INTRODUCTION
Hepatitis C virus (HCV) is a positive-strand RNA virus of the Flaviviridae family that primarily infects hepatocytes and can lead to liver cirrhosis, hepatocellular carcinoma, liver failure, and liver failure–related death.1,2 Chronic HCV infection affects nearly 170 million people worldwide.3 At present, a vaccine for HCV does not exist. Until recently, the standard of care used to treat HCV included a combination of pegylated interferon-α (PEG-IFN) and ribavirin (RBV).4 Both of these drugs are general inhibitors of viral infection but only 50% or fewer of patients infected with HCV genotype 1 and treated with PEG-IFN plus RBV achieve a sustained virologic response (SVR), which is defined as the absence of detectable virus at the end of therapy and 6 months later.5 Research in understanding the mechanisms of HCV replication has led to the development of direct-acting antiviral agents (DAAs) that target specific aspects of the HCV lifecycle.6
The, NS3-4A protease, the NS5B RNA-dependent RNA polymerase (RdRp), and the NS5A protein have been extensively studied as drug targets.7–10 Some examples of NS3-4A protease inhibitors include telaprevir, boceprevir, TMC435, MK-7009, and RG7227 (reviewed in Ciesek and colleagues7). Monotherapy with telaprevir, a specific peptidomimetic inhibitor of NS3-4A, has been found to be very effective in suppressing viral loads (nearly 3–4 logs); however, some patients, especially those infected with genotype 1a, exhibit rapid viral breakthrough and the emergence of resistant virus.11–13 Similarly, boceprevir monotherapy has showed potent antiviral activity.14 DAAs targeting NS5B include the nucleoside/nucleotide inhibitors (such as NM283, R1626, R7128 and IDX184, reviewed in Thompson and colleagues10 and Rong and Perelson15) and non-nucleoside inhibitors (such as R803, HCV371 and HCV086, reviewed in Thompson and colleagues10 and Rong and Perelson15), which act respectively by direct targeting the catalytic site and via indirect allosteric mechanisms.
DAA-based monotherapy can result in viral breakthrough because of development of drug resistance.16–18 One mechanism to overcome resistance is to combine DAAs with PEG-IFN+RBV. In the case of the protease inhibitors boceprevir and telaprevir, this approach has been highly effective, yielding SVR rates of approximately 70%, and these combinations have been approved for patient use.8,19
Understanding the key mechanisms of action of DAA-based therapy is essential for the development of more-potent drugs and in designing effective IFN-free regimes. Previously, mathematical modeling of HCV infection under therapy has been successful in deciphering the modes of action of IFN, PEG-IFN, and PEG-IFN+RBV by studying the viral dynamics of HCV RNA decline under therapy.20–23 Mathematical models fit to patient data provided key parameter estimates, such as the HCV half-life, the virus clearance rate, and infected cell life spans, which have been crucial in understanding the disease. Only a few mathematical models have been used to fit DAA treatment data. In this review, we outline the current mathematical models for HCV dynamics under IFN-based and DAA therapy, and future directions.
MODELING IFN ANTIVIRAL THERAPY
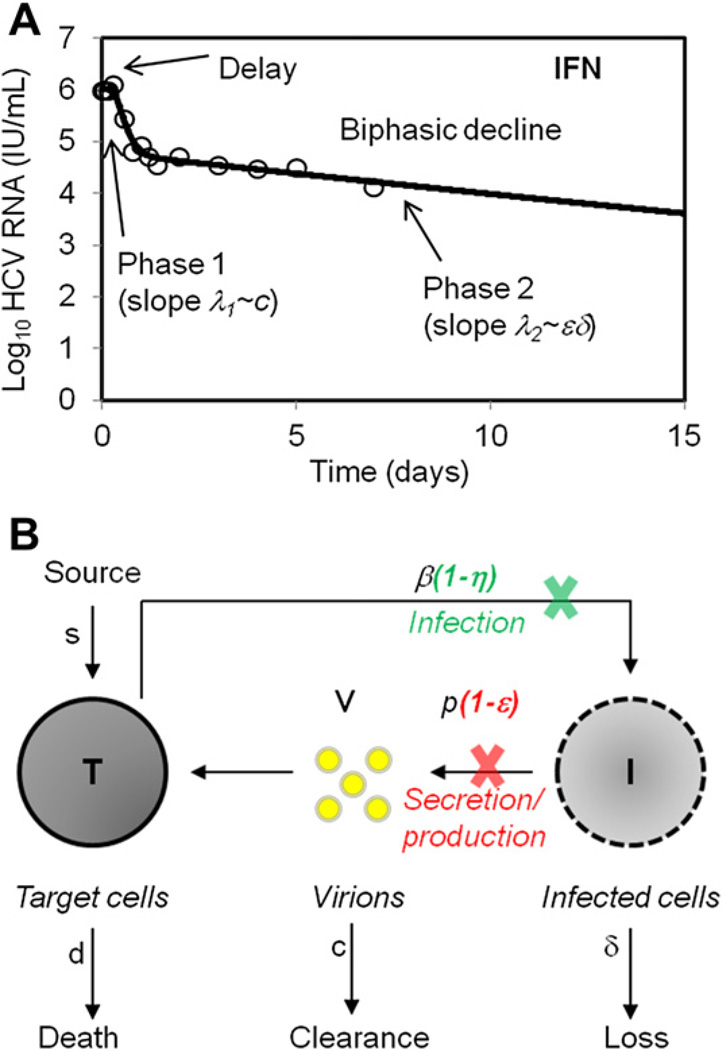
A mathematical model, developed by Neumann and colleagues,20 explained the biphasic HCV RNA decline observed during high-dose daily IFN treatment (Fig. 1A). According to the model, IFN blocked viral production from infected cells with an effectiveness, ε, leading to a rapid reduction in viral load as existing virus was cleared from the circulation and not effectively replaced because of the block in virion production. This rapid first phase was then followed by a slower second phase that reflected the loss of infected cells, which because of lower loads were not effectively replaced by de novo infections. Because the parameters in this model have gained widespread use, it is worthwhile to present them in the context of a schematic version of the model (see Fig. 1B).
Fig. 1.
Viral decline during treatment with IFN. (A) A biphasic HCV RNA decline during IFN-α treatment of chronic HCV. (B) Schematic representation of the standard viral dynamic model. Treatment may block vRNA production with effectiveness ε, and viral infection by a factor η. T and I represent target and infected cells, respectively, and V represents virus in serum. Target cells are created and die with constant rates s and d, respectively, and are infected by virus, V, with rate constant β. Infected cells, I, are lost with rate constant δ and virus, V, is cleared from serum with rate constant c.
