The role of HAN in regulating floral development was examined in Arabidopsis. HAN represses hundreds of genes, especially genes involved in hormone responses and floral organ specification. Overexpression of HAN represses the expression of HAN and other GATA3 family genes (HANL2, GNC, and GNL), forming a negative regulatory loop, and thus serves as a key repressor regulating floral development.
Abstract
Plant inflorescence meristems and floral meristems possess specific boundary domains that result in proper floral organ separation and specification. HANABA TARANU (HAN) encodes a boundary-expressed GATA3-type transcription factor that regulates shoot meristem organization and flower development in Arabidopsis thaliana, but the underlying mechanism remains unclear. Through time-course microarray analyses following transient overexpression of HAN, we found that HAN represses hundreds of genes, especially genes involved in hormone responses and floral organ specification. Transient overexpression of HAN also represses the expression of HAN and three other GATA3 family genes, HANL2 (HAN-LIKE 2), GNC (GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM-INVOLVED), and GNL (GNC-LIKE), forming a negative regulatory feedback loop. Genetic analysis indicates that HAN and the three GATA3 family genes coordinately regulate floral development, and their expression patterns are partially overlapping. HAN can homodimerize and heterodimerize with the three proteins encoded by these genes, and HAN directly binds to its own promoter and the GNC promoter in vivo. These findings, along with the fact that constitutive overexpression of HAN produces an even stronger phenotype than the loss-of-function mutation, support the hypothesis that HAN functions as a key repressor that regulates floral development via regulatory networks involving genes in the GATA3 family, along with genes involved in hormone action and floral organ specification.
INTRODUCTION
Flower formation is a fundamental feature of angiosperm plants and has attracted intensive study in the past decades. Flowers arise from a specialized structure in the shoot tip called the shoot apical meristem (SAM), which comprises a pool of stem cells that continuously divide and replenish themselves (Fletcher, 2002). Despite the fact that flowers display an enormous diversity of morphology in different plants, most flowers have four types of floral organs arranged in concentric whorls, specifically sepals in whorl 1, petals in whorl 2, stamens in whorl 3, and carpels in whorl 4. For a particular plant species, the floral organ number, size, shape, and the relative spatial position are generally fixed.
In Arabidopsis thaliana, floral organ identity is specified by the combinatorial actions of three classes of genes, termed A, B, and C, that act in developing floral meristems. A function (provided in part by the APETALA1 and APETALA2 genes) determines sepal identity; B function (provided by the APETALA3 and PISTILLATA genes), along with A function, determines petal identity; B function plus C (provided by the AGAMOUS gene) determines stamen identity; and C function determines carpel identity (Bowman et al., 1991; Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). In addition to organ identity genes, floral architecture is also affected by genes that function in meristem activity or/and boundary formation. For example, the CLAVATA genes (CLV1, CLV2, and CLV3), PERIANTHIA (PAN), WUSCHEL (WUS), SHOOT MERISTEMLESS (STM), and UNUSUAL FLORAL ORGANS (UFO) largely function in the meristem to regulate floral organ number and shape. Mutation of CLV or PAN results in increased floral organ number; by contrast, mutation of WUS, STM, or UFO generates flowers with reduced numbers of organs (Long et al., 1996; Clark et al., 1997; Mayer et al., 1998; Chuang et al., 1999; Fletcher et al., 1999; Durfee et al., 2003). On the other hand, SUPERMAN, RABBIT EARS (RBE), CUP-SHAPED COTYLEDON (CUC), PETAL LOSS, and BLADE-ON-PETIOLE (BOP), whose expression is mainly in the meristem-organ boundary regions, the organ-organ boundary regions, or both, function by establishing proper boundaries to regulate organ number, shape, separation, and relative position (Sakai et al., 1995; Aida et al., 1997; Vroemen et al., 2003; Brewer et al., 2004; Takeda et al., 2004; Hepworth et al., 2005).
HANABA TARANU (HAN) is expressed at the boundaries between meristem and organ primordia and at the boundaries between different floral organs (Zhao et al., 2004). HAN knockouts display small, flat SAMs, fused sepals, and reduced numbers of floral organs, whereas HAN overexpression leads to delayed plant growth, disturbed cell division, and loss of meristem activity (Zhao et al., 2004). HAN also plays an important role in proembryo boundary formation through regulation of auxin flux and establishing cotyledon identity during embryogenesis in Arabidopsis (Nawy et al., 2010; Kanei et al., 2012). The gene expression domain and function of HAN are not conserved between monocots and dicots. In the grass family, HAN homologs such as TASSEL SHEATH (TSH) in maize (Zea mays), NECK LEAF1 (NL1) in rice (Oryza sativa), and THIRD OUTER GLUME (TRD) in barley (Hordeum vulgare) are expressed similarly in the cells of the suppressed bract, and loss of function of TSH/NL1/TRD results in bract outgrowth (Wang et al., 2009; Whipple et al., 2010).
HAN belongs to a family of 30 members in Arabidopsis that code for GATA3–type transcription factors that have a single zinc finger domain (Zhao et al., 2004; Bi et al., 2005). GATA factors were initially named for their ability to bind the consensus DNA sequence WGATAR (W = T or A; R = G or A) (Lowry and Atchley, 2000). In animals, GATA factors have been shown to act in development, differentiation, and cell proliferation (Patient and McGhee, 2002), whereas fungal GATA factors participate in nitrogen metabolism, circadian regulation, mating-type switching, and light-regulated photomorphogenesis (Scazzocchio, 2000). In plants, GATA factors have been identified to regulate both developmental processes and responses to environmental stimuli, such as light signaling, circadian rhythms, photoperiodic control of flowering, seed germination, brassinosteroid signaling, lateral root founder cell specification, and stress responses (Putterill et al., 1995; Teakle et al., 2002; Sugimoto et al., 2003; Liu et al., 2005; De Rybel et al., 2010; Luo et al., 2010). GNC (for GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM-INVOLVED) and GNL (for GNC-LIKE) are two GATA transcription factors that belong to the same subfamily II as HAN in Arabidopsis (Reyes et al., 2004). GNC and GNL have been shown to mediate nitrogen metabolism, chlorophyll biosynthesis, and glucose sensitivity (Bi et al., 2005). Furthermore, GNC and GNL are directly repressed by floral homeotic genes APETALA3 and PISTILLATA during flower development and are important repressors of gibberellin signaling that regulate germination, greening, flowering time, and leaf elongation (Bi et al., 2005; Mara and Irish, 2008; Richter et al., 2010). However, little is known about the potential interactions between plant GATA family members, and little if anything is known about the underlying mechanism by which boundary-expressed HAN regulates floral organ development.
Here, we show that transient overexpression of HAN causes large-scale gene repression, especially repression of genes that are involved in hormone responses and floral organ specification. Induction of HAN also leads to negative autoregulation and repression of three additional GATA3 family genes: HAN-LIKE2 (HANL2), GNC, and GNL. Genetic analyses indicate that HAN and other GATA3 family genes coordinately regulate sepal separation, petal number, silique length, and stamen and embryo development. Transcripts of HANL2, GNC, and GNL have similar accumulation patterns that are partially overlapping with the expression pattern of HAN. We further show that HAN can homodimerize and heterodimerize with GATA3 family proteins and that HAN directly binds to its own promoter and the promoter of GNC in vivo. Our results suggest that HAN may function as a key repressor that regulates floral development via regulatory networks involving genes in the GATA3 family, hormone action, and floral organ specification.
RESULTS
Genome-Wide Transcription Analyses upon Inducible Overexpression of HAN
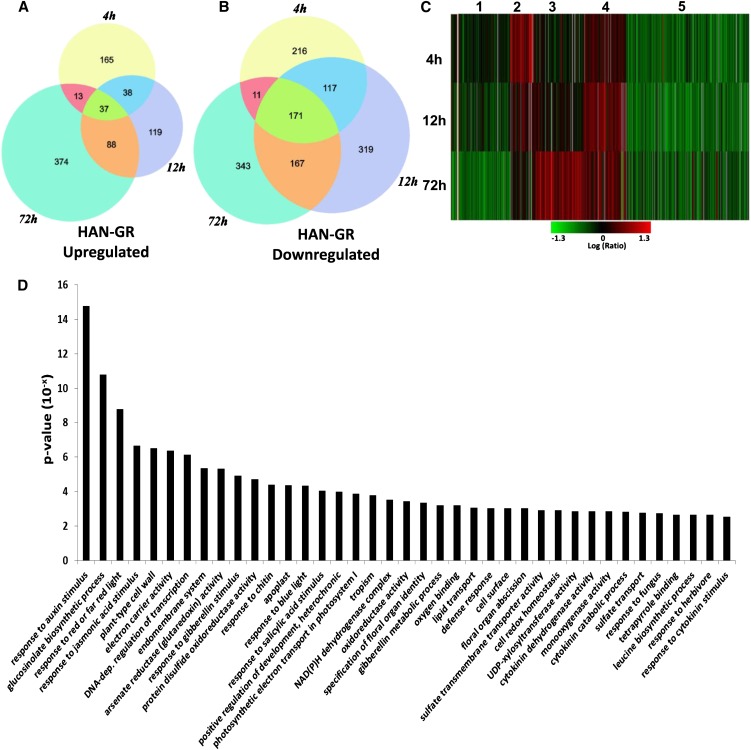
To dissect the molecular function of HAN, we performed time-course transcriptome analysis using a full-genome microarray, after the transient induction of HAN activity in p35S:HAN-GR plants, the same line as previously described (Zhao et al., 2004). Briefly, the p35S:HAN-GR line was constructed by inserting the full-length cDNA of HAN (no stop codon) into the pGreen 0229 vector containing a 2X35S promoter, a glucocorticoid receptor (GR) fragment (conferring resistance to dexamethasone [DEX]), and a nopaline synthase terminator (Zhao et al., 2004). Plants were treated with 10 μM DEX, and inflorescences containing flower buds from stages 1 to 9 were collected for microarray assays from plants at 4, 12, and 72 h after the initial DEX treatments and corresponding mock treatments. We identified 2074 genes that showed significantly differential expression (P < 0.001 and fold change > 2 or < −2) under DEX treatment in at least one of the three time points (q < 0.009) (see Supplemental Data Set 1 online). Although the numbers of upregulated genes and downregulated genes were similar after 72 h of induction, downregulated genes outnumbered upregulated genes significantly at the early time points of HAN induction. The numbers of upregulated genes were 253 and 282, and the numbers of downregulated genes were 515 and 774 after 4- and 12-h treatments, respectively. Moreover, there are many more shared downregulated genes than upregulated genes (88 shared up versus 444 shared down) (Figures 1A and 1B). To further understand the dynamic trend of these RNA abundances after HAN induction, we clustered genes with similar expression patterns across the three time points. As shown in Figure 1C, genes that showed differential expression in at least one time point can be grouped into five clusters. Genes in clusters 1 and 5 were downregulated in the HAN induction samples compared with the controls, whereas genes in the other three clusters were upregulated. Cluster 1 genes are repressed, and cluster 3 genes are activated only at 72 h, suggesting that these are probably late effects of HAN induction. Cluster 2 genes were induced only at 4 h and later returned to their normal expression levels, indicating that they may be stress-responsive genes. By contrast, clusters 4 and 5 likely reveal the specific mechanisms of HAN-mediated transcriptional regulation. Cluster 4 genes were weakly upregulated at 4 h, reached highest induction at 12 h, and returned to weak upregulation at 72 h. Cluster 5 genes were greatly repressed at 4 h and maintained this repression later on, suggesting that these genes may be the direct targets of HAN. Cluster 5 is the biggest group and accounts for over 40% of all the differentially expressed genes (Figure 1C; see Supplemental Data Set 1 online), implicating HAN as a master repressor that downregulates the transcription of a large numbers of genes.
