Abstract
Background
We tested the hypothesis that genes involved in the alcohol oxidation pathway modify the association between alcohol intake and breast cancer.
Methods
Subjects were women aged 55–74 at baseline from the screening arm of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Incident breast cancers were identified through annual health surveys. Controls were frequency matched to cases by age and year of entry into the trial. A self-administered food frequency questionnaire queried frequency and usual serving size of beer, wine or wine coolers and liquor. Three SNPs in genes in the alcohol metabolism pathway were genotyped: alcohol dehydrogenase 2, alcohol dehydrogenase 3 and CYP2E1.
Results
The study included 1041 incident breast cancer cases and 1070 controls. In comparison to non-drinkers, the intake of any alcohol significantly increased the risk of breast cancer, and this risk increased with each category of daily alcohol intake, (OR=2.01, 95% CL=1.14, 3.53) for women who drank three or more standard drinks per day. Stratification by genotype revealed significant gene/environment interactions. For the ADH1B gene, there were statistically significant associations between all levels of alcohol intake and risk of breast cancer (all OR>1.34 and all lower CL >1.01), while for women with the GA or AA genotype, there were no significant associations between alcohol intake and risk of breast cancer.
Conclusion
Alcohol intake, genes involved in alcohol metabolism and their interaction increase the risk of breast cancer in post-menopausal women.
Impact
This information could be useful for primary care providers to personalize information about breast cancer risk reduction.
Keywords: breast cancer, alcohol, metabolizing enzyme, genetics, risk factors
Breast cancer is the most common cancer diagnosed in women in the US, with an estimated 230,480 female breast cancer cases and 39,520 deaths in the year 2011. [1] While breast cancer incidence increased from the 1980s, due primarily to an increase in mammography utilization, death rates from breast cancer have fallen due to earlier detection and improved treatment options. The consistent results of epidemiologic studies suggesting a positive association between moderate alcohol intake and increased risk of breast cancer [2] have led the National Cancer Institute (www.nci.nih.gov) and the American Cancer Society (www.cancer.org) to list alcohol as a modifiable risk factor for breast cancer. Epidemiologic studies published prior to this 2001 review indicated relative risk estimates as high as 4.0 for high versus low alcohol consumption (most commonly defined as highest versus lowest tertile of intake), although the majority of studies found more moderate relative risk estimated in the range of 1.2 to 2.6. Given the fact that alcohol intake is so common, the population attributable risk for alcohol intake as a risk factor for breast cancer is potentially quite high. Several potential mechanisms have been proposed for the increased risk of breast cancer with alcohol consumption. It has been suggested that alcohol intake increases circulating levels of estrogen [3–4] or that the production of reactive oxygen species during alcohol metabolism induces DNA damage that results in breast cancer. [5]
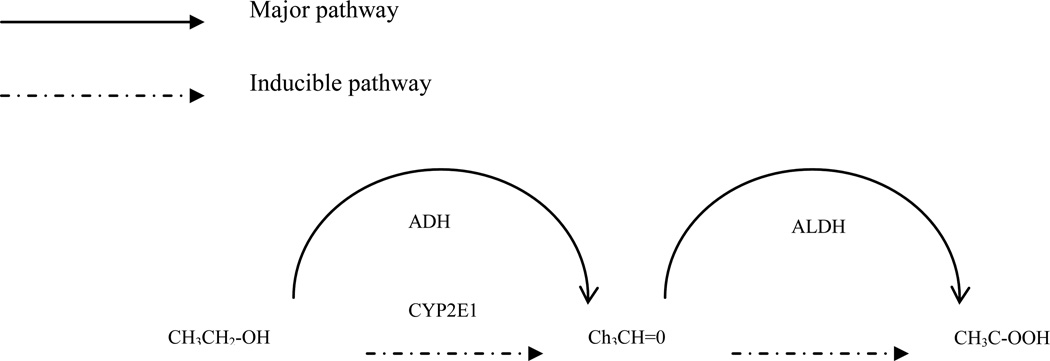