In the model, target cells, T, are infected by virus, V, a process characterized by the rate constant β. Infected cells, I, are lost via a first-order process at rate δ per infected cell. This loss represents cell death as well as possible “cure” through loss of replicative intermediates. Infected cells produce new virus at rate p per cell, and free virions are assumed to be cleared at rate c per virion. Target cells are assumed to be generated at a rate s and die at a rate d per cell. Later models allow target cells to proliferate.24 Drug, in this case, IFN, was assumed to act in 2 possible ways: to block new infections with an effectiveness η and/or to block virion production with an effectiveness ε. These effectiveness parameters were assumed to take on values between 0 and 1, with 0 meaning having no effect and 1 meaning 100% effective.
For IFN treatment, fitting patient data under the assumption that the target cell level remained constant for the 14 days of therapy studied, showed that IFN is mainly responsible for blocking virion production, whereas the effect on reducing infection was found to be less significant and difficult to estimate.20 Because the effect of η on viral load was insignificant compared with the effect of blocking production, η is set to zero in many models. In the Neumann and colleagues20 model, it was assumed that the dose of drug was high enough that the drug effectiveness could be assumed to be constant over the course of treatment. Thus, this model is sometimes called the constant effectiveness (CE) model.25
The CE model, when fitted to HCV RNA data after initiation of IFN treatment, provided an estimate of the mean virion clearance rate c of approximately 6 d−1,20 corresponding to a mean viral half-life in serum of about 2.7 hours. Estimates for ε and δ have been found to be dependent on the HCV genotype, ethnicity, polymorphism in the interleukin 28 B (IL28B), baseline viral load, baseline inducible protein 10 (IP-10) levels, and histologic factors,26–30 but for patients with genotype 1, high daily doses of 10 or 15 million international units (MIU) of standard IFN lead to effectiveness estimates of ε of approximately 0.95.20
The mathematical form of the “standard” model shown in Fig. 1B is valid when the constant drug effectiveness assumption holds, such as in the case of high-dose daily administration of IFN. However, in cases such as PEG-IFN therapy, in which the drug is administered once a week, the concentration of drug initially increases and then decreases over the dosing interval, resulting in fluctuations in HCV RNA levels.25,31,32 Under these circumstances, the CE assumption fails, and such conditions are better described by a varying effectiveness (VE) model based on the pharmacokinetic/pharmacodynamic (PK/PD) characteristics of the drugs used.25 In the VE model, the drug effectiveness is considered to vary over time. These models have been reviewed in Shudo and colleagues25 and Guedj and colleagues.33
THE PRESENT: MODELING HCV RNA DECLINE DURING DAA-BASED THERAPY
Modeling HCV RNA Decline During Telaprevir-Based Therapy
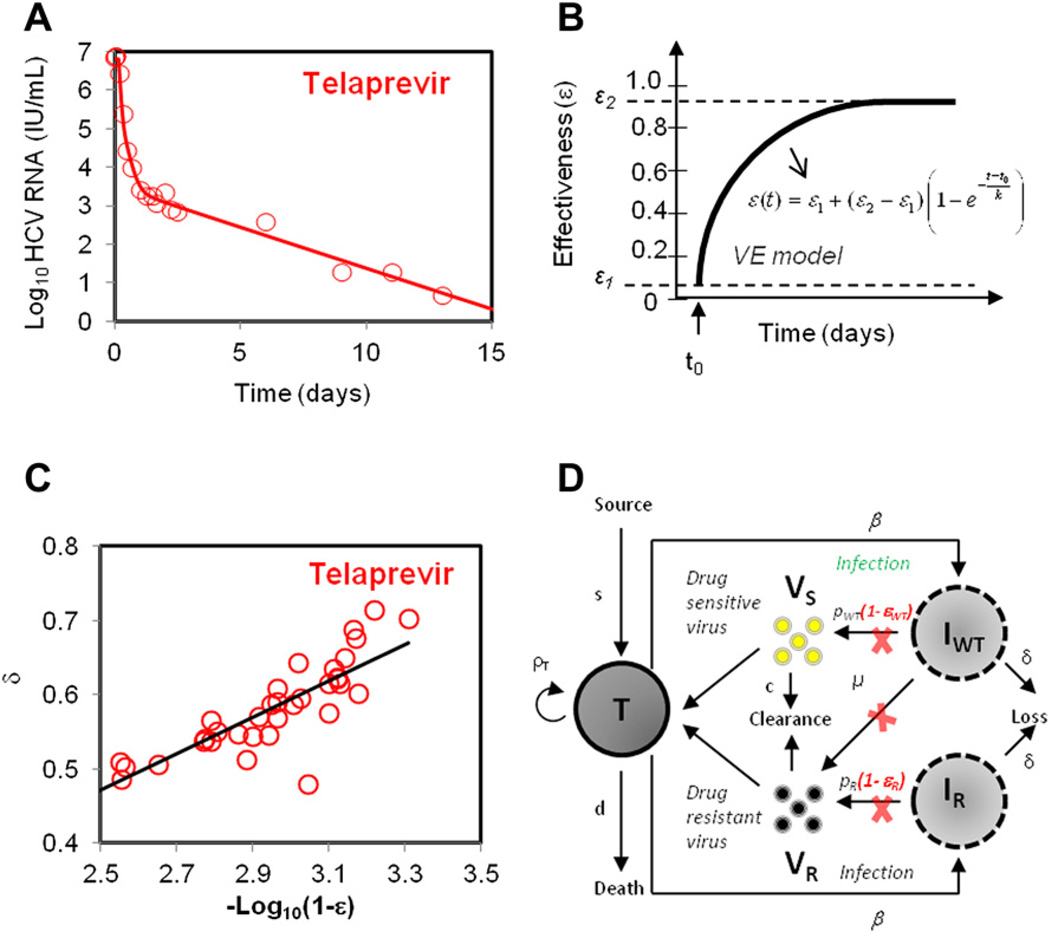
During treatment with telaprevir (TVR), an HCV NS3-4A protease inhibitor, a biphasic viral decline is also observed (Fig. 2A). Mathematical models when fit to data from 44 patients treated with TVR, showed a more rapid second-phase decline than seen previously with IFN-based therapies.34 To quantify the HCV dynamics, Adiwajaya and colleagues34 used a version of the standard (CE) model to fit the early phase of treatment (first 3 days), a period presumed short enough to justify neglecting the influence of resistant variants (see Fig. 1B). The model estimated the median infected cell clearance rate to be δ = 1.2 d−1, which was roughly 10 times higher than that reported for IFN-based treatment. Additionally, the median δ for TVR monotherapy was more than twice that of TVR+PEG-IFN-α2a+RBV treatment. This is also 3 to 4 times higher than that reported for another protease inhibitor BILN-2061. On the other hand, estimates for the plasma virion clearance rate, c, were found to be similar to that reported for PEG-IFN-α2a+RBV and BILN-2061–based regimens, and about twofold higher compared with IFN-α2b–based and PEG-IFN-α2a–based regimen.35
Fig. 2.