Figure 1.
Genome-Wide Transcription Analysis upon Time-Course Induction of HAN in the Floral Buds of 35S-HAN-GR Arabidopsis.
(A) and (B) Venn diagrams of differentially expressed genes that were significantly upregulated (A) or downregulated (B) (P value < 0.001, fold change >2 or <−2) 4, 12, and 72 h after DEX induction of HAN.
(C) Clustering displays of expression ratios (DEX treatment versus mock-treated control) for genes that are differentially expressed in at least one of the time points (4, 12, or 72 h). Red color indicates upregulation and green indicates downregulation.
(D) GO terms that are significantly enriched (P < 0.003) in cluster 5. GO terms were sorted based on P value.
Quantitative RT-PCR Corroboration of Differentially Expressed Genes under Transient Overexpression of HAN
To verify the differentially expressed genes identified by microarray, we performed quantitative RT-PCR (qRT-PCR) using independently generated DEX-treated versus mock-treated p35S:HAN-GR flowers (Table 1). Among the 20 genes we tested, 16 genes showed the same differential expression in the DEX-treated sample, and the other four genes also displayed the same patterns as those observed in microarray analysis, although the quantitative fold change in the qRT-PCR experiment was smaller than a twofold cutoff. The qRT-PCR and microarray data exhibited close agreement (Pearson correlation coefficient was 0.80, P = 1.4E-05) in the fold change between DEX-treated and mock-treated samples, indicating that the microarray data are reliable.
Table 1. qRT-PCR Corroboration of Differentially Expressed Genes under Transient Overexpression of HAN.
| Gene ID | Gene Name | P Value | Microarray DEX, Fold Change | qRT-PCR Fold Change | han-1 Fold Changea |
|---|---|---|---|---|---|
| At1g74890 | ARR15 | 4.1E-26 | −3.40b | −4.92 ± 1.20 | −1.86 ± 0.19 |
| At1g66350 | RGL1 | 0.0E+00 | −3.80b | −3.23 ± 0.43 | −1.39 ± 0.08 |
| At3g23050 | IAA7 | 0.0E+00 | −2.10b | −1.68 ± 0.55 | −1.70 ± 0.06 |
| At3g10000 | EMBRYO SAC DEVELOPMENT ARREST31 (EDA31) | 5.4E-07 | −5.20b | −7.06 ± 1.29 | −1.40 ± 0.03 |
| At4g32980 | ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) | 3.5E-09 | −2.30b | −5.04 ± 1.23 | −4.39 ± 1.24 |
| At1g70510 | KNAT2 | 4.1E-14 | −2.40b | −3.57 ± 1.25 | −1.00 ± 0.16 |
| At5g11060 | KNAT4 | 0.0E+00 | −3.90b | −2.24 ± 0.30 | −4.22 ± 0.03 |
| At1g24260 | SEP3 | 2.5E-20 | −2.20b | −2.21 ± 0.15 | −2.19 ± 0.33 |
| At1g69530 | ATEXPA1 | 0.0E+00 | −3.90b | −2.52 ± 0.55 | −1.45 ± 0.06 |
| At1g01470 | LATE EMBRYOGENESIS ABUNDANT PROTEIN14 (LEA14) | 3.3E-19 | 2.90b | 2.10 ± 0.23 | −2.60 ± 0.53 |
| At1g51950 | IAA18 | 8.0E-07 | −2.00c | −2.04 ± 0.27 | −4.15 ± 0.67 |
| At5g13220 | JAZ10 | 1.3E-10 | −2.60c | −6.85 ± 0.90 | −4.92 ± 0.19 |
| At2g34600 | JAZ7 | 4.9E-26 | −3.30c | −2.81 ± 0.65 | −5.82 ± 1.40 |
| At2g45190 | ABNORMAL FLORAL ORGANS (AFO) | 1.4E-35 | −2.70c | −5.95 ± 0.10 | −2.64 ± 0.38 |
| At3g26790 | FUS3 | 4.8E-10 | −4.80c | −1.11 ± 0.37 | −2.36 ± 0.53 |
| At2g21650 | MATERNAL EFFECT EMBRYO ARREST3 (MEE3) | 2.2E-05 | −3.20c | −7.11 ± 1.31 | 2.23 ± 0.03 |
| At1g70210 | CYCD1;1 | 0.0E+00 | −3.70c | −5.37 ± 1.99 | −1.46 ± 0.06 |
| At1g59940 | ARR3 | 1.3E-07 | 4.00c | 4.46 ± 0.86 | −1.83 ± 0.17 |
| At3g48100 | ARR5 | 5.7E-31 | 4.00c | 1.70 ± 0.11 | −1.19 ± 0.06 |
| At5g06070 | RBE | 7.8E-09 | 2.10c | 1.60 ± 0.37 | −2.67 ± 0.02 |
Fold change presented as relative abundance of transcripts in han1/wild-type Ler floral buds.
Fold change at 4 h interval
Fold change at 72 h interval
HAN Regulates Flower Development via Hormone Action and Floral Organ Regulatory Networks
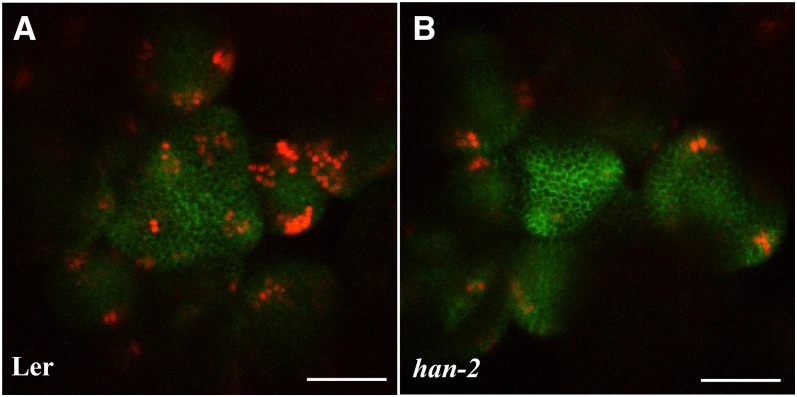
To analyze the functions of the differentially expressed genes upon HAN induction, we performed Gene Ontology (GO) term enrichment analysis for each cluster. In addition to the metabolic and stress-related pathways that were identified in all clusters, hormone action and floral organ regulators are significantly enriched in cluster 5 (Figure 1D, Tables 2 and 3; see Supplemental Figure 1 online). The most significantly enriched GO term is “response to auxin stimulus” (P < 10−14). Accordingly, lists of three well-known groups of early auxin-responsive genes including the auxin/indole-3-acetic acid (Aux/IAA), the small auxin-up RNA (SAUR), and the GH3 gene families are differentially expressed upon HAN overexpression (Wang et al., 2008) (Figure 1D, Table 2). IAAs and SAURs are generally repressed, while GH3s are induced by HAN overexpression. For example, the expression of IAA16 and SAUR68 decreases 5.3- and 4.4-fold, respectively, and the expression of GH3 family genes YADOKARI1 (YDK1) and GH3.5 increases 3.2- and 3.6-fold, respectively, upon 4 h DEX-induced HAN treatment (Table 2). We verified the repression of IAA16 and the induction of GH3.5 by quantitative PCR (see Supplemental Table 1 online). The expression of IAA16 showed no change in han mutants versus wild-type plants, while GH3.5 RNA is reduced in the HAN null allele han-1 (8.8-fold decrease) (see Supplemental Table 1 online), suggesting that GH3.5 but not IAA16 may be a direct target of HAN. To further verify whether HAN indeed regulates auxin action during flower development, we introduced the auxin response reporter pDR5rev:3XVENUS-N7 together with the auxin transport marker pPIN1:PIN1-GFP (for green fluorescent protein) to han-2 mutant plants (Heisler et al., 2005). The fluorescence from the pDR5rev:3XVENUS-N7 reporter is greatly reduced in the inflorescence meristems (IMs) of the han-2 mutant plants, especially at the floral primordia, compared with those in Landsberg erecta (Ler). By contrast, the pPIN1:PIN1-GFP reporter shows no appreciable change in fluorescence in han-2 (Figure 2). Consistent with our microarray data, live imaging results reveal that HAN mediates auxin response/signaling but not auxin transport during flower development.
Table 2. Examples of hormone action genes that are differentially expressed in the 35S-HAN-GR flowers in DEX-treated samples vs. mock-treated controls.
