAC proteins functioning as TRDs are highly variable with frequent occurrence of regions rich in Ser, Thr, Pro, Gln, or acidic residues (Olsen et al., 2005; Jensen et al., 2010).
Bioinformatic and experimental studies have revealed that the C-terminal TRDs of NAC TFs are intrinsically disordered with larger apparent molecular weights, depletion of order-promoting residues, and enrichment of disorder-promoting residues (Jensen et al., 2010). They also have potential capability of disorder-to-order transition through α-helical folding driven by trifluoroethanol (TFE), a reagent that mimics the environment of protein interaction (Kjaersgaard et al., 2011). A motif search of the entire Arabidopsis NAC family revealed that the structurally folded NAC domains of most NAC TFs share five or six highly conserved sequence motifs occurring as either α-helices or β-strands, whereas the intrinsically disordered C-terminal TRDs contain many subgroup-specific conserved motifs, usually spanning between 10 and 20 amino acid residues (Jensen et al., 2010). Despite their variable sequences, many of these subgroup-specific conserved motifs have a common feature of being dominated by polar and charged residues with highly conserved hydrophobic or aromatic residues embedded in the polar matrix. Similar arrangements were also found in the barley (Hordeum vulgare) NAC TFs (Kjaersgaard et al., 2011) and, more widely, in the plant-specific GRAS protein family (see next section). Some of these subgroup-specific conserved motifs have been reported to be important for transactivational properties and protein interactions (Taoka et al., 2004; Ko et al., 2007; Kjaersgaard et al., 2011).
Interestingly, we have observed from both GRAS protein family members (Sun et al., 2011) and NAC TFs that the subgroup-specific conserved motifs seem to be correlated with the functional specificity of each subgroup. Each subgroup of NAC TFs possesses some similar motifs in the TRDs for its specific functions, possibly through interacting with specific proteins of transcriptional complexes using either one or more of these subgroup-specific motifs. This observation is supported by existing experimental evidence. For example, ANAC019 (for ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 19) from subgroup III-3 of the Arabidopsis NAC TFs, a positive regulator of abscisic acid signaling during germination and early seedling development, requires its characteristic TRD for abscisic acid signal regulation (Jensen et al., 2010). CUC1 (for CUP-SHAPED COTYLEDON1), a member of subgroup II-3 of the Arabidopsis NAC TFs, relies on its specific W motif in the TRD for transactivational properties and loses the ability to promote adventitious shoot formation when its TRD is substituted by a potent TRD from a protein outside of the NAC TF family. This suggests that certain structural features of the TRD of NAC TFs are necessary for the function of the CUC group of NAC TFs (Taoka et al., 2004). Hv-NAC013, a barley NAC TF and homolog of subgroup II-3 of the Arabidopsis NAC TFs, uses its LP motif in the TRD as a major component of its transactivational properties. It also interacts with the RST domain of barley radical-induced cell death 1 (Hv-RCD1) via its C terminus, which contains specific YF and RR motifs that are distant from the LP motif (Kjaersgaard et al., 2011). By contrast, ANAC012 from subgroup II-1 of the Arabidopsis NAC TFs, a negative regulator of xylem fiber development, contains LP and WQ motifs in the TRD, and the WQ motif rather than the LP motif is responsible for its transactivational properties (Ko et al., 2007). Hv-NAC005, a barley NAC TF and homolog of subgroup III-2 of the Arabidopsis NAC TFs, contains no LP and WQ motifs but does have other specific motifs and shows neither transactivational properties nor interactions with Hv-RCD1. Therefore, although Hv-NAC005 and Hv-NAC013 both are upregulated by senescence in barley (Kjaersgaard et al., 2011), they likely are involved in plant senescence through distinct interactions with transcriptional complexes. These examples illustrate that functional specificity correlates with specific motifs located within the disordered TRDs.
bZIP Family
As the second largest family of eukaryotic dimerizing TFs, the bZIPs regulate eukaryotic genes associated with a wide range of functions, such as development, metabolism, circadian rhythm, and responses to stress and radiation. Like other TFs, bZIPs have a modular structure (Figure 2B) that generally includes a bZIP domain (i.e., DBD) and an intrinsically disordered activation domain (i.e., TRD) and has been observed in bZIPs including Arabidopsis HY5 (Yoon et al., 2006), human c-Fos (Campbell et al., 2000), and Coix lacryma Opaque-2 (Moreau et al., 2004). bZIP monomers make specific contacts with cognate DNA half-sites using their basic regions; thus, bZIPs form 2:1 complexes with DNA in which the basic regions form continuous α-helices with the C-terminal Leucine zipper regions (Ellenberger et al., 1992). Both sequence and experimental analyses have shown that the Leucine zipper regions always exist in a highly ordered α-helix, while the basic regions, in the absence of DNA, populate an ensemble of highly dynamic transient structures with substantial helical character (Bracken et al., 1999). The basic regions can be completely structured (e.g., ATF4; Podust et al., 2001), contain a certain amount of helical secondary structure (e.g., HY5; Yoon et al., 2006), or be completely disordered (e.g., Opaque-2; Moreau et al., 2004). Experimental evidence has suggested that, in complex with DNA, the basic regions uniformly form α-helical conformations (Hollenbeck et al., 2002). A study of conformational ensembles for 15 different bZIPs proposed that the basic regions of the bZIPs have quantifiable preferences for α-helical conformations in their unbound monomeric forms and this helicity varies from one basic region to another despite significant sequence similarity of the bZIP domains. Intramolecular interactions between basic regions and an eight-residue segment directly N-terminal to the basic regions are the primary modulators of helicity in the basic regions (Das et al., 2012).
The bZIP HY5 from Arabidopsis positively regulates plant photomorphogenesis through light-dependent regulation of transcription from promoters that contain a G-box, one of several light-responsive elements. A study of full-length HY5 indicated that the N-terminal 77 amino acids are crucial for interacting with the E3 ubiquitin ligase COP1 and interaction of HY5 with G-box elements is mediated by the bZIP domain. Therefore, HY5 has two functional domains, the C-terminal domain for DNA binding and the regulatory N-terminal domain for interacting with COP1 (Ang et al., 1998). Structural studies of full-length HY5 have revealed that the protein is largely disordered under physiological conditions. The N-terminal domain (amino acids 1 to 77) is largely disordered with some local residual secondary structure that can be induced by TFE to fold as an α-helix (Yoon et al., 2006). On the other hand, the basic region of HY5 has been shown to acquire a significant amount of helical secondary structure but lacks a well-folded tertiary structure that may be subsequently acquired through DNA binding. The Leucine zipper region exhibits a well-defined coiled-coil structure (Yoon et al., 2006).