Mathematical modeling of HCV kinetics during treatment with protease inhibitor telaprevir. (A) A biphasic viral decline of measured HCV RNA during treatment with telapre-vir+IFN is observed. (B) The effectiveness of drug varying over time using VE models. (C) The rate of loss of infected liver cells is linearly correlated with log-transformed drug effectiveness, ε2. (D) Schematic representation of 2-strain viral dynamic model for drug-sensitive (VS) and drug-resistant virus (VR); ρT is the proliferation rate of target cells, pS and pR are the production rate of drug-sensitive and drug-resistant virus strains respectively. Drug effectiveness in reducing viral production from drug-sensitive and drug-resistant strains is denoted by εS and εR respectively, and µ is the rate of mutation from drug-sensitive to drug-resistant strain. ([D] Adapted from Rong L, Dahari H, Ribeiro RM, et al. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med 2010;2(30):30ra32; with permission.)
Guedj and Perelson36 fit the same patient data using a VE model, instead of the CE model to account for time taken for TVR concentration in plasma to reach maximum levels. Treatment effectiveness ε was allowed to vary over time, as shown in Fig. 2B, where ε1 and ε2 are the initial and final treatment effectiveness (ε2 >ε1), respectively, and k is rate of change of effectiveness. Interestingly, no significant difference was found for drug effectiveness parameters (k, ε1 and ε2) and viral dynamics when comparing TVR+PEG-IFN-α2a with TVR monotherapy. The model estimated the initial treatment effectiveness of ε1 = 0.974, defined as the effectiveness at the first time a viral load decline could be discerned, and a significantly higher final treatment effectiveness of ε2 = 0.999. The mean infected cell loss rate was estimated to be δ = 0.58 d−1 for patients receiving monotherapy and δ = 0.57 d−1 for patients receiving combination therapy (TVR+PEG-IFN), which was significantly lower than that estimated by the CE model (δ = 1.2 d−1).34 Both CE34 and VE36 models indicated that TVR exhibits a rapid second-phase decrease when compared with IFN-based treatments.
Typically, the second-phase decline is attributed to infected cell death δ. Interestingly, a linear correlation between δ and the log-transformed treatment effectiveness (1–ε2) was found (see Fig. 2C), indicating that high effectiveness not only generated a more profound first-phase decline but also a faster second-phase decline. As a result, the time required for achieving less than 1 virion in the extracellular volume or less than 1 infected cell in the liver, was predicted to be considerably shortened with TVR as compared with IFN-based treatments. The VE model predicted that in fully compliant patients with no evidence of drug resistance, the threshold of less than 1 virion could be achieved within 7 and 8 weeks for 95% and 99% of patients respectively. Although this time frame is significantly shorter than currently recommended, the emergence of drug resistance could further extend the needed treatment duration.
Viral Breakthrough During Telaprevir Monotherapy
Viral breakthrough hinders the applicability of current protease inhibitors as monotherapy agents.11,17,37,38 This is primarily because of the high replication rate of HCV and the high error rate of HCV RdRp (estimated as 2.5 × 10−5 per base per generation39). Because DAAs target specific structural properties of HCV proteins, often a single point mutation is sufficient to confer resistance.15 For example, the single mutations T54A, V36A/M, R155K/T, and A156V/T confer different degrees of TVR resistance.40 Some studies have monitored the selection and kinetics of drug-resistant HCV variants.11–13,17,40–42 A 14-day monotherapy treatment with TVR showed that all 4 treated patients with genotype 1a developed resistance,13 with approximately 5% to 20% of the viral pool consisting of drug-resistant variants as early as day 2 of treatment.
Because of the importance of understanding drug resistance to DAAs, mathematical models have been used to quantify the emergence and buildup of drug-resistant variants.29,37,43 Rong and colleagues37 used a model (see Fig. 2D) that considered 2 viral strains, drug sensitive and drug resistant, to track the development of drug resistance during TVR treatment. The model assumed low levels of drug-resistant variants existed before the start of the treatment, with their levels determined by “mutation-selection balance” (ie, the HCV RdRp error rate and the fitness of the variant).44 The high frequency of drug-resistant variants detected in the patients with genotype 1a on day 2 of therapy could be explained by the profound first-phase decline in the drug-sensitive population, in essence revealing the preexisting drug-resistant variants. The rapid growth of the resistant variants observed after day 2 required “replication space.” In the model, this replication space was provided by the proliferation of target cells. There are other possible sources of replication space, such as growth in cells already infected by drug-sensitive virus or the loss of an IFN-induced antiviral state in cells previously protected by the endogenous IFN response. We expect that as viral loads decline because of DAA therapy, endogenous IFN responses will decline as well. Whether this occurs sufficiently rapidly to explain the observed growth of the resistant variants remains to be resolved.
The Rong and colleagues’37 model that included only 2 viral strains was purposely kept simple. More complex models that kept track of multiple resistant variants, each with different fitness and different drug sensitivity, were developed by Adiwijaya and colleagues43 and Rong and colleagues.45 The Adiwijaya and colleagues’ model43 was fit to data from telaprevir clinical trials and explained the dynamics of the various resistant variants observed in the patient data. The Rong and colleagues’ model45 studied from a more mathematical perspective the properties of multistrain systems and explored the role of mutation versus preexistence of resistant strains and the specific viral properties that determine variant frequency. Both models showed that the mere presence of resistant variants does not mean that they will grow and cause viral rebound. The fitness and drug sensitivity of the variants, as well as the properties of the species they are competing with within the patient, may all be needed to predict patient outcome. A more practical approach to the drug-resistance problem is to use combination therapy that provides a high barrier to resistance. Thus, modeling has also focused on determining the viral kinetic properties and modes of action of other classes of antiviral agents.
