Abstract
Background
Adjuvant chemotherapy improves survival among patients with stage III colon cancer, but no reliable molecular predictors of outcome have been identified.
Methods
We evaluated loss of chromosomal material (also called loss of heterozygosity or allelic loss) from chromosomes 18q, 17p, and 8p; cellular levels of p53 and p21WAF1/CIP1 proteins; and microsatellite instability as molecular markers. We analyzed tumor tissue from 460 patients with stage III and high-risk stage II colon cancer who had been treated with various combinations of adjuvant fluorouracil, leucovorin, and levamisole to determine the ability of these markers to predict survival.
Results
Loss of heterozygosity at 18q was present in 155 of 319 cancers (49 percent). High levels of microsatellite instability were found in 62 of 298 tumors (21 percent), and 38 of these 62 tumors (61 percent) had a mutation of the gene for the type II receptor for transforming growth factor β1 (TGF-β1). Among patients with microsatellite-stable stage III cancer, five-year overall survival after fluorouracil-based chemotherapy was 74 percent in those whose cancer retained 18q alleles and 50 percent in those with loss of 18q alleles (relative risk of death with loss at 18q, 2.75; 95 percent confidence interval, 1.34 to 5.65; P=0.006). The five-year survival rate among patients whose cancer had high levels of microsatellite instability was 74 percent in the presence of a mutated gene for the type II receptor for TGF-β1 and 46 percent if the tumor did not have this mutation (relative risk of death, 2.90; 95 percent confidence interval, 1.14 to 7.35; P=0.03).
Conclusions
Retention of 18q alleles in microsatellite-stable cancers and mutation of the gene for the type II receptor for TGF-β1 in cancers with high levels of microsatellite instability point to a favorable outcome after adjuvant chemotherapy with fluorouracil-based regimens for stage III colon cancer.
Colorectal cancer is the second most common cause of death from cancer in the United States.1 Postoperative adjuvant chemotherapy improves the outcome in stage III (Dukes’ stage C) colon cancer and is now widely accepted as standard therapy.2,3 Many patients with stage II (Dukes’ stage B) disease are considered to be at high risk for recurrence and receive adjuvant therapy, although its benefit in such cases is uncertain. Markers that reliably predict survival are needed.2,4,5
The sequence of genetic alterations leading to colorectal cancer usually begins with the inactivation of the pathway involving the adenomatous polyposis coli tumor-suppressor gene and β-catenin.6,7 Subsequent changes often include the loss of portions of chromosomes, termed loss of heterozygosity or allelic loss, or the loss of whole chromosomes.8 In about 15 percent of cases of sporadic colorectal cancers, there are insertions or deletions of nucleotides within repeated sequences of DNA, termed microsatellite instability, due to defective repair of mismatched nucleotides.9–11 Tumors with high levels of microsatellite instability are characteristic of the hereditary non-polyposis colorectal cancer syndrome, but in most cases such tumors are sporadic.11–13 Neoplasms with high levels of microsatellite instability accumulate mutations in microsatellites within the coding region of certain genes,10,11,14 but loss of chromosomes is rare.8
Some of these genetic alterations are prognostic markers in colorectal cancer.15 Loss of heterozygosity at chromosome 18q indicates a poor prognosis.15–24 Other alterations that have been found to have prognostic value are allelic loss at chromosomes 17p, 1p, 3p, 4p, 5q, or 8p; changes in the levels of certain gene products, including the DCC (deleted in colorectal cancer) protein, p53, and p27Kip1; mutation of the ras gene; and increased expression of genes involved in fluoropyrimidine metabolism.15,25 In addition, colorectal cancers with high levels of microsatellite instability metastasize less often and have a better prognosis than microsatellite-stable cancers.15,26–33
Molecular alterations also have the potential to predict survival after chemotherapy.34–38 We examined a panel of molecular markers (listed in the Glossary) in specimens of colon cancer from patients enrolled by the Eastern Cooperative Oncology Group in two National Cancer Institute Gastrointestinal Intergroup clinical trials of adjuvant chemotherapy with fluorouracil-based regimens.
Glossary of Molecular Markers.
| Marker | Abnormality or Abnormal Gene* | Functions of Wild-Type Gene Product | Reported Prognostic or Predictive Value in Colorectal Cancer |
|---|---|---|---|
| Loss of heterozygosity at chromosome 18q | DCC | Netrin-1 receptor; caspase substrate in apoptosis; cell adhesion molecule; tumor suppression | Adverse prognostic marker |
| Smad4 (DPC4, MADH4) | Nuclear transcription factor in TGF-β1 signaling; regulation of angiogenesis; activator of WAF1 promoter; downstream mediator of Smad2; tumor suppression | Adverse prognostic marker | |
| Smad2 (MADH2, JV18) | Endodermal differentiation; interacts with SKI protein | None | |
| Loss of heterozygosity at chromosome 17p | p53 (TP53) | Transcription factor; regulator of cell-cycle progression after cellular stress, of apoptosis, of gene expression, and of DNA repair; tumor suppression | Adverse prognostic marker |
| Loss of heterozygosity at chromosome 8p | Unknown | Unknown | Adverse prognostic marker |
| High labeling index for p53 protein | p53 (TP53) | Same as for loss of heterozygosity at chromosome 17p | Adverse prognostic marker, adverse predictive marker |
| Increased labeling index for p21WAF1/CIP1 protein | WAF1 (CIP1, SDI1) | Cyclin-dependent kinase inhibitor; controller of cell-cycle progression | Favorable predictive marker |
| Microsatellite instability | Consequence of abnormal genes in mismatch-repair family | Repair of nucleotide mismatches | Favorable prognostic marker, favorable predictive marker |
| Mutation in gene for type II receptor for TGF-β1 | TGF-β1 RII | Receptor for signaling in TGF-β1 pathway; inhibitor of colonic epithelial proliferation | None |
| Mutation in BAX gene | BAX | Proapoptosis | Adverse prognostic marker |
Alternative terms are given in parentheses.
METHODS
Patients
Specimens from 516 eligible patients enrolled in two randomized trials of adjuvant chemotherapy for colon carcinoma were studied. These patients had stage III cancer (Dukes’ stage C, with lymph-node metastasis) or high-risk stage II cancer (Dukes’ stage B2, with colonic obstruction, adherence to or invasion of adjacent organs, or tumor perforation and with en bloc resection of all visible disease, including regional peritoneal metastases). In one trial (Eastern Cooperative Oncology Group protocol E2284 [National Cancer Institute Gastrointestinal Intergroup INT 0035]), three treatments — fluorouracil plus levamisole, levamisole alone, and surgery alone — were compared.39 Patients in this trial were enrolled from February 1985 to October 1987, and the median duration of follow-up in surviving patients was 9.0 years. In the other trial (protocol E2288 [INT 0089]), four treatments — low-dose leucovorin plus fluorouracil, high-dose leucovorin plus fluorouracil, levamisole plus fluorouracil, and low-dose leucovorin plus fluorouracil plus levamisole — were compared.40 Patients were enrolled from August 1988 to July 1992, and the median duration of follow-up in surviving patients was 4.8 years.