TFs function in DNA binding, molecular recognition, and transcriptional activation in various biological processes. TFs are often involved in complicated and flexible protein interaction networks with members of other TF families and/or signaling proteins. For example, human tumor repressor p53 (Figure 1D) is a key player in a large signaling network for activating the expression of genes in response to cell cycle progression, apoptosis, DNA repair, and cellular stress. This remarkable protein regulates more than 150 genes and binds to over 100 protein partners (Oldfield et al., 2008). NAC TFs have been experimentally demonstrated to interact, as either transcriptional regulators or subjects of regulation, with a large number of other TFs that are involved in plant development in response to both biotic and abiotic stress (Olsen et al., 2005). NAC TFs have also been shown to interact with many signaling proteins, such as PASTICCINO1 for control of plant cell division (Smyczynski et al., 2006) and RCD1 involved in plant senescence (Kjaersgaard et al., 2011). Similarly, HY5 likely functions as part of a large multiprotein complex in vivo (Hardtke et al., 2000). In addition to interacting with COP1, HY5 also interacts with the CIRCADIAN CLOCK ASSOCIATED1 protein, which is necessary for normal circadian clock regulation in Arabidopsis (Andronis et al., 2008). The intrinsically disordered C-terminal activation domain (i.e., TRD) of the human bZIP c-Fos interacts directly with the TFs TATA-box binding protein, CREB binding protein, and Smad3 and activates transcription in a diverse range of cellular processes (Campbell et al., 2000). These studies suggest a broad functional and structural repertoire of the TF families that often function as integrators of crosstalk between different pathways. The intrinsic disorder of TFs, either in almost the entire protein or in TRDs, enables them to act as hubs in protein interaction networks and sensors of a wide variety of different intracellular and extracellular stimuli, providing strong support to the hypothesis that the global or local structural flexibility of TFs contribute to their functional diversity.
The binding plasticity mediated by MoRFs enables TFs to bind to multiple targets with high specificity. For example, p53 uses different motifs in the disordered regions to bind to multiple partners at the same time, and one motif is even able to bind to different partners by adopting different secondary structures (Oldfield et al., 2008). NAC TFs contain many functionally important, subgroup-specific conserved motifs in their disordered TRDs. These motifs mostly overlap with predicted MoRFs and may be recognized by both specific and general proteins of the transcriptional apparatus (Jensen et al., 2010). The propensity of binding-induced folding of disordered TRDs has been confirmed in the case of barley NAC TFs (Kjaersgaard et al., 2011), suggesting a potential role of MoRFs in the function of NAC TFs. We suggest here that a rigid segment in HY5 (residues from 43 to 58 within the disordered N-terminal domain) shows characteristics typical of MoRFs, namely, the propensity to interact with other proteins and undergo a disorder-to-order transition. This is supported by experiments showing that the N-terminal domain of HY5 undergoes α-helical folding induced by TFE (Yoon et al., 2006), and this potential MoRF overlaps with the region in HY5 (residues from 25 to 60) that binds to COP1 (Hardtke et al., 2000). Potential MoRFs have also been suggested to exist in the basic regions of 15 different bZIPs (Das et al., 2012). Since one of the major functional advantages of IDPs is their ability to interact specifically with multiple molecular targets, it is not surprising that intrinsically disordered TFs are prime candidates for being the crucial hub proteins or building blocks upon which flexible networks have evolved to exert fine-tuned transcriptional control over signaling pathways, through specific binding-induced folding of various MoRFs in the TRDs (Figure 2B).
SIGNAL TRANSDUCTION PROTEINS
GRAS Protein Family
The plant-specific GRAS (for GIBBERELLIC ACID INSENSITIVE, REPRESSOR of GAI, and the SCARECROW) proteins play critical and diverse roles in plant development, signaling, and transcriptional coactivation and act as integrators of signals from multiple plant growth regulatory and environmental inputs (Figure 2C). GRAS proteins have been divided into 10 subfamilies named either after one of their members or after a common motif (Sun et al., 2011). Most GRAS proteins possess a variable N-terminal domain (N-domain) and a highly conserved C-terminal domain (GRAS domain) present in the entire GRAS family. In the GRAS domain, Leucine-rich regions I (LRI) and II (LRII) flank the VHIID motif to form a LRI-VHIID-LRII pattern present in most GRAS proteins. It has been experimentally confirmed for many GRAS proteins that the LRI-VHIID-LRII pattern or individual motifs within the pattern are used for interactions with protein partners (Cui et al., 2007; Fode et al., 2008; Hirsch et al., 2009; Hou et al., 2010). Since the Leucine-rich regions were found to be involved in various types of transcriptional coactivation (Heery et al., 1997), it was speculated that the GRAS proteins act as transcriptional coactivators by either blocking or enhancing the transcriptional activity of their partners through the highly conserved LRI-VHIID-LRII pattern or entire GRAS domain (Sun et al., 2012a).
In contrast with the widely conserved motifs in the GRAS domains, subfamily-restricted motifs are conserved within the N-domains of almost all of the subfamilies, although the sequences of the N-domains are highly variable between the subfamilies (Sun et al., 2011). Like those in the disordered C-terminal TRDs of NAC TFs, these subfamily-restricted motifs in the N-domains of GRAS proteins share a common pattern: repeated hydrophobic or aromatic residues forming the framework for the conserved motifs. The repeated hydrophobic or aromatic residues of the conserved motifs in the N-domains of DELLA subfamily proteins interact with the GA-bound receptor GID1 and thereby play a critical role in perceiving GA signals to regulate plant development (Murase et al., 2008; Sun et al., 2010). Such repeated hydrophobic or aromatic residues in the N-domains of other GRAS subfamilies have been proposed to play a similar role in binding to their interacting partners (Sun et al., 2011). The N-domains of GRAS proteins accommodating these subfamily-restricted motifs are intrinsically disordered and undergo binding-induced folding upon binding to their partners. They constitute a plant-specific unfoldome and act as signal receivers to initiate key molecular recognition events in plant development, whereas structurally folded GRAS domains are involved in regulatory transcriptional coactivation interactions with other proteins (Sun et al., 2011).
Each of the GRAS subfamilies influences different aspects of plant growth and development through signaling processes and transcriptional coactivation in response to various phytohormones or stress-induced signals. Members within the same subfamily play similar regulatory roles, but in response to distinct signals, or play roles in crosstalk with many different signaling pathways in plant development by interacting with key proteins in these pathways. GRAS proteins are functionally required to accommodate different partners, and the intrinsically disordered nature of the N-domains facilitates the function of GRAS proteins as hub proteins and allows them to perform key roles in integrating multiple developmental and environmental signals (Sun et al., 2012a). As discussed previously, IDRs are able to efficiently recognize different partners in signaling via MoRF–partner interactions. All of the predicted MoRFs of GRAS proteins fall exclusively in the intrinsically disordered N-domains and most nest within the subfamily-restricted conserved motifs, representing potential protein–protein binding sites. Such MoRFs have been experimentally confirmed in the case of the DELLA subfamily (Sun et al., 2010). The N-domains of GRAS proteins bear one or more MoRFs as molecular baits to hook multiple interacting partners in various molecular recognition events in signaling processes. The structural information from intrinsic disorder-based MoRFs could guide future experiments to understand the mechanism of signaling and regulation for the entire GRAS family (Sun et al., 2012a).
