A high-throughput imaging platform and custom algorithm are presented for the quantitative analysis of complex plasmodesmata (PD) formation. Complex PDs are shown to be derived from simple PDs in a pattern that is accelerated when leaves undergo the sink–source transition. This approach will facilitate further large-scale analyses of the endogenous and exogenous factors that influence PD formation.
Abstract
Plasmodesmata (PD) form tubular connections that function as intercellular communication channels. They are essential for transporting nutrients and for coordinating development. During cytokinesis, simple PDs are inserted into the developing cell plate, while during wall extension, more complex (branched) forms of PD are laid down. We show that complex PDs are derived from existing simple PDs in a pattern that is accelerated when leaves undergo the sink–source transition. Complex PDs are inserted initially at the three-way junctions between epidermal cells but develop most rapidly in the anisocytic complexes around stomata. For a quantitative analysis of complex PD formation, we established a high-throughput imaging platform and constructed PDQUANT, a custom algorithm that detected cell boundaries and PD numbers in different wall faces. For anticlinal walls, the number of complex PDs increased with increasing cell size, while for periclinal walls, the number of PDs decreased. Complex PD insertion was accelerated by up to threefold in response to salicylic acid treatment and challenges with mannitol. In a single 30-min run, we could derive data for up to 11k PDs from 3k epidermal cells. This facile approach opens the door to a large-scale analysis of the endogenous and exogenous factors that influence PD formation.
INTRODUCTION
Plasmodesmata (PD) are specialized membrane structures that form intercellular communication channels between cells. In recent years, there has been growing evidence that PDs may traffic a range of substances from small solutes to large protein and RNA complexes, including viral genomes (Roberts and Oparka, 2003; Maule et al., 2011; Burch-Smith and Zambryski, 2012). The picture emerging depicts the PD pore as a dynamic structure that modulates transport in response to a number of endogenous and exogenous cues (Ueki and Citovsky, 2011; Burch-Smith and Zambryski, 2012).
PDs exist in a number of discrete forms whose functions have not been fully determined. Primary PDs are laid down at cytokinesis during the formation of the cell plate and arise by entrapment of an endoplasmic reticulum strand that later forms the desmotubule of the pore. These PDs have a simple architecture; that is, they exist as single linear pores (Hepler, 1982; Faulkner et al., 2008). During development, secondary PDs may form to keep pace with wall expansion (Ehlers and Kollmann, 2001). These secondary pores arise postcytokinetically across the mature cell wall, either in association with existing pores (Faulkner et al., 2008) or entirely de novo (Ehlers and Kollmann, 2001). From electron microscope images alone, it is impossible to tell whether a PD pore is primary or secondary, as these terms refer strictly to the ontogenetic deposition of PD (Ehlers and Kollmann, 2001). However, many of the PDs that arise in the later growth of an organ have a complex, branched structure, with several pores leading into a shared wall cavity (Ding and Lucas, 1996; Ehlers and Kollmann, 2001; Roberts and Oparka, 2003; Faulkner et al., 2008). There has been considerable debate as to whether complex PDs are modifications of existing simple (primary) PDs or whether they arise de novo in areas that were not previously occupied by primary PDs (critically reviewed in Burch-Smith and Zambryski, 2012). Regardless of their precise origin, in leaf tissues, complex PDs supersede the simple PDs laid down at cytokinesis (Ding et al., 1992; Oparka et al., 1999; Roberts et al., 2001).
In an extensive study of the expanding basal cell wall of the tobacco (Nicotiana tabacum) trichome (Faulkner et al., 2008), we found that new PD pores were inserted immediately adjacent to the original primary PD to form paired PDs. With continued expansion, multiple twinning events gave rise to primary pit fields, with the original primary PDs acting as templates for the formation of new PD pores. In this model, new PDs arise in direct connection with existing pores rather than randomly across the mature cell wall (Faulkner et al., 2008). Although primary and secondary PDs cannot be distinguished unequivocally, it is possible to distinguish simple PDs from complex PD using appropriate genetic markers. Fortuitously, a unique feature of complex PDs is their ability to be targeted by viral movement proteins (MPs; Lucas, 2006), which appear to accumulate specifically in the central cavity of the shared pore (Ding et al., 1992; Fitzgibbon et al., 2010). However, this may not be a universal feature of all MPs. For example, the MP of tobacco mosaic virus (TMV) has been shown recently to target both simple and complex PDs in Arabidopsis thaliana (Burch-Smith et al., 2011). By contrast, the MP of potato leafroll virus (MP17) is thought to target only complex PDs (Burch-Smith et al., 2011; Hofius et al., 2001), thus providing a suitable marker for the appearance of the complex PDs in the leaf. We therefore used the MP17-GFP (for green fluorescent protein) tag to identify the progression of complex PD formation during development of the Arabidopsis leaf epidermis.
It is now well established that PD may change their size exclusion limit (SEL) in response to a number of intrinsic and extrinsic cues (reviewed in Burch-Smith and Zambryski, 2012), providing a means of controlling the nature of molecules that pass between cells. The formation of complex PDs is associated with a decrease in PD conductance (Oparka et al., 1999), and the selective deposition of complex PDs at specific wall interfaces may provide an additional means of controlling communication between cells. For example, complex PDs appear very early during development at the interface of specialized cell types (Roberts et al., 2001; Faulkner et al., 2008). During the sink–source transition in tobacco leaves, the PD population changes from predominantly simple to predominantly branched, with a concomitant decrease in the SEL of the remaining PD (Oparka et al., 1999). Studies using leaves bombarded with a 2xGFP (54 kiloDalton) construct have shown that between 30 and 46% of cells in sink leaves (but only 2 to 9% in source leaf cells) are able to traffic this probe (Kobayashi et al., 2005). To date, very little is known about the conversion of simple to complex forms of PDs and the factors that regulate this remarkable structural alteration within the cell wall. Combined confocal and electron microscopy of tobacco leaves undergoing the sink–source transition suggests that the alteration from simple to complex PDs may occur rapidly (Roberts et al., 2001). At the end of the sink–source transition, the leaf contains predominantly complex PDs that must function in solute transport throughout the remainder of its life.
One of the major obstacles in studying the dynamic conversions of PD is their small size and inaccessibility. To date, most studies of PD frequencies and distributions have required painstaking serial sectioning and electron microscopy followed by calculations of densities between shared wall interfaces (Ehlers and van Bel, 2010). We set out to devise a high-throughput, live-imaging approach that would allow robust, accurate quantification of the appearance of complex PDs in the developing leaf epidermis. To do this, we used a spinning disc confocal microscope (Opera) to observe intact Arabidopsis leaves in multiwell plates (Salomon et al., 2010). We developed image analysis algorithms to detect PDs in different wall interfaces and generated large-scale data sets for the appearance of complex PD during leaf development. To facilitate this study, we crossed a transgenic Arabidopsis line expressing MP17-GFP (Hofius et al., 2001) with a second line expressing the PD marker PLASMODESMATA LOCALISED PROTEIN1-monomeric RED FLOURESCENT PROTEIN (PDLP1-mRFP) which labels all PD (Bayer et al., 2008). This allowed us to examine the appearance of complex PDs and their spatial relationship to existing simple PDs.
Our results reveal that complex PD formation occurs in a highly regulated pattern between individual cell types and within specific regions of the cell wall. We show also that alterations in leaf carbon balance, imposed by source removal, induce a rapid insertion of complex PDs and an accelerated sink–source transition. Finally, we reveal that the insertion of complex PDs is sensitive to a range of stresses imposed during early leaf development. Our high-throughput approach for studying PD numbers in intact tissues establishes an analysis framework for complex PD formation and opens the door to the development of genetic screens that will identify factors influencing the formation of complex PD in plants.
RESULTS
MP17-GFP Selectively Targets Complex PDs in Arabidopsis Leaves
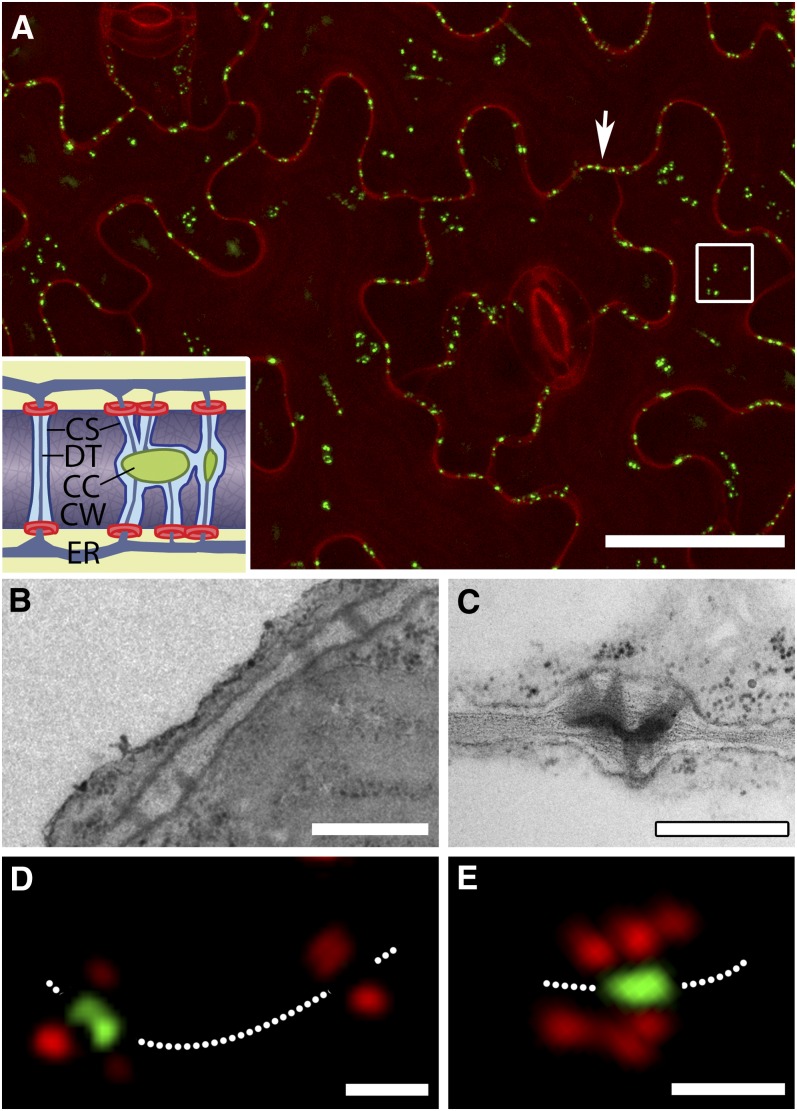
We infiltrated leaves of transgenic plants expressing MP17-GFP with propidium iodide, a cell wall stain, to highlight the positions of PD in the epidermis (Figure 1A; Hofius et al., 2001). Under the confocal microscope, we identified two distinct populations of PDs, an anticlinal population that connected epidermal cells and a periclinal population that connected the lower epidermis to the mesophyll (Figure 1A). For a quantitative, high-throughput analysis to be effective, it was essential to confirm that MP17 targets only complex PDs. To do this, we took the first six leaves of plants expressing MP17-GFP and aligned them on a microscope slide. Confocal imaging revealed that the GFP signal first appeared at the tip of leaf 2 with a basipetal progression of PD labeling in subsequent leaves. We cut small pieces of leaf from those regions expressing or not expressing GFP and prepared them for electron microscopy. Regions of leaf not expressing GFP contained only simple PDs (Figure 1B), while those expressing GFP showed exclusively complex PDs with a characteristic central cavity (Figure 1C). Next, we used super-resolution imaging (Fitzgibbon et al., 2010) to examine the epidermis of MP17-GFP plants stained with callose antibody to identify PD orifices. We found that MP17-GFP was located exclusively in the central cavities of complex PD, spatially separated from the callose signals (Figures 1D and 1E). These data confirm that MP17 is a bona fide marker for complex PDs with shared central cavities.