Modeling HCV Kinetics with Mericitabine (RG7128)
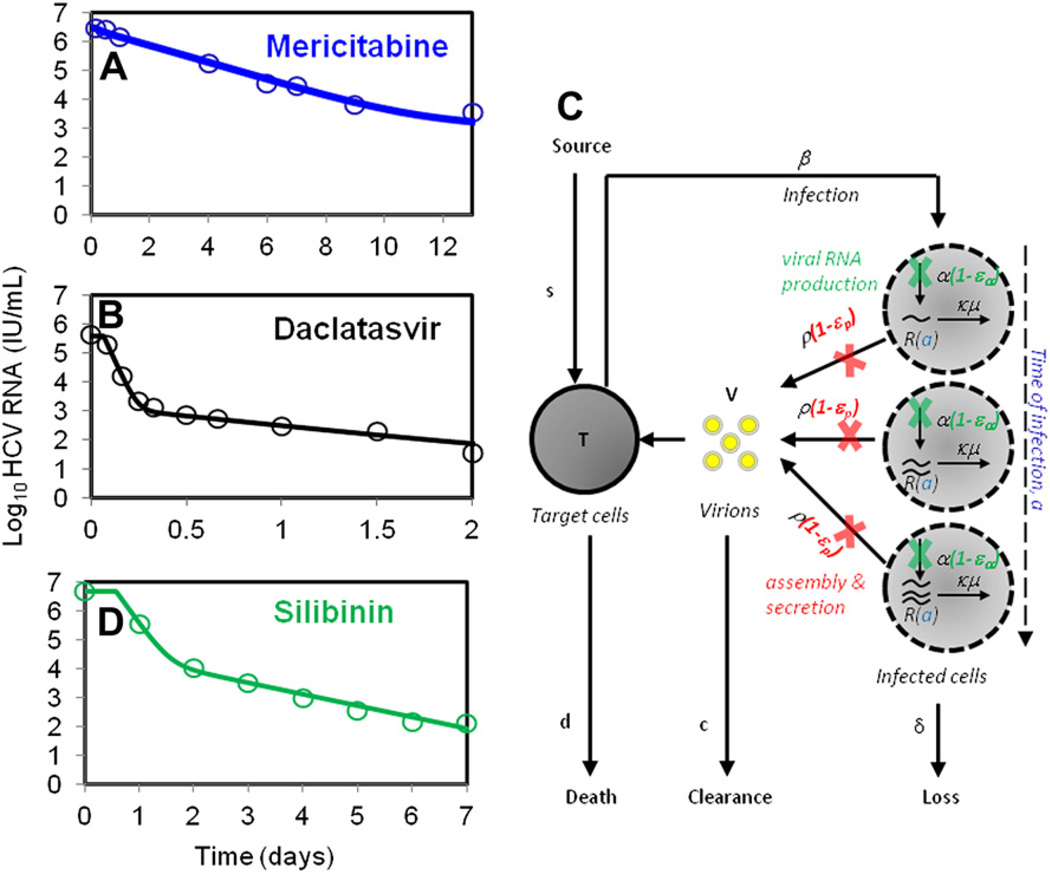
Mericitabine is an oral cytidine nucleoside that exhibits a strong antiviral effectiveness against HCV RNA polymerase across all genotypes.46 On cellular uptake, mericitabine is converted to cytidine monophosphate, which is further phosphorylated intracellularly to the active cytidine triphosphate and uridine triphosphate forms. Motivated by the need of the active triphosphates to accumulate intracellularly, the variable effectiveness model was used by Guedj and colleagues47 to fit data from 32 IFN-treatment experienced patients infected by HCV-genotype 1 (Fig. 3A). In this study, mericitabine was given for 14 days to 4 groups of patients at dosages of 750 mg once daily (750 mg every day), 1500 mg once daily (1500 mg every day), 750 mg twice daily (750 mg twice a day), and 1500 mg twice daily (1500 mg twice a day).
Fig. 3.
Viral decline during treatment with various DAAs and an improved mathematical model of HCV kinetics. (A, B) A monophasic decline of measured HCV RNA with mericitabine monotherapy (A), and a biphasic decline with daclatasvir (B) is observed. (C) Schematic representation of the modified model for fitting early HCV viral load decline during treatment with daclatasvir. Here, intracellular HCV RNA (vRNA) is denoted by R; production, degradation, and assembly/secretion occur at rates α, µ, and ρ, respectively. Treatment may block vRNA production with effectiveness εα, and/or virion assembly/secretion with effectiveness εs and/or enhance the degradation rate (µ) of vRNA by a factor κ. (D) A biphasic viral decline of measured HCV RNA during treatment with silibinin monotherapy is observed.
The effectiveness of mericitabine was found to increase during treatment in a dose-dependent manner, with mean final effectiveness ε2 = 0.86 (for 750 mg every day) and ε2 = 0.94 (for 1500 mg every day), and a significantly higher mean effectiveness for the twice-a-day groups, ε2 = 0.98 (for 750 mg twice a day) and ε2 = 0.998 (for 1500 mg twice a day). Not surprisingly, the rate at which the treatment effectiveness built up (parameter k in equation shown in Fig. 2B) was found to be significantly higher for patients in the twice-a-day dosing groups than in the patients given drug once a day. With twice-a-day dosing it took 2.9 days on average for 90% of the final effectiveness to be attained and 6.5 days to reach 99% of the final effectiveness.47 Because of the slow buildup of effectiveness, all patients treated with mericitabine showed a slow viral decline within the first 4 days of treatment when compared with patients treated with IFN, a protease inhibitor, or an NS5A inhibitor.34,36 In patients who needed additional time to reach high levels of antiviral effectiveness, there was a slower initial rate of viral decline, with 12 patients exhibiting a single phase of monotonic decline, rather than the typical biphasic decline usually observed with IFN or protease inhibitor therapy. The mean value of δ was 0.023 d−1, with no significant differences among the various treatment groups. This value is much lower than that reported for IFN-based therapy and may be biased, as all these patients failed prior IFN-based therapy and hence may have slow second-phase declines. Interestingly, unlike other DAAs, viral breakthrough was not reported at the end of treatment. This suggests that, even though mericitabine may act slowly via a monophasic manner, it could be combined with other DAAs to develop effective treatments.