Thus, of the seven cohorts in these two trials, five received fluorouracil-based chemotherapy. We studied survival in relation to the presence or absence of molecular markers in available tumor specimens from 460 patients in the five cohorts that received fluorouracil. There were no statistically significant differences in outcome among the patients in these five cohorts. The study population was representative of all the patients in the two clinical trials; the only statistically significant difference between patients included in this study and those not included was the frequency of regional metastases (6 percent vs. 9 percent, P=0.04) (Table 1).
Table 1.
Principal Characteristics of the Patients Enrolled in the Two Clinical Trials, According to Whether the Tumor Was Analyzed in the Current Study.*
| Characteristic | Protocol E2284 | Protocol E2288 | ||
|---|---|---|---|---|
| TUMOR ANALYZED (N=85) | TUMOR NOT ANALYZED (N=331) | TUMOR ANALYZED (N=431) | TUMOR NOT ANALYZED (N=3106) | |
| Regional metastases — no. of patients (%)† | ||||
| No | 81 (95) | 316 (95) | 406 (94) | 2825 (91) |
| Yes | 4 (5) | 14 (4) | 25 (6) | 281 (9) |
| Probability of disease-free survival at 5 yr —% | 48 | 51 | 58 | 59 |
| Probability of overall survival at 5 yr — % | 59 | 60 | 65 | 65 |
Because of rounding, not all percentages total 100. There were no significant differences between the patients whose tumors were analyzed in this study and those whose tumors were not analyzed according to the following variables: sex, race (white, black, or other), age (≤65 or >65 years), site of the cancer (cecum and ascending colon, transverse colon and flexures, descending colon, sigmoid colon, or multiple primary sites), extent of spread (submucosa only, muscular wall, serosa, or contiguous structures), differentiation of carcinoma (well, moderately, or poorly differentiated), stage of disease (stage II or stage III), obstruction (present or absent), tumor size (≤5 or >5 cm), or survival rates.
The difference in the frequency of regional metastases between patients enrolled in protocol E2288 who were included in this study of molecular markers and those who were not included in this study was significant (P=0.04 after stratification according to protocol). Data were missing for 1 patient with a regional metastasis in the subgroup of 331 patients from protocol E2284 whose tumor was not analyzed.
Analysis of Tumor Specimens
Formalin-fixed, paraffin-embedded specimens were obtained through the Eastern Cooperative Oncology Group Pathology Coordinating Office. A data base of information on the patients was maintained at the Eastern Cooperative Oncology Group Statistical Center. Laboratory analysis of tumor specimens was performed without knowledge of the patients’ clinical data. Microdissection of tumor tissue and of non-neoplastic control tissue, when available, and preparation of DNA were performed as previously described.14,16,32,41
Microsatellite Markers
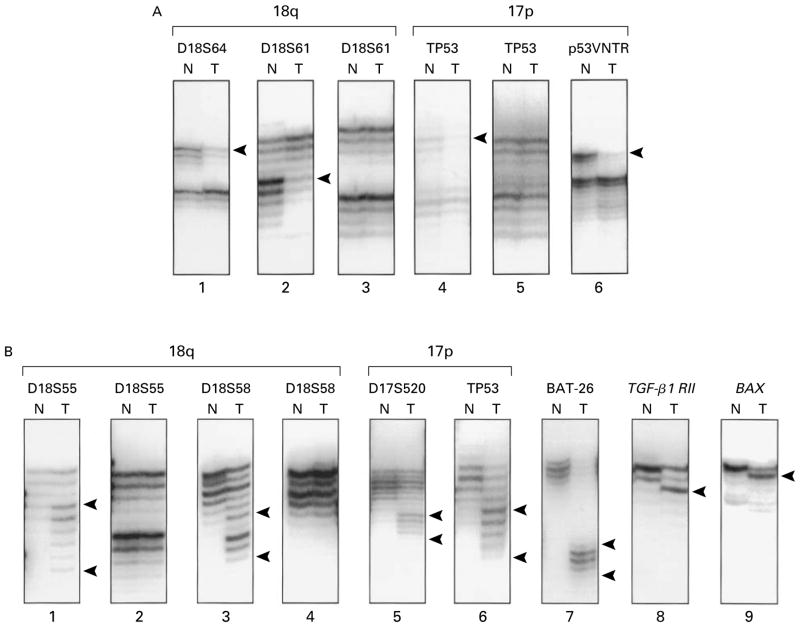
The microsatellite analysis depended on the type of tissue available (Fig. 1). In the case of 298 tumor specimens for which non-neoplastic control tissue was also available, allelic losses from chromosomes 18q, 17p, and 8p were evaluated with polymorphic markers, and microsatellite instability was determined with eight dinucleotide and two mononucleotide markers (additional information is available with the full text of the article at www.nejm.org).14,16,32,42 The 218 tumor specimens for which insufficient control tissue was available were tested for microsatellite instability with two mononucleotide markers that are rarely polymorphic and that do not require control tissue for evaluation. The mononucleotide repeat in the BAX gene14 was analyzed in cancers with high levels of microsatellite instability.
Figure 1. Autoradiographs of Molecular Markers.
Panel A shows an analysis of allelic deletions on chromosomes 18q (lanes 1, 2, and 3) and 17p (lanes 4, 5, and 6). N denotes non-neoplastic mucosa and T tumor tissue. The carcinomas in lanes 1, 2, 4, and 6 have allelic loss in tumor DNA, as indicated by bands with reduced intensity (arrowheads) as compared with the intensity of matched control DNA from non-neoplastic mucosa. Panel B shows an analysis of microsatellite instability and frame-shift mutations in genes with repeated sequences in coding regions. The tumors in lanes 1, 3, 5, and 6 have allelic shifts (flanked by arrowheads) in dinucleotide markers on chromosomes 18q and 17p, in comparison with the matched control DNA from non-neoplastic mucosa. The carcinoma in lane 7 has shifts (flanked by arrowheads) in the BAT-26 polyadenine tract in the fifth intron of the hMSH2 gene. The tumors in lanes 8 and 9 have mutations (arrowheads) in the polyadenine tract of the gene for the type II receptor for TGF-β1 and in the polydeoxyguanosine tract in the proapoptotic BAX gene, respectively.