Figure 1.
Leaf Epidermal PDs Viewed by Confocal and Super-Resolution Microscopy.
(A) Surface view of the epidermis stained with propidium iodide reveals PDs labeled by MP17-GFP in both anticlinal (arrow) and periclinal (boxed region) interfaces. The inset shows a diagram of simple (left) and complex (right) PDs, showing the location of MP17 (green) in the central cavity of complex PDs. Callose collars are shown in red. CC, central cavity; CS, cytoplasmic sleeve; CW, cell wall; DT, desmotubule; ER, endoplasmic reticulum. Bar = 30 μm.
(B) Image of simple PDs in the cell wall connecting epidermal cells in a region of leaf not labeled with MP17-GFP. Bar = 0.5 μm.
(C) Image of a complex PDs from a region of leaf expressing MP17-GFP. Note that multiple branches lead into a shared central cavity. Bar = 0.5 μm.
(D) and (E) Super-resolution images of PDs acquired using three-dimensional structured illumination microscopy. MP17 is associated exclusively with central cavities of complex PDs, identified by callose collars (red). A simple PD with two callose collars (right in [D]) shows no labeling in the central region of the pore. In (E), several callose collars surround a single shared central cavity. The dotted line depicts the orientation of the cell wall. Bars = 0.5 µm.
Conversion of Simple to Complex PDs
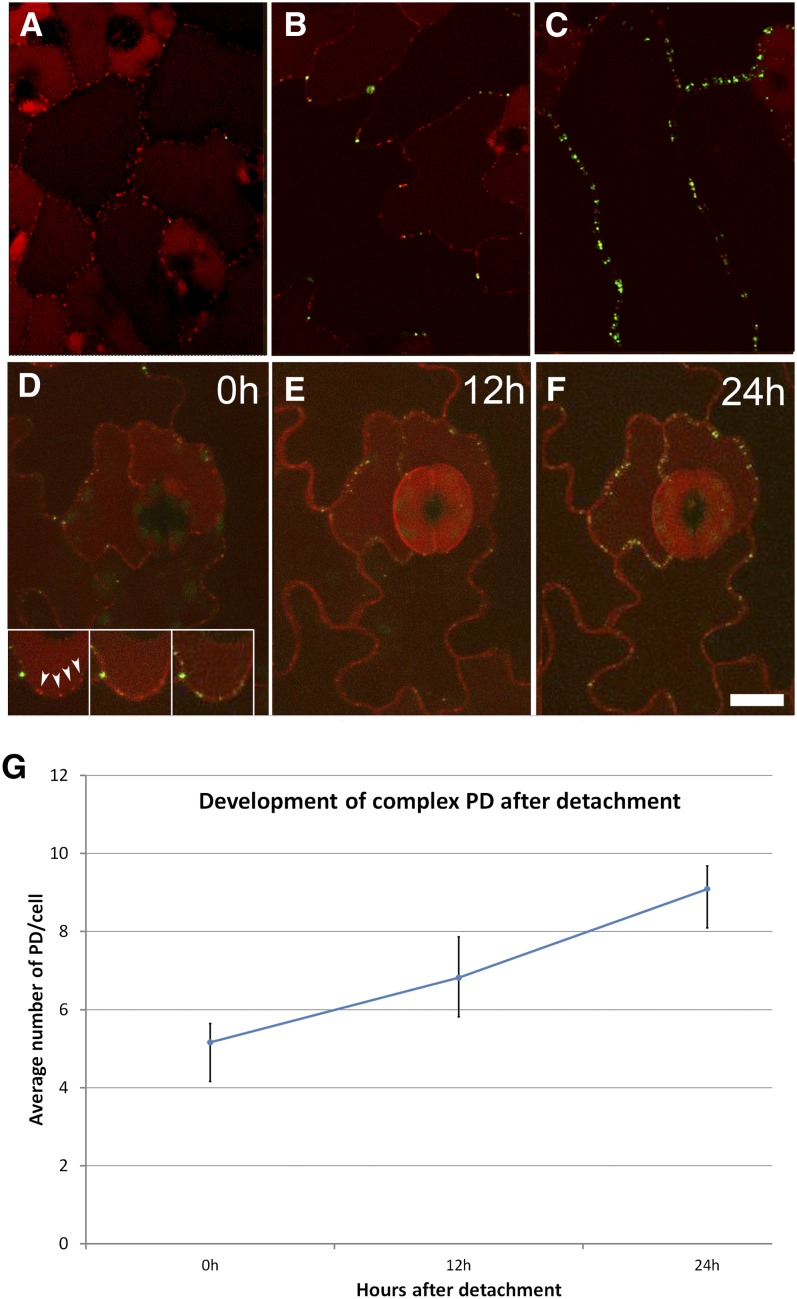
To study the conversion of simple to branched PDs, we used a stable cross between PDLP1-mRFP and MP17-GFP transgenic lines. PDLP1 has been shown to label all PDs, both simple and complex (Bayer et al., 2008), while MP17 selectively labels only complex PDs (Hofius et al., 2001; this study). Although older leaves (leaves 5 and 6) showed nonspecific cell wall labeling with mRFP (Figures 2D to 2F), we found PDLP1-mRFP to be a suitable marker for detecting the simple PDs in young leaves. When we examined the first emerged leaf of these plants, all PDs were labeled with PDLP1-mRFP (Figure 2A). In leaf 2, both PDLP1-mRFP and MP17-GFP signals were detected, indicating the appearance of complex PDs (Figure 2B), and in leaf 3, the proportion of MP17-GFP–labeled PDs increased further (Figure 2C). To study in detail the locations of new complex PDs, we used time-lapse imaging of detached leaves maintained in observation chambers. The chamber system developed by Chan et al. (2007) was adapted to study the temporal insertion of complex PDs in the same cells over time. This was constructed using a standard cover slip and a section of breathable membrane, with a ring of double-sided adhesive forming a spacer between (see Supplemental Figure 1 online). This system allowed leaves to be maintained and imaged repeatedly for ∼72 h. Using observation chambers, we followed the progression of simple-to-complex PD formation and found that many of the new MP17-GFP signals appeared at the same locations as existing PDLP1-mRFP signals, producing yellow overlap signals (shown around a guard cell complex in Figures 2D to 2F). The inset of Figure 2D shows detail of an individual pavement cell wall observed over a period of 24 h, indicating the spatial overlay between red and green signals. With young detached leaves (leaves 1 and 2), we noticed that the conversion of simple to complex PDs occurred rapidly. New complex PDs could be visualized within 1 h of leaf detachment and were formed continuously at an average rate of 0.18 PDs per cell per hour (Figure 2G). In the areas observed, the overall numbers of PDs increased two- to threefold within 48 h.
Figure 2.
Dual Labeling of PDs with PDLP1-mRFP (All PDs) and MP17-GFP (Complex PDs Only) Allows Imaging of the Conversion of Simple to Complex PDs.
(A) PDs at the base of leaf 1 show exclusively simple PDs (red). Vacuolar fluorescence is due to nonspecific accumulation of mRFP.
(B) The first appearance of complex PDs (green) occurs at three-way junctions and epidermal lobes.
(C) In older leaves, the PD signals become yellow as complex PDs supersede simple PDs in the epidermis.
(D) to (F) Time-lapse imaging of an anisocytic complex over a 24-h period showing that complex PDs arise at sites occupied by existing simple PDs. The boxed region shows detail of an epidermal lobe in which simple PDs (red) are converted to complex PDs (yellow). Progression of complex PD numbers in epidermal cells in leaf 1 over a 24-h period following its detachment from the plant. Bar = 10 μm.
(G) Increase in complex PD numbers following leaf detachment. Graph shows data averaged from a range of leaf ages and sizes.