Alcohol is primarily eliminated in the liver through enzymatic oxidation (see Figure). [6] Alcohol dehydrogenase is the enzyme responsible for the first step and aldehyde dehydrogenase is the enzyme involved in the second step. The microsomal ethanol-oxidizing system (CYP2E1) becomes important in alcohol metabolism as alcohol concentration rises or in chronic alcohol use. Tolerant drinkers may increase their clearance of alcohol through induction of the CYP2E1 pathway. Alcohol dehydrogenase is generally the rate-limiting step in alcohol oxidation and the gene is known to be polymorphic, with a minor allele frequency of 0.06 in Caucasians. Researchers have shown that ADH2 [7] and ADH3 [8] genotype modify the association between alcohol consumption and breast cancer risk, especially in pre-menopausal women8. Furthermore, class I alcohol dehydrogenase has been found to be highly expressed in normal human mammary epithelium but not in invasive breast cancer, perhaps suggesting a tumor suppressor role for the gene. [9] Aldehyde dehydrogenase is not polymorphic in Caucasian populations. A significant interaction was reported in Korean women between Cytochrome P450 2E1 (CYP2E1) and alcohol intake for developing breast cancer (OR=1.9 for the interaction between genotype and alcohol intake, p for interaction=0.043). [10] CYP2E1 has been shown to be expressed in normal and breast tissue [11–13], with contradictory evidence as to whether the gene is up-regulated or down-regulated in breast cancer.
Figure 1.
Ethanol oxidation. ADH=alcohol dehydrogenase, ALDH=aldehyde dehydrogenase
These previously published data on alcohol metabolism genes and breast cancer support the hypothesis tested in the current study that genes involved in the alcohol oxidation pathway that remove alcohol from circulation are important in the development of breast cancer and that they modify the association between alcohol intake and breast cancer incidence.
Methods
Subjects in this study were women randomly assigned to the screening arm of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial, a multi-center trial to determine the efficacy of screening versus usual care to reduce mortality from the four cancers. Details of the PLCO trial have been published previously. [14] Briefly, more than 150,000 men and women aged 55–74 without existing PLCO cancers were enrolled between 1993 and 2001 at one of ten screening centers. All subjects provided written informed consent and the project was approved by the institutional review board at the National Cancer Institute and at each of the individual sites. Additional approval was obtained from the IRBs at the Marshfield Clinic and the National Cancer Institute for the current nested case-control study.
All study subjects were asked to complete a self-administered baseline questionnaire that included questions on demographic factors, medical history and health-related behaviors. Of the 77,376 women enrolled in the trial, the 38,660 women randomly assigned to the screening arm were asked to complete a dietary history questionnaire at baseline; 31,411 women (81%) completed the questionnaire. The PLCO food frequency questionnaire (FFQ) was designed to be self-administered and to characterize usual food intake over the previous 12 months (http://www.cancer/gov/prevention/plco/DQX.pdf). This 16-page questionnaire queried the frequency of consumption of 60 fruits and vegetables. The serving size (small, medium, large) and frequency of consumption of 77 individual food items in the grains, meats, dairy, beverages and ‘other’ groups were also queried. Frequency and usual serving size was queried for beer, wine or wine coolers and liquor separately. Usual method for food preparation of a number of foods and use of multi-vitamins and other supplements was also queried. A cognitive approach [15–17] was used to develop wording and formatting that would optimize comprehensibility, ease of completion, and participation. Daily dietary and supplemental folate intake was quantified and high folate intake, due to supplemental intake, (as well as moderate alcohol intake) was found to increase risk of breast cancer in this cohort. [18] Finally, two questions related to usual number of hours currently and at the age of 40 that a person is engaged in vigorous activities per week.