CRY Protein Family
Cryptochromes (CRYs) represent another example of intrinsic disorder playing an essential role in plant signaling. Found in plants, animals, and bacteria, CRYs have an N-terminal domain sharing sequence similarity to photolyases. CRYs are flavoproteins that catalyze the repair of UV light–damaged DNA but do not have photolyase activity (Sancar, 2003). Both plant and animal CRYs possess a variable C-terminal domain, ranging from 30 to 250 amino acids beyond their photolyase homology regions (PHRs; ∼500 amino acids) and mediate light signal transduction (Cashmore, 2003). Despite their diverse functions, photolyases and CRYs preserve a common structural fold in their PHRs as well as having a similar dependence on flavin adenine dinucleotide and, therefore, a similar light-driven intramolecular electron transfer mechanism to initiate signaling (Zoltowski et al., 2011). The signaling mechanisms of CRYs are often related to specific protein–protein interactions between CRYs and their associated effectors (Figure 2D).
Arabidopsis CRYs (CRY1 and CRY2) interact with phytochromes in the regulation of photomorphogenic development and floral initiation and in the molecular circuitry of the circadian clock (Li and Yang, 2007). CRYs also regulate inhibition of hypocotyl growth and induction of flowering in response to blue light through the light-dependent inhibition of COP1, which results in accumulation of TFs to initiate the photomorphogenic program (Lin and Shalitin, 2003). Furthermore, At-CRYs act together with the blue light receptor phototropins to mediate blue light regulation of stomatal opening (Li and Yang, 2007), which is also regulated by the interaction between CRYs and COP1, a repressor of stomatal opening (Mao et al., 2005). Yeast two-hybrid and coimmunoprecipitation studies have shown that CRYs interact directly with COP1 through the C-terminal domain of the CRYs in a light-independent manner (Wang et al., 2001; Yang et al., 2001). Inhibition of COP1 function was achieved by regulating the conformation or accessibility of the C-terminal domain to COP1 in a light-dependent manner (Yang et al., 2000), suggesting that the signaling mechanism of CRYs is driven by a light-dependent conformational rearrangement in the C-terminal domain. In an analogous animal system, the Drosophila melanogaster cryptochrome (dCRY) regulates the circadian clock by targeting the TIMELESS protein (TIM) for ubiquitin-mediated degradation in the presence of light (Koh et al., 2006). Again, the C-terminal domain of dCRY is essential for light-dependent interaction between dCRY and TIM, whereas dCRY with its C-terminal domain removed exhibits light-independent interaction with TIM, indicating that a light-modulated interaction between the C-terminal domain and the PHR of dCRY is critical for proper regulation of the circadian clock (Busza et al., 2004; Dissel et al., 2004).
The recombinant C-terminal domains from both animal and plant CRYs have been experimentally demonstrated to be intrinsically disordered in solution (Partch et al., 2005). These C-terminal domains interact directly with their PHRs, inducing stable tertiary structure in the C-terminal domains. Upon light exposure, a light-dependent structural rearrangement occurs in the latter part of the C-terminal domain of At-CRY1 that may subsequently be accessible and bind to COP1, resulting in the inhibition of COP1 function. This provides experimental evidence that intrinsic disorder can be involved in signaling mechanisms of the CRYs in response to light (Partch et al., 2005). In further support of this light-dependent conformational change of the disordered C-terminal domain, time-resolved protein conformational changes in full-length At-CRY1 reveal that light-dependent modulation of the interaction between the PHR and the C-terminal domain is critical for At-CRYs to transform a blue light signal into protein–protein interactions and for the subsequent biological activity of At-CRY1 (Kondoh et al., 2011). In darkness, the PHR and the C-terminal domain form a metastable tertiary structure by interdomain interaction. A conformational change in full-length At-CRY1 is dependent on light activation of the PHR through flavin reduction by intraprotein electron transfer. This conformational switch triggers the dissociation of the C-terminal domain from the PHR in full-length At-CRY1. Subsequently, this dissociation leads to exposure of the previously buried interfaces in both the PHR and the C-terminal domain. These newly exposed segments act as binding sites for other molecules or effectors, thereby activating signals in the photomorphogenic pathways (Kondoh et al., 2011). These proposed signaling mechanisms of CRYs are further supported by the crystal structure of full-length dCRY (Zoltowski et al., 2011). Compared with the long C-terminal domain of At-CRY1 (∼200 amino acids), dCRY has a shorter C-terminal domain (∼40 amino acids) that is loosely folded and packed against the PHR in its crystallized state. Most of the residues in the C-terminal domain of dCRY take turn and loop conformations with a helical tail formed by the last 10 residues. The C-terminal helical tail docks in the analogous groove that binds DNA substrates in the Drosophila photolyase. Importantly, the light-driven electron transfer in the PHR results in a reduced conformation of flavin adenine dinucleotide, which subsequently influences the extensive interactions between the binding pocket in dCRY and the C-terminal residues. In fact, two independent dCRY molecules in a crystallographic asymmetric unit within the crystal of full-length dCRY display significant variation in binding mode of the helical tail, reflecting the structural flexibility of the C-terminal domain, which is loosely bound in the crystal structure.
The C-terminal domains of CRYs use their intrinsic disorder nature to assure their binding promiscuity and plasticity, which are required for efficient recognition of multiple partners and provide diverse binding sites for different partners (Figure 2D). Similar to other IDPs discussed in this article, we propose that the disordered C-terminal domains of CRYs may carry out binding-induced folding to make high-specificity/low-affinity reversible interactions with various partners; in darkness, they may fold in a way that allows binding to the PHRs and upon light exposure be released and fold in another way to bind to various effectors, such as COP1. Disorder predictions on a set of animal and plant CRYs have shown some short and rigid segments distributed within the disordered C-terminal domains of most of CRYs analyzed (Partch et al., 2005). These short rigid segments constitute potential MoRFs acting as nucleation sites for binding-induced folding. Among them, a potential α-MoRF corresponding to a short and rigid segment (residues 530 to 540) in the C-terminal domain of full-length dCRY has been verified to fold as an α-helix in binding to its PHR domain (Zoltowski et al., 2011). Interestingly, it seems likely that the different lengths of the C-terminal domains of At-CRYs and dCRY render them capable of regulating their signaling differently. At-CRYs appear to use their longer C-terminal domains to interact directly with COP1 (Wang et al., 2001; Yang et al., 2001) or other effectors, though it cannot be excluded that other molecules may bind to the active site of the PHRs preoccupied by the C-terminal domains. By contrast, dCRY dissociates its short C-terminal domain from the PHR so that effectors such as TIM can bind to the PHR domain in a light-dependent manner (Busza et al., 2004; Dissel et al., 2004). Although the signaling end points for At-CRYs and dCRY are different, light-dependent C-terminal domain switching appears to be a common feature for CRYs in regulating signal transduction to downstream effectors.