Allelic Loss at Chromosomes 18q, 17p, and 8p
Allelic loss was determined by examining autoradiographs of DNA amplified by the polymerase chain reaction (PCR). Allelic loss was defined as a reduction in the intensity of the autoradiograph of one of the two alleles in the amplified tumor DNA to at least 50 percent of the level of DNA in non-neoplastic control tissue.14,16,42 Of the 298 cases for which control DNA was available, the results for 18q were interpretable in 279 (94 percent), for 17p in 223 (75 percent), and for 8p in 201 (67 percent). Tumors with high levels of microsatellite instability infrequently lose chromosomes, including 18q, 17p, and 8p,8,14 and were therefore categorized as having no allelic loss; these consisted of 40 additional tumors with no loss at 18q, 102 with no loss at 17p, and 102 with no loss at 8p.
The p53 and p21WAF1/CIP1 Proteins
Immunohistochemical analysis was performed as previously described.43,44 Three categories of p53 staining were defined for statistical analysis with the use of labeling-index cutoff points of 40 percent and 5 percent after quantitation by computer-assisted image analysis.43 After initial evaluation of immunohistochemical results for p21WAF1/CIP1, the labeling index was estimated and categorized as greater than 30 percent, 20 to 30 percent, 10 to 19 percent, 5 to 9 percent, and less than 5 percent of nuclei. Of the 516 analyzed tumors, results were interpretable for 445 (86 percent).
Classification of Microsatellite Instability
Changes in the electrophoretic mobility of DNA amplified by PCR were used to assess microsatellite instability.14,16,32,42 The number of markers with altered allelic sizes and the number of technically satisfactory markers were recorded for each tumor. In evaluating the 298 carcinomas for which control DNA was available, we ascertained microsatellite instability with the use of the interpretable markers among eight dinucleotide markers and two polyadenine tracts. Tumors with a shift in at least two markers and at least 30 percent of the interpretable markers were classified as having high levels of microsatellite instability, in accordance with international criteria.11 A low level of microsatellite instability was defined as a shift in only one dinucleotide marker. In this study, tumors with low levels of microsatellite instability were categorized as microsatellite-stable tumors.11
All the tumors with a shift in a mononucleotide marker had high levels of microsatellite instability when examined with the complete panel of markers, as reported previously.45,46 Therefore, in the 218 cases without control DNA that were evaluated with two mono-nucleotide markers, a shift in a marker was considered to indicate high levels of microsatellite instability.
Statistical Analysis
Cases with missing results were included in all analyses that did not involve the missing data. The Cochran–Mantel–Haenszel (stratum-adjusted Pearson’s chi-square) test and Pearson’s chi-square test were used to analyze associations among categorical variables.47 Analysis of variance was applied to data on age and tumor size. Survival curves were estimated by the method of Kaplan and Meier,48 and differences were assessed by means of the stratified log-rank test.49 Proportional-hazards regression models were used for multivariable comparisons of time-to-event end points.50 All computations were performed with SAS software (version 6.12, SAS Institute, Cary, N.C.). All P values were calculated with two-sided tests of significance.
RESULTS
We first present the genetic abnormalities in the colon cancers we studied and then relate these molecular findings to the survival of the patients after adjuvant chemotherapy.
Genetic Alterations
Loss of heterozygosity at chromosome 18q was observed in 155 of 319 cancers (49 percent). Of these 155 specimens, 143 (92 percent) had loss of all the analyzed 18q markers. Loss of heterozygosity at 17p was found in 166 of 325 tumors (51 percent). Of 309 tumors, 254 (82 percent) had allelic loss from both 18q and 17p or from neither (P<0.001 for concordance between the status of 18q and 17p). Loss of 8p alleles was found in 95 of 303 tumors (31 percent).
Of 445 cancers, 205 (46 percent) had a high labeling index for p53 protein, a finding consistent with a mutation of the p53 gene.51 In 204 of 288 tumors (71 percent), a high p53 labeling index was associated with allelic loss from 17p and a low index was associated with retention of 17p (P<0.001 for the concordance between p53 labeling index and 17p status). The p21WAF1/CIP1 protein was detected in 211 of 445 tumors (47 percent). In 276 of 445 cancers (62 percent), there was an inverse relation between p53 and p21WAF1/CIP1 labeling (P<0.001).
Of the 298 tumor specimens we evaluated for microsatellite instability, 62 (21 percent) had high levels of microsatellite instability. Low levels of microsatellite instability were found in 28 of 298 cancers (9 percent), which were categorized as the microsatellite-stable tumors.
Of the 218 tumor specimens for which control DNA was not available, high levels of microsatellite instability were found in 40 specimens (18 percent). Of the 516 specimens in the entire study that could be analyzed, 102 (20 percent) were classified as having high levels of microsatellite instability and 227 (44 percent) as having microsatellite stability; in 187 specimens (36 percent) the results of evaluation were indeterminate or unsatisfactory.
High levels of microsatellite instability and a high labeling index for p53 protein were found in 24 of 90 tumor specimens (27 percent), whereas 106 of 202 tumor specimens (52 percent) with microsatellite stability had a high p53 labeling index (P<0.001). Among the tumor specimens that had high levels of microsatellite instability and that were evaluated with dinucleotide and mononucleotide markers, mutation of the gene for the type II receptor for transforming growth factor β1 (TGF-β1) was present in 38 of 62 specimens (61 percent). None of the tumor specimens with low levels of microsatellite instability or with microsatellite stability had such a mutation. Mutation of the BAX gene was present in 22 of 60 cancers (37 percent) with high levels of microsatellite instability.
Survival Analysis
Among the 460 patients who were treated with fluorouracil-based chemotherapy, female sex, less advanced stage of disease, and the absence of regional metastases were significant favorable predictors of five-year disease-free survival and five-year overall survival after chemotherapy (Table 2). Younger age (≤65 years) and the presence of a well-differentiated adenocarcinoma, as compared with a moderately or poorly differentiated tumor, were also favorable predictors of five-year overall survival.
Table 2.