A self-administered health survey was mailed annually to all study subjects to ascertain any diagnoses of cancer in the previous year. Incident breast cancers were identified through these annual health surveys, through state cancer registries and death certificates, and from physician reports. Pathology reports were requested for all cases and 73% of breast cancer cases were confirmed through medical record review. All incident breast cancer cases diagnosed between 1993 and June 30, 2005 were eligible for inclusion in the present study. Controls had to have no report of breast cancer as of June 30, 2005 and were frequency matched to the breast cancer cases by age of entry into the trial (four 5-year age intervals) and year of entry into the trial (two categories: prior to 1997, 1997+). Participants were excluded if they did not complete a baseline questionnaire or an annual study update, their dietary questionnaire was regarded as unacceptable, they had breast cancer prior to study enrollment, they had no DNA sample or necessary consent was not obtained. Ten controls had genotyping results that suggested sample handling errors, and were excluded.
Genotyping was conducted at the Core Genotyping Facility at the National Cancer Institute (http://cgf.nci.nih.gov/home.cfm). TaqMan™ assays were purchased from Applied Biosystems, Inc. (Foster City, CA). Three SNPs were genotyped: 1) alcohol dehydrogenase 2 (rs1229984/ABI # C_2688467_20), 2) alcohol dehydrogenase 3 (rs698/ABI# C_26457410_10) and CYP2E1 (rs2031920/custom made assay).
Using Cox proportional hazards models, the main analyses involved computing relative risks (RR) and 95% confidence intervals (CI) for breast cancer incidence by quintile or other category of average daily alcohol intake. All statistical tests using Cox proportional hazards models were based on the Wald-F test using the SUDAAN software program (SUDAAN Release 9.0, Research Triangle Institute, Research Triangle Park, North Carolina) for analysis of weighted data from sample surveys. These weights allow inference back to the original distribution of cases and controls in the entire PLCO cohort. Stratified analyses were performed to assess the interaction of genotype and alcohol intake with risk of breast cancer. A two-sided p-value <0.05 was considered to be statistically significant.
Results
The study included 1041 incident breast cancer cases and 1070 controls. In the breast cancer cases with available information, 768 were invasive and 203 were in-situ. A comparison of baseline characteristics between breast cancer cases and controls is presented in Table 1. Despite the limited frequency matching, the cases and controls are similar in terms of demographics (age, race and education level). Age at menarche and menopause, ever used oral contraceptives, BMI at the time of enrollment, mean weight at the age of 20 and average weekly hours of physical activity were also not significantly different between cases and controls. There was a significant linear trend between age at birth of first child and number of live births, with breast cancer cases having their first child at older ages and having fewer children (both p<0.01). Personal history of benign breast disease was significantly higher in breast cancer cases than controls (37.9% versus 25.3%, p<0.001). Twenty percent of the breast cancer cases reported a first degree relative with breast cancer, in comparison with 16% of the controls (p=0.0295). The mean caloric intake and the mean level of folate from diet and supplements were higher in breast cancer cases than controls, although did not reach statistical significance (both p=0.05).