INTRINSIC DISORDER UNDERLIES VARIOUS MODES OF ACTION
Ample evidence indicates that intrinsic disorder is a critical factor for plant protein function in various responsive, regulatory, and signaling processes and it often underlies their modes of action through its effects on binding promiscuity and plasticity. In addition to entirely disordered IDPs (e.g., dehydrins), partly disordered IDPs (e.g., modular TFs, regulatory GRAS, or CRY proteins) contain both structurally folded domains and intrinsically disordered domains. The physical proximity of these contrasting domains in partly disordered IDPs allows conformational signals to be easily transduced from the disordered to the ordered portions of the proteins. A common feature of both entirely and partly disordered IDPs is that their IDRs actively participate in, and sometimes dominate, the protein–protein interactions between the IDPs and partners through binding-induced folding. The IDP studies so far have produced sufficient data for proposing various binding modes for plant IDPs. Whether entirely or partly disordered, plant IDRs commonly bear potential MoRFs and phosphorylation sites for controlling and regulating protein interactions. They may serve to initiate or mediate the interactions and molecular recognition events. The resultant bound conformation could be characterized by either regular secondary or irregular structures. Given this premise, we present three potential modes of action for plant IDPs (Figure 3).
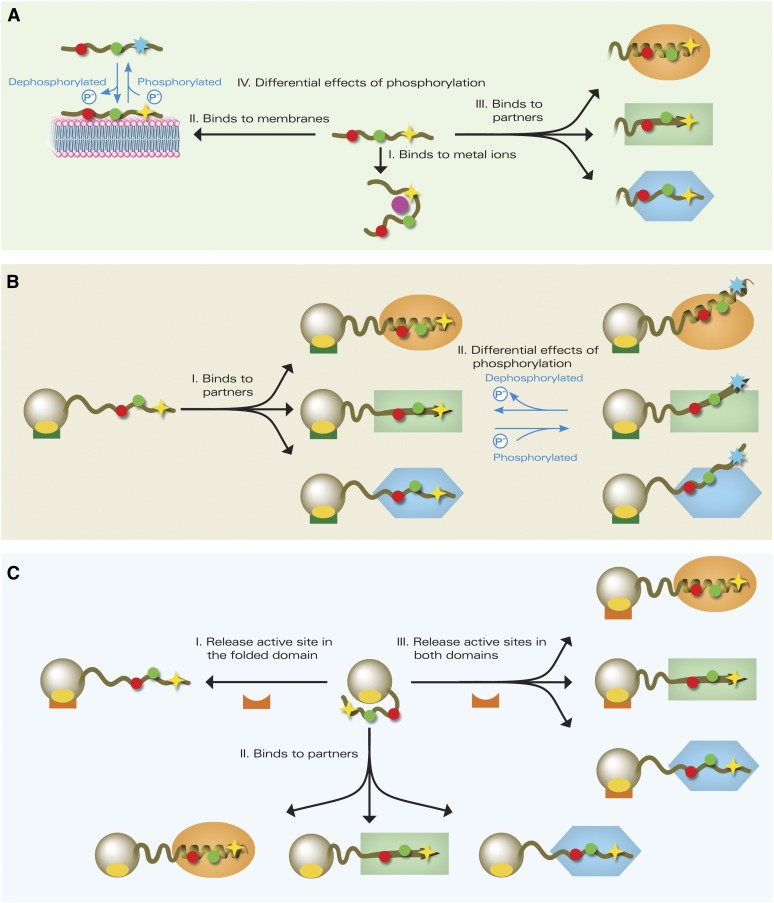
Figure 3.
Proposed Modes of Action of Plant IDPs.
The IDPs and IDRs are shown as olive coils, with filled red and green circles representing potential MoRFs; four- and seven-point stars represent the potential and phosphorylated sites, respectively. Orange ovals, light-green rectangles, and blue hexagons represent different protein partners with which IDRs fold into α-helix, β-strand, and irregular structure, respectively.
(A) An entirely disordered IDP binds to a metal ion, shown as a pink-filled circle (I), the membrane (II), and protein partners (III). Phosphorylation can induce differential binding. In this case, binding to membrane is prevented by phosphorylation (IV).
(B) A modular IDP with an IDR binds to protein partners, while the folded domain, shown as an olive sphere, binds to another target shown as a green block (I) or differential binding is induced by phosphorylation. In this case, binding to the partners being weakened by phosphorylation is shown (II).
(C) A disordered domain with an intramolecular masking interaction releases the active site (yellow oval) in the folded domain for another partner, shown as an orange block (I), binds to protein partners and exposes the active site in the folded domain (II) or releases active sites in both the disordered domain and the folded domain for interaction with various partners (III).
In the case of entirely disordered IDPs, they could bind metal ions (Figure 3A, I), membrane (Figure 3A, II), and protein partners in response to abiotic stress, resulting in three potential ordered conformations (Figure 3A, III). Phosphorylation (Figure 3A, IV) adds a further layer of binding complexity. Different lines of experimental evidence support these potential modes of binding (Table 2). In contrast with the entirely disordered IDPs, the ordered domains in the partly disordered IDPs generally constitute a functional platform, e.g., binding specific DNA sequences (TFs), interacting with proteins in transcriptional coactivation regulation (GRAS proteins) or hosting light-driven intramolecular electron transfer for initiating light signaling (CRYs). The intrinsically disordered domains generally play diverse roles in signal perception and/or molecular recognition through interacting with partners from various signaling processes. Depending on whether or not there are intramolecular interactions between the ordered domain and the disordered domain, these IDPs could be further divided into two subclasses in terms of binding modes. Intrinsically disordered domains without noticeable intramolecular interactions might play a role in consecutive regulatory mechanisms and cooperate with the ordered domain by interacting with other proteins (Figure 3B, I), e.g., most TFs that generally show structural and functional modularity). This model can also accommodate modulation by differential binding associated with changes in phosphorylation (Figure 3B, II). Experimental studies on some plant TFs have supported this type of binding (Table 2). The involvement of intrinsically disordered domains in intramolecular interactions with their own ordered domains provides another avenue for intrinsic disorder to mediate functionality of plant IDPs. In this case, the disordered domain could release an active site in the ordered domain (Figure 3C, I); the disordered domain could also be released from the ordered domain to bind to partners for signal perception (Figure 3C, II); alternatively, the release of the disordered domain could free the active sites in both ordered and disordered domains for interaction with partners in signaling processes (Figure 3C, III). These potential modes of binding have some experimental support (Table 2). Analogously to that described above, phosphorylation also has the potential to affect access to active sites and partner binding sites, which is not shown in the figure for the sake of simplicity.