Viral Kinetics During Treatment with the NS5A Inhibitor Daclatsvir (BMS-79002)
The first-in-class inhibitor of the nonstructural NS5A protein of HCV, daclatasvir, is very effective and can decrease HCV RNA levels by as much as 3 orders of magnitude within 12 hours postdosing (see Fig. 3B)48; however, the mechanism(s) of action of daclatasvir remain unclear. To understand the action of daclatasvir, Guedj and colleagues (submitted for publication) developed a multilevel model, which accounted for the dynamics of intracellular genomic viral RNA (vRNA) so as to incorporate the steps of intracellular viral replication directly targeted by DAAs (see Fig. 3C).
In this model, vRNA was assumed to be produced at a constant rate α, removed at a constant rate µ, and exported into the circulation as virions at rate ρ per vRNA. The model incorporates the age of an infected cell, denoted a, which is defined as the time elapsed postinfection. As time after infection increases, vRNA should accumulate in the infected cell and this new model keeps track of this. The model allows for 3 possible effects of daclatasvir: (1) blocking of vRNA production with effectiveness εα, (2) blocking vRNA assembly into virions and/or secretion with effectiveness εp, and (3) enhancement of vRNA degradation (denoted as µ) by a factor κ>1. The model was fit to patient data from 5 subjects receiving a single dose of 10 or 100 mg of daclatasvir. The model was also fit to data from 20 patients on IFN therapy, and revealed that daclatasvir appeared to have significant activity in blocking both viral assembly/secretion and vRNA replication, whereas IFN mainly affected vRNA replication (Guedj and colleagues, submitted for publication). Further, the first-phase decay observed with daclatasvir was very rapid and suggested that HCV may have a half-life as short as 45 minutes. This fast decay can be deduced from the HCV RNA decay shown in Fig. 1 of Gao and colleagues,48 without the use of any modeling, and is comparable to the half-life reported for HIV.49
Modeling the Viral Kinetics During Treatment with Silibinin
Silibinin, the active component of Legalon SIL, has exhibited significant antiviral activity against HCV genotypes 1 and 4. SVR has been reported in a patient treated with SIL monotherapy for 1 week, followed by 15 weeks of PEG+RBV. Despite its success, the mode of action of SIL is not clearly understood. Ahmed-Belkacem and colleagues,50 based on in vitro studies, showed that the mode of action of SIL was inhibiting the HCV NS5B-RdRp, whereas Wagoner and colleagues,51,52 based on in vivo data, suggested silibinin blocks infection and cell-to-cell spread.
Guedj and colleagues27 modeled HCV RNA kinetics measured (once per day) in 25 patients, treated for 1 week with SIL monotherapy, with dosages of 10, 15, and 20 mg/kg, as reported in Ferenci and colleagues.53 Using the standard model to fit the data yielded a dose-dependent effectiveness (ε = 0.69 for the 10 + 15 mg/kg group and ε = 0.91 for the 20 mg/kg group). The infected cell loss rate was estimated at δ = 0.91 d−1, which is higher than that reported for IFN-based therapy.20,34 Interestingly, the data indicated that 40% of patients showed a monophasic viral RNA decline (see Fig. 3D). A higher infected cell loss rate was estimated for monophasic patients (δ = 1.1 d−1) compared with patients with a biphasic decline (δ = 0.83 d−1), although both groups showed a similar viral decline rate between days 2 and 7. A model assuming SIL inhibits viral infection as well as viral production estimated an effectiveness in blocking infection of η = 0.6 and yielded a statistically improved fit to patient data. This analysis suggested that SIL potentially acts via both these modes in vivo, with blocking viral production being the more pronounced mode of action, consistent with experimental data.50–52,54
Impact of Viral Kinetic Modeling on Therapeutics and Drug Development
The insights gained from viral kinetic modeling have contributed both to improving our understanding of the treatment of patients with HCV, as well as to the development of new antivirals. The Neumann and colleagues’20 model for IFN, described previously, was extended by Dahari and colleagues24 and further applied to the prediction of treatment outcomes. Snoeck and colleagues55 applied HCV viral kinetic modeling to a large database of more than 2000 patients with clinical outcomes who were treated with PEG-IFN-α-2a and RBV, using nonlinear mixed effects modeling. Introducing the concept of a “cure boundary,” the investigators were able to describe and predict therapeutic outcomes with a high degree of accuracy. This model has further been used to simulate therapeutic outcomes of alternative PEG-IFN-α-2a dosing regimens and in special patient populations, facilitating clinical trial design and support of dose recommendations for drug labeling. This PEG-IFN-α-2a–based model has been further extended to incorporate the protease inhibitor danoprevir, and qualified to simulate clinical trial outcomes of triple combination therapy (PEG-IFN-α-2a plus danoprevir/ritonavir and RBV) regimens including different danoprevir doses and treatment durations (Roche, unpublished data).
Viral kinetic modeling has also been used to facilitate the design of early-phase clinical trials, both as a translational tool to bridge preclinical data to human trials, and to generate hypotheses for optimizing dosing regimens of DAAs, which can then be evaluated in patients. For example, the varying effectiveness of the viral kinetic model for mericitabine, described previously,47 has led to the hypothesis that a short lead-in dosing strategy of nucleoside polymerase inhibitors, when given in combination with DAAs with a low barrier to resistance, may enhance the ability of the nucleoside to protect against resistance. This strategy is currently being tested in 2 clinical trials involving both mericitabine and GS-7977. The results of the trials will determine whether the benefit of this approach translates into a clinically meaningful difference.
An example of a translational use of HCV viral kinetic modeling is provided by the Toll-like receptor 7 (TLR7) agonist PF-04878691.56 In this example, an HCV viral kinetic model was used to predict the antiviral activity of a TLR7 agonist, based on the observed level of stimulation of the IFN system with various dosing regimens of PF-0487691 in animals. Translational pharmacokinetic-pharmacodynamic modeling facilitated the selection of the first human doses and the estimation of the likely human effective dose.56 Once healthy volunteer data were available, the model was updated with human clinical data and used to support a go/no-go decision with regard to further clinical development.57
HCV viral kinetic modeling has also been applied to drug discovery and early drug development. Reddy and colleagues58 described the identification of key parameters associated with the clinical potency of HCV protease inhibitors and non-nucleoside polymerase inhibitors. These model-based relationships were used to optimize the nomination of compounds for consideration to move into clinical development, and to facilitate the design of phase I clinical trials, by improving the prediction of clinically effective doses.