Accelerated Formation of Complex PDs Is Accompanied by a Premature Sink–Source Transition
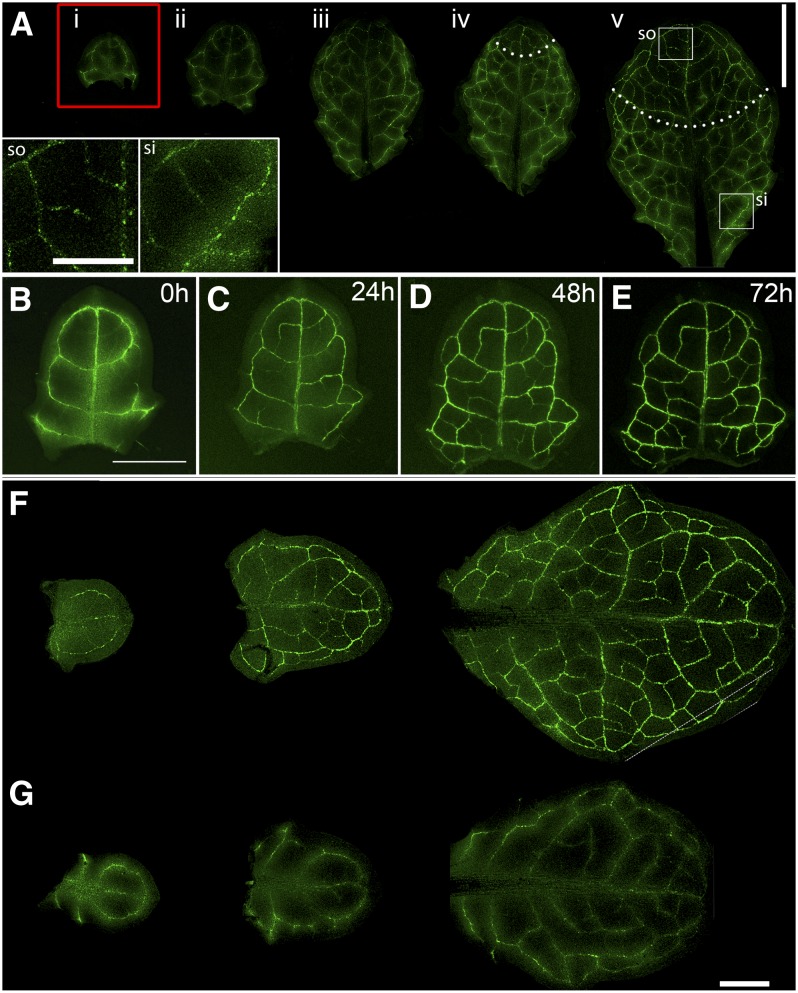
Because the bulk appearance of complex PD has been linked to the sink–source transition (Oparka et al., 1999), we tested whether leaf detachment, as described above, also influenced the onset of the sink–source transition. To do this, we used an Arabidopsis line expressing the Suc transporter, AtSUC2-GFP, a reporter expressed in the companion cells of the exporting phloem (Imlau et al., 1999). In these plants, GFP unloading is observed in the major veins of sink leaves, with subsequent appearance of the GFP in companion cells active in phloem loading, resulting in a restricted localization of GFP to the minor veins (Imlau et al., 1999; Wright et al., 2003). In attached control leaves, we observed the sink–source transition to occur characteristically in leaves 4 and 5 (Figure 3A). In these leaves, the veins in the apical part of the leaf were beginning to show discrete companion cell labeling (Figure 3A), indicative of phloem loading. In comparison, when we detached the first emerged leaf and floated it on water, we observed a rapid sink–source transition within 24 h after detachment, similar to the rapid formation of complex PDs observed in detached leaves (Figures 2D and 2E). At this point, the leaf was <2 mm in length (Figures 3B to 3E). In control leaves, the sink–source transition had not yet commenced (cf. Figure 3A). We were able to produce identical results to these by removing all of the leaves on the plant except for the last three emerged leaves. In this case, also, the remaining leaves showed an accelerated sink–source transition relative to control plants (Figures 3F and 3G). These source–sink manipulations suggested that complex PD formation is influenced by leaf carbon balance, possibly the supply of Suc from exporting leaves.
Figure 3.
Leaf Detachment Is Accompanied by an Accelerated Sink–Source Transition.
(A) The sink–source transition on an intact plant is revealed using the At-SUC2 promoter to drive GFP expression. Leaves i to iii show diffuse vein labeling indicative of GFP unloading (Imlau et al., 1999; Wright et al., 2003). Leaves iv and v show the onset of loading in the apical part of leaf, indicated by restriction of GFP to the companion cells (areas above dotted lines). The insets show the source (so) and sink (si) regions of leaf (v). Bar = 2 cm.
(B) to (E) Accelerated sink–source transition in leaf i after its detachment from the plant. Restriction of GFP to developing veins is seen after 24 h (C) and progresses as new veins mature ([D] and [E]). Bar = 0.5 cm.
(F) and (G) Removal of all leaves on the plant except leaves i to iii also accelerates the sink–source transition.
(F) Punctate vein labeling, indicative of phloem loading, is apparent in leaves i to iii when all other leaves are removed.
(G) Control leaves from an untreated plant continue to show diffuse GFP labeling, indicative of phloem unloading (cf. [A], i to iii). Bar = 0.5 cm.
Anisocytic Complexes Display an Early Appearance of Complex PDs
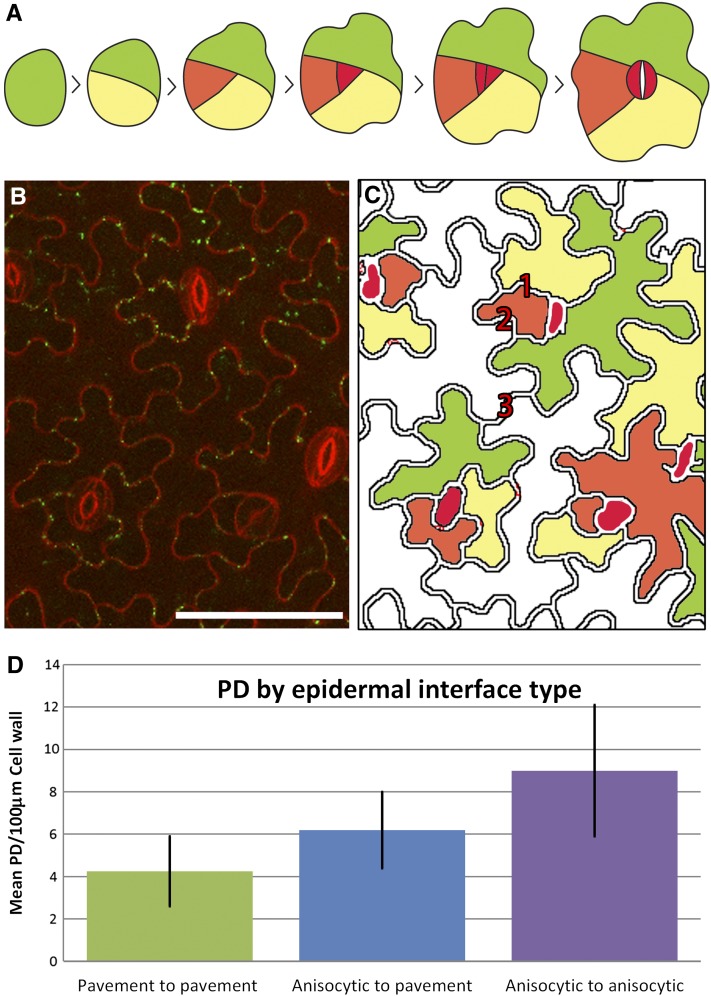
It is well known that maturing guard cells become symplasmically isolated at maturity due to a loss of PDs in their walls (Wille and Lucas, 1984). However, we noted that cells developmentally related to guard cells also showed different PD development patterns compared with pavement cells. In Arabidopsis, stomatal complexes are formed by progressive spiral divisions of meristemoid initials, resulting in each guard cell pair being surrounded by three monoclonal cells termed an anisocytic complex (Nadeau and Sack, 2002; Figure 4A). When we examined developing epidermal cells in the confocal microscope, we found that the anisocytic complexes showed conspicuous formation of complex PDs (Figures 4B and 4C; see also Figures 2D to 2F). We confirmed this observation by counting the number of complex PDs in the anticlinal walls of different cell types of the epidermis. Cells of the anisocytic complex developed complex PDs at a faster rate, and to a higher overall density, than other epidermal cells, with interfaces between anisocytic cells developing the highest overall densities in the epidermis (Figure 4D).
Figure 4.
Appearance of Complex PDs in Anisocytic Complexes.
(A) Asymmetric division of a single epidermal precursor (green) gives rise to two daughter cells, the smaller of which (yellow) undergoes further asymmetric division to give rise to a cell (orange) that will form the precursor of the meristemoid cell (red). Division of this cell gives rise to the guard cells.
(B) Early appearance in the leaf epidermis of complex PDs (green) occurs in anisocytic complexes adjacent to guard cells. Bar = 50 μm.
(C) Representation, based on (A), of the relative age of cells in the anisocytic complexes. (1) Anisocytic-anisocytic interface, (2) anisocytic-pavement interface, and (3) pavement-pavement interface. The oldest pavement epidermal cells are shown in white.
(D) Densities of complex PDs associated with different interfaces of the anisocytic complex.