Table 2. Experimental Evidence Supporting the Proposed Modes of Binding of Plant IDPs.
| Proposed Modes of Binding | Experimental Evidence | Reference |
|---|---|---|
| Entirely disordered IDPs bind to: | (I) Dehydrins bind metal ions using a specific motif containing His residues. | Heyen et al. (2002); Hara et al. (2005) |
| Metal ions (Figure 3A, I) | Ts-DHN-1 and -2 undergo disorder-to-order transition upon binding to zinc ion to form mainly β-strand. | Rahman et al. (2011b) |
| Membrane (Figure 3A, II) | (II) The K-segments of DHN1 adopt α-helices upon binding to model membranes. | Koag et al. (2003, 2009) |
| Ts-DHN-1 and -2 and LEA18 bind to model membranes and fold mainly into β-strands. | Rahman et al. (2010); Hundertmark et al. (2011) | |
| Protein partners (Figure 3A, III) | (III) Lj-IDP1 effectively protects model enzymes against stress-induced inactivation and shows propensity of folding into α-helix. | Haaning et al. (2008) |
| Differential effects of phosphorylation (Figure 3A, IV) | (IV) Phosphorylation has differential effects on interactions between IDPs and partners. It can activate, enhance, or prevent binding of dehydrins to metal ions and membranes. | Alsheikh et al. (2005); Rahman et al. (2011a); Eriksson et al. (2011) |
| Partly disordered plant IDPs without intramolecular interaction between the disordered and ordered domains bind to protein partners (Figure 3B, I) | (I) The subgroup-specific conserved motifs in the disordered TRDs of NAC TFs most likely serve as molecular recognition sites and interact with specific and general proteins of the transcriptional apparatus. | Taoka et al. (2004); Jensen et al. (2010); Kjaersgaard et al. (2011) |
| The disordered N-terminal domain of HY5 interacts with COP1 to negatively regulate the level and activity of HY5. | Ang et al. (1998); Hardtke et al. (2000) | |
| Differential effects of phosphorylation (Figure 3B, II) | (II) Phosphorylation can either promote or obstruct the interactions between the disordered domains and their partners. Phosphorylation at several sites enables Sic1 (an inhibitor of a cyclin dependent kinase) to bind to Cdc4 (a subunit of an ubiquitin ligase). Conversely, phosphorylation weakens binding between the disordered N-terminal domain of HY5 and COP1. | Mittag et al. (2010); Hardtke et al. (2000) |
| Partly disordered plant IDPs with intramolecular interaction between the disordered and ordered domains Disordered domain releases an active site of the ordered domain (Figure 3C, I) | (I) dCRY dissociates its disordered C-terminal domain from the PHR so that TIM can bind to the PHR domain of dCRY preoccupied by the C-terminal domain for its subsequent ubiquitin-mediated degradation. Here, the short C-terminal domain of dCRY contributes to regulation of the circadian clock by determining availability of the active site in the PHR domain to TIM. | Busza et al. (2004); Dissel et al. (2004); Koh et al. (2006) |
| Disordered domains fold alternatively for binding to protein partners (Figure 3C, II) | (II) Light-driven release of the C-terminal domain of At-CRYs from the PHR domain results in direct interaction between the C-terminal domain of CRY and COP1, which initiates the downstream photomorphogenic program. Here, the long C-terminal domain of CRY contributes to plant photomorphogenesis by alternative folding to directly bind to COP1. | Yang et al. (2000, 2001); Wang et al. (2001); Partch et al. (2005) |
| The release of disordered domains free the active sites of both ordered and disordered domains (Figure 3C, III) | (III) Light-driven dissociation of the C-terminal domain from the PHR of full-length At-CRY1 leads to exposure of the previously buried interaction sites in both the PHR and the C-terminal domain, indicating the potential simultaneous interactions of different partners or effectors with both the PHR and the C-terminal domain of CRY1. | Kondoh et al. (2011) |
| The disordered N-domains of DELLA subfamily of GRAS proteins were suggested to mask the active site in the GRAS domain that is responsible for interaction with the F-box proteins to target DELLA proteins for 26S proteasomal degradation in response to GA signal. Upon perceiving GA signal, the DELLA motif (α-MoRFs) and the VHYNP motif (ι-MoRFs) in the N-domains of DELLA proteins fold and bind to the GA receptor GID1, which may make the active site in the GRAS domain accessible to the F-box proteins. | Murase et al. (2008); Sun et al. (2010); Sheerin et al. (2011) |
FUTURE PERSPECTIVES
The plant protein families discussed above show that plant IDPs possess crucial cellular functions to respond to changing physiological conditions. The binding promiscuity and plasticity of plant IDPs confer upon them a unique predisposition to play critical roles in plant protein interaction networks and many, if not most, biological processes. For ordered proteins, a great deal of our understanding of their molecular mechanisms derives from studies of the roles of active sites or protein interfaces. By analogy, insight into the molecular mechanism of IDPs or IDRs may well be driven in the future by our understanding of MoRFs and other disorder-based binding motifs. We speculate that the analysis of MoRFs or other disorder-based binding motifs should be a practical tool for delineating the potential binding sites of plant IDPs that are crucial for their interactions with specific partners. This would facilitate the development of a conceptual framework for the role of specific segments within IDRs to reveal how coupled binding and folding is used in protein interactions. As revealed in both GRAS proteins and the NAC TFs, MoRFs often nest within the subfamily (or subgroup)–restricted conserved motifs. These subfamily (or subgroup)–restricted motifs, existing as hydrophobic or aromatic residue repeats within the long polar regions, appear to represent a general model adopted by many plant IDPs. These may provide guidance for experimental design involving mutational analysis in concert with pull-down experiments and other interaction-based screening assays, leading to efficient strategies for discovering new interacting proteins in signaling pathways. We postulate that these approaches will contribute to elucidating the molecular mechanisms of key plant proteins that are either IDPs themselves or interact with IDPs.
Acknowledgments
We thank Margaret Sunde and Andrew Kralicek for their critical reading of the manuscript and useful comments. We also thank the scientific editors and reviewers of this manuscript for their helpful suggestions during the preparation of this article. We apologize to colleagues whose work could not be cited due to space limitations. This work was supported by core CRI funding from MBIE provided to the Plant and Food Research Institute of New Zealand (Grant P/312003/01).
AUTHOR CONTRIBUTIONS
X.S. and E. H. R. conceived the initial idea for the review, carried out the relevant literature research and drafted the article. V.N.U. and W.T.J. revised the article and provided important content. All authors edited and approved the final version of the article.