Disease-free and Overall Survival at Five Years in Relation to Clinical and Pathological Characteristics of Patients with Stage II or III Colon Cancer Treated with Fluorouracil-Based Adjuvant Chemotherapy.*
| Characteristic | No. of Patients | Disease-free Survival at 5 yr | P Value† | Overall Survival at 5 yr | P Value† |
|---|---|---|---|---|---|
| % | % | ||||
| Age | 0.19 | 0.04 | |||
| ≤65 yr | 248 | 62 | 71 | ||
| 65 yr | 212 | 55 | 61 | ||
| Sex | 0.01 | 0.009 | |||
| Female | 195 | 66 | 72 | ||
| Male | 265 | 54 | 62 | ||
| Differentiation of carcinoma‡ | 0.17§ | 0.01§ | |||
| Well | 55 | 65 | 77 | ||
| Moderate | 302 | 59 | 62 | ||
| Poor | 95 | 57 | 60 | ||
| Stage of disease | <0.001 | <0.001 | |||
| II (Dukes’ stage B) | 105 | 70 | 80 | ||
| II (Dukes’ stage C) | 355 | 54 | 63 | ||
| Regional metastases | <0.001 | <0.001 | |||
| No | 432 | 61 | 69 | ||
| Yes | 28 | 29 | 36 |
The data were stratified according to the protocol in which the patient was enrolled. There were no significant differences in survival rates with respect to race (white, black, or other), site of the cancer (cecum and ascending colon, transverse colon and flexures, descending colon, sigmoid colon, or multiple primary sites), extent of spread (submucosa only, muscular wall, serosa, or contiguous structures), obstruction (present or absent), tumor size (≤5 or >5 cm), or protocol (protocol E2284 or protocol E2288).
P values were calculated by the log-rank test.
Data were missing for eight patients.
The P value is for the linear trend.
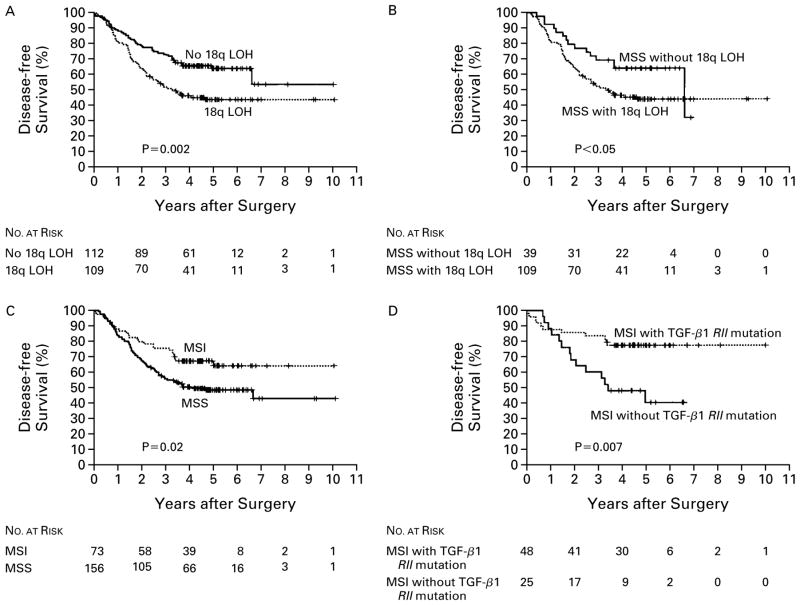
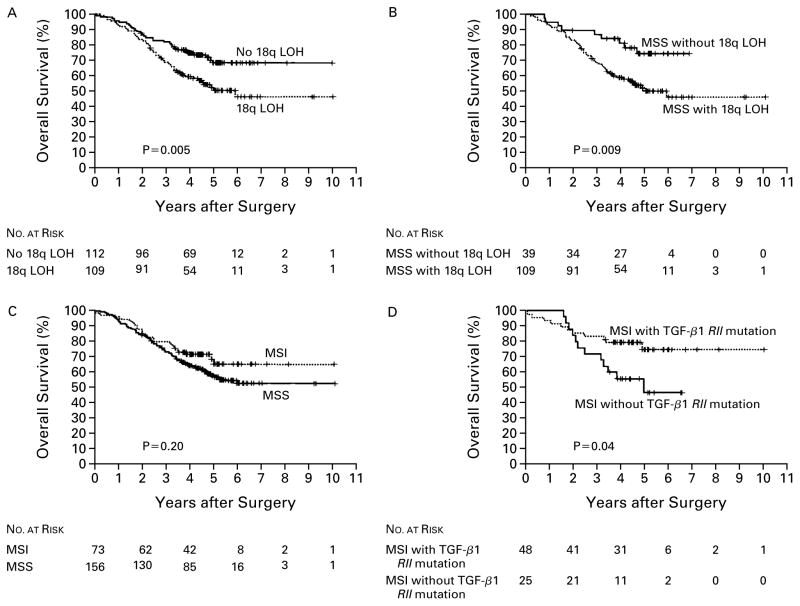
Because the efficacy of adjuvant chemotherapy is more firmly established for patients with stage III cancer than for those with stage II cancer, survival among those with stage II disease was analyzed separately. Of the molecular markers we tested, the status of 18q was significantly associated with both five-year disease-free survival and five-year overall survival after chemotherapy among patients with stage III cancer (Table 3). Patients with tumors that retained 18q had a five-year disease-free survival rate of 64 percent, as compared with 44 percent among those with loss of heterozygosity at 18q (P=0.002) (Fig. 2 and Table 3). The corresponding five-year overall survival rates were 69 percent with retention of 18q alleles and 50 percent with allelic loss at 18q (P=0.005) (Fig. 3 and Table 3). The 18q status also had predictive value in an analysis of the subgroup of patients with microsatellite-stable stage III carcinoma.
Table 3.
Disease-free and Overall Survival at Five Years in Relation to Molecular Markers in Patients with Stage III Colon Cancer Treated with Fluorouracil-Based Adjuvant Chemotherapy.*
| Molecular Marker | No. of Patients | Probability of Disease-Free Survival at 5 yr | P Value† | Probability of Overall Survival at 5 yr | P Value† |
|---|---|---|---|---|---|
| % | % | ||||
| Allelic status of 18q | 0.002 | 0.005 | |||
| No loss | 112 | 64 | 69 | ||
| Loss | 109 | 44 | 50 | ||
| Allelic status of 18q in microsatellite-stable tumors | 0.05 | 0.009 | |||
| No loss | 39 | 67 | 74 | ||
| Loss | 109 | 44 | 50 | ||
| Microsatellite status | 0.02 | 0.20 | |||
| High levels of instability | 73 | 64 | 68 | ||
| Stability | 156 | 49 | 56 | ||
| Status of gene for type II receptor for TGF-β1 | 0.002 | 0.06 | |||
| Mutation | 48 | 79 | 74 | ||
| No mutation | 280 | 50 | 59 | ||
| Status of gene for type II receptor for TGF-β1 in tumors with high levels of microsatellite instability | 0.007 | 0.04 | |||
| Mutation | 48 | 79 | 74 | ||
| No mutation | 25 | 40 | 46 | ||
| Status of BAT-26 marker | 0.02 | 0.15 | |||
| Shift | 61 | 69 | 70 | ||
| No shift | 269 | 50 | 59 |
The data were stratified according to the protocol in which the patient was enrolled. There were no statistically significant differences in survival according to the presence or absence of allelic loss from chromosome 17p; the presence or absence of allelic loss from chromosome 8p; the presence or absence of an increased labeling index for p53 protein; the activation or inactivation of the p53 pathway; the presence or absence of p21WAF1/CIP1 protein; the number of shifted dinucleotide markers; and the status of the BAX gene (mutant or wild-type).