| Fold Change |
|||||
| Gene ID | Gene Name | 4h | 12h | 72h | Cluster |
| Auxin response | |||||
| At1g19220 | ARF19 (AUXIN RESPONSE FACTOR 19) | –2.4 | –1.8 | –2.1 | 5 |
| At3g04730 | IAA16 (INDOLE-3-ACETIC ACID INDUCIBLE16) | –5.3 | –3.2 | –2.0 | 5 |
| At1g51950 | IAA18 (INDOLE-3-ACETIC ACID INDUCIBLE18) | –1.5 | –2.0 | –2.0 | 5 |
| At3g17600 | IAA31 (INDOLE-3-ACETIC ACID INDUCIBLE31) | –2.0 | –2.3 | –1.9 | 5 |
| At2g01200 | IAA32 (INDOLE-3-ACETIC ACID INDUCIBLE32) | –2.0 | –2.5 | –1.5 | 5 |
| At1g52830 | IAA6 (INDOLE-3-ACETIC ACID INDUCIBLE6) | –2.0 | –2.1 | 3.5 | 5 |
| At3g23050 | IAA7 (INDOLE-3-ACETIC ACID INDUCIBLE7) | –2.1 | –3.0 | –2.0 | 5 |
| At1g29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | –4.4 | –4.5 | –3.3 | 5 |
| At5g63160 | BT1 (BTB AND TAZ DOMAIN PROTEIN 1) | –1.8 | –2.3 | –2.0 | 5 |
| At4g37390 | YDK1 (YADOKARI-1) | 3.2 | 2.0 | –1.5 | 2 |
| At3g07390 | AIR12 (AUXIN-INDUCED PROTEIN12) | 2.5 | 1.2 | 1.1 | 2 |
| At4g27260 | GH3.5 (Auxin-responsive GH3 family protein) | 3.6 | 3.4 | 1.7 | 4 |
| At1g12980 | DRN( DORNROSCHEN) | 1.1 | 3.3 | 1.6 | 4 |
| Jasmonate response | |||||
| At1g32640 | MYC2 (bHLH DNA-binding family protein) | –1.7 | –2.2 | –1.3 | 5 |
| At3g17860 | JAZ3 (JASMONATE-ZIM-DOMAIN PROTEIN 3) | –1.9 | –2.3 | –1.5 | 5 |
| At1g17380 | JAZ5 (JASMONATE-ZIM-DOMAIN PROTEIN 5) | –1.3 | –2.2 | –1.2 | 5 |
| At2g34600 | JAZ7 (JASMONATE-ZIM-DOMAIN PROTEIN 7) | –2.7 | –2.7 | –3.3 | 5 |
| At5g13220 | JAZ10 (JASMONATE-ZIM-DOMAIN PROTEIN 10) | –2.4 | –2.5 | –2.6 | 5 |
| At4g23600 | CORI3 (CORONATINE INDUCED 3) | –2.7 | –5.2 | –20.5 | 5 |
| At3g45140 | LOX2 (LIPOXYGENASE 2) | –2.0 | –6.4 | –7.7 | 5 |
| At5g47220 | ERF2 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2) | –1.5 | –3.1 | –1.4 | 5 |
| Gibberellin response | |||||
| At1g14920 | GAI (GA INSENSITIVE) | –2.3 | –2.9 | –2.1 | 5 |
| At1g66350 | RGL1 (RGA-LIKE1) | –3.8 | –3.9 | –3.1 | 5 |
| At5g15230 | GASA4 (GAST1 PROTEIN HOMOLOG 4) | –2.6 | –5.2 | –2.8 | 5 |
| At5G59780 | MYB59 (MYB DOMAIN PROTEIN 59) | –2.2 | –3.8 | –1.9 | 5 |
| At5g61420 | MYB28 (MYB DOMAIN PROTEIN 28) | –2.0 | –3.8 | –3.8 | 5 |
| At4g36410 | UBC17 (UBIQUITIN CONJUGATING ENZYME 17) | –3.6 | –3.1 | –1.6 | 5 |
| At1g68320 | MYB62 (MYB DOMAIN PROTEIN 62) | –1.8 | –2.2 | –3.4 | 5 |
| Cytokinin response | |||||
| At1g74890 | ARR15 (RESPONSE REGULATOR 15) | –3.4 | –3.2 | –2.6 | 5 |
| At1g59940 | ARR3 (RESPONSE REGULATOR 3) | –1.1 | 1.2 | 4.0 | 3 |
| At3g48100 | ARR5 (RESPONSE REGULATOR 5) | –1.8 | 1.6 | 4.0 | 3 |
| At5g62920 | ARR6 (RESPONSE REGULATOR 6) | –1.1 | 2.8 | 3.3 | 3 |
| At1g19050 | ARR7 (RESPONSE REGULATOR 7) | –4.7 | –1.9 | –1.4 | 5 |
| At5g56970 | CKX3 (CYTOKININ OXIDASE 3) | –4.1 | –5.0 | –16.5 | 5 |
| At3g16360 | AHP4 (ARABIDOPSIS HISTIDINE-CONTAINING PHOSPHOTRANSFER FACTOR 4) | –5.0 | –3.6 | –3.9 | 5 |
Table 3. Examples of developmental regulators that are differentially expressed in the 35S-HAN-GR flowers in DEX-treated samples vs. mock-treated controls.
| Fold Change |
|||||
| Gene ID | Gene Name | 4h | 12h | 72h | Cluster |
| Flower development | |||||
| At2g41370 | BOP2 (BLADE ON PETIOLE2) | –1.5 | –2.0 | –2.1 | 5 |
| At3g57130 | BOP1 (BLADE ON PETIOLE1) | –2.0 | –2.0 | –1.3 | 5 |
| At4g32980 | ATH1 (ARABIDOPSIS THALIANA HOMEOBOX GENE 1) | –2.3 | –3.2 | –1.7 | 5 |
| At1g24260 | SEP3 (SEPALLATA3) | –2.2 | –2.1 | –1.9 | 5 |
| At2g45190 | AFO (ABNORMAL FLORAL ORGANS) | –2.4 | –3.9 | –2.7 | 5 |
| At1g70510 | KNAT2 (KNOTTED-LIKE2) | –2.4 | –2.5 | –1.5 | 5 |
| At5g11060 | KNAT4 (KNOTTED-LIKE4) | –3.9 | –2.3 | –1.6 | 5 |
| At4g08150 | BP (BREVIPEDICELLUS) | –2.2 | –2.8 | –1.1 | 5 |
| At1g68480 | JAG (JAGGED) | –1.8 | –1.7 | –2.1 | 5 |
| At5g67180 | TOE3 (TARGET OF EAT 3) | –2.3 | –2.3 | –1.8 | 5 |
| At1g69490 | NAP (NAC-LIKE ACTIVATED BY AP3/PI) | 2.4 | 2.3 | 2.7 | 4 |
| At5g06070 | RBE (RABITT EARS) | 1.5 | 2.6 | 2.1 | 4 |
| At1g76420 | CUC3 (CUP-SHAPED COTYLEDON 3) | 1.4 | 3.4 | 1.8 | 4 |
| Reproductive development | |||||
| At3g10000 | EDA31 (EMBRYO SAC DEVELOPMENT ARREST 31) | –5.2 | –6.2 | –4.1 | 5 |
| At2g21650 | MEE3 (MATERNAL EFFECT EMBRYO ARREST 3) | –1.8 | –2.5 | –3.2 | 5 |
| At4g21330 | DYT1 (DYSFUNCTIONAL TAPETUM 1) | –1.5 | –2.3 | –3.1 | 5 |
| At1g01470 | LEA14 (LATE EMBRYOGENESIS ABUNDANT PROTEIN) | 2.9 | 4.2 | 2.1 | 4 |
| At4g28520 | CRU3 (CRUCIFERIN 3) | –1.2 | 2.0 | –3.6 | 1 |
| At3g26790 | FUS3 (FUSCA3) | –1.0 | 1.3 | –4.8 | 1 |
| Positive regulation of development | |||||
| At1g53230 | TCP3 (TCP family transcription factor 3) | –1.6 | –2.4 | –1.8 | 5 |
| At3g02150 | TCP13 (TCP family transcription factor 13) | –2.9 | –1.8 | –1.2 | 5 |
| At3g15030 | TCP4 (TCP family transcription factor 4) | –1.8 | –2.7 | –2.2 | 5 |
| At5g60970 | TCP5 (TCP family transcription factor 5) | –2.0 | –1.9 | –1.5 | 5 |
| Cell division & expansion | |||||
| At1g70210 | CYCD1;1 (CYCLIN DELTA-1) | –1.9 | –2.0 | –3.7 | 1 |
| At1g69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | –3.9 | –1.7 | –2.0 | 5 |
| At1g20190 | ATEXPA11 (ARABIDOPSIS THALIANA EXPANSIN A11) | –2.7 | –2.2 | –1.3 | 5 |
| At5g56320 | ATEXPA14 (ARABIDOPSIS THALIANA EXPANSIN A14) | –1.6 | –2.4 | –2.6 | 5 |
| At3g29030 | ATEXPA5 (ARABIDOPSIS THALIANA EXPANSIN A5) | –2.5 | –2.4 | –1.2 | 5 |
| GATA3 family | |||||
| At3g50870 | HAN (HANABA TARANU) | –1.2 | –1.6 | –2.0 | 1 |
| At5g56860 | GNC (GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM-INVOLVED) | –2.6 | –5.3 | –6.6 | 5 |
| At4g36620 | HANL2 (HAN-LIKE2) | –3.3 | –3.2 | –3.2 | 5 |
| At4g26150 | GNL (GNC LIKE) | 1.5 | –4.9 | –5.1 | 5 |
Figure 2.
Auxin Response Is Greatly Compromised in han-2 Mutant Plants.
pDR5rev:3XVENUS-N7 (red) and pPIN1:PIN1-GFP (green) markers in wild-type Ler (A) and han-2 mutant (B) IMs. Images are representative of 10 to 20 plants grown under the same environmental conditions. Bars = 50 µm.
Genes that are involved in responses to other hormones, such as jasmonic acid, gibberellin, and cytokinin, are significantly enriched in cluster 5 as well (Figure 1D). For example, the jasmonate signaling genes that encode jasmonate ZIM domain proteins (JAZ3, 5, 7, and 10), MYC2, and CORONATINE INDUCED1 (CORI3) are significantly repressed upon HAN induction (Table 2) (Fonseca et al., 2009). The DELLA family genes GA INSENSITIVE (GAI ) and RGA-LIKE1 (RGL1) in the gibberellin signaling and RESPONSE REGULATOR15 (ARR15) and ARABIDOPSIS HISTIDINE-CONTAINING PHOSPHOTRANSFER FACTOR4 (AHP4) in the cytokinin two-component signaling pathway are also largely downregulated upon transient HAN overexpression (Table 2) (Hirano et al., 2008; To and Kieber, 2008). Therefore, HAN seems to function as a negative regulator that mediates multiple hormone response and/or signaling pathways.
In addition, the expression of genes encoding many well-known developmental regulators are significantly altered upon HAN induction, including flower developmental genes, such as BOP1 and 2, RBE, CUC3, BREVIPEDICELLUS (BP), KNOTTED-LIKE2 (KNAT2), and SEPALLATA3 (SEP3), and reproductive development genes, such as FUSCA3 (FUS3) and DYSFUNCTIONAL TAPETUM1 (DYT1), indicating that HAN regulates floral organ and embryo development through the interaction with known developmental regulators (Table 3) (Pelaz et al., 2000; Pautot et al., 2001; Vroemen et al., 2003; Takeda et al., 2004; Tsuchiya et al., 2004; Zhao et al., 2004; Hepworth et al., 2005; Hibara et al., 2006; Zhang et al., 2006). In addition, transient HAN overexpression greatly represses the expression of HAN itself and several homologous genes in the same family (Table 3), suggesting a negative regulatory feedback. Therefore, we performed a more detailed characterization of this group of genes.