References
- Alsheikh M.K., Svensson J.T., Randall S.K. (2005). Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant Cell Environ. 28: 1114–1122 [Google Scholar]
- Andronis C., Barak S., Knowles S.M., Sugano S., Tobin E.M. (2008). The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 1: 58–67 [DOI] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A.A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadó P., Blanchard L., Timmins P., Marion D., Ruigrok R.W.H., Blackledge M. (2005). A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc. Natl. Acad. Sci. USA 102: 17002–17007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher V., Buitink J., Lin X., Boudet J., Hoekstra F.A., Hundertmark M., Renard D., Leprince O. (2010). MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant Cell Environ. 33: 418–430 [DOI] [PubMed] [Google Scholar]
- Boudet J., Buitink J., Hoekstra F.A., Rogniaux H., Larré C., Satour P., Leprince O. (2006). Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol. 140: 1418–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C., Carr P.A., Cavanagh J., Palmer A.G., III (1999). Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: Implications for the entropy of association with DNA. J. Mol. Biol. 285: 2133–2146 [DOI] [PubMed] [Google Scholar]
- Busza A., Emery-Le M., Rosbash M., Emery P. (2004). Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 304: 1503–1506 [DOI] [PubMed] [Google Scholar]
- Campbell K.M., Terrell A.R., Laybourn P.J., Lumb K.J. (2000). Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry 39: 2708–2713 [DOI] [PubMed] [Google Scholar]
- Cashmore A.R. (2003). Cryptochromes: Enabling plants and animals to determine circadian time. Cell 114: 537–543 [PubMed] [Google Scholar]
- Cheng Y.G., Oldfield C.J., Meng J.W., Romero P., Uversky V.N., Dunker A.K. (2007). Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 46: 13468–13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard T. (2011). Structural biology: Breaking the protein rules. Nature 471: 151–153 [DOI] [PubMed] [Google Scholar]
- Ciceri P., Gianazza E., Lazzari B., Lippoli G., Genga A., Hoscheck G., Schmidt R.J., Viotti A. (1997). Phosphorylation of Opaque2 changes diurnally and impacts its DNA binding activity. Plant Cell 9: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T.J. (1997). Dehydrins: A commonality in the response of plants to dehydration and low temperature. Physiol. Plant. 100: 291–296 [Google Scholar]
- Cui H.C., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Czikkel B.E., Maxwell D.P. (2007). NtGRAS1, a novel stress-induced member of the GRAS family in tobacco, localizes to the nucleus. J. Plant Physiol. 164: 1220–1230 [DOI] [PubMed] [Google Scholar]
- Das R.K., Crick S.L., Pappu R.V. (2012). N-terminal segments modulate the α-helical propensities of the intrinsically disordered basic regions of bZIP proteins. J. Mol. Biol. 416: 287–299 [DOI] [PubMed] [Google Scholar]
- Davey N.E., Haslam N.J., Shields D.C., Edwards R.J. (2010). SLiMFinder: A web server to find novel, significantly over-represented, short protein motifs. Nucleic Acids Res. 38 (Web Server issue): W534–W539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R.B., Tanabe S., Koshioka M., Mitsui T., Itoh H., Ueguchi-Tanaka M., Matsuoka M., Kaku H., Shibuya N., Minami E. (2004). Two rice GRAS family genes responsive to N -acetylchitooligosaccharide elicitor are induced by phytoactive gibberellins: Evidence for cross-talk between elicitor and gibberellin signaling in rice cells. Plant Mol. Biol. 54: 261–272 [DOI] [PubMed] [Google Scholar]
- Disfani F.M., Hsu W.-L., Mizianty M.J., Oldfield C.J., Xue B., Dunker A.K., Uversky V.N., Kurgan L. (2012). MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics 28: i75–i83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S., Codd V., Fedic R., Garner K.J., Costa R., Kyriacou C.P., Rosato E. (2004). A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 7: 834–840 [DOI] [PubMed] [Google Scholar]
- Dosztányi Z., Mészáros B., Simon I. (2009). ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 25: 2745–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Brown C.J., Lawson J.D., Iakoucheva L.M., Obradović Z. (2002). Intrinsic disorder and protein function. Biochemistry 41: 6573–6582 [DOI] [PubMed] [Google Scholar]
- Dunker A.K., et al. (2001). Intrinsically disordered protein. J. Mol. Graph. Model. 19: 26–59 [DOI] [PubMed] [Google Scholar]
- Dunker A.K., Oldfield C.J., Meng J.W., Romero P., Yang J.Y., Chen J.W., Vacic V., Obradovic Z., Uversky V.N. (2008). The unfoldomics decade: An update on intrinsically disordered proteins. BMC Genomics 9 (Suppl 2): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T.E., Brandl C.J., Struhl K., Harrison S.C. (1992). The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein-DNA complex. Cell 71: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Eriksson S.K., Kutzer M., Procek J., Gröbner G., Harryson P. (2011). Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 23: 2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst H.A., Olsen A.N., Larsen S., Lo Leggio L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode B., Siemsen T., Thurow C., Weigel R., Gatz C. (2008). The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20: 3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi S., Hosoda K., Homma K., Gojobori T., Nishikawa K. (2011). Binary classification of protein molecules into intrinsically disordered and ordered segments. BMC Struct. Biol. 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M., Simon I., Friedrich P., Tompa P. (2004). Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Fuxreiter M., Tompa P., Simon I. (2007). Local structural disorder imparts plasticity on linear motifs. Bioinformatics 23: 950–956 [DOI] [PubMed] [Google Scholar]
- Gould C.M., et al. (2010). ELM: The status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 38 (Database issue): D167–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaning S., Radutoiu S., Hoffmann S.V., Dittmer J., Giehm L., Otzen D.E., Stougaard J. (2008). An unusual intrinsically disordered protein from the model legume Lotus japonicus stabilizes proteins in vitro. J. Biol. Chem. 283: 31142–31152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M., Fujinaga M., Kuboi T. (2004). Radical scavenging activity and oxidative modification of citrus dehydrin. Plant Physiol. Biochem. 42: 657–662 [DOI] [PubMed] [Google Scholar]
- Hara M., Fujinaga M., Kuboi T. (2005). Metal binding by citrus dehydrin with histidine-rich domains. J. Exp. Bot. 56: 2695–2703 [DOI] [PubMed] [Google Scholar]
- Hara M., Terashima S., Fukaya T., Kuboi T. (2003). Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 217: 290–298 [DOI] [PubMed] [Google Scholar]
- Hara M., Terashima S., Kuboi T. (2001). Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol. 158: 1333–1339 [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C., Oldfield C.J., Ji F., Klitgord N., Cusick M.E., Radivojac P., Uversky V.N., Vidal M., Iakoucheva L.M. (2006). Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Wang K.J., Liu Y.L., Xue B., Uversky V.N., Dunker A.K. (2009). Predicting intrinsic disorder in proteins: An overview. Cell Res. 19: 929–949 [DOI] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven E., Hoare S., Parker M.G. (1997). A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387: 733–736 [DOI] [PubMed] [Google Scholar]