Insertion of Complex PDs in Pavement Cells
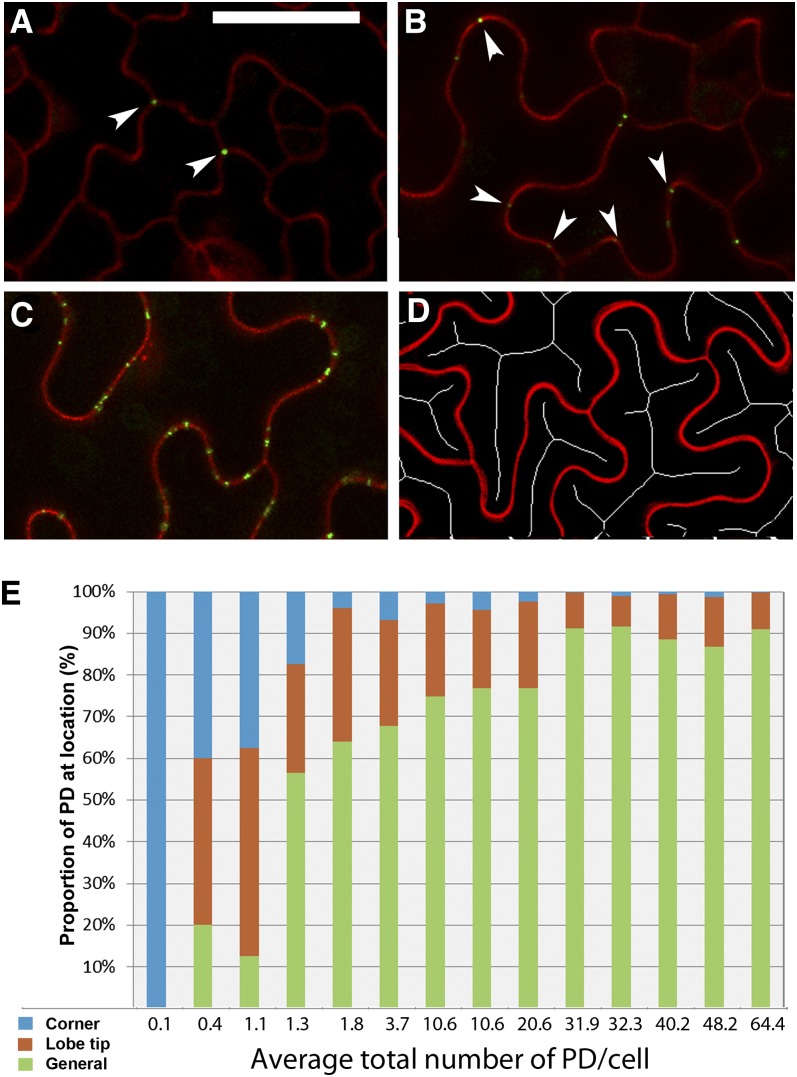
Detailed observations revealed that the insertion of complex PDs is not random within individual cells. In pavement cells, the first complex PDs appeared at the three-way junctions between cells. These complex PDs occurred early in leaf development, when the cells were still relatively unexpanded (Figure 5A). The next complex PDs appeared at the lobe tips of the expanding pavement cells (Figures 5B and 5C). Using ImageJ analysis software, we reduced the interior of the cells to skeleton lines (Xiong et al., 2010) to identify the lobe tips of the cells (Figure 5D). We were then able to allocate PDs to lobed and nonlobed regions in pavement cells of differing sizes. Following their appearance at lobe tips, complex PDs were inserted in nonlobed regions around the cell periphery, leading eventually to all regions of the cell wall containing complex PDs (as in Figure 1A). Figure 5E shows the proportion of complex epidermal PDs at different cellular locations as a function of increasing complex PD numbers, emphasizing the early appearance of complex PDs at the cell corners and lobe tips.
Figure 5.
Progression of Complex PD Formation in Pavement Epidermal Cells.
(A) The first complex PDs appear at the three-way junctions between epidermal cells (arrowheads). Bar = 20 μm for (A) to (C).
(B) Subsequent PDs appear at the lobed tips of pavement epidermal cells (arrowheads).
(C) With continued development of the epidermis, complex PDs appear in the intervening wall regions between lobes (general PD labeling).
(D) Skeleton cell profiles, generated using ImageJ, are used to determine the positions of lobes.
(E) Quantitative assessment of the appearance of complex PDs at three-way junctions, lobe tips, and general regions, respectively, as a function of increasing complex PD numbers in a sample of leaves.
High-Throughput Imaging of Complex PD Formation
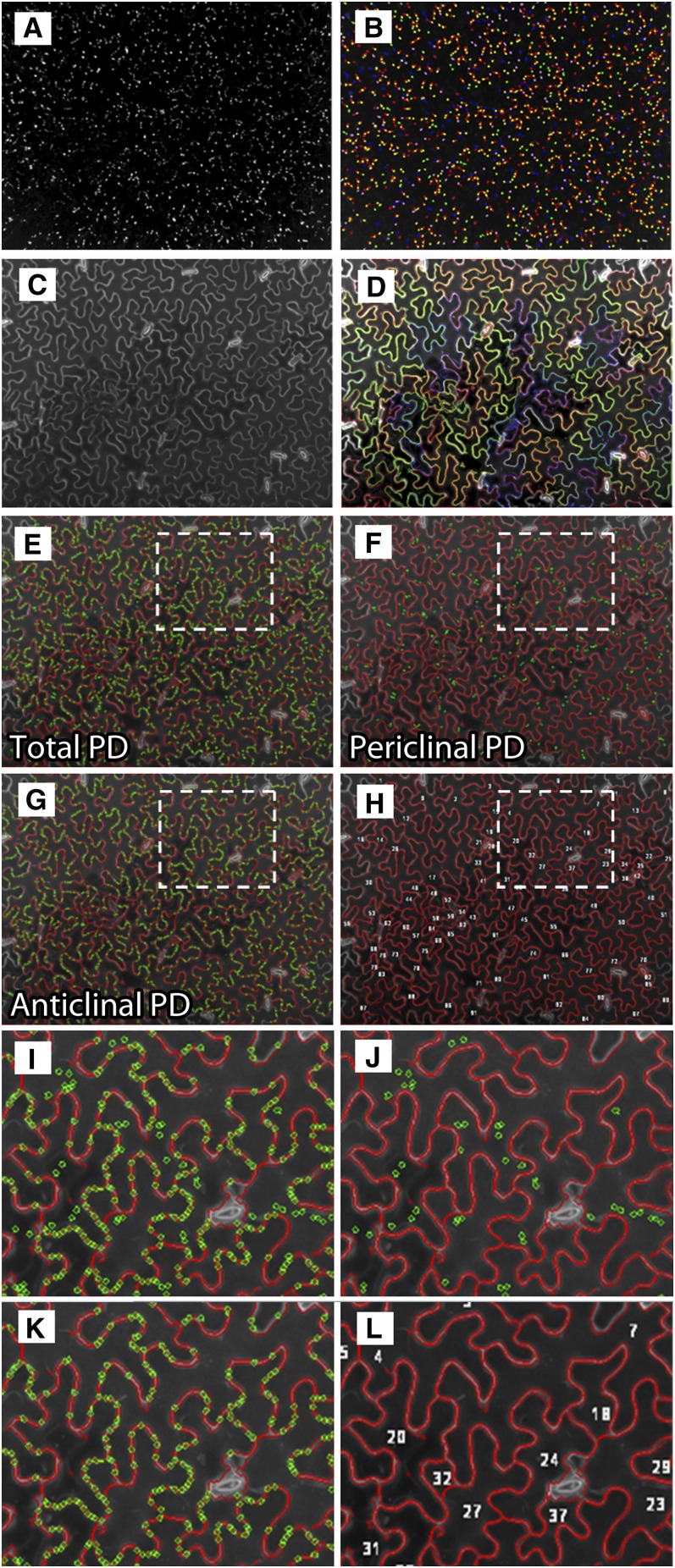
While the above observations provided data on the locations of complex PD, it was not possible to determine accurately the total numbers of complex PDs inserted into leaves of increasing age using manual microscopy, as the inherent variability in leaf maturation rates necessitated unfeasibly large samples sizes. To do this, we chose the Opera high-content screening system (Perkin-Elmer) to perform high-throughput confocal imaging. Two cameras in the Opera system were used during image acquisition that captured two consecutive image series of 21 to 35 Z planes with a spacing of 1 μm. One series (the PD channel; MP17-GFP) contained spot-like PD signals and the other one (the cell wall channel; propidium iodide) included information on the jigsaw-shaped epidermal cells. We developed a complex algorithm called PDQUANT that provided an automated image processing workflow that detected and quantified PD candidates, identified cell boundaries, and segmented immature cells from older cells (Figure 6). Full details of the PDQUANT workflow are provided in Methods and in Supplemental Figures 2 and 3 online. Once the analysis pipeline had been established, a single automated run of 60 leaves took ∼30 min. To ensure good coverage, we imaged nine separate (432 × 328 μm) fields on each leaf, each containing between 30 and 200 epidermal cells (mean 94), depending on the area of the leaf, allowing us to reconstruct the spatial distribution of complex PDs during leaf development. We also attempted to image simple PDs labeled with PDLP1-mRFP using the Opera system. However, this was not possible because in older leaves (leaves 4 to 6) the mRFP signal became increasingly associated with the cell walls in a nonspecific manner (see above), interfering with the computational spot detection program developed to recognize PD punctae. Thus, for high-throughput imaging, we concentrated our efforts on the transgenic line expressing MP17-GFP. The method potentially allows for calculation of PD/area values. However, in the epidermal-mesophyll interface PD/area, values are misleading as the area of contact between the two cell layers is broken by airspaces, so data are presented in the form of more biologically relevant PD/cell values.
Figure 6.
The High-Throughput Analysis Workflow for Detecting PDs and Segmenting Jigsaw-Shaped Cells Using PDQUANT.
(A) A maximum intensity projection was used to merge 21 to 35 optical Z planes (image sequences in the PD channel) into a two-dimensional pseudo-image for PD signals.
(B) A set of PD candidates detected from PD images, which are presented by randomly colored circles.
(C) A maximum intensity projection was used to merge 21 to 35 optical Z planes (image sequences in the cell channel) into a two-dimensional pseudo-image for cell boundary signals.
(D) Jigsaw-shaped cells are segmented and cell boundaries are highlighted by colored lines.
(E) Detected cell boundaries (colored red) are overlapped with recognized PDs (colored green). Cells with unsuitable size (<200 μm), contrast, and location (e.g., small cells attached to the image border) are discarded.