THE FUTURE: MODELING COMBINATION THERAPY OF DAAS AND ROLE OF INTRACELLULAR FACTORS DURING DAA TREATMENT
Currently DAA therapy with telaprevir or boceprevir involves concomitant administration of PEG-IFN and RBV, for both treatment-naïve and treatment-experienced patients. Although significant improvement in the treatment outcomes have been observed, the underlying limitation of tolerability and efficacy issues associated with IFN-based treatment highlight the need to switch to DAA combination therapy in the future. In the first of its kind study, INFORM-1 provided proof-of-concept that combination of 2 different DAAs can result in successful treatment outcome.59 In this study, 2 different DAAs, danoprevir, a NS3/4A protease inhibitor,60 and mericitabine, a nucleoside inhibitor, were administered to patients without concomitant use of IFN or RBV. Danoprevir monotherapy has shown potent activity against HCV60; however, similar to other protease inhibitors, has shown emergence of viral breakthrough. The INFORM-1 study showed that with combination therapy a significant 5-log reduction in viral load over a period of 14 days could be achieved, in addition to no viral breakthrough being reported. The success of INFORM-1 has led to several phase-2 trials that have combined various DAAs (summarized in Gane61).
So far, mathematical models for IFN-free DAA combination therapy have not been reported. Although the conceptual framework of the biphasic model has brought valuable insights into the origin of the typical viral decline during standard high-dose daily IFN and has been fairly successful in explaining DAA+IFN-based treatments, extensions of this model are needed to understand other observed patterns of viral decline and the response to DAAs. Especially because DAAs act via targeting specific mechanisms during HCV replication, the current models need to be refined to include the pertinent biology. There is still lack of information about hepatocyte kinetics as well as understanding of the intracellular viral RNA replication dynamics. Dahari and colleagues62 developed a mathematical model describing the dynamics of intracellular HCV replication, providing the first step toward developing a detailed model for HCV replication that can serve as an important tool for evaluating new antivirals. An important aspect of combination therapy is to optimize the number and type of DAAs to achieve high rates of SVR as well as reduced treatment duration.
Additional advances are also needed in the development of viral kinetic models that evaluate the role of host factors that have been shown to influence treatment outcome, such as cyclophilin A,63 IP-10,64 and IL28B polymorphism.65 Until now, most of the DAA-based modeling efforts have been focused on protease inhibitors and NS5A and NS5B nucleoside inhibitors. Mathematical models describing viral kinetics during treatment with entry inhibitors, such as cyanovirin-N,66 and non-nucleoside inhibitors of HCV RdRp, such as setrobuvir,67 BI 2027127,68 and GS-6620,69 or host factor inhibitors, such as alisporivir,70 have yet to be explored.
SUMMARY
The future goal for HCV therapy is an IFN-free treatment, reduced treatment duration, and higher treatment effectiveness for all genotypes, for both treatment-naïve and treatment-experienced patients. To achieve these goals, viral kinetic models that incorporate new drug classes, resistance, and combination therapy should be developed. This may allow us to further improve the therapeutics of HCV by applying these models in a way that both enhances the development of new drugs and further optimizes the use of approved therapies. In the past, mathematical modeling of HCV kinetics has allowed for estimation of crucial parameters associated with treatment and provided relevant biological explanations for HCV kinetics during primary infection as well as during antiviral therapy. These models have also been applied in an attempt to improve the success rate and efficiency of drug development, and to optimize the use of currently approved therapies. In the future, mathematical modeling will certainly continue to serve as an important tool to quantitatively assess and develop an improved understanding of HCV therapy.
KEY POINTS.
The future goal for hepatitis C virus (HCV) therapy is an interferon-free treatment, reduced treatment duration, and higher treatment effectiveness for all genotypes, for both treatment-naïve and treatment-experienced patients.
To achieve these goals, viral kinetic models that incorporate new drug classes, resistance, and combination therapy should be developed.
This may allow us to further improve the therapeutics of HCV by applying these models in a way that both enhances the development of new drugs and further optimizes the use of approved therapies.
In the past, mathematical modeling of HCV kinetics has allowed for estimation of crucial parameters associated with treatment and provided relevant biological explanations for HCV kinetics during primary infection as well as during antiviral therapy.
These models have also been applied in an attempt to improve the success rate and efficiency of drug development, and to optimize the use of currently approved therapies.
In the future, mathematical modeling will certainly continue to serve as an important tool to quantitatively assess and develop an improved understanding of HCV therapy.
ACKNOWLEDGMENTS
This work was performed under the auspices of the US Department of Energy under contract DE-AC52-06NA25396, and supported by NIH grants AI028433, P20-RR018754, R34-HL109334, AI078881, the National Center for Research Resources and the Office of Research Infrastructure Programs through grant 8R01-OD011095-21 (ASP), and Roche Inc. We also acknowledge the Los Alamos National Laboratory LDRD (Laboratory Directed Research and Development) Program for providing partial funding for A.C.