Table 1.
Comparison of baseline characteristics between breast cancer cases and controls
| Variable | Controls, n=1070 |
Cases, n=1041 |
p-value from Wald F or t-test |
|---|---|---|---|
| Race White, non-Hispanic (n, %) Black non-Hispanic (n, %) Hispanic (n, %) Asian (n, %) Other (n, %) |
966, 90.3 39, 3.6 13, 1.2 44, 4.1 8, 0.8 |
950, 91.3 41, 3.9 8, 0.8 35, 3.4 7, 0.7 |
0.65 |
| Mean age in yrs at baseline (SD) | 62.5 (5.1) | 62.5 (5.0) | <0.001 |
| Education level Less than high school (n, %) Graduated high school (n, %) Some post high school (n, %) College graduate (n, %) |
65, 6.1 281, 26.3 380, 35.5 344, 32.2 |
54, 5.2 238, 23.0 361, 34.7 388, 37.3 |
0.14 |
| Age at menarche <12 (n, %) 12–13 (n, %) 14+ (n, %) |
209, 19.5 586, 54.8 275, 25.7 |
207, 19.9 574, 55.1 260, 25.0 |
0.91 |
| Age at menopause <45 (n, %) 45–49 (n, %) 50–54 (n, %) 55+ (n, %) |
282, 26.4 258, 24.1 391, 36.5 139, 13.0 |
264, 25.4 222, 21.3 407, 39.1 148, 14.2 |
0.70 |
| Mean BMI at baseline (SD) | 26.9 (5.4) | 27.1 (5.2) | 0.53 |
| Mean BMI at age 20 y (SD) | 21.2 (2.8) | 21.1 (2.7) | 0.66 |
| Hours per week of vigorous physical activity 0–1 (n, %) 1–4 (n, %) 4–5 (n, %) Unknown (n, %) |
315, 29.4 415, 38.8 228, 21.3 112, 10.5 |
312, 30.0 457, 43.9 183, 17.6 89, 8.6 |
0.03 |
| Smoking status at baseline Never smoker (n, %) Current smoker (n, %) Former smoker (n, %) |
636, 59.4 91, 8.5 343, 32.1 |
565, 54.3 90, 8.7 386, 37.1 |
0.10 |
| Age at birth of first child <19 (n, %) 20–24 (n, %) 25–29 (n, %) 30+ (n, %) No children (n, %) |
170, 15.9 536, 50.1 207, 19.4 71, 6.6 86, 8.0 |
150, 14.4 444, 42.7 247, 23.7 92, 8.8 108, 10.4 |
<0.001 |
| Number of live births None (n, %) 1–2 (n, %) 3–4 (n, %) 5+ (n %) |
86, 8.0 326, 30.5 436, 40.8 222, 20.8 |
110, 10.6 354, 34.0 420, 40.4 157, 15.1 |
0.008 |
| Ever used oral contraceptives (n, %) | 580, 54.2 | 574, 55.1 | 0.86 |
| Hormone replacement therapy at baseline Never user (n, %) Current user (n %) Former user (n, %) |
319, 29.8 561, 52.4 190, 17.8 |
275, 26.4 632, 60.7 134, 12.9 |
0.0006 |
| Personal history of benign breast disease (n, %) | 271, 25.3 | 394, 37.9 | <0.001 |
| Number (%) with a first degree relative with breast cancer | 173, 16.2 | 205, 19.7 | 0.03 |
| Mean food energy in kcal from diet excluding alcohol (SD) | 1695.0 (606.0) | 1762.6 (671.5) | 0.0598 |
| Mean folate from diet and supplements in mcg/day (SD) | 596.1 (344.0) | 642.7 (367.2) | 0.0525 |
Significant primary effects were observed for both alcohol intake and the alcohol metabolizing genes for breast cancer incidence (Table 2). In comparison to non-drinkers, the intake of any alcohol significantly increased the risk of breast cancer, and this risk increased with each category of daily alcohol intake, with an odds ratio of 2.01 (95% CL=1.14, 3.53) from the minimally adjusted model for women who drank three or more standard drinks per day. The results from the minimally adjusted models were similar to the fully adjusted models.