Repression of GATA3 Family Genes upon Inducible Overexpression of HAN
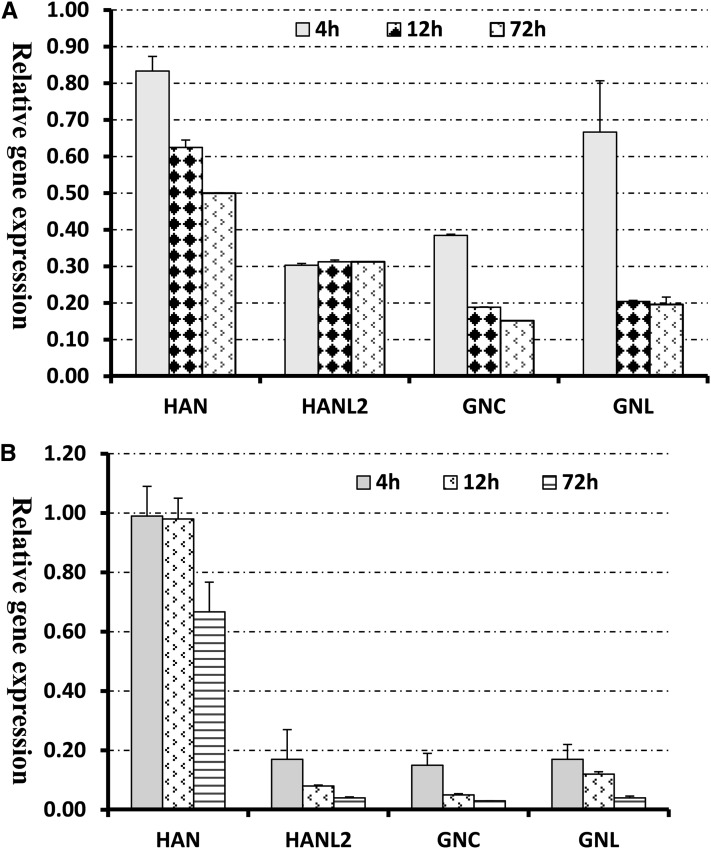
Induction of HAN with DEX leads to a progressive reduction of endogenous HAN RNA. After 72 h of DEX treatment, the accumulation of HAN transcripts decreases by half compared with the mock-treated plants (Figure 3A, Table 3). Moreover, induction of HAN significantly represses the expression of three GATA3 family genes: HANL2, GNC, and GNL, which belong to the same subfamily II as HAN (Reyes et al., 2004). For simplicity, the term GATA3 family genes will hereafter refer to HANL2, GNC, and GNL collectively. HANL1 and HANL2 are the two close homologs of HAN, which contain the HAN motif in Arabidopsis, and their biological function is unknown (Zhao et al., 2004). After 4 h of DEX treatment, transcripts of HANL2 decrease by 3.3-fold and subsequently remain at this level, while there is no change in the level of HANL1 transcripts (Table 3). Similarly, HAN induction reduces the levels of GNC and GNL transcripts as well. After 72 h of DEX treatment, the accumulation of GNC and GNL transcripts is only 15 and 20% of those in the mock treatments (Table 3). To validate our results and control for potential cross-hybridization between family members in the microarray, we performed quantitative real-time RT-PCR. As shown in Figure 3B, HAN overexpression from a transgene indeed results in a progressive repression of RNA accumulation from the endogenous gene and a rapid negative regulation of HANL2, GNC, and GNL. For example, upon 4 h of DEX treatment, HAN expression has not changed, whereas transcripts of HANL2, GNC, and GNL decrease by 5.8-, 6.6-, and 5.8-fold, respectively. Upon 72 h DEX treatment, HAN expression is reduced to 70% of its starting value, similar to the microarray results, while HANL2, GNC, and GNL transcripts decrease 25-, 33-, and 25-fold respectively compared with the mock-treated samples.
Figure 3.
Transient Induction of HAN Represses the Transcription of HAN and the Three GATA3 Family Genes HANL2, GNC, and GNL.
(A) Microarray analyses of the expression of HAN and GATA3 family genes in the floral buds of the 35S-HAN-GR line upon DEX treatment. Data are presented as the ratios of expression levels in the DEX-treated samples versus mock-treated samples. Four biological replicates were used for the microarray hybridization.
(B) Transcription analyses by qRT-PCR using independently generated RNA samples. Three biological replicates were used for each time point, and all samples were normalized according to the expression level of Actin2. Error bars represent the se between biological replicates.
Interactions between HAN and GATA3 Family Genes
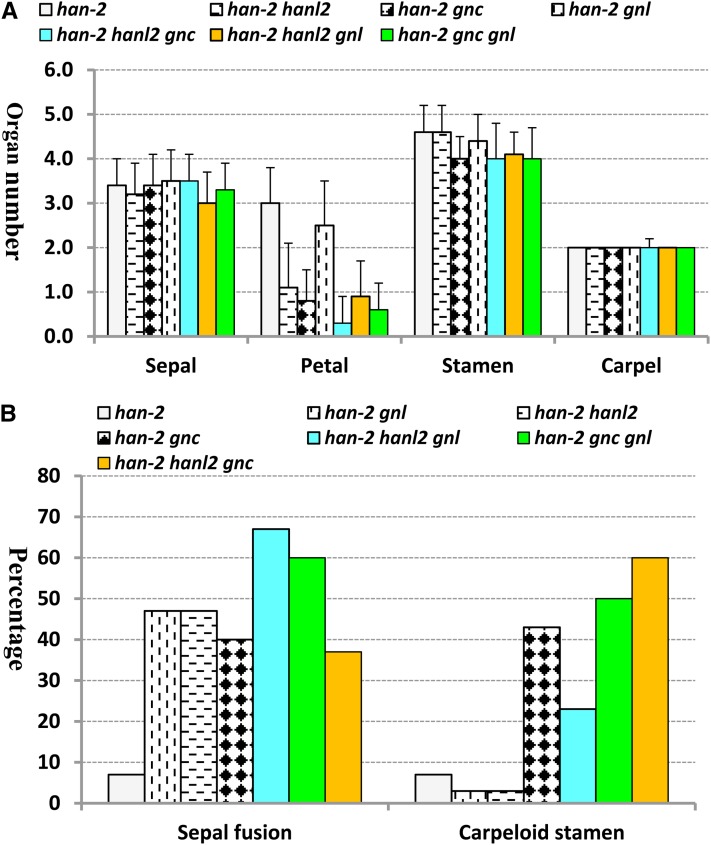
To investigate the genetic interactions between HAN and the GATA3 family genes identified in the microarray experiments, we used a weak allele of HAN (han-2) and T-DNA insertion lines of HANL2 (SALK_138626), GNC (SALK_001778), and GNL (SAILK_003995) to generate all possible combinations of double and triple mutants (Bi et al., 2005). Although single mutants and any combination of double mutants of GNC, GNL, and HANL2 had no evident floral developmental phenotypes, double and triple mutants of HAN with GNC, GNL, and HANL2 showed progressive and synergistic effects on sepal fusion, petal number, fertility defects, and carpel abnormality (Figures 4 and 5). The single han-2 mutant has reduced silique length, decreased fertility, and fewer sepals, petals, and stamens, and it displays occasional sepal fusion and carpelloid stamens (Zhao et al., 2004). The silique length in the double mutants (han-2 hanl2, han-2 gnc, and han-2 gnl) is decreased ∼20%, and seed yields are reduced (see Supplemental Figure 2 online). In the triple mutants (han-2 hanl2 gnc, han-2 hanl2 gnl, and han-2 gnc gnl), the siliques exhibit almost no expansion or elongation, and the plants are almost sterile (Figures 4B and 4C; see Supplemental Figure 2 online). Floral organ development is even more severely affected in the double and triple mutants. The most obvious defect is the reduction of petal number. For example, the single han-2 mutant usually has three petals, while the average petal number is 1.1 and 0.3 in the han-2 hanl2 double and han-2 hanl2 gnc triple mutant plants, respectively (Figures 4D and 5A). Moreover, there are increases in the frequency of sepal fusion and carpelloid stamens in plants with combined mutations in HAN with GATA3 family genes. The frequency of sepal fusion in the han-2 single mutant is 7%, while it increases to 47, 47, and 40% in the double mutants han-2 gnl, han-2 hanl2, and han-2 gnc, respectively, and 67, 60, and 37% in the triple mutants han-2 hanl2 gnl, han-2 gnc gnl, and han-2 hanl2 gnc, respectively (Figure 5B). By contrast, stamen/carpel development seems to be mainly regulated by GNC and HAN, double mutations of GNC and HAN result in an over 12-fold increase in the occurrence of carpelloid stamens compared with han-2 single mutants, whereas in the han-2 hanl2 or han-2 gnl double mutant plants, the frequency of carpelloid stamens is about the same as in the han-2 single mutant (Figure 5B). There is no phenotypic difference between homozygous triple mutants and plants with homozygous mutations in HAN, homozygous for a second member, and heterozygous for a third member, suggesting functional redundancy among HANL2, GNC, and GNL.
Figure 4.
Floral Phenotypes of Mutations in HAN and GATA3 Family Genes.
(A) Representative image of a han-2 single mutant plant and double mutant plants of HAN and GATA3 family genes.
(B) Representative image of a han-2 single mutant plant and triple mutants of HAN and GATA3 family genes.
(C) and (D) Silique length (C) and floral morphology (D) are progressively more defective in higher combinations of mutations of HAN and GATA3 family genes.
Bars = 1 cm in (A) and (B) and 1 mm in (C) and (D).
Figure 5.
Quantification of Floral Abnormality in GATA3 Family Mutants.
(A) The average floral organ numbers of 30 flowers counted from han-2 single mutant and all of the combinatory double and triple mutants between HAN and GATA3 family genes. Error bars represent the sd.
(B) The frequency of sepal fusion and carpelloid stamens calculated from 30 flowers of each mutant background.
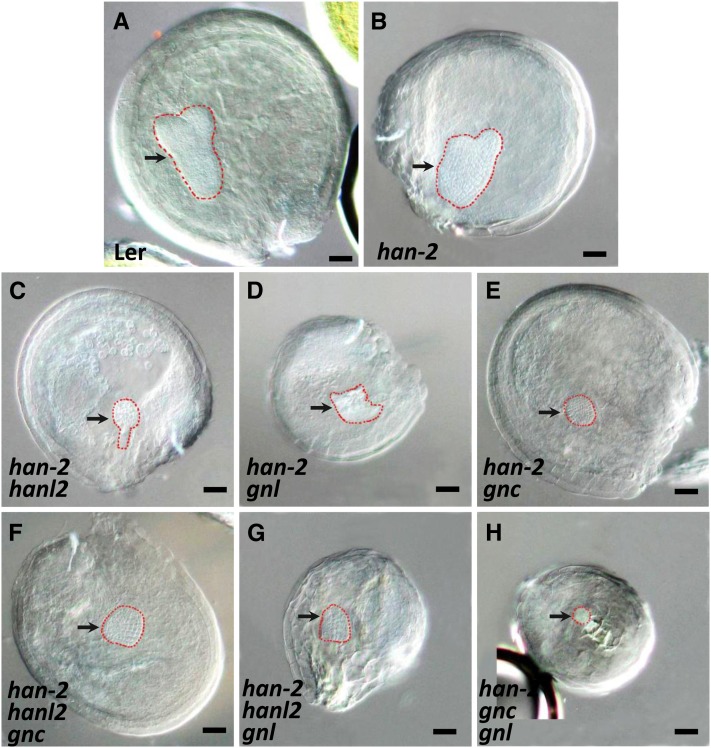
We also compared embryo development in the han-2 single mutant with development in double and triple mutants. HAN has previously been shown to function during Arabidopsis embryo development and is required for proper proembryo boundary position (Zhao et al., 2004; Nawy et al., 2010). As shown in Figure 6, han-2 embryo development displays some degree of abnormality, such as unequal or partially fused cotyledons, along with slightly disorganized embryo shape and boundary positions. In the double or triple mutants of HAN and GATA3 family genes, embryo development is severely disrupted. Most embryos prematurely terminated at the globular stage, after forming a cluster of cells, either in a round or irregular shape (Figure 6). Moreover, HAN and GNL may be involved in general plant growth, since the plant height and size decreased greatly when the plants bore combinations of HAN and GNL mutations (Figures 4A and 4B).
Figure 6.
Embryo Development Is Defective in Double and Triple Mutants of GATA3 Family Genes.