- Heyen B.J., Alsheikh M.K., Smith E.A., Torvik C.F., Seals D.F., Randall S.K. (2002). The calcium-binding activity of a vacuole-associated, dehydrin-like protein is regulated by phosphorylation. Plant Physiol. 130: 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E.D. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck J.J., McClain D.L., Oakley M.G. (2002). The role of helix stabilizing residues in GCN4 basic region folding and DNA binding. Protein Sci. 11: 2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.L., Lee L.Y.C., Xia K.F., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Hughes S., Graether S.P. (2011). Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 20: 42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M., Dimova R., Lengefeld J., Seckler R., Hincha D.K. (2011). The intrinsically disordered late embryogenesis abundant protein LEA18 from Arabidopsis thaliana modulates membrane stability through binding and folding. Biochim. Biophys. Acta 1808: 446–453 [DOI] [PubMed] [Google Scholar]
- Hundertmark M., Hincha D.K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A., Cao D.N., Cheng H., Wen Z.L., Peng J.R. (2005). Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J. 44: 88–99 [DOI] [PubMed] [Google Scholar]
- Iakoucheva L.M., Brown C.J., Lawson J.D., Obradović Z., Dunker A.K. (2002). Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323: 573–584 [DOI] [PubMed] [Google Scholar]
- Iakoucheva L.M., Radivojac P., Brown C.J., O’Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. (2004). The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail A.M., Hall A.E., Close T.J. (1999). Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol. 120: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Sasaki A., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Hasegawa Y., Minami E., Ashikari M., Matsuoka M. (2005). Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46: 1392–1399 [DOI] [PubMed] [Google Scholar]
- Jarsch I.K., Ott T. (2011). Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol. Plant Microbe Interact. 24: 7–12 [DOI] [PubMed] [Google Scholar]
- Jaspard E., Macherel D., Hunault G. (2012). Computational and statistical analyses of amino acid usage and physico-chemical properties of the twelve late embryogenesis abundant protein classes. PLoS ONE 7: e36968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Johnston S.A., Zavortink M.J., Debouck C., Hopper J.E. (1986). Functional domains of the yeast regulatory protein GAL4. Proc. Natl. Acad. Sci. USA 83: 6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaersgaard T., Jensen M.K., Christiansen M.W., Gregersen P., Kragelund B.B., Skriver K. (2011). Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J. Biol. Chem. 286: 35418–35429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Yang S.H., Park A.H., Lerouxel O., Han K.H. (2007). ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 50: 1035–1048 [DOI] [PubMed] [Google Scholar]
- Koag M.C., Fenton R.D., Wilkens S., Close T.J. (2003). The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 131: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koag M.C., Wilkens S., Fenton R.D., Resnik J., Vo E., Close T.J. (2009). The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 150: 1503–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Zheng X.Z., Sehgal A. (2006). JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312: 1809–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh M., Shiraishi C., Müller P., Ahmad M., Hitomi K., Getzoff E.D., Terazima M. (2011). Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J. Mol. Biol. 413: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs D., Kalmar E., Torok Z., Tompa P. (2008). Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.H., Yang H.Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83: 94–101 [DOI] [PubMed] [Google Scholar]
- Liang S., Li L., Hsu W.-L., Pilcher M.N., Uversky V., Zhou Y., Dunker A.K., Meroueh S.O. (2009). Exploring the molecular design of protein interaction sites with molecular dynamics simulations and free energy calculations. Biochemistry 48: 399–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.T., Shalitin D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Lisse T., Bartels D., Kalbitzer H.R., Jaenicke R. (1996). The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineum displays no defined three-dimensional structure in its native state. Biol. Chem. 377: 555–561 [DOI] [PubMed] [Google Scholar]
- Liu J.G., Perumal N.B., Oldfield C.J., Su E.W., Uversky V.N., Dunker A.K. (2006). Intrinsic disorder in transcription factors. Biochemistry 45: 6873–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Zhang Y.C., Sang Y., Li Q.H., Yang H.Q. (2005). From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín, M., and Ott, T. (2012). Phosphorylation of intrinsically disordered regions in remorin proteins. Front. Plant Science 3: 86. [DOI] [PMC free article] [PubMed]
- McCubbin W.D., Kay C.M., Lane B.G. (1985). Hydrodynamic and optical properties of the wheat-germ Em protein. Can. J. Biochem. Cell Biol. 63: 803–811 [Google Scholar]
- Mittag T., Kay L.E., Forman-Kay J.D. (2010). Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 23: 105–116 [DOI] [PubMed] [Google Scholar]
- Moreau V.H., da Silva A.C., Siloto R.M.P., Valente A.P., Leite A., Almeida F.C.L. (2004). The bZIP region of the plant transcription factor opaque-2 forms stable homodimers in solution and retains its helical structure upon subunit dissociation. Biochemistry 43: 4862–4868 [DOI] [PubMed] [Google Scholar]
- Mouillon J.M., Eriksson S.K., Harryson P. (2008). Mimicking the plant cell interior under water stress by macromolecular crowding: Disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 148: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon J.M., Gustafsson P., Harryson P. (2006). Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol. 141: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nylander M., Svensson J., Palva E.T., Welin B.V. (2001). Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 45: 263–279 [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Cheng Y., Cortese M.S., Brown C.J., Uversky V.N., Dunker A.K. (2005). Comparing and combining predictors of mostly disordered proteins. Biochemistry 44: 1989–2000 [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Meng J., Yang J.Y., Yang M.Q., Uversky V.N., Dunker A.K. (2008). Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 9 (suppl. 1): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Partch C.L., Clarkson M.W., Ozgür S., Lee A.L., Sancar A. (2005). Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44: 3795–3805 [DOI] [PubMed] [Google Scholar]
- Patil A., Nakamura H. (2006). Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 580: 2041–2045 [DOI] [PubMed] [Google Scholar]
- Podust L.M., Krezel A.M., Kim Y. (2001). Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J. Biol. Chem. 276: 505–513 [DOI] [PubMed] [Google Scholar]
- Popova A.V., Hundertmark M., Seckler R., Hincha D.K. (2011). Structural transitions in the intrinsically disordered plant dehydration stress protein LEA7 upon drying are modulated by the presence of membranes. Biochim. Biophys. Acta 1808: 1879–1887 [DOI] [PubMed] [Google Scholar]
- Radivojac P., Iakoucheva L.M., Oldfield C.J., Obradovic Z., Uversky V.N., Dunker A.K. (2007). Intrinsic disorder and functional proteomics. Biophys. J. 92: 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman L.N., Bamm V.V., Voyer J.A.M., Smith G.S.T., Chen L., Yaish M.W., Moffatt B.A., Dutcher J.R., Harauz G. (2011b). Zinc induces disorder-to-order transitions in free and membrane-associated Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2: A solution CD and solid-state ATR-FTIR study. Amino Acids 40: 1485–1502 [DOI] [PubMed] [Google Scholar]
- Rahman L.N., Chen L., Nazim S., Bamm V.V., Yaish M.W., Moffatt B.A., Dutcher J.R., Harauz G. (2010). Interactions of intrinsically disordered Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 with membranes - Synergistic effects of lipid composition and temperature on secondary structure. Biochem. Cell Biol. 88: 791–807 [DOI] [PubMed] [Google Scholar]