Table 2.
Relation of alcohol intake and alcohol metabolism genotype to breast cancer incidence
| Daily alcohol intake or alcohol metabolism genotype |
Controls N, % |
Cases N, % |
Minimally adjusted hazard ratio (95%CL)* |
Fully adjusted hazard ratio (95% CL)** |
|---|---|---|---|---|
| Daily servings of alcohol | ||||
| None (n, %) | 202, 18.9 | 146, 14.0 | 1.00 | 1.00 |
| >0–0.99 (n, %) | 604, 56.5 | 604, 58.0 | 1.34 (1.04, 1.73) | 1.31 (1.01, 1.71) |
| 1.00–1.99 (n, %) | 80, 7.5 | 99, 9.5 | 1.64 (1.12, 2.39) | 1.54 (1.04, 2.28) |
| 2.00–2.99 (n, %) | 32, 3.0 | 43, 4.1 | 1.77 (1.05, 2.97) | 1.75 (1.02, 3.00) |
| 3.00+ (n, %) | 25, 2.3 | 35, 3.4 | 2.01 (1.14, 3.53) | 2.00 (1.11, 3.61) |
| Alcohol servings/day (quintile) | ||||
| 0–<0.001 | 202, 18.9 | 146, 14.0 | 1.00 | 1.00 |
| 0.001–<0.032 | 262, 24.5 | 243, 23.3 | 1.28 (0.96, 1.71) | 1.27 (0.94, 1.71) |
| 0.032–<0.111 | 200, 18.7 | 217, 20.9 | 1.41 (1.05, 1.90) | 1.38 (1.01, 1.89) |
| 0.111–<0.672 | 172, 16.1 | 176, 16.9 | 1.35 (0.99, 1.85) | 1.31 (0.95, 1.82) |
| 0.672+ | 107, 10.0 | 145, 13.9 | 1.82 (1.30, 2.56) | 1.74 (1.22, 2.47) |
| ADH1B genotype | ||||
| GG | 960, 89.7 | 917, 88.1 | 1.00 | 1.00 |
| GA | 80, 7.5 | 91, 8.7 | 1.41 (1.00, 1.99) | 1.31 (0.91, 1.87) |
| AA | 26, 2.4 | 26, 2.5 | 1.46 (0.58, 3.67) | 1.58 (0.59, 4.23) |
| GA or AA | 106, 9.9 | 117, 11.2 | 1.41 (1.00, 1.99) | 1.31 (0.92, 1.88) |
| ADH1C genotype | ||||
| GG | 164, 15.3 | 173, 16.6 | 1.05 (0.81, 1.37) | 1.04 (0.79, 1.37) |
| GA | 465, 43.5 | 427, 41.0 | 0.87 (0.71, 1.06) | 0.86 (0.70, 1.05) |
| AA | 423, 39.5 | 425, 40.8 | 1.00 | 1.00 |
| CYP2E1 genotype | ||||
| CC | 974, 91.0 | 957, 91.9 | 1.00 | 1.00 |
| CT | 70, 6.5 | 64, 6.2 | 0.88 (0.59, 1.31) | 0.81 (0.54, 1.23) |
| TT | 4, 0.4 | 7, 0.7 | 1.91 (0.57, 6.39) | 1.70 (0.49, 5.94) |
| CT or TT | 74, 6.9 | 71, 6.8 | 0.95 (0.66, 1.37) | 0.87 (0.60, 1.28) |
adjusted for age at baseline, year of baseline, race and ethnicity
adjusted for age at baseline, year of baseline, race and ethnicity, age at menarche, age at menopause, parity, age at first live birth, family history of breast cancer, personal history of benign breast disease
Stratification by genotype of the association between alcohol intake and breast cancer incidence revealed significant gene/environment interactions (Table 3). For the ADH1b gene, there were statistically significant associations between all levels of alcohol intake and risk of breast cancer (all OR>.34 and all lower CL >1.01), while for women with the GA or AA genotype, there were no significant associations between alcohol intake and risk of incident breast cancer, with some suggestion that alcohol intake may be protective. For the ADH1C gene, the association between various levels of alcohol intake and risk of incident breast cancer were similar across genotypes. Although there is some suggestion that the association between alcohol intake and risk of incident breast cancer may vary by genotype for the CYP2E1 gene, the results for the CT and TT genotype are not precise due to the relatively small sample size for those genotypes.
Table 3.