Compared with the wild type (A), the han-2 single mutant (B) showed slightly defective embryos with unequal cotyledons and disordered cellular organization. Embryo development in the double ([C] to [E]) or triple mutants ([F] to [H]) of HAN and GATA3 family genes are mostly terminated prematurely at the globular stage, showing only a cluster of cells without proper embryo shape or organization. Arrows indicate the position of embryos, which are outlined with red, dashed lines. Bars = 50 µm.
Similar Transcript Accumulation Patterns of HANL2, GNC, and GNL
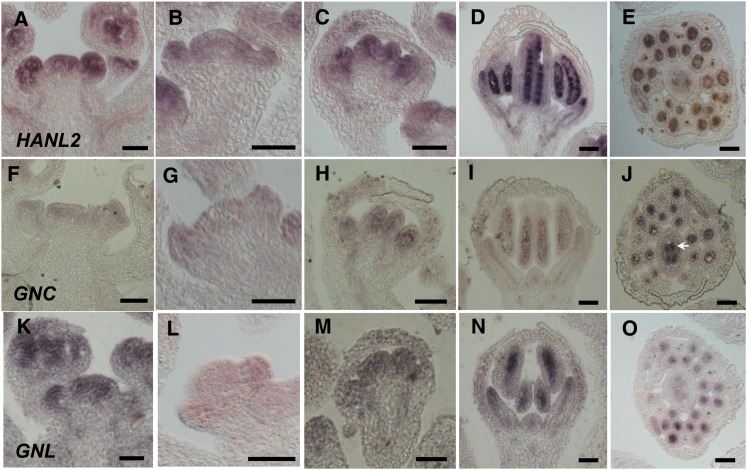
We next examined the expression patterns of HANL2, GNC, and GNL during flower development by in situ hybridization. We examined the specificity of our probes by hybridizing the HANL2 probe to flowers of a hanl2 null mutant, SALK_138626 (Bi et al., 2005). As shown in Supplemental Figure 3 online, there is almost no signal in the hanl2 mutant background, showing that the probe reflects the in vivo transcript accumulation of GATA3 family genes. HANL2 is expressed throughout the IM and early stages of floral primordia (stages 1 to 3) (Figures 7A and 7B). From stage 6 onward, HANL2 is largely restricted to the inner three whorls, specifically in the petals, stamens, and carpels (Figures 7C and 7D). Within the stamens, the strongest expression of HANL2 is detected in the anther locules as well as in the vascular strands, whereas in carpels, the strongest expression is limited to ovules (Figure 7E). The expression patterns of GNC and GNL overlap with that of HANL2, confirming that the three GATA3 family genes may share similar functions. Transcripts of GNC and GNL are first detected in the IM and young flower buds. As the flowers develop (from stage 6 onward), GNC and GNL signals are mostly limited to the inner three whorls, which is consistent with data from previous studies (Mara and Irish, 2008) (Figures 7F to 7I and 7K to 7N). Transverse sections showed that GNC and GNL expression is mainly detectable in the anther locules, vascular strands, and ovules (Figures 7J and 7O). However, the signal intensity of HANL2, GNC, and GNL are different, with HANL2 having the strongest, GNL the intermediate, and GNC the weakest expression in all of the tissues that we examined (Figure 7). Given that transcripts of HAN are detected at the boundaries between the meristem and lateral organ primordia, the lateral and basal regions of carpels, and the anther locules and vascular strands (Zhao et al., 2004), HAN partially overlaps with the expression patterns of HANL2, GNC, and GNL.
Figure 7.
Similar Expression Patterns of GATA3 Family Genes in the Arabidopsis Flowers, as Detected by in Situ Hybridization.
The expression domains of HANL2 ([A] to [E]), GNC ([F] to [J]), and GNL ([K] to [O]) are overlapping during wild-type flower development. Longitudinal sections of shoot apices ([A], [F], and [K]), stage 4 flowers ([B], [G], and [L]), stage 6 flowers ([C], [H], and [M]) and stage 8 flowers ([D], [I], and [N]) reveal that GATA3 family genes are expressed throughout the IM and young floral primordia (stages 1 to 3) and then limited to the inner three whorls (petal, stamen, and carpel). Transverse sections reveal that GATA3 family genes are specifically expressed in the anther locules, vascular strands, and ovules ([E], [J], and [O]). Arrow indicates the strong expression of GNC in the ovules within the ovary. Bars = 50 µm.
HAN Negatively Regulates GATA3 Transcription Factor Expression
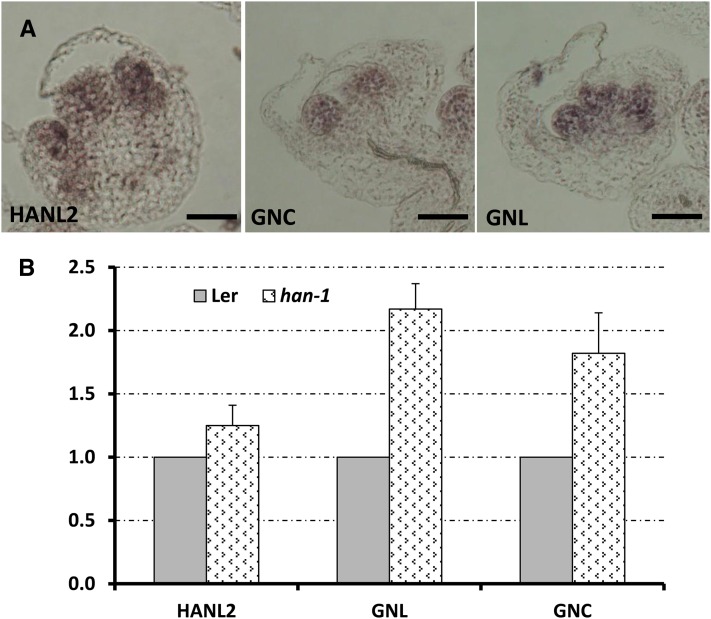
The expression data and genetic analysis imply that HAN may function as a negative regulator that modulates its own expression and that of other GATA3 family genes. To further test this possibility, we examined the expression of GATA3 family genes in the loss-of-function han-1 line (Figure 8). The expression patterns of HANL2, GNC, and GNL remain similar in the han-1 mutant background, with expression specifically restricted to the inner three whorls in the older flower primordia (Figure 8A). Quantification of the expression by qRT-PCR revealed that transcript accumulation from HANL2, GNC, and GNL increases in the han-1 floral buds compared with those of the wild type (Figure 8B), suggesting that HAN may control the expression level of its family genes to ensure proper flower development.
Figure 8.
Transcript Accumulation from GATA3 Family Genes Is Slightly Increased in the han-1 Loss-of-Function Mutant Background.
(A) In situ hybridization analyses showed that the expression patterns of GATA3 family genes (left, HANL2; middle, GNC; right, GNL) remained the same in han-1 mutant flowers. Bars = 50 µm.
(B) qRT-PCR comparison of transcript accumulation from GATA3 family genes in Ler and han-1 floral buds (stages 0 to 9). Three biological replicates were performed for each gene, and all samples were normalized to actin2. The expression level of each gene in the Ler background was set as 1. Bars represent the se.
HAN Forms Homomers and Heteromers with GATA3 Family Proteins
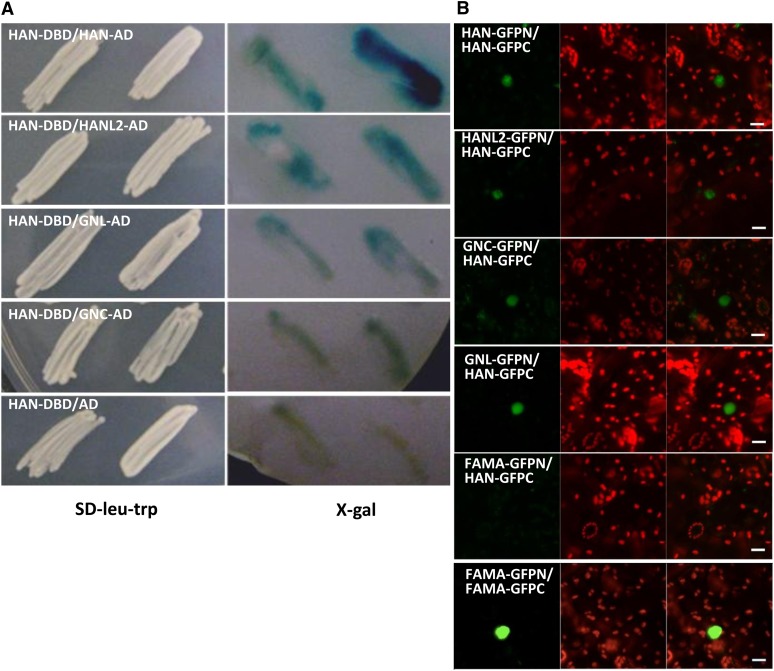
The additive phenotype of the triple mutants suggested that HAN likely works with its family members to control floral development. To examine the physical interaction between these proteins, we performed a yeast two-hybrid assay. In the X-Gal colony lift assay, HAN interacts with HAN and HANL2 strongly (strong blue color) and also interacts with GNL and GNC (moderate blue color), compared with the combination with HAN and empty vector only (Figure 9), suggesting that GATA3 family proteins physically interact. To confirm the interactions between HAN and the GATA3 family proteins in vivo, we also performed a bimolecular fluorescence complementation (BiFC) experiment. HAN interacted with itself and with HANL2, GNC, and GNL (Figure 9; see Supplemental Figure 4 online), but not with FAMA, an unrelated bHLH protein that had been shown to form dimers with bHLH family proteins (Ohashi-Ito and Bergmann, 2006).
Figure 9.
HAN and GATA3 Family Proteins Interact in Yeast Two-Hybrid Assays and in BiFC.
(A) Yeast two-hybrid assays. Bait constructs express HAN fused with the GAL4 DNA binding domain (DBD). Prey constructs express HAN, HANL2, GNC, or GNL fused with the GAL4 activation domain (AD). Empty prey constructs expressing the GAL4 activation domain alone serve as a negative control. Left panel shows yeast patches expressing both constructs derived from independent transformed colonies, which were streaked onto SD-Leu-Trp selection plates. Right panel indicates the X-Gal–based colony lift yeast two-hybrid assay. Blue color indicates the cumulative β-galactosidase activity caused by the activation of the lacZ reporter gene, which is activated by the physical interaction between HAN and GATA3 family proteins. At least two independent experiments were performed, and the result of one representative experiment is shown.
(B) BiFC interactions between HAN and GATA3 family proteins in transiently transformed Nicotiana benthamiana leaves. For each picture, a positive interaction is indicated by GFP fluorescence (green) in nuclei (left panel), the tobacco cells are indicated by chlorophyll autofluorescence (red) (middle panel), and the two merged channels are also shown (right panel). The label HAN-GFPN represents HAN fused with the N-terminal half of GFP in frame, with similar labels used for the other constructs. Representative images of different combinations, including HAN-GFPN with HAN-GFPC, HANL2-GFPN with HAN-GFPC, GNC-GFPN with HAN-GFPC, GNL-GFPN with HAN-GFPC, FAMA-GFPC with HAN-GFPC (negative control), and FAMA-GFPN with FAMA-GFPC (positive control) (Ohashi-Ito and Bergmann, 2006), are shown. All pictures were taken using the same settings, and each interaction was confirmed three times with independent infiltrations. Bars = 20 µm.