P values were calculated by the log-rank test.
Figure 2. Disease-free Survival According to the Analysis of Molecular Markers in Patients with Stage III Colon Cancer Treated with Postoperative Adjuvant Chemotherapy with Fluorouracil-Based Regimens.
The rate of disease-free survival was significantly higher in patients whose tumor had no loss of heterozygosity (LOH) at chromosome 18q than in those who did have allelic loss at 18q (Panel A); this was also true in the subgroup of patients with microsatellite-stable tumors (MSS) (Panel B). Patients whose cancers had high levels of microsatellite instability (MSI) had a higher rate of survival than those with microsatellite-stable cancers (Panel C). In the subgroup of those with high levels of MSI, those whose cancer had a mutation in the gene for the type II receptor for TGF-β1 (TGF-β1 RII) had a higher rate of disease-free survival than those without this mutation (Panel D). P values were calculated by the log-rank test.
Figure 3. Overall Survival According to the Analysis of Molecular Markers in Patients with Stage III Colon Cancer Treated with Postoperative Adjuvant Chemotherapy with Fluorouracil-Based Regimens.
The rate of overall survival was significantly higher in patients whose cancer had no loss of heterozygosity (LOH) at chromosome 18q (Panel A); this was also true in the subgroup of patients with microsatellite-stable tumors (MSS) (Panel B). The rate of survival in patients whose cancers had high levels of microsatellite instability (MSI) was not significantly different from that in patients with microsatellite-stable tumors (Panel C). Patients with high levels of microsatellite instability and a mutation of the gene for the type II receptor for TGF-β1 (TGF-β1 RII) had a greater rate of survival than those without this mutation (Panel D). P values were calculated by the log-rank test.
Mutation of the gene for the type II receptor for TGF-β1, a specific indicator of high levels of micro-satellite instability, was marginally associated with improved five-year overall survival (Table 3). High levels of microsatellite instability and alteration of the BAT-26 marker were also moderately associated with improved disease-free survival at five years (P=0.02 for both associations). Among the patients who had stage III cancer with both high levels of microsatellite instability and mutation of the gene for the type II receptor for TGF-β1, the rate of disease-free survival at five years was 79 percent, as compared with 40 percent among those whose tumors had high levels of micro-satellite instability and no mutation of this gene (P= 0.007) (Fig. 2 and Table 3). The corresponding rates of overall survival at five years were 74 percent and 46 percent (P=0.04) (Fig. 3 and Table 3).
There was no relation between survival after treatment with a particular regimen and the presence of any of the molecular markers. No marker had predictive value in the analysis of 121 patients with stage II cancer, possibly because of the small sample.
In proportional-hazards regression models that were adjusted for sex, age, the extent of spread, the presence or absence of regional metastases, and the presence or absence of obstruction, several variables —allelic loss at 18q, microsatellite stability, absence of mutation of the gene for the type II receptor for TGF-β1, and absence of an allelic shift in BAT-26 —were each independently associated with an increased risk of recurrence (Table 4). In models in which multiple markers were analyzed, microsatellite-stable cancers had a higher relative risk of recurrence than cancers with high levels of microsatellite instability and mutation of the gene for the type II receptor for TGF-β1 (relative risk, 2.60; 95 percent confidence interval, 1.36 to 4.95; P=0.004). After adjustment for multiple markers, loss of 18q alleles remained an indicator of recurrence and death (Table 4); in contrast, allelic loss at 17p was not predictive either in univariate analysis or after such adjustments (data not shown).
Table 4.
Relative Risks of Recurrence and Death in Patients with Stage III Colon Cancer Treated with Fluorouracil-Based Postoperative Adjuvant Chemotherapy, According to Proportional-Hazards Regression Models.
| Model | Relative Risk of Recurrence (95% CI) | P Value | Relative Risk of Death (95% CI) | P value |
|---|---|---|---|---|
| Clinical and pathological characteristics | ||||
| Male sex | 1.65 (1.19–2.30) | 0.003 | 1.71 (1.19–2.47) | 0.004 |
| Age (per year) | 1.01 (0.99–1.02) | 0.33 | 1.02 (1.00–1.04) | 0.04 |
| Extent of spread (serosa or contiguous structures vs. submucosa or muscular wall) | 1.38 (1.15–1.67) | <0.001 | 1.45 (1.19–1.77) | <0.001 |
| Regional metastatic implants | 3.53 (2.07–6.03) | <0.001 | 3.77 (2.17–6.57) | <0.001 |
| Obstruction | 1.38 (0.96–1.99) | 0.09 | 1.31 (0.87–1.97) | 0.20 |
| Single molecular markers* | ||||
| Allelic loss at 18q | 2.07 (1.37–3.12) | <0.001 | 2.04 (1.30–3.20) | 0.002 |
| Allelic loss at 18q in microsatellite-stable tumor | 2.01 (1.13–3.57) | 0.02 | 2.75 (1.34–5.65) | 0.006 |
| Microsatellite instability | 1.72 (1.09–2.72) | 0.02 | 1.39 (0.85–2.27) | 0.18 |
| Mutation of gene for type II receptor for TGF-β1 | 2.40 (1.29–4.48) | 0.006 | 1.64 (0.88–3.09) | 0.12 |
| Mutation of gene for type II receptor for TGF-β1 in tumor with high levels of microsatellite instability | 3.58 (1.40–9.17) | 0.008 | 2.90 (1.14–7.35) | 0.03 |
| Allelic shift in BAT-26 marker | 1.80 (1.11–2.93) | 0.02 | 1.51 (0.90–2.54) | 0.12 |
| Multiple markers† | ||||
| Microsatellite stability (vs. high levels of microsatellite instability and mutation of the gene for type II receptor for TGF-β1) | 2.60 (1.36–4.95) | 0.004 | 1.83 (0.95–3.52) | 0.07 |
| Allelic loss at 18q | 1.95 (1.24–3.07) | 0.004 | 1.92 (1.17–3.16) | 0.01 |
This analysis involved the use of multivariate regression models after adjustment for clinical and pathological characteristics but not the other molecular markers. In these analyses, neither the status of the BAX gene (wild-type vs. mutant) nor the number and percentage of shifted dinucleotide markers had a significant relation to the relative risk of recurrence or the relative risk of death.