REFERENCES
- 1.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5):S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436(7053):933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 3.Davis GL, Albright JE, Cook SF, et al. Projecting future complications of chronic hepatitis C in the United States. Liver Transpl. 2003;9(4):331–338. doi: 10.1053/jlts.2003.50073. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Lawitz EJ, Shiffman ML, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361(6):580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 5.Abdelmalek MF, Firpi RJ, Soldevila-Pico C, et al. Sustained viral response to interferon and ribavirin in liver transplant recipients with recurrent hepatitis C. Liver Transpl. 2004;10(2):199–207. doi: 10.1002/lt.20074. [DOI] [PubMed] [Google Scholar]
- 6.TenCate V, Sainz BJ, Cotler SJ, et al. Potential treatment options and future research to increase hepatitis C virus treatment response rate. Hepat Med. 2010;2:125–145. doi: 10.2147/HMER.S7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciesek S, von Hahn T, Manns MP. Second-wave protease inhibitors: choosing an heir. Clin Liver Dis. 2011;15(3):597–609. doi: 10.1016/j.cld.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Hézode C, Forestier N, Dusheiko G, et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360(18):1839–1850. doi: 10.1056/NEJMoa0807650. [DOI] [PubMed] [Google Scholar]
- 9.Pawlotsky JM. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53(5):1742–1751. doi: 10.1002/hep.24262. [DOI] [PubMed] [Google Scholar]
- 10.Thompson A, Patel K, Tillman H, et al. Directly acting antivirals for the treatment of patients with hepatitis C infection: a clinical development update addressing key future challenges. J Hepatol. 2009;50(1):184–194. doi: 10.1016/j.jhep.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 11.De Meyer SM, Dierynck I, Ghys A, et al. Similar incidence of virological failure and emergence of resistance with or without a lead-in: results of a telaprevir Phase 3 study in patients who did not achieve SVR with prior Peg-IFN/RBV treatment. Antivir Ther. 2011;16(4):A25. [Google Scholar]
- 12.Hiraga N, Imamura M, Abe H, et al. Rapid emergence of telaprevir resistant hepatitis C virus strain from wildtype clone in vivo. Hepatology. 2011;54(3):781–788. doi: 10.1002/hep.24460. [DOI] [PubMed] [Google Scholar]
- 13.Kieffer TL, Sarrazin C, Miller JS, et al. Telaprevir and pegylated interferon-alpha- 2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46(3):631–639. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 14.Malcolm BA, Liu R, Lahser F, et al. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother. 2006;50(3):1013–1020. doi: 10.1128/AAC.50.3.1013-1020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rong L, Perelson AS. Treatment of hepatitis C virus infection with interferon and small molecule direct antivirals: viral kinetics and modeling. Crit Rev Immunol. 2010;30(2):131–148. doi: 10.1615/critrevimmunol.v30.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartels DJ, Zhou Y, Zhang EZ, et al. Natural prevalence of hepatitis C virus variants with decreased sensitivity to NS3/4A protease inhibitors in treatment-naive subjects. J Infect Dis. 2008;198(6):800–807. doi: 10.1086/591141. [DOI] [PubMed] [Google Scholar]
- 17.Lin C, Gates CA, Rao BG, et al. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J Biol Chem. 2005;280(44):36784–36791. doi: 10.1074/jbc.M506462200. [DOI] [PubMed] [Google Scholar]
- 18.Nettles RE, Gao M, Bifano M, et al. Multiple ascending dose study of BMS-790052, a nonstructural protein 5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1. Hepatology. 2011;54(6):1956–1965. doi: 10.1002/hep.24609. [DOI] [PubMed] [Google Scholar]
- 19.Poordad F, McCone J, Bacon BR, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364(13):1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282(5386):103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 21.Perelson AS, Neumann AU, Markowitz M, et al. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271(5255):1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 22.Dixit NM, Layden-Almer JE, Layden TJ, et al. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432(7019):922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 23.Perelson AS, Essunger P, Cao YZ, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387(6629):188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 24.Dahari H, Lo A, Ribeiro RM, et al. Modeling hepatitis C virus dynamics: liver regeneration and critical drug efficacy. J Theor Biol. 2007;247(2):371–381. doi: 10.1016/j.jtbi.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shudo E, Ribeiro RM, Perelson AS. Modeling HCV kinetics under therapy using PK and PD information. Expert Opin Drug Metab Toxicol. 2009;5(3):321–332. doi: 10.1517/17425250902787616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahari H, Guedj J, Perelson A, et al. Hepatitis C viral kinetics in the era of direct acting antiviral agents and interleukin-28B. Curr Hepat Rep. 2011;10(3):214–227. doi: 10.1007/s11901-011-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guedj J, Dahari H, Pohl RT, et al. Understanding silibinin’s modes of action against HCV using viral kinetic modeling. J Hepatol. 2012;56:1019–1024. doi: 10.1016/j.jhep.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro RM, Layden-Almer J, Powers KA, et al. Dynamics of alanine aminotrans-ferase during hepatitis C virus treatment. Hepatology. 2003;38(2):509–517. doi: 10.1053/jhep.2003.50344. [DOI] [PubMed] [Google Scholar]
- 29.Guedj J, Neumann AU. Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010;267(3):330–340. doi: 10.1016/j.jtbi.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 30.Guedj H, Guedj J, Negro F, et al. The impact of fibrosis and steatosis on early viral kinetics in HCV genotype 1 infected patients treated with PEG-IFN-alfa-2a and ribavirin. J Viral Hepat. 2012;19(7):1365–2893. doi: 10.1111/j.1365-2893.2011.01569.x. [DOI] [PubMed] [Google Scholar]
- 31.Welling PG. Pharmacokinetics: processes and mathematics. ACS (American Chemical Society) Monograph. 1986;185 XIV+290P. [Google Scholar]
- 32.Talal AH, Ribeiro RM, Powers KA, et al. Pharmacodynamics of PEG-IFN alpha differentiate HIV/HCV coinfected sustained virological responders from nonresponders. Hepatology. 2006;43(5):943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 33.Guedj J, Rong L, Dahari H, et al. A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010;17(12):825–833. doi: 10.1111/j.1365-2893.2010.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adiwijaya BS, Hare B, Caron PR, et al. Rapid decrease of wild-type hepatitis C virus on telaprevir treatment. Antivir Ther. 2009;14(4):591–595. [PubMed] [Google Scholar]
- 35.Hinrichsen H, Benhamou Y, Wedemeyer H, et al. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology. 2004;127(5):1347–1355. doi: 10.1053/j.gastro.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Guedj J, Perelson AS. Second-phase hepatitis C virus RNA decline during telaprevir-based therapy increases with drug effectiveness: implications for treatment duration. Hepatology. 2011;53(6):1801–1808. doi: 10.1002/hep.24272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rong L, Dahari H, Ribeiro RM, et al. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010;2(30):30ra32. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong X, Bogen S, Chase R, et al. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 2008;77(3):177–185. doi: 10.1016/j.antiviral.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Ribeiro RM, Li H, Wang S, et al. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: estimates of the in vivo mutation rate. PLoS Pathog. 2012;8(8):e1002881. doi: 10.1371/journal.ppat.1002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarrazin C, Kieffer TL, Bartels D, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132(5):1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 41.Kieffer TL, De Meyer S, Bartels DJ, et al. Clinical virology findings from treatment-naive and treatment-experienced genotype 1 HCV patients receiving telaprevir/ peginterferon/ribavirin in Phase 3 clinical trials. Antivir Ther. 2011;16(4):A27. [Google Scholar]