(F) and (G) PDs overlaid with cell boundaries are categorized into the anticlinal (epidermal-epidermal) PD group, and those overlapped with cell interiors are categorized into the periclinal (epidermal-mesophyll) PD group. PDs not overlaid with any cells are discarded from the analysis.
(H) Detected cells are treated as image objects. They are labeled together with calculated features, such as size, two-dimensional coordinates, and the number of PDs per cell.
(I) to (L) High-magnification views of recognized cell boundaries showing all PDs (I), periclinal PDs (J), anticlinal PDs (K), and cell identities (L).
Complex PD Insertion in the First Six Emerged Leaves
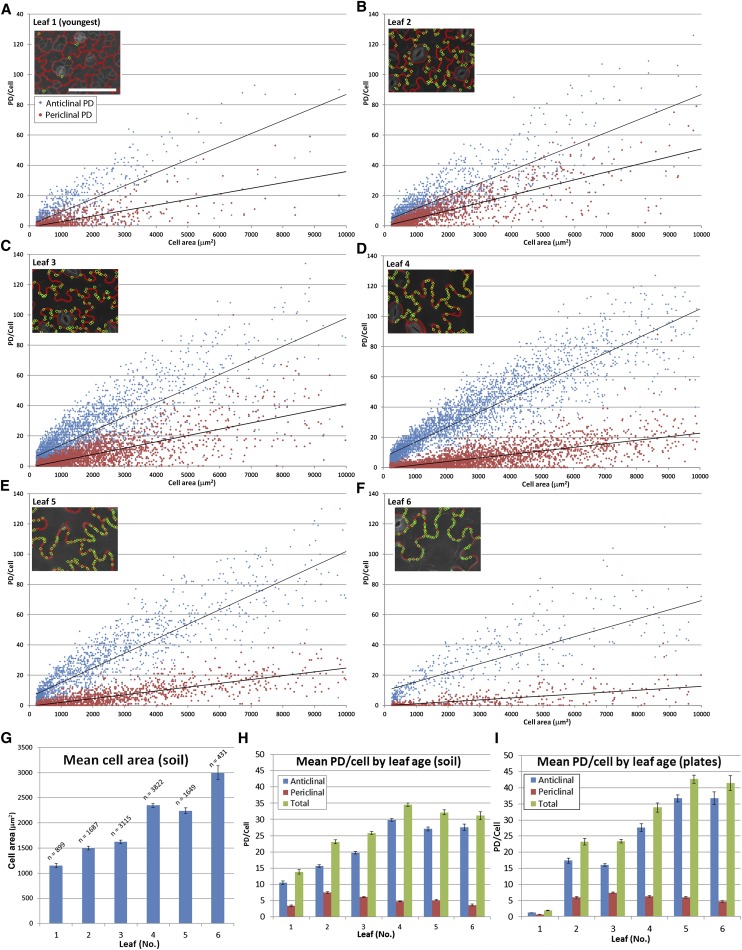
Large-scale data sets for the insertion of total complex PDs in the anticlinal and periclinal walls of the epidermis are shown in Figure 7, revealing a linear relationship between complex PDs and cell size for the first six emerged leaves. As the leaves aged, a greater number of cells displayed maximal numbers of PDs, leveling off at ∼35 complex PDs per cell (Figure 7H). By contrast, the mean cell area was continued to increase up to leaf 6 (Figure 7G). The combined data for leaves 1 to 6, grown on soil and agar, are shown in Figures 7H and 7I. Note that the increase in anticlinal complex PD numbers as a function of cell size increases until approximately leaf 5 and becomes constant thereafter. The leaves of plants grown on agar showed an apparent delay in the onset of complex PD formation compared with soil-grown plants by about one leaf position, with leaf 1 showing significantly fewer complex PDs than the equivalent leaf position on soil-grown plants (cf. Figures 7H and 7I).
Figure 7.
Large-Scale Analysis of the Insertion of Complex PDs in the Arabidopsis Leaf Epidermis Using PDQUANT.
(A) to (F) Data for the first six emerged leaves. PD numbers and area for individual cells in both anticlinal (red) and periclinal (blue) epidermal walls are shown, with cells under 200 μm not analyzed. The insets show representative images from PDQUANT showing locations of individual complex PDs in the cell wall.
(G) Data for mean cell area of the first six leaves.
(H) Mean number of complex PDs at different epidermal interfaces for the first six leaves for plants grown on soil.
(I) Mean number of PDs at different epidermal interfaces for the first six leaves for plants grown on agar.
The data for periclinal interfaces connecting epidermal cells to the overlying mesophyll are also shown in Figure 7. In this case, the interfaces showed a gradual but significant decrease in total complex PD numbers from leaf 2 to 6, indicating a loss of PDs from this interface (Figures 6H and 6I). This loss may be due to the separation of some mesophyll cells from the overlying epidermis due to turgor-driven cell expansion (Roberts et al., 2001).
Complex PD Formation Is Influenced by Exogenous Stresses
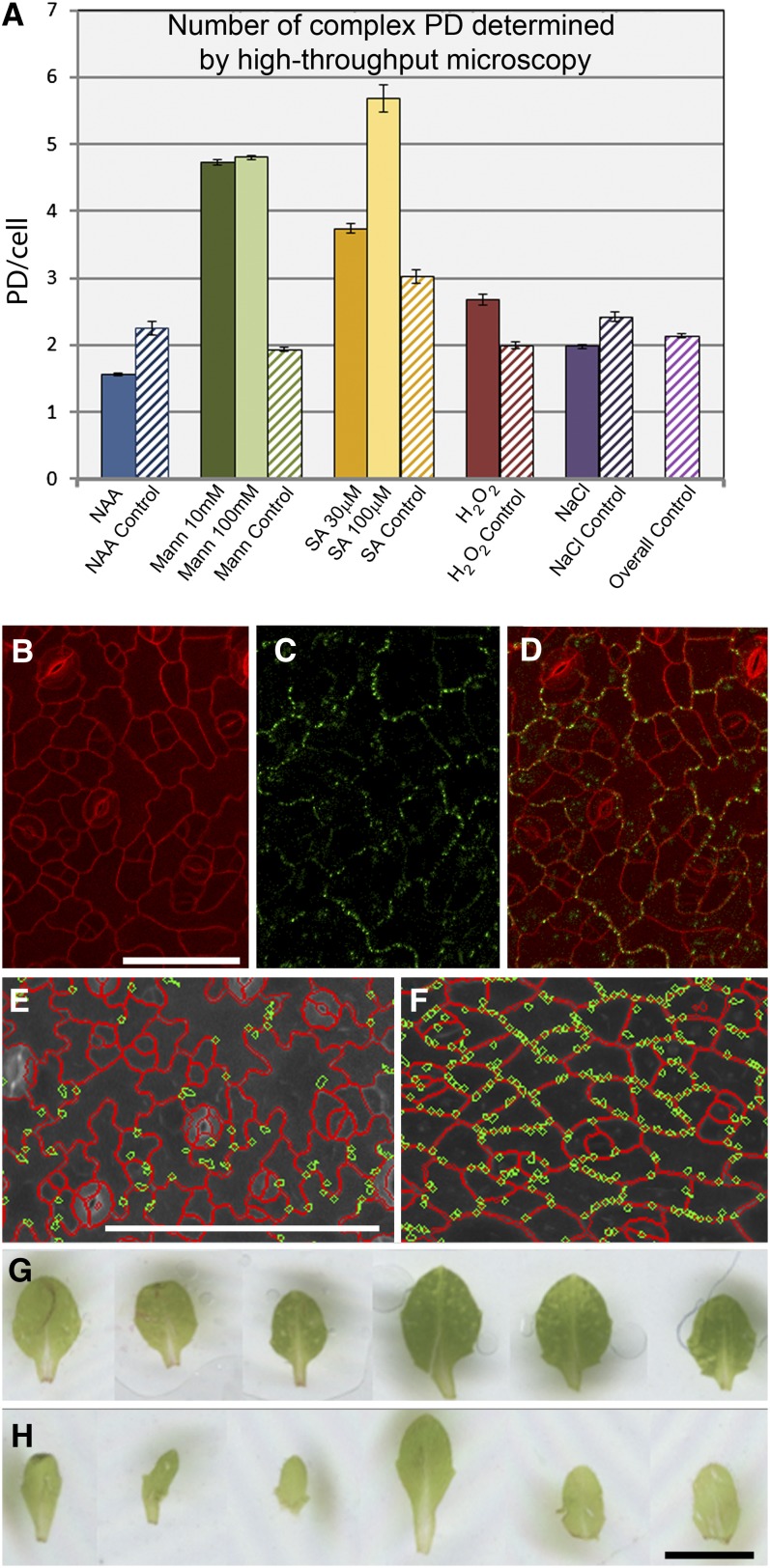
Having established a baseline pattern for complex PD formation in attached leaves (Figure 7), we examined the effects of a range of exogenous factors on the appearance of complex PDs. For these experiments, we transferred established seedlings to media plates containing supplements to simulate various growth challenges and allowed them to continue growth for 5 d. We used treatments that had previously been shown to alter the structure and function of PDs in a range of cell types (reviewed in Burch-Smith and Zambryski, 2012). These included salt stress (sodium chloride), osmotic stress (mannitol), an auxin (NAA), (naphthalene-1-acetic acid [NAA]) and defense-related treatments (hydrogen peroxide [H2O2] and salicylic acid [SA]; Figure 8A).
Figure 8.
Response of Complex PD Formation in Anticlinal Epidermal Interfaces to Exogenous Stresses.
(A) Numbers of complex PDs, derived using PDQUANT, in leaf 2 of plants subjected to a range of treatments (mannitol, 10 and 100 mM; H2O2, 1 mM; NaCl, 25 mM; NAA, 50 nM; SA 30 and 100 μm) and their respective controls that were performed in parallel to the treatment experiments. The “overall control” shows the average from all of the parallel control experiments combined. Mannitol and SA treatments show a significant increase (P < 0.01) in complex PD numbers.