This analysis involved the use of multivariate regression models after adjustment for clinical and pathological characteristics and the other molecular markers. Microsatellite stability as compared with high levels of microsatellite stability and presence of the wild-type gene for the type II receptor for TGF-β1 had no significant relation to the relative risk of recurrence or the relative risk of death. The labeling index for p53 protein (>40 percent vs. ≤40 percent), the presence or absence of p21WAF1/CIP1 protein, and the allelic status of 17p (loss vs. no loss) had no significant effect on the relative risk of recurrence or the relative risk of death.
DISCUSSION
In this study, we showed that the status of chromosome 18q in tumors with microsatellite stability and of the gene for the type II receptor for TGF-β1 in tumors with high levels of microsatellite instability could be used to predict the likelihood of survival in patients with stage III colon cancer who received fluorouracil-based adjuvant chemotherapy. We do not know whether these markers reflect resistance or sensitivity to fluorouracil or inherent differences in the biologic characteristics of the tumors.
In several15–24 but not all15,29,52,53 previous studies, loss of heterozygosity at 18q was an indicator of a poor prognosis in patients with stage II cancer, patients with stage III cancer, or both groups. This loss usually involves the DCC gene, but there are numerous other genes in the deleted region. The product of the DCC gene is the netrin-1 receptor, which guides the migration of neuronal axons.54–57 In colon cancer, loss of DCC is associated with metastasis and an adverse prognosis.58–61 If it does not bind to netrin-1, the DCC protein triggers apoptosis.62 For this reason, loss of DCC as a result of loss of 18q could impair apoptosis, thereby conferring resistance to chemotherapy. The absence of an association between survival and loss of heterozygosity at 8p or 17p suggests that loss of heterozygosity at 18q is a specific marker for survival and not simply a reflection of generalized chromosomal instability.
Our study confirmed the concordance between allelic loss at chromosome 18q and allelic loss at 17p.16 Although alteration of p53 is a plausible predictive marker,15,33,34,63–69 we found no significant relation between survival and the status of the p53 gene or p53 protein. Another study, however, found a higher rate of seven-year survival after adjuvant therapy with fluorouracil and levamisole in patients who had cancer without increased levels of p53 protein than in those who had cancer with increased p53 levels.36 The explanation for these discrepant results in patients in the same clinical trial is not apparent.
The p21WAF1/CIP1 protein is a downstream effector of the p53 protein,70,71 and we found an inverse relation between p53 and p21WAF1/CIP1 in colon cancer, as has been reported previously.72,73 Despite the importance of p21WAF1/CIP1 for in vitro responses to chemotherapeutic agents35 and the report that increased levels of p21WAF1/CIP1 were associated with chemosensitivity of metastatic colorectal cancer,74 pretreatment levels of this protein were not related to survival in our study.
A mutation of the gene that encodes the type II receptor for TGF-β1 in cancers with high levels of microsatellite instability was associated with a favorable outcome, but the mechanism of this effect is uncertain. High levels of microsatellite instability improve the prognosis15,26–33 and may also increase the likelihood of survival after chemotherapy.37,38 Because cancers with high levels of microsatellite instability usually retain 18q alleles, loss of heterozygosity in such tumors is unlikely to be a determinant of outcome after adjuvant chemotherapy. The TGF-β1 pathway inhibits tumor proliferation by blocking the cell cycle late in the G1 (gap 1) phase,75,76 so continued proliferation due to mutation of the gene for the type II receptor for TGF-β1 could increase susceptibility to chemotherapy. However, colon-cancer cell lines that are deficient in mismatch-repair activity and that have high levels of microsatellite instability are relatively resistant to fluorouracil in vitro.77
We find that specific molecular markers in resected stage III colon cancer can be used to predict survival after adjuvant fluorouracil-based regimens. Prospective studies are needed to determine whether newer chemotherapeutic agents, such as irinotecan78 and oxaliplatin, would benefit patients with stage III cancer whose tumors have molecular markers associated with a reduced efficacy of fluorouracil-based regimens. Our study is a first step toward the goal of individualized cancer treatment based on the molecular characteristics of the tumor.
Acknowledgments
Supported by grants (CA60100, CA21115, CA62924, and CA23318) from the National Institutes of Health.
We are indebted to Drs. Bert Vogelstein, Kenneth W. Kinzler, and James Eshleman for advice; to Dr. Asif Rashid for assistance; and to Nancy Folker and Cheryl Willis for secretarial support.
References
- 1.Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Galanis E, Alberts SR, O’Connell MJ. New adjuvant therapy for colon cancer: justified hope or commercial hype. Surg Oncol Clin North Am. 2000;9:813–23. [PubMed] [Google Scholar]
- 3.Macdonald JS. Adjuvant therapy of colon cancer. CA Cancer J Clin. 1999;49:202–19. doi: 10.3322/canjclin.49.4.202. [DOI] [PubMed] [Google Scholar]
- 4.O’Connell MJ, Schaid DJ, Ganju V, Cunningham J, Kovach JS, Thibodeau SN. Current status of adjuvant chemotherapy for colorectal cancer: can molecular markers play a role in predicting prognosis? Cancer. 1992;70(Suppl):1732–9. doi: 10.1002/1097-0142(19920915)70:4+<1732::aid-cncr2820701614>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Chung DC. Molecular prognostic markers and colorectal cancer: the search goes on. Gastroenterology. 1998;114:1330–2. doi: 10.1016/s0016-5085(98)70441-x. [DOI] [PubMed] [Google Scholar]
- 6.Laurent-Puig P, Blons H, Cugnenc PH. Sequence of molecular genetic events in colorectal tumorigenesis. Eur J Cancer Prev. 1999;8(Suppl 1):S39–S47. [PubMed] [Google Scholar]
- 7.Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854–65. doi: 10.1053/gast.2000.16507. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 9.Prolla TA. DNA mismatch repair and cancer. Curr Opin Cell Biol. 1998;10:311–6. doi: 10.1016/s0955-0674(98)80005-7. [DOI] [PubMed] [Google Scholar]
- 10.Jiricny J, Nystrom-Lahti M. Mismatch repair defects in cancer. Curr Opin Genet Dev. 2000;10:157–61. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 11.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 12.Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–11. [PubMed] [Google Scholar]