- Rahman L.N., Smith G.S.T., Bamm V.V., Voyer-Grant J.A.M., Moffatt B.A., Dutcher J.R., Harauz G. (2011a). Phosphorylation of Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 facilitates cation-induced conformational changes and actin assembly. Biochemistry 50: 9587–9604 [DOI] [PubMed] [Google Scholar]
- Riera M., Figueras M., López C., Goday A., Pagès M. (2004). Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc. Natl. Acad. Sci. USA 101: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Obradovic Z., Li X.H., Garner E.C., Brown C.J., Dunker A.K. (2001). Sequence complexity of disordered protein. Proteins 42: 38–48 [DOI] [PubMed] [Google Scholar]
- Russouw P.S., Farrant J., Brandt W., Lindsey G.G. (1997). The most prevalent protein in a heat-treated extract of pea (Pisum sativum) embryos is an LEA group I protein; Its conformation is not affected by exposure to high temperature. Seed Sci. Res. 7: 117–123 [Google Scholar]
- Sancar A. (2003). Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103: 2203–2237 [DOI] [PubMed] [Google Scholar]
- Schulz, G.E. (1979). Nucleotide binding proteins. In Molecular Mechanism of Biological Recognition, M. Balaban, ed (New York: Elsevier/North-Holland), pp. 79–94. [Google Scholar]
- Seo P.J., Kim M.J., Park J.Y., Kim S.Y., Jeon J., Lee Y.H., Kim J., Park C.M. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Sheerin D.J., Buchanan J., Kirk C., Harvey D., Sun X., Spagnuolo J., Li S., Liu T., Woods V.A., Foster T., Jones W.T., Rakonjac J. (2011). Inter- and intra-molecular interactions of Arabidopsis thaliana DELLA protein RGL1. Biochem. J. 435: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih M.D., Lin S.C., Hsieh J.S., Tsou C.H., Chow T.Y., Lin T.P., Hsing Y.I.C. (2004). Gene cloning and characterization of a soybean (Glycine max L.) LEA protein, GmPM16. Plant Mol. Biol. 56: 689–703 [DOI] [PubMed] [Google Scholar]
- Smyczynski C., Roudier F., Gissot L., Vaillant E., Grandjean O., Morin H., Masson T., Bellec Y., Geelen D., Faure J.D. (2006). The C terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. J. Biol. Chem. 281: 25475–25484 [DOI] [PubMed] [Google Scholar]
- Soulages J.L., Kim K., Arrese E.L., Walters C., Cushman J.C. (2003). Conformation of a group 2 late embryogenesis abundant protein from soybean. Evidence of poly (L-proline)-type II structure. Plant Physiol. 131: 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulages J.L., Kim K., Walters C., Cushman J.C. (2002). Temperature-induced extended helix/random coil transitions in a group 1 late embryogenesis-abundant protein from soybean. Plant Physiol. 128: 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jones W.T., Rikkerink E.H.A. (2012a). GRAS proteins: The versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 442: 1–12 [DOI] [PubMed] [Google Scholar]
- Sun, X., Jones, W.T., and Uversky, V.N. (2012b). Applications of bioinformatics and experimental methods to intrinsic disorder-based protein-protein interactions. In Protein Engineering, P. Kaumaya, ed (Rijeka, Croatia: InTech), pp. 181–206. [Google Scholar]
- Sun X., Xue B., Jones W.T., Rikkerink E., Dunker A.K., Uversky V.N. (2011). A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 77: 205–223 [DOI] [PubMed] [Google Scholar]
- Sun X.L., et al. (2010). N-terminal domains of DELLA proteins are intrinsically unstructured in the absence of interaction with GID1/gibberellic acid receptors. J. Biol. Chem. 285: 11557–11571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K., Yanagimoto Y., Daimon Y., Hibara K., Aida M., Tasaka M. (2004). The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J. 40: 462–473 [DOI] [PubMed] [Google Scholar]
- Thalhammer A., Hundertmark M., Popova A.V., Seckler R., Hincha D.K. (2010). Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. Biochim. Biophys. Acta 1798: 1812–1820 [DOI] [PubMed] [Google Scholar]
- Tolleter D., Hincha D.K., Macherel D. (2010). A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim. Biophys. Acta 1798: 1926–1933 [DOI] [PubMed] [Google Scholar]
- Tolleter D., Jaquinod M., Mangavel C., Passirani C., Saulnier P., Manon S., Teyssier E., Payet N., Avelange-Macherel M.H., Macherel D. (2007). Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell 19: 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27: 527–533 [DOI] [PubMed] [Google Scholar]
- Tompa P., Bánki P., Bokor M., Kamasa P., Kovács D., Lasanda G., Tompa K. (2006). Protein-water and protein-buffer interactions in the aqueous solution of an intrinsically unstructured plant dehydrin: NMR intensity and DSC aspects. Biophys. J. 91: 2243–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P., Kovacs D. (2010). Intrinsically disordered chaperones in plants and animals. Biochem. Cell Biol. 88: 167–174 [DOI] [PubMed] [Google Scholar]
- Triezenberg S.J. (1995). Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5: 190–196 [DOI] [PubMed] [Google Scholar]
- Tunnacliffe, A., Hincha, D., Leprince, O., and Macherel, D. (2010). LEA proteins: Versatility of form and function. Dormancy and resistance in harsh environments. In E. Lubzens, J. Cerda, and M. Clark, eds (Berlin/Heidelberg: Springer), pp. 91–108. [Google Scholar]
- Tunnacliffe A., Wise M.J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94: 791–812 [DOI] [PubMed] [Google Scholar]
- Uversky V.N. (2010). The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010: 568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N., Gillespie J.R., Fink A.L. (2000). Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41: 415–427 [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Oldfield C.J., Dunker A.K. (2005). Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 18: 343–384 [DOI] [PubMed] [Google Scholar]
- Vacic V., Oldfield C.J., Mohan A., Radivojac P., Cortese M.S., Uversky V.N., Dunker A.K. (2007). Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 6: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wells M., Tidow H., Rutherford T.J., Markwick P., Jensen M.R., Mylonas E., Svergun D.I., Blackledge M., Fersht A.R. (2008). Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. USA 105: 5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkers W.F., McCready S., Brandt W.F., Lindsey G.G., Hoekstra F.A. (2001). Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim. Biophys. Acta 1544: 196–206 [DOI] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. (1999). Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293: 321–331 [DOI] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. (2009). Linking folding and binding. Curr. Opin. Struct. Biol. 19: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Dunker A.K., Uversky V.N. (2012). Orderly order in protein intrinsic disorder distribution: Disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 30: 137–149 [DOI] [PubMed] [Google Scholar]
- Xue B., Li L., Meroueh S.O., Uversky V.N., Dunker A.K. (2009). Analysis of structured and intrinsically disordered regions of transmembrane proteins. Mol. Biosyst. 5: 1688–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q., Tang R.H., Cashmore A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q., Wu Y.J., Tang R.H., Liu D.M., Liu Y., Cashmore A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yao Q., Gao J., Bollinger C., Thelen J.J., Xu D. (2012). Predicting and analyzing protein phosphorylation sites in plants using musite. Front. Plant Sci. 3: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M.K., Shin J., Choi G., Choi B.S. (2006). Intrinsically unstructured N-terminal domain of bZIP transcription factor HY5. Proteins 65: 856–866 [DOI] [PubMed] [Google Scholar]
- Zoltowski B.D., Vaidya A.T., Top D., Widom J., Young M.W., Crane B.R. (2011). Structure of full-length Drosophila cryptochrome. Nature 480: 396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]