HAN Directly Binds to Its Own Promoter and the GNC Promoter in Vivo
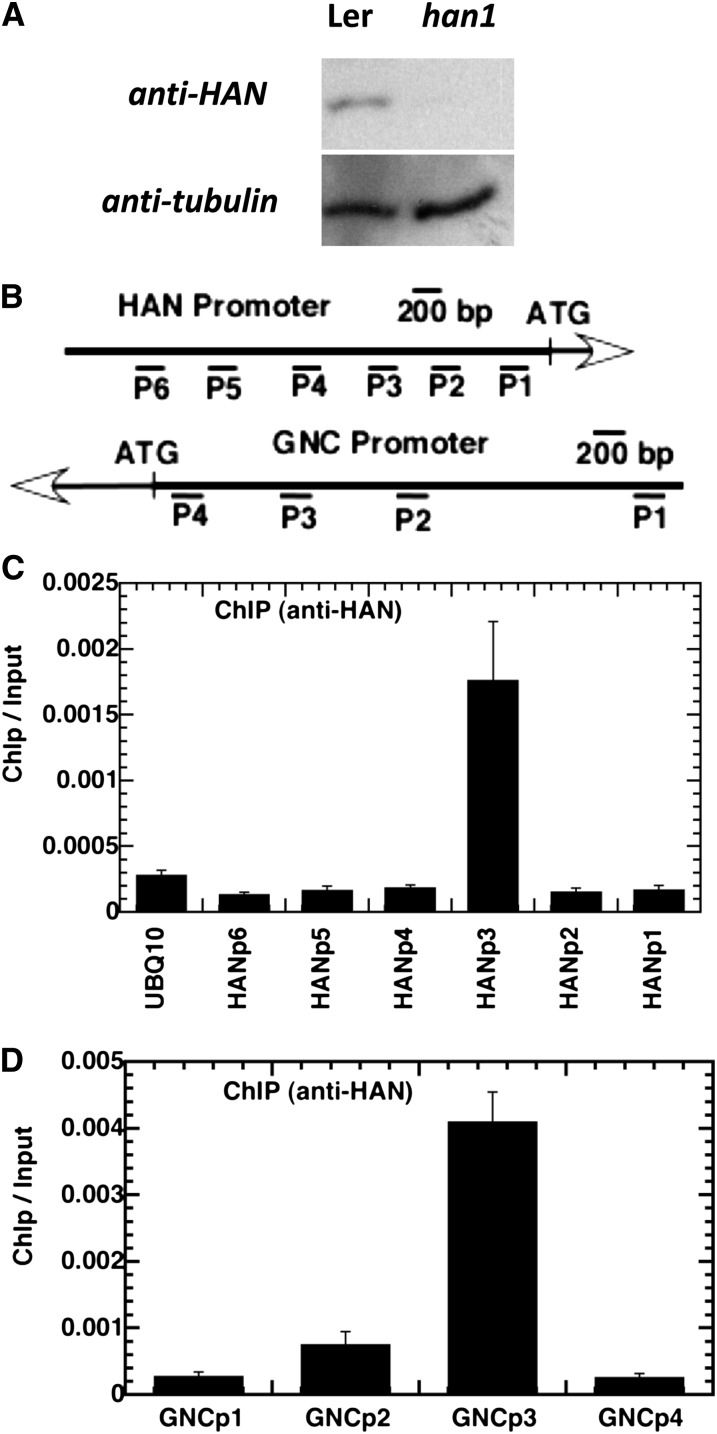
To investigate the mechanism by which HAN repressed its own expression and that of other members of the HAN-like gene family, we performed chromatin immunoprecipitation (ChIP) experiments with anti-HAN antibody, followed by quantitative PCR analysis of the precipitated genomic DNA. We first confirmed the specificity of the anti-HAN antibody we generated. In protein gel blot analysis, the anti-HAN antibody recognized a 32-kD band in the protein extracts from wild-type flowers, but not in the protein extracts from han1 null mutant flowers, suggesting that the anti-HAN antibody is highly specific (Figure 10A). The various amplicons used for the ChIP analyses are shown in Figure 10B, which covered different segments of the 5′ 2.5 kb of the HAN and GNC promoters. Amplicon 3 (HANp3), which spans the region from −977 to −735 bp in the HAN promoter, was significantly enriched, when normalized to a control using anti-actin antibody (Figure 10C). The ChIP/input ratio for amplicon 3 (HANp3) increases sixfold above a negative control amplicon from the UBQ10 promoter in two independent biological replicates (Figure 10C). By contrast, all other amplicons from the HAN promoter were not enriched compared with the UBQ10 amplicon, which demonstrated the specificity of the assay. Amplicon 3 (GNCp3) in the GNC promoter, which spans the region from −763 to −656 in the GNC promoter, was also significantly enriched, and the enrichment is up to 17-fold after normalization (Figure 10D). Amplicon 3 from −977 to −735 of the HAN promoter and amplicon 3 from the GNC promoter both contain a putative GATA consensus binding site for GATA3 family transcription factors.
Figure 10.
ChIP Analyses Indicate That HAN Directly Associates with Its Own Promoter and with That of GNC.
(A) Protein gel blot analyses of the specificity of binding of anti-HAN antibody in wild-type (Ler) and han1 floral buds.
(B) Schematic diagram of the amplicons located in the HAN and GNC promoters used for ChIP analysis.
(C) and (D) HAN (C) and GNC (D) chromatin regions associated with HAN protein. Quantitative data from real-time PCR show the relative enrichment value for each amplicon immunoprecipitation with anti-HAN antibody normalized to the control, in which the anti-actin antibody was used. The data presented as ChIP/input ratio were from two independent ChIP analyses of wild-type plants and were calculated for each amplicon using the following equation: ChIP/input = 2(Ct(MOCK) − Ct(HAN-ChIP))/2(Ct(MOCK) − Ct(INPUT)). Error bars represent the se from different biological replicates.
DISCUSSION
HAN is a boundary-expressed GATA3 transcription factor that regulates meristem organization, flower development, and cell division (Zhao et al., 2004). The molecular mechanisms by which it performs these functions are unknown. In this study, we found that transient induction of HAN represses many more genes than that are induced, and genes that function in hormone responses and flower organ specification are significantly enriched among the differentially expressed genes upon HAN induction (Figure 1). Moreover, HAN induction leads to autorepression and negative regulation of the related GATA3 family genes HANL2, GNC, and GNL (Figure 3). Genetic analyses reveal interactions between HAN and GATA3 family members in the regulation of sepal separation, petal number, silique length, and stamen and embryo development (Figures 4 to 6). HANL2, GNC, and GNL have similar expression patterns that are partially overlapping with the expression domain of HAN (Figure 7) (Zhao et al., 2004). Furthermore, we showed that the expression of GATA3 family members is slightly increased in the null mutant han-1 (Figure 8) and that HAN can form homo- and heteromultimers with GATA3 family members (Figure 9). Finally, we show that HAN protein binds to GATA-containing elements in the HAN and GNC promoters (Figure 10). The experiments together indicate that HAN may be a repressor that modulates the spatial and temporal expression patterns of itself and other GATA3 family genes, and this protein may also regulate genes involved in hormone actions and floral organ specification to control flower development.
Autoregulation May Be a Conserved Feature for GATA Transcription Factors
The data from microarray and real-time RT-PCR analyses indicate that HAN negatively regulates its own expression (Figure 3). ChIP data suggest that this regulation is direct, with HAN binding to its own promoter (Figure 10). These data suggest a mechanism that balances HAN protein and gene expression levels through a negative feedback loop. Similarly, GATA3 family members GNC and GNL have been reported to trigger a homeostatic mechanism that controls their transcript abundance, which mediates proper GA signaling and responses, including germination, greening, leaf elongation growth, and flowering time (Richter et al., 2010). Furthermore, GATA2, a member of the subfamily I of Arabidopsis GATA factors, has also been shown to repress endogenous GATA2 expression in overexpression lines (Reyes et al., 2004; Luo et al., 2010). GATA2 can form a desensitizing mechanism during the light response by binding to its own promoter to feedback inhibit its transcription (Luo et al., 2010). In vertebrates, GATA factors have been shown to be involved in self-association and autoregulation as well (Crossley et al., 1995; Bates et al., 2008), suggesting that autoregulation may be a conserved feature of GATA factors. HAN also can interact with itself and several GATA3 family proteins in yeast and during a BiFC assay in transiently transformed tobacco (Nicotiana tabacum) cells (Figure 9; see Supplemental Figure 4 online).
GATA3 Family Genes Are Targets of HAN and Function Redundantly with HAN during Flower Development
Among the 30 members of the family of Arabidopsis GATA transcription factors, only mutations in the han gene have previously been shown to cause developmental defects in flowers (Reyes et al., 2004; Zhao et al., 2004; Bi et al., 2005). Although GNC and GNL have been shown to be directly repressed by the floral homeotic genes APETALA3 and PISTILLATA, single mutants or gnc gnl double mutants do not show any visible flower abnormalities (Mara and Irish, 2008). Since hanl2 gnc or hanl2 gnl also do not show flower defects, we proposed that GNC, GNL, and HANL2 are functionally redundant with HAN, and their functions are largely masked by HAN activity in the wild type, since HAN directly represses and confines the expression of these genes during floral development. However, mutations in these genes can greatly enhance the phenotype of a weak han allele, namely, a sensitized background with reduced HAN meristem and boundary activities (Figures 4 to 6) (Zhao et al., 2004). Moreover, our genetic data suggest that GATA3 family members have distinct and redundant functions, in which HANL2 and GNL contribute mainly to sepal separation and petal number, while GNC participates in stamen and carpel development in addition to sepal separation and petal number (Figures 4 and 5). The more abundant expression of GNC than HANL2 and GNL in stamens and carpels (arrow in Figure 7J) is consistent with the prominent function of GNC in stamen/carpel separation. The level of gene transcript accumulation also displays similarity as well as specificity in HAN null allele han-1 and han-2 hanl2 gnc triple mutant plants (see Supplemental Table 1 online). For example, TCP family transcription factor4 (TCP4) RNA exhibits a reduction in both han-1 and han-2 hanl2 gnc, with a greater level of reduction in han-1 than in han-2 hanl2 gnc. Similarly, the level of GH3.5 RNA is ninefold decreased in the han-1 mutant flowers, whereas it remains unchanged in han-2 hanl2 gnc. The level of CORI3 mRNA, however, is fourfold decreased in the han-1 mutant, but it is threefold increased in han-2 hanl2 gnc, suggesting that HAN and GATA3 family genes only share partial downstream targets.