(B) to (D) Appearance of epidermal cells from plants grown on mannitol. Lobing is absent in the pavement cells (B), and complex PDs (C) are restricted to the interfaces of the oldest epidermal cells. (D) shows the merged images. Bar = 50 μm.
(E) to (F). PDQUANT images of control (E) and 100 mM mannitol treated (F) leaves, showing detected cells (red) and PD (green). Scale = 100 μm. (G) to (H) Mannitol treatment results in narrower leaves (G) compared to control plants (H). Scale = 2 mm.
As above, leaves from the treated plants were placed in 96-well plates and complex PDs counted using the Opera system. A summary of the results is shown in Figure 8A. In these experiments, all leaves were stained with propidium iodide for 10 min and placed immediately into the observation chambers to minimize increases in PD numbers due to leaf detachment. Of the treatments tested, two showed major effects on the development of complex PDs. The first was SA, a long-range signal produced during plant defense responses (Spoel and Dong, 2012). SA produced significantly increased numbers (P < 0.01) of complex PDs at both high (100 μm) and low concentrations (30 μm), with values from >800 cells (100 µM SA) and >4500 cells (30 µM SA) compared with values from >1500 cells from the parallel control population. The second was mannitol, which has been used in a wide range of studies to promote osmotic stress (Schulz, 1995). Mannitol significantly increased the numbers (P < 0.01) of complex PDs at both high (100 mM) and low (10 mM) concentrations, with values from >19,000 cells (100 mM mannitol) and >22,000 cells (10 mM mannitol) compared with values from >9000 cells from the parallel control. However, in the case of the high concentration, increased complex PD numbers were accompanied by a major change in leaf and cellular morphology. Leaves from plants grown on 100 mM mannitol grew to similar lengths as the controls but were significantly narrower (Figures 8E and 8F), and the epidermal cells were uniformly square or oblong shaped rather than jigsaw shaped (Figures 8B to 8D). Furthermore, mannitol induced a marked difference in the distribution of PDs in these cells, with complex PDs forming predominantly in larger epidermal cells surrounding smaller dividing cells (Figures 8B to 8D). These morphological effects were absent in the low mannitol concentration, although the increase in complex PD numbers persisted (Figure 8A). These data suggest that development of complex PDs is influenced by a number of exogenous cues associated with stress-related pathways. Equivalent data sets are provided for periclinal PDs from the same samples in Supplemental Figure 4 online; in all cases, the response of periclinal PDs was similar to that of anticlinal PDs. For all treatments the number of PDs observed was smaller than equivalent values for plate-grown plants in the experiment shown in Figure 7I. We attribute this to the fact that the seedlings in the treatment experiment were used at a younger stage (four leaves instead of eight; see Methods).
To determine whether these treatments influenced the final numbers of complex PDs, we examined the same leaf over an extended time treatment on SAs (from 5 d extended to 14 d). At the end of the treatment period, the tagged leaf had become the sixth emerged leaf, at which stage complex PD numbers would normally have plateaued (Figure 7). These leaves did not show a significant increase (P > 0.5) in complex PD numbers relative to control plants (see Supplemental Figure 5 online), suggesting that the rate of insertion of complex PDs had been stimulated by SA, rather than the total number of complex PDs produced.
DISCUSSION
Our high-throughput, live-imaging approach to studying the development of PDs is amenable to plants that express suitable markers for the detection of PDs. Unfortunately, an exclusive marker for simple PDs has not yet been isolated. Thus, we were dependent on fluorescent reporters that detected either all PDs (e.g., PDLP1; Thomas et al., 2008) or complex PDs alone (MP17). Using the Opera confocal system, we have shown that it is possible to obtain large-scale data sets for the insertion of complex PDs during leaf development and, using observation chambers, to examine the spatial relationship of these PDs to existing simple PDs using a transgenic line expressing both the above PD markers. Our data show that many complex PDs arise at the same positions as existing simple PDs, confirming our previous study of the leaf trichome base (Faulkner et al., 2008). It seems likely that many of these new pores became connected to a shared wall cavity, although we were not able to image this process here. In observation chambers, detached leaves showed a rate of complex PD insertion of ∼0.2 PDs/cell/hour. These data suggest a remarkable remodeling of the cortical endoplasmic reticulum and a corresponding structural modification of the cell wall to accommodate the new pores of the complex PD. We should stress that the insertion of new PD channels adjacent to existing PDs is distinct from the formation of secondary PDs that arise de novo across graft unions. In the latter, the new PD pores may arise in regions of wall where PDs were not previously present (Ehlers and Kollmann, 2001).
It was apparent that the onset of complex PD formation occurs very early in leaf development. In intact plants, we were able to observe the first complex PDs in the second emerged leaf. It was previously thought that complex PDs were absent from embryonic tissues. However, the Arabidopsis embryo contains between 5 and 9% complex PDs, and it is now clear that complex PDs may form between some cell types very early in embryo development. Two genes, INCREASED SIZE EXCLUSION LIMIT1 (ISE1) and ISE2, were discovered in a screen for Arabidopsis embryos that showed enhanced dextran movement (Kobayashi et al., 2007). Recently, it was shown that the loss of these genes increased the formation of secondary PDs (seen as profiles of twinned and branched PDs) in the embryo, with an increase in GFP trafficking in plants silenced for the ISE genes (Burch-Smith and Zambryski 2010). This is unusual in that formation of branched (complex) PDs is usually associated with a downregulation of PD SEL (Itaya et al., 1998; Oparka et al., 1999; Crawford and Zambryski, 2001; reviewed in Roberts and Oparka, 2003). It is not clear if the increased branched PD numbers in the embryo observed by Burch-Smith and Zambryski (2010) were maintained through to leaf maturity.
In our experiments, we noted that the appearance of complex PDs was accelerated rapidly following leaf detachment, as was the sink–source transition, suggesting that leaf carbon balance may play an important role in the conversion of simple to complex forms of PDs. Our data suggest that a signal is received from older leaves that maintains PDs in the simple architecture because sink leaves, in the absence of any other leaves on the plant, also showed accelerated complex PD formation. The nature of this signal clearly deserves further study. Extensive studies on detached leaves conducted in the early twentieth century established that the response of most leaves to detachment is not death, but survival (reviewed in Yarwood, 1946), with detached leaves of many species able to regenerate shoots and roots and become independent, viable plantlets. Our work suggests that this survival response is just as strong in immature leaves, as even the youngest leaves undergo a precocious sink–source transition upon detachment that allows them to become self-sufficient. It is significant that even the smallest leaves, which have almost no complex PDs at detachment, use the available resources to produce complex PDs.
With an aim of carrying out automated high-throughput image analysis, we needed to address three technical issues: (1) how to quantify punctate PD signals, (2) how to segment and measure jigsaw-shaped plant cells, and (3) how to differentiate anticlinal and periclinal PDs. A number of automated methods have been developed for extracting quantifiable cell-scale data from confocal images (Zanella et al., 2010). To quantify spot-like membrane compartments and trafficking within and between cells, an endomembrane script was developed previously (Salomon et al., 2010). To perform cell segmentation and cell network analysis, additional methods (Butenuth, 2008; Zanella et al., 2010) and software tools (Kamentsky et al., 2011; Sethuraman et al., 2011; Pound et al., 2012) were introduced to the plant science research community. We found that many of the above methods performed reasonably well on cell unilayers but were exceptionally inaccurate when applied to intact leaves. In addition, we found that many open source software tools (e.g., FIJI/ImageJ, ICY, CellSeT, and CellProfiler) either did not support images (flex files) generated by the Opera system or could not efficiently batch process flex files. Because of that, we developed our own analysis pipeline on the basis of the Acapella image analysis software platform (version 2.0; Perkin-Elmer). Our script, PDQUANT, provided an automated image processing workflow (see Supplemental Figure 2A online) that detected and quantified PD candidates, identified cell boundaries, and segmented immature cells from older cells.
Using multititer plates, in a single run, we were able to derive data for up to 3k epidermal cells, detecting ∼11k complex PDs in the process. This approach allowed us to collect new data for complex PDs during leaf development and to examine the factors that influence their development. Our data show that increasing numbers of complex PDs occur in the epidermis up to the 4th emerged leaf. After this, complex PD numbers remained constant, suggesting that by leaf 5, the full quota of complex PDs had been reached. Exogenously applied stresses appeared to affect the rate of insertion of complex PDs rather than the final number per cell. It will be interesting to determine if maximal PD numbers per cell are genetically determined.
PDs in immature tissues generally allow diffusional passage of relatively large macromolecules (Oparka et al., 1999; Crawford and Zambryski, 2001; Imlau et al., 1999; Kim et al., 2005). As simple PDs predominate in newly formed leaves, the symplasm of these tissues can be viewed as syncytial, to the extent that simple PDs allow relatively nonspecific passage of molecules (Oparka et al., 1999). The onset of complex PD formation, exemplified by MP17 accumulation, appears to mark the concomitant formation of symplasmic domains in which communication between cells, and groups of cells, becomes restricted (Lucas and Lee, 2004). It is likely that the judicial placement of complex PDs at specific cell interfaces, or within regions of individual cells, may reduce intercellular communication at these points. In stomatal complexes, the PDs that connect the guard cells to the epidermis become truncated, isolating the stomatal complex from surrounding cells and allowing solute transporters to become active (Wille and Lucas, 1984). This may be important for correct stomatal patterning. In the chorus mutant of Arabidopsis, Guseman et al., (2010) found that decreased callose accumulation at PDs was related to an increase in symplasmic connectivity, leading to aberrant stomatal patterning.