- 42.Kieffer T, Zhou Y, Zhang E, et al. Evaluation of viral variants during a Phase 2 study (PROVE2) of telaprevir with peginterferon alfa-2a and ribavirin in treatment-naive HCV genotype I-infected patients. Hepatology. 2007;46(4):862A. [Google Scholar]
- 43.Adiwijaya BS, Herrmann E, Hare B, et al. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput Biol. 2010;6(4):e1000745. doi: 10.1371/journal.pcbi.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domingo E. Biological significance of viral quasispecies. Viral Hepatitis Rev. 1996;2:247–261. [Google Scholar]
- 45.Rong L, Ribeiro RM, Perelson AS. Modeling quasispecies and drug resistance in hepatitis C patients treated with a protease inhibitor. Bull Math Biol. 2012;74:1789–1817. doi: 10.1007/s11538-012-9736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy R, Rodriguez-Torres M, Gane E, et al. Antiviral activity, pharmacokinetics, safety and tolerability of R7128, a novel nucleoside HCV RNA polymerase inhibitor, following multiple, ascending, oral doses in patients with HCV genotype 1 infection who have failed prior interferon therapy. Hepatology. 2007;46(4):862A–863A. [Google Scholar]
- 47.Guedj J, Dahari H, Shudo E, et al. Hepatitis C viral kinetics with the nucleoside polymerase inhibitor mericitabine (RG7128) Hepatology. 2012;55:1030–1037. doi: 10.1002/hep.24788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao M, Nettles RE, Belema M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465(7294):96–108. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramratnam B, Bonhoeffer S, Binley J, et al. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet. 1999;354(9192):1782–1785. doi: 10.1016/S0140-6736(99)02035-8. [DOI] [PubMed] [Google Scholar]
- 50.Ahmed-Belkacem A, Ahnou N, Barbotte L, et al. Silibinin and related compounds are direct inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Gastroenterology. 2010;138(3):1112–1122. doi: 10.1053/j.gastro.2009.11.053. [DOI] [PubMed] [Google Scholar]
- 51.Wagoner J, Morishima C, Graf TN, et al. Differential in vitro effects of intravenous versus oral formulations of silibinin on the HCV life cycle and inflammation. PLoS One. 2011;6(1):e16464. doi: 10.1371/journal.pone.0016464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagoner J, Negash A, Kane OJ, et al. Multiple effects of silymarin on the hepatitis C virus lifecycle. Hepatology. 2010;51(6):1912–1921. doi: 10.1002/hep.23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferenci P, Scherzer TM, Kerschner H, et al. Silibinin is a potent antiviral agent in patients with chronic hepatitis C not responding to pegylated interferon/ribavirin therapy. Gastroenterology. 2008;135(5):1561–1567. doi: 10.1053/j.gastro.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 54.Polyak SJ, Morishima C, Shuhart MC, et al. Inhibition of T-cell inflammatory cytokines, hepatocyte NF-kappa B signaling, and HCV infection by standardized silymarin. Gastroenterology. 2007;132(5):1925–1936. doi: 10.1053/j.gastro.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 55.Snoeck E, Chanu P, Lavielle M, et al. A comprehensive hepatitis C viral kinetic model explaining cure. Clin Pharmacol Ther. 2010;87(6):706–713. doi: 10.1038/clpt.2010.35. [DOI] [PubMed] [Google Scholar]
- 56.Benson N, de Jongh J, Duckworth JD, et al. Pharmacokinetic-pharmacodynamic modeling of alpha interferon response induced by a Toll-like 7 receptor agonist in mice. Antimicrob Agents Chemother. 2010;54(3):1179–1185. doi: 10.1128/AAC.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones HM, Chan PL, van der Graaf PH, et al. Use of modelling and simulation techniques to support decision making on the progression of PF-04878691, a TLR7 agonist being developed for hepatitis C. Br J Clin Pharmacol. 2012;73(1):77–92. doi: 10.1111/j.1365-2125.2011.04047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy MB, Morcos PN, Le Pogam S, et al. Pharmacokinetic/pharmacodynamic predictors of clinical potency for hepatitis C virus nonnucleoside polymerase and protease inhibitors. Antimicrob Agents Chemother. 2012;56(6):3144–3156. doi: 10.1128/AAC.06283-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gane EJ, Roberts SK, Stedman CA, et al. Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1): a randomised, double-blind, placebo-controlled, dose-escalation trial. Lancet. 2010;376(9751):1467–1475. doi: 10.1016/S0140-6736(10)61384-0. [DOI] [PubMed] [Google Scholar]
- 60.Forestier N, Larrey D, Guyader D, et al. Treatment of chronic hepatitis C patients with the NS3/4A protease inhibitor danoprevir (ITMN-191/RG7227) leads to robust reductions in viral RNA: a phase 1b multiple ascending dose study. J Hepatol. 2011;54(6):1130–1136. doi: 10.1016/j.jhep.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Gane E. Future hepatitis C virus treatment: interferon-sparing combinations. Liver Int. 2011;31:62–67. doi: 10.1111/j.1478-3231.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- 62.Dahari H, Ribeiro RM, Rice CM, et al. Mathematical modeling of subgenomic hepatitis C virus replication in Huh-7 cells. J Virol. 2007;81(2):750–760. doi: 10.1128/JVI.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang F, Robotham JM, Nelson HB, et al. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82(11):5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lagging M, Romero AI, Westin J, et al. IP-10 predicts viral response and therapeutic outcome in difficult-to-treat patients with HCV genotype 1 infection. Hepatology. 2006;44(6):1617–1625. doi: 10.1002/hep.21407. [DOI] [PubMed] [Google Scholar]
- 65.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helle F, Wychowski C, Vu-Dac N, et al. Cyanovirin-N Inhibits hepatitis C virus entry by binding to envelope protein glycans. J Biol Chem. 2006;281(35):25177–25183. doi: 10.1074/jbc.M602431200. [DOI] [PubMed] [Google Scholar]
- 67.Lawitz E, Rodriguez-Torres M, DeMico M, et al. Antiviral activity of ANA598, a potent non-nucleoside polymerase inhibitor, in chronic hepatitis C patients. J Hepatol. 2009;50:S384. [Google Scholar]
- 68.Larrey D, Lohse AW, de Ledinghen V, et al. Rapid and strong antiviral activity of the non-nucleosidic NS5B polymerase inhibitor BI 207127 in combination with peginterferon alfa 2a and ribavirin. J Hepatol. 2012;57:39–46. doi: 10.1016/j.jhep.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Ray AS, Feng JY, Wang T, et al. 1233 GS-6620: A liver targeted nucleotide prodrug with potent pan-genotype anti-hepatitis C virus activity in vitro. J Hepatol. 2011;54:S487. [Google Scholar]
- 70.Coelmont L, Hanoulle X, Chatterji U, et al. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a Cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS One. 2010;5(10):e13687. doi: 10.1371/journal.pone.0013687. [DOI] [PMC free article] [PubMed] [Google Scholar]