HAN May Function as a Transcription Repressor That Triggers a Feedback Mechanism
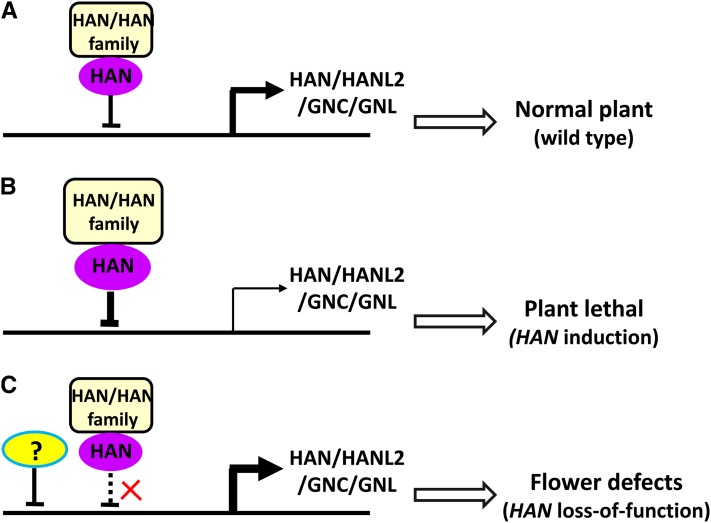
Our microarray and qRT-PCR data showed that overexpression of HAN leads to reduced expression of HAN and GATA3 family genes (Figure 3), whereas in the han-1 null mutants, transcripts of GATA3 family genes HANL2, GNC, and GNL are slightly increased (Figure 8), supporting the notion that HAN may act as a transcriptional repressor during flower development. HAN is likely a negative regulator of cell growth as well, which is supported by the fact that many cell cycle genes and expansin genes, such as CYCLIN DELTA-1 (CYCD1;1) and ARABIDOPSIS THALIANA EXPANSIN A1 (ATEXPA1), are significantly repressed upon HAN overexpression (Table 3). Furthermore, plants with overaccumulation of HAN protein, such as 35S:HAN or DEX-treated 35S:HAN-GR lines, have reduced growth (Zhao et al., 2004). Taken together, the evidence suggests a model for HAN function (Figure 11), where in wild-type plants, HAN interacts with itself and the genes in the GATA3 family, which feeds back on the transcription of HAN, HANL2, GNC, and GNL to allow their expression at levels that are appropriate for the production of plants with normal development (Figure 11A). Transient overexpression of HAN will greatly enhance the transcriptional repression, resulting in reduced expression of HAN and GATA3 family genes. Such reduction is so deleterious to plant development that these plants die after ∼10 d of DEX treatment (Zhao et al., 2004) (Figure 11B). In the han mutant background, on the other hand, reduced HAN protein levels result in weaker transcriptional repression and higher HANL2, GNC, and GNL expression (Figures 8 and 11C). However, the transcriptional reduction in overexpression lines, with only slight increases in the han null mutant background, suggest that there is an alternate repressor that functions independently of HAN to modulate the expression of GATA3 family genes, which in turn leads to plants with floral developmental defects (Figure 11C). Our quantitative PCR data support this model (Table 3). Gene expression patterns are similar in han-1 and DEX-treated 35S:HAN-GR lines, suggesting that transient overexpression of HAN mimics the loss-of-function line han1 through self-repression. Moreover, the quantitative RNA level changes are generally larger in the DEX-treated plants than in han-1 (Table 3), which is consistent with the stronger phenotypic effect observed in the overexpression of HAN than in han-1 plants.
Figure 11.
A Model for HAN Function in Arabidopsis.
(A) In the wild-type plant, HAN interacts with itself and with its family genes in a negative feedback loop, which decreases the transcription of HAN, HANL2, GNC, and GNL to produce the moderate level of expression necessary for normal plant development.
(B) Transient overexpression of HAN enhances transcriptional repression, which results in substantially reduced expression of endogenous HAN and GATA3 family genes and therefore has deleterious effects on plant development.
(C) In the han mutant background, reduced HAN protein levels result in weakened transcriptional repression and higher HANL2, GNC, and GNL expression. The levels are not much higher than in wild-type plants, which may indicate that an alternative (and unknown) repressor is triggered to down-modulate the expression of GATA3 family genes, producing a plant with floral developmental defects, but not complete deregulation of the GATA3 family genes.
HAN Regulates Flower Development via Multiple Regulatory Networks
Transient induction of HAN results in the repression of a large number of genes involved in auxin response, jasmonic acid signaling, gibberellin signaling, cytokinin response, floral organ specification, reproductive regulation, and GATA3 family genes as well (Figures 1 and 3). Biochemical analyses indicate that HAN can directly interact with itself and GATA3 family members, both at the protein level and at the DNA level (Figures 9 and 10), and the expression domains of HAN and GATA3 family members are partially overlapping. Considering that GATA3 family genes GNC and GNL mediate many similar pathways, including gibberellin signaling, cytokinin response, light response, nitrogen metabolism, sugar sensing, and chlorophyll biosynthesis, and that GNC and GNL regulate genes in the AP3/PI pathway (Bi et al., 2005; Naito et al., 2007; Mara and Irish, 2008; Richter et al., 2010), HAN may function partially through GNC and GNL in boundary regions to control hormone signaling and nutrient distribution, thus ensuring proper flower separation and specification. It is also possible that HAN directly regulates hormonal response genes in the boundary regions, which in turn affect the floral organ number, size, and position. Given the complex interaction between auxin, jasmonate, gibberellin, and cytokinin during plant development, further study is needed to elucidate which, if any, hormone pathway is directly regulated by HAN (Khan and Stone, 2007; Moubayidin et al., 2009; Peng, 2009; Bishopp et al., 2011). HAN may control flower development via direct interactions with well-known floral organ regulators as well (Table 3). BOP, RBE, and CUC3 have overlapping expression with HAN in the boundary regions, and BP and KNAT2 share expression domain with HAN at the lower halves of SAMs and in carpels, respectively, while FUS3 has an overlapping expression region with HAN in the embryo, indicating the possibility that the floral and embryo defects seen in the han mutant plants result in part from changes in the expression of these developmental regulators (Pautot et al., 2001; Vroemen et al., 2003; Takeda et al., 2004; Tsuchiya et al., 2004; Zhao et al., 2004; Hepworth et al., 2005; Hibara et al., 2006) . Future work should examine how boundary-expressing HAN interacts with hormone signaling and floral organ specification pathways during flower development.
METHODS
Plant Materials and Growth Conditions
Plants used for microarray experiments were 35S:HAN-GR lines in the Ler background. The han-1 and han-2 alleles are in the Wassilewskija and Ler ecotypes, respectively (Zhao et al., 2004). The mutant lines of HANL2 (SALK_138626), GNC (SALK_001778), and GNL (SALK_003995) are in the Columbia background. Double and triple mutant plants between HAN and GATA3 family genes with the er mutation were chosen for genetic and morphological analyses. All of the phenotypes were confirmed from multiple segregation lines, which rule out the effects of the ecotype. Plants were grown in a soil:vermiculite:perlite mixture under continuous illumination with a light intensity range of 80 to 100 µmol·m–2·s–1 at 20°C.
Sample Collection and Microarray Analysis
Arabidopsis thaliana inflorescences containing flower buds from stages 1 to 9 were collected for microarray analysis 0, 4, 12, and 72 h after 10 µM DEX treatment. DEX solution was made and applied every 24 h as previously described (Wellmer et al., 2006). Total RNA was extracted with TRIZOL (Invitrogen) and purified with an RNeasy kit (Qiagen). Isolated RNA was assessed for integrity using an Agilent 2100 Bioanalyzer. RNA samples from mock- and DEX-treated plants at each time point were cohybridized, and labeling dyes were swapped between replicates to reduce dye-related bias. Four biological replicates were used for microarray hybridization. Microarray labeling and hybridization were performed as described (Wellmer et al., 2004, 2006). Microarrays were scanned with a GenePix 4200A scanner (Axon Instruments), and raw data were analyzed using the Resolver gene expression data analysis system version 4.0 (Rosetta Biosoftware) as described previously (Wellmer et al., 2004, 2006). Briefly, we first removed spots that were flagged during data acquisition by Genepix software or had intensities in both channels below zero after background subtraction. We then normalized the signal intensities as described (Schadt et al., 2001). To calculate P values, we combined additive and multiplicative error components in both channels and loaded the resultant ratio profiles into the Resolver system. Analysis within the Resolver system was performed at the sequence level as described (Stoughton and Dai, 2002). If multiple data points corresponded to the same gene, their values were combined using a weighted scheme such that the feature with the lowest error was given the greatest weight. The P values calculated with Resolver were adjusted with the Benjamini and Hochberg procedure using the Bioconductor multtest package (http://www.bioconductor.org/packages/bioc/stable/src/contrib/html/multtest.html). The data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE38658.
GO Term Enrichment Analysis
We clustered genes with similar expression patterns using the clustering algorithm in Resolver as described previously (Sugimoto et al., 2010). To determine which categories of genes were enriched in each cluster, we tested for enrichment of GO terms using GOEAST software (Zheng and Wang, 2008) with default parameters except for the use of algorithms to eliminate local dependencies between GO terms (Alexa et al., 2006).
Live Imaging of IMs
All imaging was performed using a Zeiss LSM 510 Meta confocal microscope with a ×40 water-dipping objective lens. Similar settings of laser power and filters were used for imaging of GFP/VENUS combination as previously described (Heisler et al., 2005). Zeiss LSM software was used for reconstructing the Z-stacks into a projection view.
Quantitative Real-Time RT-PCR
qRT-PCR analyses were first performed on cDNA synthesized from independently generated samples that were either mock- or DEX-treated for 4, 12, 72, or 9 d, using 2XsensiMix SYBR Mastermix (Bioline). Then, qRT-PCR analyses were compared using stage 0 to 9 flower buds of Ler (control), han-1, han2, and han-2 hanl2 gnc mutant backgrounds. Three biological replicates were used, upon which three technical replicates were performed. Actin2 was used as a control to normalize the expression data. Fold change was calculated as 2ΔΔCt (cycle threshold), and standard deviation was calculated among three biological replicates. The gene-specific primers are listed in Supplemental Table 2 online.
In Situ Hybridization
Tissue fixation and in situ hybridizations were performed as described (Zhang et al., 2007) with minor modifications. Western Blue plus 1 mM tetramisole was used instead of 5-Bromo-4-Chloro-3-Indolyl Phosphate/Nitroblue tetrazolium as the substrate solution to reduce the hybridization background. In situ probes were synthesized by PCR amplification of cDNA using gene-specific primers containing T7 and SP6 RNA polymerase binding sites. Antisense probes were generated using T7 RNA polymerase, and sense probes were made using SP6 RNA polymerase. The gene primer pairs are listed in Supplemental Table 2 online.
Embryo Clearing
Similar developmental stages of immature seeds were cleared with Hoyer’s clearing buffer (2.5 g gum arabic, 33.3 g chloral hydrate, and 1.66 g glycerol in 10 mL water) and examined with a differential interference contrast microscope as described (Lukowitz et al., 2004).