Relation of alcohol intake to breast cancer incidence, stratified by genotype
| Gene | Genotype | Average daily alcohol intake (drinks per day) |
Minimally adjusted OR (95%CL)* |
Fully adjusted OR (95% CL)** |
|---|---|---|---|---|
| ADH1B | GG | None (n, %) >0–0.99 (n, %) 1.00–1.99 (n, %) 2.00–2.99 (n, %) 3.00+ (n, %) |
1.00 1.34 (1.02, 1.76) 1.80 (1.20, 2.68) 1.91 (1.12, 3.28) 2.25 (1.25, 4.05) |
1.00 1.29 (0.97, 1.72) 1.66 (1.09, 2.52) 1.88 (1.07, 3.29) 2.22 (1.19, 4.13) |
| GA or AA | None (n, %) >0–0.99 (n, %) 1.00–1.99 (n, %) 2.00–2.99 (n, %) 3.00+ (n, %) |
1.00 0.95 (0.39, 2.30) 0.53 (0.12, 2.45) 0.24, 0.02, 3.58) 0.00 |
1.00 0.90 (0.33, 2.46) 0.51 (0.06, 4.55) 0.30 (0.01, 8.77) 0.00 |
|
| ADH1C | GG | None (n, %) >0–0.99 (n, %) 1.00–1.99 (n, %) 2.00–2.99 (n, %) 3.00+ (n, %) |
1.00 1.71 (0.92, 3.19) 1.29 (0.49, 3.39) 1.24 (0.33, 4.63) 2.11 (0.33, 4.63) |
1.00 1.68 (0.81, 3.49) 1.11 (0.36, 3.41) 1.69 (0.42, 6.75) 2.14 (0.31, 14.91) |
| GA | None (n, %) >0–0.99 (n, %) 1.00–1.99 (n, %) 2.00–2.99 (n, %) 3.00+ (n, %) |
1.00 0.99 (0.66, 1.47) 1.50 (0.86, 2.64) 1.85 (0.77, 4.46) 1.06 (0.47, 2.39) |
1.00 1.03 (0.66, 1.58) 1.29 (0.71, 2.36) 1.59 (0.61, 4.14) 1.01 (0.43, 2.41) |
|
| AA | None (n, %) >0–0.99 (n, %) 1.00–1.99 (n, %) 2.00–2.99 (n, %) 3.00+ (n, %) |
1.00 1.57 (1.05, 2.36) 1.69 (0.87, 3.28) 1.72 (0.78, 3.79) 5.31 (1.76, 16.01) |
1.00 1.59 (1.03, 2.46) 1.80 (0.87, 3.72) 1.87 (0.80, 4.36) 4.13 (1.19, 14.26) |
|
| CYP2E1 | CC | None (n, %) >0–0.99 (n, %) 1.00–1.99 (n, %) 2.00–2.99 (n, %) 3.00+ (n, %) |
1.00 1.32 (1.01, 1.71) 1.77 (1.19, 2.63) 1.61 (0.93, 2.79) 1.96 (1.09, 3.54) |
1.00 1.29 (0.98, 1.70) 1.70 (1.13, 2.57) 1.61 (0.91, 2.85) 1.96 (1.06, 3.65) |
| CT or TT | None (n, %) >0–0.99 (n, %) 1.00–1.99 (n, %) 2.00–2.99 (n, %) 3.00+ (n, %) |
1.00 1.25 (0.43, 3.59) 0.34 (0.06, 1.90) 3.20 (0.50, 20.36) 0.56 (0.03, 10.36) |
1.00 0.86 (0.20, 3.74) 0.05 (0, 0.95) 5.28 (0.64, 43.41) 0.46 (0.01, 15.71) |
adjusted for age at baseline, year of baseline, race and ethnicity
adjusted for age at baseline, year of baseline, race and ethnicity, age at menarche, age at menopause, parity, age at first live birth, family history of breast cancer, personal history of benign breast disease
To assess the potential bias induced by self-selection of subjects into low levels of alcohol intake due to alcohol metabolizing genotype, a cross-tab of alcohol intake and genotype was calculated (Table 4). For the two highest categories of average daily alcohol intake, there is some indication that subjects with the GA or AA genotype for ADH1B do not consume this level of alcohol and are more likely to abstain from alcohol (20.2% versus 15.9% for subjects with the GG genotype).
Table 4.
Alcohol intake by alcohol metabolism genotype
| Alcohol servings per day |
ADH1B | ADH1C | CYP2E1 | |||
|---|---|---|---|---|---|---|
| GG n (%) |
GA or AA n (%) |
GG or GA n (%) |
AA n (%) |
CC n (%) |
CT or TT n (%) |
|
| None | 298 (15.9) | 45 (20.2) | 204 (16.6) | 140 (16.5) | 313 (16.2) | 29 (20.0) |
| >0–0.99 | 1071 (57.1) | 133 (59.6) | 695 (56.6) | 496 (58.5) | 1112 (57.6) | 75 (51.7) |
| 1.00–1.99 | 167 (8.9) | 12 (5.4) | 119 (9.7) | 57 (6.7) | 163 (8.4) | 14 (9.7) |
| 2.00–2.99 | 72 (3.8) | 3 (1.4) | 38 (3.1) | 34 (4.0) | 67 (3.5) | 8 (5.5) |
| 3.00+ | 57 (3.0) | 2 (0.9) | 38 (3.1) | 21 (2.5) | 55 (2.9) | 3 (2.1) |
Discussion
We examined the role of alcohol intake, genes in the alcohol metabolism pathway and their interaction in the development of incident breast cancer in post-menopausal women. As has been observed in many previous observational studies [2], women with incident breast cancer in this study had a significantly higher daily alcohol intake than controls, with increasing risk observed along with increasing average daily alcohol intake.