The insertion of complex PDs showed a characteristic pattern in the pavement cells of the epidermis, first appearing at three-way cell junctions of undifferentiated cells, followed by their appearance in anisocytic complexes. Within the pavement epidermal cells, complex PDs appeared at the tips of the lobes before being inserted into the intervening areas of these cells. The significance of these patterns is not yet clear. During epidermal development, significant cytoskeletal reorganizations occur in pavement cells (Panteris and Galatis, 2005). It is possible that the early conversion of simple to complex PDs at this site, associated with downregulation of PD conductance, may allow more stringent control of cytoskeletal signaling events between epidermal cells. The question remains as to whether the appearance of complex PDs is always accompanied by a downregulation of PD SEL. Answering this will require a major correlative study between complex PD insertion and transport fluxes between individual cells. However, we propose that the rapid insertion of complex PDs in the leaf may mark a shift from unselective to selective protein transport, allowing increasing control of the molecules that pass between cells. As macromolecular trafficking occurs routinely through complex PDs, exemplified by the passage of viral genomes (Lucas, 2006), this indicates that complex PDs require an interaction with unique chaperones to permit selective macromolecular trafficking (Ueki and Citovsky, 2011). Indeed, at the base of the trichome, PD-mediated transport of the transcription factor KNOTTED1 has been shown recently to require interaction with specific chaperonins to permit movement (Xu et al., 2011).
In addition to subcellular regulation by specific classes of proteins, it appears that PD SEL may be sensitive to a range of extrinsic cues that influence the permeability of PDs. Closure of PDs by callose deposition during viral pathogenesis is common (reviewed in Burch-Smith and Zambryski, 2012); recently, cellular energy levels and reactive oxygen species (ROS; H2O2 treatment) have been implicated in regulation of PD function (Benitez-Alfonso et al., 2011; Burch-Smith et al., 2011). Furthermore, the site of ROS production (e.g., chloroplast versus mitochondrion) may differentially regulate specific cellular targets that in turn affect PD permeability. While ROS may influence the gating capabilities of complex PDs (Benitez-Alfonso et al., 2011; Burch-Smith and Zambryski, 2012), we found no evidence in leaves treated with H2O2 that increased ROS levels led to an accelerated appearance of complex PDs. By contrast, SA had a marked effect on complex PD formation. The premature onset of complex PD formation in response to SA acid may be a specific response to pathogen stress, allowing the plant to downregulate cell–cell communication rapidly in response to invasion, reducing the nonselective movement of pathogen-induced macromolecules between host cells. The response of complex PD formation to mannitol stress is less easily explained. Application of mannitol was shown to increase the PD SEL in roots (Schulz, 1995), and we found here that the rate of insertion of complex PDs was greater in mannitol-treated than in control plants. In addition, mannitol induced the formation of complex PDs at specific epidermal cell interfaces. This suggests that osmotic stress may also influence the spatial distribution of complex PDs. The significance of this is unclear, but our data suggest that complex PD insertion may be sensitive to both positional and stress-related cues.
Overall, our approach offers a new route to studying the formation of complex PDs, an area that has been sadly neglected in plant developmental biology. As PDs are central to cellular development (Burch-Smith et al., 2011), the positioning and distribution of simple and complex PDs may play important roles during cell differentiation in plants. This is especially true when morphogen gradients require to be established between cells within specific niches (Burch-Smith et al., 2011). The major onset of complex PD formation during early leaf development offers an opportunity to search for mutants that are specifically affected in the formation of complex PDs, thereby unraveling the function of these unique structures.
METHODS
Growth Conditions
For Arabidopsis thaliana grown on soil, seeds were vernalized for 48 h at 4°C and grown in controlled climate growth chambers (21°C, 50% humidity, 16 h/day illumination) and watered twice a week.
For Arabidopsis grown on plates, seeds were surface sterilized by immersion in 70% ethanol for 2 min and 100% bleach for 10 min and sown on prepared Petri dishes of solid Murashige and Skoog (MS) medium (4.9 g/L MS salts, 3 g/L Suc, and 10 g/L Difco agar, pH 5.8). Plates were stratified for 2 d at 4°C and then placed on angled racks under a light bench with 16-h/day illumination.
In the treatment experiments, plants were grown on plates as above for 5 d and then transferred with fine forceps to prepared plates of solid MS supplemented with the following treatments: NAA (50 nM), mannitol (10 and 100 mM), SA (30 and 100 μm), H2O2 (1 mM), and NaCl (25 mM). Seedlings were grown for a further 5 or 10 d before the third leaf was detached and imaged. For the 5-d treatment experiments, this meant that the plants were at the four-leaf stage and that the third leaf was the second youngest leaf on the plant. For each treatment experiment, a control was performed in parallel where plants were transferred to media plates without treatments.
Confocal Microscopy
For confocal imaging, leaves were infiltrated with 100 μg/mL propidium iodide. Tissues were mounted on slides and imaged using a Leica SP2 confocal laser scanning microscope with a ×63 water dipping lens (Leica Microsystems). GFP was excited using a 488-nm laser and mCherry using a 593-nm laser. For the confocal experiments, cell periphery and PD number were quantified using ImageJ.
Transmission Electron Microscopy
For transmission electron microscopy, tissue sections were cut using a clean razor and fixed for 3 h in 3% glutaraldehyde in 0.1 M sodium cacodylate, washed for 3 × 20 min in 0.1 M sodium cacodylate, postfixed for 1 h in 1% osmium tetroxide in 0.1 M sodium cacodylate, and dehydrated in a graded ethanol series Tissues were then infiltrated with Agar100 epoxy resin (Agar Scientific) for 1 h. The tissue in Agar 100 was then placed into capsules and the resin polymerized in an oven at 58°C for 48 h. Embedded tissues were then sectioned, stained with uranyl acetate and lead citrate, and imaged using a Philips BioTwin transmission electron microscope.
Structured Illumination Microscopy and Immunofluorescence
Structured illumination microscopy imaging and immunofluorescence were both performed exactly as described by Fitzgibbon et al. (2010).
Maintenance of Detached Leaves
Observation chambers were adapted from Chan et al. (2007). Leaves were detached using a clean razor and mounted in a drop of sterile water in a chamber formed by a standard cover slip and a section of breathable membrane (Lumox Folie 25-μm film; In Vitro Systems and Services) separated by a circle of double-sided adhesive (SecureSeal Adhesive Sheet; Grace Bio-labs) and mounted in a custom-made plastic frame (see Supplemental Figure 1 online). Chambers were maintained horizontally on a light bench with 16-h/day illumination.
Leaf Removal Experiments
Mature pAtSUC2
GFP plants at the 15- to 16-leaf stage were selected. All leaves but the youngest visible three were removed using tweezers. Plants were then imaged immediately (0 h) or returned to the growth chamber and imaged 12 or 24 h after leaf removal.
High-Throughput Microscopy
High-throughput confocal imaging was performed using the spinning disc automated Opera microscope (Perkin-Elmer Cellular Technologies) as described by Salomon et al. (2010). Excitation of the samples was performed at 488 nm for GFP and 561 nm for propidium iodide; the emission spectrum for GFP was captured using the 540/575 band-pass filter; and RFP emission was detected using the 600/40 band-pass filter. The exposure time was set to 40 ms. Leaves were prepared in 96-well plates with an optical glass bottom (Greiner), and a ×20 water immersion objective was used for taking images of a consecutive series of 31 planes with a distance of 1 μm. The collected image data were further analyzed using Acapella software (Perkin-Elmer Cellular Technologies).
Quantitative Analysis of Complex PD Insertion
Based on the earlier developed endomembrane script (Salomon et al., 2010), we refined the spot detection algorithm and embedded it in the PDQUANT script so that PD signals (see Supplemental Figure 2B online) could be detected and quantified. PDs were initially detected on MP17-GFP channel images and then filtered according to their size, roundness, intensity, and contrast (in comparison to their surrounding areas) (see Supplemental Figure 2C online). To image cell walls, we infiltrated the leaves under vacuum with 100 μg/mL propidium iodide. While processing cell wall signals, PDQUANT first discarded out-of-focus images from the analysis (see Supplemental Figure 2C online). After that, cell channel images were merged (using a maximum intensity projection) and then filtered to reduce noise through a median filter (see Supplemental Figure 2D online) and a convolution filter (see Supplemental Figure 2G online). On the filtered images, a complex masking method was applied to identify regions with high-intensity/contrast values (see Supplemental Figure 2E online). Generated masks were screened based on their intensity, size, and contrast values. After removing unsuitable masks, the remaining masked objects were retained in the analysis, which represented the potential cell boundaries between epidermal cells (see Supplemental Figures 2F and 2H online). Where image quality was poor (e.g., many areas were out of the focal plane on merged images), only clearly preserved cell boundaries were used in this process.
Cell boundaries, identified from the two approaches above, were combined (see Supplemental Figure 3A online) and inverted to represent the cells of interest (see Supplemental Figure 3B online). An image mask was applied to the inverted image that generated a range of masks to segment the monochrome image into regions representing recognized cell candidates (see Supplemental Figure 3C online). Subsequently, cell candidates were classified into three relative size groups (large, medium, and small) to facilitate segmentation. Merged cell candidates were split, and candidates under 200 μm (where pavement cells were difficult to distinguish from artifacts and guard cells) were removed. The resulting cell segmentation was refined using an erosion technique, which filled gaps between and within cells. Cells with unsuitable contrast (too bright or too dim) and position (e.g., touching the image border) were discarded (see Supplemental Figure 3D online). The remaining borders of recognized cells were treated as cell boundaries (see Supplemental Figure 3E online). Once segmented, the cells of the three different size classes were combined into a single image for further analysis.