Yeast Two-Hybrid and BiFC Assays
Yeast transformation and X-Gal–based β-galactosidase assays were performed following the manufacturer’s instructions. Full-length cDNAs for HAN, HANL2, GNC, and GNL were cloned into pENTR/D/TOPO and then Gateway cloned to pDEST32 and pDEST22 through the LR reaction (Invitrogen). The bait and prey vectors were transformed to yeast strain MaV203, and three single transformed colonies were picked and streaked onto yeast peptone dextrose adenine plates for the X-Gal colony lift assay, as described in the yeast protocols handbook from Clontech. For BiFC experiments, full-length HAN, HANL2, GNL, and GNC cDNA (without stop codon) Gateway clones were recombined into vectors containing each half of GFP (N or C terminus) to generate the fusion proteins (such as HAN-GFP N terminus) in frame (Walter et al., 2004). Two plasmids for testing the specific interaction were cotransformed into Nicotiana benthamiana leaves through Agrobacterium tumefaciens infiltration as previously described (Lavy et al., 2002). The tobacco leaves were imaged on a Zeiss LSM 510 Meta confocal microscope 2 d after infiltration. GFP signals in nuclei (which demonstrate the physical interaction) and chlorophyll autofluorescence signals (which indicate tobacco cells) were detected at the same time from different detection channels. FAMA constructs, which have been shown to form homodimers, were included as positive controls for specificity (Ohashi-Ito and Bergmann, 2006).
Protein Expression and Antibody Preparation
Full-length HAN cDNA was cloned into the pET28-a vector to express 6xHIS-HAN protein in Escherichia coli RosettaBlue competent cells (Novagen). The recombinant fusion protein was purified using Ni-NTA agarose beads (Qiagen) and was used to generate polyclonal HAN antisera in rabbits by the Strategic Biosolution Company.
Protein Gel Blot Analysis
Floral buds were ground in liquid nitrogen, and proteins were extracted using a plant total protein extraction kit (Sigma-Aldrich). SDS-PAGE, blotting, and detection were performed as described (Zhang et al., 2005), with HAN or tubulin antibodies at dilutions of 1:12,000 or 1:3000, respectively, and anti-rabbit or anti-mouse secondary antibodies at a dilution of 1:2000 (Amersham). The HAN antibody recognizes the native HAN specifically as a 32-kD band (Figure 10A).
ChIP
The association of HAN with the HAN and GNC promoters was investigated in planta using ChIP, followed by a quantitative real-time PCR approach, as described (Bowler et al., 2004; Sawa et al., 2007; Zhou et al., 2009) with some modifications. In general, 2 g of shoot apex from Ler wild-type plants was harvested and fixed with 1% formaldehyde under vacuum. Nuclei were isolated and lysed, and chromatin was sheared to an average size of 500 bp by sonication. The sonicated chromatin served as input or positive control. Immunoprecipitations were performed with anti-HAN antibody and using anti-actin antibody (Promega) as a control. The precipitated DNA was isolated and purified and served as a template for PCR. Quantitative PCR was performed as described for real-time PCR analysis. The degree of enrichment of the HAN or GNC promoter fragments was presented as the ChIP/input ratio, normalized to the antiactin control. The value was calculated as for each amplicon using following the equation 2(Ct(MOCK) − Ct(HAN-ChIP))/2(Ct(MOCK) − Ct(INPUT)). The primer pairs used in ChIP-PCR are listed in Supplemental Table 2 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative databases under the following accession numbers: HAN (AT3G50870), HANL2 (AT4G36620), GNC (AT5G56860), and GNL (AT4G26150).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene Ontology Term Enrichment (P < 0.003) in Cluster 1, Cluster 2, Cluster 3, and cluster 4 of Differentially Expressed Genes.
Supplemental Figure 2. Seed Yield Is Greatly Reduced in the Combinatory Mutants of HAN and GATA3 Family Genes.
Supplemental Figure 3. In Situ Hybridization of HANL2 in hanl2 Mutant Flowers.
Supplemental Figure 4. Negative Controls for Interactions between HAN and GATA3 Family Proteins in BiFC Experiments in Transiently Transformed Nicotiana benthamiana Leaves.
Supplemental Table 1. Gene Expression Patterns of Five Non-GATA HAN Targets in Mutant Plants and in the Transient Overexpression of HAN Plants.
Supplemental Table 2. Primer Information Used in This Study.
Supplemental Data Set 1. List of Genes That Are Differentially Expressed under Transient Overexpression of HAN.
Acknowledgments
We thank members of the Meyerowitz lab for the discussion and technique help. We thank Xuemei Chen and Xigang Liu for help with the in situ hybridization, Wolfgang Lukowitz for communicating unpublished information, Arnavaz Garda for technical assistance, Adrienne Roeder for sharing constructs before publication, and Adrienne Roeder, Kaoru Sugimoto, An Yan, and Zachary Nimchuk for critical reading and comments on the article. This work was supported by National Institutes of Health Grant 1R01 GM086639 to E.M.M., by the National Basic Research of China 973 Program 2012CB113900 and National Natural Science Foundation of China 31171399 to X.Z., and by a California Institute of Technology Gosney Postdoctoral Fellowship to Y.Z.
AUTHOR CONTRIBUTIONS
X.Z. and Y.Z. conceived and performed most of the experiments and wrote the article along with E.M.M. L.D. did the real-time PCR for data verification. Z.W. and R.L. performed the microarray data analyses.
Glossary
- SAM
shoot apical meristem
- DEX
dexamethasone
- qRT-PCR
quantitative RT-PCR
- GO
Gene Ontology
- Ler
Landsberg erecta
- GFP
green fluorescent protein
- IM
inflorescence meristem
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
References
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenführer J., Lengauer T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Bates D.L., Chen Y., Kim G., Guo L., Chen L. (2008). Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J. Mol. Biol. 381: 1292–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y.M., Zhang Y., Signorelli T., Zhao R., Zhu T., Rothstein S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Bishopp A., Benková E., Helariutta Y. (2011). Sending mixed messages: Auxin-cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 14: 10–16 [DOI] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Brewer P.B., Howles P.A., Dorian K., Griffith M.E., Ishida T., Kaplan-Levy R.N., Kilinc A., Smyth D.R. (2004). PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131: 4035–4045 [DOI] [PubMed] [Google Scholar]
- Chuang C.F., Running M.P., Williams R.W., Meyerowitz E.M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13: 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Crossley M., Merika M., Orkin S.H. (1995). Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15: 2448–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Durfee T., Roe J.L., Sessions R.A., Inouye C., Serikawa K., Feldmann K.A., Weigel D., Zambryski P.C. (2003). The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 8571–8576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J.C. (2002). Shoot and floral meristem maintenance in Arabidopsis. Annu. Rev. Plant Biol. 53: 45–66 [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hepworth S.R., Zhang Y., McKim S., Li X., Haughn G.W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K., Karim M.R., Takada S., Taoka K., Furutani M., Aida M., Tasaka M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Ueguchi-Tanaka M., Matsuoka M. (2008). GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13: 192–199 [DOI] [PubMed] [Google Scholar]
- Kanei M., Horiguchi G., Tsukaya H. (2012). Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development 139: 2436–2446 [DOI] [PubMed] [Google Scholar]
- Khan S., Stone J.M. (2007). Arabidopsis thaliana GH3.9 in auxin and jasmonate cross talk. Plant Signal. Behav. 2: 483–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Bracha-Drori K., Sternberg H., Yalovsky S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.P., Koizuka N., Martin R.C., Nonogaki H. (2005). The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 44: 960–971 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lowry J.A., Atchley W.R. (2000). Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 50: 103–115 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D., Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Luo X.M., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19: 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara C.D., Irish V.F. (2008). Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Moubayidin L., Di Mambro R., Sabatini S. (2009). Cytokinin-auxin crosstalk. Trends Plant Sci. 14: 557–562 [DOI] [PubMed] [Google Scholar]
- Naito T., Kiba T., Koizumi N., Yamashino T., Mizuno T. (2007). Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Nawy T., Bayer M., Mravec J., Friml J., Birnbaum K.D., Lukowitz W. (2010). The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell 19: 103–113 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient R.K., McGhee J.D. (2002). The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 12: 416–422 [DOI] [PubMed] [Google Scholar]
- Pautot V., Dockx J., Hamant O., Kronenberger J., Grandjean O., Jublot D., Traas J. (2001). KNAT2: Evidence for a link between knotted-like genes and carpel development. Plant Cell 13: 1719–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Peng J. (2009). Gibberellin and jasmonate crosstalk during stamen development. J. Integr. Plant Biol. 51: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Reyes J.C., Muro-Pastor M.I., Florencio F.J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134: 1718–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R., Behringer C., Müller I.K., Schwechheimer C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Medrano L.J., Meyerowitz E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203 [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. (2000). The fungal GATA factors. Curr. Opin. Microbiol. 3: 126–131 [DOI] [PubMed] [Google Scholar]
- Schadt, E.E., Li, C., Ellis, B., and Wong, W.H. (2001). Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell Biochem. Suppl. 37: 120–125. [DOI] [PubMed]
- Stoughton, R.S., and Dai, H. (2002). Statistical combining of cell expression profiles. U.S. Patent 6351712.
- Sugimoto K., Jiao Y., Meyerowitz E.M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Takeda S., Hirochika H. (2003). Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible Myb gene NtMyb2. Plant J. 36: 550–564 [DOI] [PubMed] [Google Scholar]
- Takeda S., Matsumoto N., Okada K. (2004). RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131: 425–434 [DOI] [PubMed] [Google Scholar]
- Teakle G.R., Manfield I.W., Graham J.F., Gilmartin P.M. (2002). Arabidopsis thaliana GATA factors: Organisation, expression and DNA-binding characteristics. Plant Mol. Biol. 50: 43–57 [DOI] [PubMed] [Google Scholar]
- To J.P., Kieber J.J. (2008). Cytokinin signaling: Two-components and more. Trends Plant Sci. 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Nambara E., Naito S., McCourt P. (2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37: 73–81 [DOI] [PubMed] [Google Scholar]
- Vroemen C.W., Mordhorst A.P., Albrecht C., Kwaaitaal M.A., de Vries S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schutze K., Batistic O., Weckermann K., Nake C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H., Tian C.-e., Duan J., Wu K. (2008). Research progresses on GH3s, one family of primary auxin-responsive genes. Plant Growth Regul. 56: 225–232 [Google Scholar]
- Wang L., Yin H., Qian Q., Yang J., Huang C., Hu X., Luo D. (2009). NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Res. 19: 598–611 [DOI] [PubMed] [Google Scholar]
- Weigel D., Meyerowitz E.M. (1994). The ABCs of floral homeotic genes. Cell 78: 203–209 [DOI] [PubMed] [Google Scholar]
- Wellmer F., Alves-Ferreira M., Dubois A., Riechmann J.L., Meyerowitz E.M. (2006). Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F., Riechmann J.L., Alves-Ferreira M., Meyerowitz E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C.J., Hall D.H., DeBlasio S., Taguchi-Shiobara F., Schmidt R.J., Jackson D.P. (2010). A conserved mechanism of bract suppression in the grass family. Plant Cell 22: 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang X., Li X., Marshall J.B., Zhong C.X., Dawe R.K. (2005). Phosphoserines on maize CENTROMERIC HISTONE H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell 17: 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Madi S., Borsuk L., Nettleton D., Elshire R.J., Buckner B., Janick-Buckner D., Beck J., Timmermans M., Schnable P.S., Scanlon M.J. (2007). Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Medrano L., Ohashi K., Fletcher J.C., Yu H., Sakai H., Meyerowitz E.M. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16: 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Wang X.J. (2008). GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 36(Web Server issue): W358–W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang X., Kang X., Zhao X., Zhang X., Ni M. (2009). SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]