The genes responsible for alcohol metabolism were selected for this study because of the metabolic pathway for alcohol (Figure). The polymorphisms in the alcohol metabolizing genes were selected because of previous studies in the medical literature. As mentioned previously, markers in the gene responsible for aldehyde dehydrogenase were not genotyped in this study because it is not polymorphic in Caucasians and therefore not relevant to this primarily Caucasian study population. It would be worth genotyping in Asian populations where the gene is found to be polymorphic.
Alcohol dehydrogenase oxidizes ethanol to acetaldehyde. The SNP selected in ADH1B (ADH2), rs1229984, is non-synonymous. It is a haplotype-tagging SNP on chromosome 4 that results in arginine to histidine change. The SNP selected in ADH1C (ADH3), rs698, is also a non-synonymous haplotype-tagging SNP on chromosome 4, resulting in isoleucine to valine change. We found differences in the magnitude and direction of the effect of alcohol intake on risk of breast cancer in the PLCO cohort by both ADH1B and ADH1C genotype, with the A allele of ADH1B appearing to be protective. A case-only study in primarily post-menopausal women in Germany demonstrated an interaction between ADH1B and alcohol intake in women with invasive breast cancer. [7] A recent case-control study reported a potential protective association between ADH1B*896G and breast cancer in women with a mean age of 57 years, more than one-quarter of whom were pre-menopausal. [19]
In women with invasive breast cancer, there was an inverse association between the ADH2 polymorphism and frequency of alcohol consumption in relation to risk of breast cancer. Similarly, we observed a protective affect for the A allele of ADH2. Three prior studies failed to find significant associations between alcohol intake and breast cancer risk in post-menopausal women when stratified by ADH3 genotype, although their data suggested an interaction which may not have been able to be detected due to lack of power. [8, 20–21]
Cytochrome P450 also oxidizes ethanol to acetaldehyde, although it is only induced by heavy drinking. The SNP selected for this study, rs2031920, located on chromosome 10, acts in the 5’ flanking region of the gene to decrease enzyme activity in drinkers. One prior study demonstrated an interaction between alcohol intake and CYP2E1 genotype and risk of breast cancer in alcohol-consuming Korean women, although their findings were not statistically significant due to small sample size for the minor allele. [10] The findings were similar regardless of menopausal status. The magnitude of the effect of current alcohol intake on breast cancer risk in our study (OR ranging from 1.32 to 1.96) was slightly greater than was observed in the Korean study (OR=1.4), perhaps because of the importance of aldehyde dehydrogenase in the Korean population.
Higher levels of acetaldehyde due to alterations in the oxidative pathway because of genetic polymorphisms may explain the association between alcohol intake and risk of breast cancer. Another potential mechanism to explain this association is a modulating effect on estrogen metabolism or receptors. [22] Interaction between alcohol intake and estrogen and progesterone (ER/PR) receptor status of the breast cancer tumor has been observed in previous studies [23–28], as well as a synergistic effect of alcohol intake and estrogen replacement therapy [29], further supporting the hypothesis that alcohol influences hormonal status to increase breast cancer risk. ER/PR status is not currently available for PLCO cohort.