Finally, the identified cells were overlapped with the detected PD candidates (see Supplemental Figure 2C online). If a putative PD candidate (green) was not overlaid with any cell boundaries (red) or within cells (areas enclosed by the cell boundaries), this PD candidate was discarded (see Supplemental Figure 3F online). PDs overlaid with cell boundaries were categorized into the anticlinal (epidermal-epidermal) PD group (see Supplemental Figure 3G online) or, if they occurred in nonanticlinal walls, as the periclinal (epidermis-mesophyll) PD group (see Supplemental Figure 3H online). Cellular traits (area, maximum width [x], maximum length [y], two-dimensional coordinates, and fluorescence signal intensity) and numbers of PDs were also calculated at this step, and the results were exported to a CSV file for further statistical analysis. The numbers of PDs (anticlinal and periclinal) were counted for every recognized cell. Each cell was given a specific numerical label, allowing further manual processing of the results if required (see Supplemental Figure 3I online).
With minor changes to the input parameters of the PDQUANT script, our analysis pipeline could be applied to cells with different sizes and shapes, for example, the small, square-shaped cells in immature leaves. In order to enable the automated analysis to run on other Acapella platforms (with different purchased modules) as well as to be translated into open-source image processing solutions (e.g., FIJI), the PDQUANT script solely uses low-level image analysis functions/modules of the Acapella image library.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Diagram Showing the Construction of a Simple Imaging Chamber for Analyzing PDs in Detached Arabidopsis Leaves.
Supplemental Figure 2. The PDQUANT Analysis Workflow for Detecting PD Candidates and Identifying Jigsaw-Shaped Cell Boundaries.
Supplemental Figure 3. The PDQUANT Analysis Workflow for Segmenting Plant Cells and Measuring PDs.
Supplemental Figure 4. Response of Complex PD Formation to Exogenous Stresses on the Periclinal Epidermal Interfaces, from the Same Data Sets as Data for Anticlinal Epidermal PDs Presented in Figure 8A.
Supplemental Figure 5. Effects of Long-Term Application of Salicylic Acid on Complex PD Numbers.
Acknowledgments
J.F. was the recipient of a postgraduate award funded by the Biotechnology and Biological Science Research Council. M.B. is funded by the Deutsche Forschungsgemeinschaft. Work in the laboratory of K.O. is funded by the Biotechnology and Biological Science Research Council. The S.R. laboratory is supported by the Gatsby Charitable Foundation.
AUTHOR CONTRIBUTIONS
J.F. designed and performed research and analyzed data. M.B. performed research and analyzed data. J.Z. designed computational tools and analyzed data. C.F. performed research. S.R. analyzed data and contributed new microscopy tools. K.O. designed research and wrote the article.
Glossary
- PD
plasmodesmata
- MP
movement protein
- GFP
green fluorescent protein
- SEL
size exclusion limit
- mRFP
monomeric red fluorescent protein
- H2O2
hydrogen peroxide
- SA
salicylic acid
- ROS
reactive oxygen species
- MS
Murashige and Skoog
References
- Bayer E., Thomas C., Maule A. (2008). Symplastic domains in the Arabidopsis shoot apical meristem correlate with PDLP1 expression patterns. Plant Signal. Behav. 3: 853–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Cilia M., San Roman A., Thomas C., Maule A., Hearn S., Jackson D. (2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Stonebloom S., Xu M., Zambryski P.C. (2011). Plasmodesmata during development: Re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma 248: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Zambryski P.C. (2010). Loss of INCREASED SIZE EXCLUSION LIMIT (ISE)1 or ISE2 increases the formation of secondary plasmodesmata. Curr. Biol. 20: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Zambryski P.C. (2012). Plasmodesmata paradigm shift: Regulation from without versus within. Annu. Rev. Plant Biol. 63: 239–260 [DOI] [PubMed] [Google Scholar]
- Butenuth M. (2008). Topology-preserving network snakes. Int. Arch. of Photogram. Rem. Sens. Spatial Inform. Sci. 37: 229–234 [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Crawford K.M., Zambryski P.C. (2001). Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 125: 1802–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Haudenshield J.S., Hull R.J., Wolf S., Beachy R.N., Lucas W.J. (1992). Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., and Lucas, W.J. (1996). Secondary plasmodesmata: Biogenesis, special functions and evolution. In Membranes: Specialised Functions in Plants. M. Smallwood, P. Knox, and D. Bowles, eds (Oxford, UK: BIOS Scientific Publishers), pp. 489–506. [Google Scholar]
- Ehlers K., Kollmann R. (2001). Primary and secondary plasmodesmata: Structure, origin, and functioning. Protoplasma 216: 1–30 [DOI] [PubMed] [Google Scholar]
- Ehlers K., van Bel A.J. (2010). Dynamics of plasmodesmal connectivity in successive interfaces of the cambial zone. Planta 231: 371–385 [DOI] [PubMed] [Google Scholar]
- Faulkner C., Akman O.E., Bell K., Jeffree C., Oparka K. (2008). Peeking into pit fields: A multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell 20: 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J., Bell K., King E., Oparka K. (2010). Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol. 153: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman J.M., Lee J.S., Bogenschutz N.L., Peterson K.M., Virata R.E., Xie B., Kanaoka M.M., Hong Z.L., Torii K.U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741 [DOI] [PubMed] [Google Scholar]
- Hepler P.K. (1982). Endoplasmic reticulum in the formation of the cell plate and plasmodesmata. Protoplasma 111: 121–133 [Google Scholar]
- Hofius D., Herbers K., Melzer M., Omid A., Tacke E., Wolf S., Sonnewald U. (2001). Evidence for expression level-dependent modulation of carbohydrate status and viral resistance by the potato leafroll virus movement protein in transgenic tobacco plants. Plant J. 28: 529–543 [DOI] [PubMed] [Google Scholar]
- Imlau A., Truernit E., Sauer N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya A., Woo Y.M., Masuta C., Bao Y.M., Nelson R.S., Ding B. (1998). Developmental regulation of intercellular protein trafficking through plasmodesmata in tobacco leaf epidermis. Plant Physiol. 118: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L., Jones T.R., Fraser A., Bray M.-A., Logan D.J., Madden K.L., Ljosa V., Rueden C., Eliceiri K.W., Carpenter A.E. (2011). Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics 27: 1179–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Kobayashi K., Cho E., Zambryski P.C. (2005). Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 102: 11945–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, K., Insoon, I., Cho, E., and Zambryski, P.C. (2005). Plasmadesmata and plant morphogenesis. In Plasmodesmata: Annual Plant Reviews, K.J. Oparka, ed (Oxford, UK: Blackwell Publishing), 90–109 [Google Scholar]
- Kobayashi K., Otegui M.S., Krishnakumar S., Mindrinos M., Zambryski P. (2007). INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19: 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J. (2006). Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344: 169–184 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Lee J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5: 712–726 [DOI] [PubMed] [Google Scholar]
- Maule A.J., Benitez-Alfonso Y., Faulkner C. (2011). Plasmodesmata - Membrane tunnels with attitude. Curr. Opin. Plant Biol. 14: 683–690 [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002). Stomatal development in Arabidopsis In The Arabidopsis Book 1: e0066, doi/10.1199/tab.0066. [DOI] [PMC free article] [PubMed]
- Oparka K.J., Roberts A.G., Boevink P., Santa Cruz S., Roberts I., Pradel K.S., Imlau A., Kotlizky G., Sauer N., Epel B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 743–754 [DOI] [PubMed] [Google Scholar]
- Panteris E., Galatis B. (2005). The morphogenesis of lobed plant cells in the mesophyll and epidermis: Organization and distinct roles of cortical microtubules and actin filaments. New Phytol. 167: 721–732 [DOI] [PubMed] [Google Scholar]
- Pound M.P., French A.P., Wells D.M., Bennett M.J., Pridmore T.P. (2012). CellSeT: Novel software to extract and analyze structured networks of plant cells from confocal images. Plant Cell 24: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.G., Oparka K.J. (2003). Plasmodesmata and the control of symplastic transport. Plant Cell Environ. 26: 103–124 [Google Scholar]
- Roberts I.M., Boevink P., Roberts A.G., Sauer N., Reichel C., Oparka K.J. (2001). Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218: 31–44 [DOI] [PubMed] [Google Scholar]
- Salomon S., Grunewald D., Stüber K., Schaaf S., MacLean D., Schulze-Lefert P., Robatzek S. (2010). High-throughput confocal imaging of intact live tissue enables quantification of membrane trafficking in Arabidopsis. Plant Physiol. 154: 1096–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A. (1995). Plasmodesmal widening accompanies the short-term increase in symplasmic phloem unloading in pea root-tips under osmotic-stress. Protoplasma 188: 22–37 [Google Scholar]
- Sethuraman V., French A., Wells D., Kenobi K., Pridmore T. (2011). Tissue-level segmentation and tracking of cells in growing plant roots. Mach. Vis. Appl. 23: 639–658 [Google Scholar]
- Spoel S.H., Dong X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S., Citovsky V. (2011). To gate, or not to gate: Regulatory mechanisms for intercellular protein transport and virus movement in plants. Mol. Plant 4: 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille A.C., Lucas W.J. (1984). Ultrastructural and histochemical-studies on guard-cells. Planta 160: 129–142 [DOI] [PubMed] [Google Scholar]
- Wright K.M., Roberts A.G., Martens H.J., Sauer N., Oparka K.J. (2003). Structural and functional vein maturation in developing tobacco leaves in relation to AtSUC2 promoter activity. Plant Physiol. 131: 1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Kabacoff C., Franca-Koh J., Devreotes P.N., Robinson D.N., Iglesias P.A. (2010). Automated characterization of cell shape changes during amoeboid motility by skeletonization. BMC Syst. Biol. 4: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Wang J., Xuan Z., Goldshmidt A., Borrill P.G., Hariharan N., Kim J.Y., Jackson D. (2011). Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Yarwood C.E. (1946). Detached leaf culture. Bot. Rev. 12: 1–56 [Google Scholar]
- Zanella C., Campana M., Rizzi B., Melani C., Sanguinetti G., Bourgine P., Mikula K., Peyrieras N., Sarti A. (2010). Cells segmentation from 3-D confocal images of early zebrafish embryogenesis. IEEE Trans. Image Process. 19: 770–781 [DOI] [PubMed] [Google Scholar]