- 13.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara T, Stolker JM, Watanabe T, et al. Accumulated clonal genetic alterations in familial and sporadic colorectal carcinomas with widespread instability in microsatellite sequences. Am J Pathol. 1998;153:1063–78. doi: 10.1016/S0002-9440(10)65651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLeod HL, Murray GI. Tumour markers of prognosis in colorectal cancer. Br J Cancer. 1999;79:191–203. doi: 10.1038/sj.bjc.6690033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jen J, Kim H, Piantadosi S, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213–21. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- 17.Ookawa K, Sakamoto M, Hirohashi S, et al. Concordant p53 and DCC alterations and allelic losses on chromosomes 13q and 14q associated with liver metastases of colorectal carcinoma. Int J Cancer. 1993;53:382–7. doi: 10.1002/ijc.2910530307. [DOI] [PubMed] [Google Scholar]
- 18.Iino H, Fukayama M, Maeda Y, et al. Molecular genetics for clinical management of colorectal carcinomas: 17p, 18q, and 22q loss of heterozygosity and decreased DCC expression are correlated with the metastatic potential. Cancer. 1994;73:1324–31. doi: 10.1002/1097-0142(19940301)73:5<1324::aid-cncr2820730503>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 19.Kato M, Ito Y, Kobayashi S, Isono K. Detection of DCC and Kiras gene alterations in colorectal carcinoma tissue as prognostic markers for liver metastatic recurrence. Cancer. 1996;77(Suppl):1729–35. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1729::AID-CNCR47>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 20.Ogunbiyi OA, Goodfellow PJ, Herfarth K, et al. Confirmation that chromosome 18q allelic loss in colon cancer is a prognostic indicator. J Clin Oncol. 1998;16:427–33. doi: 10.1200/JCO.1998.16.2.427. [DOI] [PubMed] [Google Scholar]
- 21.Arai T, Akiyama Y, Yamamura A, et al. Allelotype analysis of early colorectal cancers with lymph node metastasis. Int J Cancer. 1998;79:418–23. doi: 10.1002/(sici)1097-0215(19980821)79:4<418::aid-ijc18>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 22.Lanza G, Matteuzzi M, Gafa R, et al. Chromosome 18q allelic loss and prognosis in stage II and III colon cancer. Int J Cancer. 1998;79:390–5. doi: 10.1002/(sici)1097-0215(19980821)79:4<390::aid-ijc14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Lopez E, Abad A, Font A, et al. Allelic loss on chromosome 18q as a prognostic marker in stage II colorectal cancer. Gastroenterology. 1998;114:1180–7. doi: 10.1016/s0016-5085(98)70423-8. [DOI] [PubMed] [Google Scholar]
- 24.Jernvall P, Makinen MJ, Karttunen TJ, Makela J, Vihko P. Loss of heterozygosity at 18q21 is indicative of recurrence and therefore poor prognosis in a subset of colorectal cancers. Br J Cancer. 1999;79:903–8. doi: 10.1038/sj.bjc.6690144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salonga D, Danenberg KD, Johnson M, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–7. [PubMed] [Google Scholar]
- 26.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 27.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–9. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 28.Lothe RA, Peltomaki P, Meling GI, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–52. [PubMed] [Google Scholar]
- 29.Halling KC, French AJ, McDonnell SK, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 30.Johannsdottir JT, Bergthorsson JT, Gretarsdottir S, et al. Replication error in colorectal carcinoma: association with loss of heterozygosity at mismatch repair loci and clinicopathological variables. Anticancer Res. 1999;19:1821–6. [PubMed] [Google Scholar]
- 31.Jernvall P, Makinen MJ, Karttunen TJ, Makela J, Vihko P. Microsatellite instability: impact on cancer progression in proximal and distal colorectal cancers. Eur J Cancer. 1999;35:197–201. doi: 10.1016/s0959-8049(98)00306-2. [DOI] [PubMed] [Google Scholar]
- 32.Lukish JR, Muro K, DeNobile J, et al. Prognostic significance of DNA replication errors in young patients with colorectal cancer. Ann Surg. 1998;227:51–6. doi: 10.1097/00000658-199801000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gryfe R, Kim H, Hsieh ETK, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 34.Pirollo KF, Bouker KB, Chang EH. Does p53 status influence tumor response to anticancer therapies? Anticancer Drugs. 2000;11:419–32. doi: 10.1097/00001813-200007000-00002. [DOI] [PubMed] [Google Scholar]
- 35.O’Connor PM, Jackman J, Bae I, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–300. [PubMed] [Google Scholar]
- 36.Ahnen DJ, Feigl P, Quan G, et al. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: a Southwest Oncology Group study. Cancer Res. 1998;58:1149–58. [PubMed] [Google Scholar]
- 37.Elsaleh H, Powell B, Soontrapornchai P, et al. p53 Gene mutation, microsatellite instability and adjuvant chemotherapy: impact on survival of 388 patients with Dukes’ C colon carcinoma. Oncology. 2000;58:52–9. doi: 10.1159/000012079. [DOI] [PubMed] [Google Scholar]
- 38.Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–50. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 39.Moertel CG, Fleming TR, Macdonald JS, et al. Fluorouracil plus levamisole as effective adjuvant therapy after resection of stage III colon carcinoma: a final report. Ann Intern Med. 1995;122:321–6. doi: 10.7326/0003-4819-122-5-199503010-00001. [DOI] [PubMed] [Google Scholar]
- 40.Haller DG, Catalano PJ, Macdonald JS, Mayer RJ. Fluorouracil (FU), leucovorin (LV) and levamisole (LEV) adjuvant therapy for colon cancer: five-year final report of INT-0089. Prog Proc Am Soc Clin Oncol. 1998;17:256a. abstract. [Google Scholar]
- 41.Moskaluk CA, Kern SE. Microdissection and polymerase chain reaction amplification of genomic DNA from histological tissue sections. Am J Pathol. 1997;150:1547–52. [PMC free article] [PubMed] [Google Scholar]
- 42.Wu TT, Watanabe T, Heitmiller R, Zahurak M, Forastiere AA, Hamilton SR. Genetic alterations in Barrett esophagus and adenocarcinomas of the esophagus and esophagogastric junction region. Am J Pathol. 1998;153:287–94. doi: 10.1016/S0002-9440(10)65570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baas IO, Mulder J-WR, Offerhaus GJA, Vogelstein B, Hamilton SR. An evaluation of six antibodies for immunohistochemistry of mutant p53 gene product in archival colorectal neoplasms. J Pathol. 1994;172:5–12. doi: 10.1002/path.1711720104. [DOI] [PubMed] [Google Scholar]
- 44.Moskaluk CA, Heitmiller R, Zahurak M, Schwab D, Sidransky D, Hamilton SR. p53 And p21WAF1/CIP1/SDI1 gene products in Barrett esophagus and adenocarcinoma of the esophagus and esophagogastric junction. Hum Pathol. 1996;27:1211–20. doi: 10.1016/s0046-8177(96)90317-2. [DOI] [PubMed] [Google Scholar]
- 45.Dietmaier W, Wallinger S, Bocker T, Kullman F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–56. [PubMed] [Google Scholar]
- 46.Perucho M. Correspondence re: C. R. Boland et al., A National Cancer Institute Workshop on Microsatellite Instability for Cancer Detection and Familial Predisposition: Development of International Criteria for the Determination of Microsatellite Instability in Colorectal Cancer: Cancer Res 58: 5248–5257, 1998. Cancer Res. 1999;59:249–53. [PubMed] [Google Scholar]
- 47.Agresti A. Categorical data analysis. New York: John Wiley; 1990. [Google Scholar]
- 48.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–81. [Google Scholar]
- 49.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–70. [PubMed] [Google Scholar]
- 50.Cox DR. Regression models and life-tables. J R Stat Soc [B] 1972;34:187–202. [Google Scholar]
- 51.Save V, Nylander K, Hall PA. Why is p53 protein stabilized in neoplasia? Some answers but many more questions? J Pathol. 1998;184:348–50. doi: 10.1002/(SICI)1096-9896(199804)184:4<348::AID-PATH1227>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 52.Cohn KH, Ornstein DL, Wang F, et al. The significance of allelic deletions and aneuploidy in colorectal carcinoma: results of a 5-year follow-up study. Cancer. 1997;79:233–44. [PubMed] [Google Scholar]
- 53.Carethers JM, Hawn MT, Greenson JK, Hitchcock CL, Boland CR. Prognostic significance of allelic loss at chromosome 18q21 for stage II colorectal cancer. Gastroenterology. 1998;114:1188–95. doi: 10.1016/s0016-5085(98)70424-x. [DOI] [PubMed] [Google Scholar]
- 54.Fearon ER. DCC: is there a connection between tumorigenesis and cell guidance molecules? Biochem Biophys Acta. 1996;1288:M17–M23. doi: 10.1016/0304-419x(96)00023-6. [DOI] [PubMed] [Google Scholar]
- 55.Rieger-Christ KM, Brierley KL, Reale MA. The DCC protein — neural development and the malignant process. Front Biosci. 1997;2:438–48. doi: 10.2741/a203. [DOI] [PubMed] [Google Scholar]
- 56.Keino-Masu K, Masu M, Hinck L, et al. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–85. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 57.Chan SS, Zheng H, Su MW, et al. UNC-40, a C. elegans homolog of DCC (Deleted in Colorectal Cancer), is required in motile cells responding to UNC-6 netrin cues. Cell. 1996;87:187–95. doi: 10.1016/s0092-8674(00)81337-9. [DOI] [PubMed] [Google Scholar]
- 58.Shibata D, Reale MA, Lavin P, et al. The DCC protein and prognosis in colorectal cancer. N Engl J Med. 1996;335:1727–32. doi: 10.1056/NEJM199612053352303. [DOI] [PubMed] [Google Scholar]
- 59.Reymond MA, Dworak O, Remke S, Hohenberger W, Kirchner T, Kockerling F. DCC protein as a predictor of distant metastases after curative surgery for rectal cancer. Dis Colon Rectum. 1998;41:755–60. doi: 10.1007/BF02236264. [DOI] [PubMed] [Google Scholar]
- 60.Saito M, Yamaguchi A, Goi T, et al. Expression of DCC protein in colorectal tumors and its relationship to tumor progression and metastasis. Oncology. 1999;56:134–41. doi: 10.1159/000011954. [DOI] [PubMed] [Google Scholar]
- 61.Itoh F, Hinoda Y, Ona M, et al. Decreased expression of DCC mRNA in human colorectal cancers. Int J Cancer. 1993;53:260–3. doi: 10.1002/ijc.2910530215. [DOI] [PubMed] [Google Scholar]
- 62.Mehlen P, Rabizadeh S, Snipes SJ, Assa-Munt N, Salvesen GS, Bredesen DE. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–4. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 63.Soussi T. The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann N Y Acad Sci. 2000;910:121–37. doi: 10.1111/j.1749-6632.2000.tb06705.x. [DOI] [PubMed] [Google Scholar]
- 64.North S, Hainaut P. p53 And cell-cycle control: a finger in every pie. Pathol Biol (Paris) 2000;48:255–70. [PubMed] [Google Scholar]
- 65.Smith DM, Gao G, Zhang X, Wang G, Dou QP. Regulation of tumor cell apoptotic sensitivity during the cell cycle. Int J Mol Med. 2000;6:503–7. doi: 10.3892/ijmm.6.5.503. [DOI] [PubMed] [Google Scholar]
- 66.el-Deiry WS. The p53 pathway and cancer therapy. Cancer J. 1998;11:229–36. [Google Scholar]
- 67.Bunz F, Hwang PM, Torrance C, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chu E, Copur SM, Ju J, et al. Thymidylate synthase protein and p53 mRNA form an in vivo ribonucleoprotein complex. Mol Cell Biol. 1999;19:1582–94. doi: 10.1128/mcb.19.2.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nita ME, Nagawa H, Tominaga O, et al. 5-Fluorouracil induces apoptosis in human colon cancer cell lines with modulation of Bcl-2 family proteins. Br J Cancer. 1998;78:986–92. doi: 10.1038/bjc.1998.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cox LS. Multiple pathways control cell growth and transformation: overlapping and independent activities of p53 and p21Cip/WAF1/Sdi1. J Pathol. 1997;183:132–40. doi: 10.1002/(SICI)1096-9896(199710)183:2<134::AID-PATH960>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 71.Boulaire J, Fotedar A, Fotedar R. The functions of the cdk-cyclin kinase inhibitor p21WAF1. Pathol Biol (Paris) 2000;48:190–202. [PubMed] [Google Scholar]
- 72.Doglioni C, Pelosio P, Laurino L, et al. p21/WAF1/CIP1 Expression in normal mucosa and in adenomas and adenocarcinomas of the colon: its relationship with differentiation. J Pathol. 1996;179:248–53. doi: 10.1002/(SICI)1096-9896(199607)179:3<248::AID-PATH571>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 73.Valassiadou KE, Stefanaki K, Tzardi M, et al. Immunohistochemical expression of p53, bcl-2, mdm2, and waf1/p21 proteins in colorectal adenocarcinomas. Anticancer Res. 1997;17:2571–6. [PubMed] [Google Scholar]
- 74.Cheng JD, Werness BA, Babb JS, Meropol NJ. Paradoxical correlations of cyclin-dependent kinase inhibitors p21waf1/cip1 and p27kip1 in meta-static colorectal carcinoma. Clin Cancer Res. 1999;5:1057–62. [PubMed] [Google Scholar]
- 75.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor β in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 76.Akhurst RJ, Balmain A. Genetic events and the role of TGF beta in epithelial tumour progression. J Pathol. 1999;187:82–90. doi: 10.1002/(SICI)1096-9896(199901)187:1<82::AID-PATH248>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 77.Carethers JM, Chauhan DP, Fink D, et al. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–31. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saltz LB, Cox JV, Blanke C, et al. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. N Engl J Med. 2000;343:905–14. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]