The advantages of this study include the relatively large sample size, the fact that the information about alcohol intake was collected prior to breast cancer diagnosis, and the available information about other risk factors for breast cancer. Limitations include the relative lack of ethnic diversity in the study sample and the limited sample size to assess gene/gene interactions. Also, because acetaldehyde is known to play a major role in producing unpleasant symptoms after alcohol intake, such as facial flushing, palpitations, headache, vomiting and sweating [26], women who are “poor metabolizers” may self-select into the non-drinking or lower levels of drinking groups, thus effectively decreasing the sample size of moderate or heavier alcohol drinkers and further decreasing the power to detect an association of moderate or heavy alcohol intake with breast cancer when stratified by genotype. There is some indication in the current data set that this self-selection does occur.
In conclusion, alcohol intake, genes involved in alcohol metabolism and the interaction of alcohol intake and alcohol metabolism genes increase the risk of breast cancer in post-Alcohol, genetics and breast cancer menopausal women. Further research to investigate these effects in other ethnic groups and a larger sample size to quantify gene/gene interactions are warranted. In addition, the inclusion of estrogen and progesterone receptor status would allow the investigation of potential interaction with tumor status.
Acknowledgements
Supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Department of Health and Human Services.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest in relation to this manuscript.
References
- 1.American Cancer Society. Cancer Facts and Figures 2011. Atlanta: American Cancer Society; 2010. [Google Scholar]
- 2.Singletary KW, Gapstur SM. Alcohol and breast cancer. Review of epidemiologic and experimental evidence and potential mechanisms. JAMA. 2001;286:2143–2151. doi: 10.1001/jama.286.17.2143. [DOI] [PubMed] [Google Scholar]
- 3.Ginsburg ES. Estrogen, alcohol and breast cancer risk. J Steroid Biochem Mol Biol. 1999;69:299–306. doi: 10.1016/s0960-0760(99)00047-3. [DOI] [PubMed] [Google Scholar]
- 4.Purohit V. Can alcohol promote aromatization of androgens to estrogens? A review Alcohol. 2000;20:123–127. doi: 10.1016/s0741-8329(00)00124-5. [DOI] [PubMed] [Google Scholar]
- 5.Wright RM, McManaman JL, Repine JE. Alcohol-induced breast cancer: a proposed mechanism. Free Radical Biol Med. 1999;26:348–354. doi: 10.1016/s0891-5849(98)00204-4. [DOI] [PubMed] [Google Scholar]
- 6.Goldfrank LR, Flomenbaum NE, Lewin NA, Howland MA, Hoffman RS, Nelson LS, editors. Goldfranks Toxicologic Emergencies, 7th edition. New York: Mc-Graw-Hill; 2002. [Google Scholar]
- 7.Stürmer T, Wang-Gohrke S, Arndt V, Boeing H, Kong Xkreienberg R, et al. Interaction between alcohol dehydrogenase II gene, alcohol consumption, and risk for breast cancer. Br J Cancer. 2002;87:519–523. doi: 10.1038/sj.bjc.6600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freudenheim JL, Ambrosone CB, Moysich KB, Vena JE, Graham S, Marshall JR, et al. Alcohol dehydrogenase 3 genotype modification of the association of alcohol consumption with breast cancer. Cancer Cause Control. 1999;10:369–377. doi: 10.1023/a:1008950717205. [DOI] [PubMed] [Google Scholar]
- 9.Triano EA, Slusher LB, Atkins TA, Beneski JT, Gestl SA, Zolfaghari R, et al. Class I alcohol dehydrogenase is highly expressed in normal human mammary epithelium but not in invasive breast cancer: implications for breast carcinogenesis. Cancer Res. 2003;63:3092–3100. [PubMed] [Google Scholar]
- 10.Choi J-Y, Abel J, Neuhaus T, Ko Y, Harth V, Hamaima N, et al. Role of alcohol and genetic polymorphisms of CYP2E1 and ALDH2 in breast cancer development. Pharmacogenetics. 2003;13:67–72. doi: 10.1097/00008571-200302000-00002. [DOI] [PubMed] [Google Scholar]
- 11.El-Rayes BF, Ali S, Heinbrun LK, Lababidi S, Bouwman D, Visscher D, et al. Cytochrome P450 and glutathione transferase expression in human breast cancer. Clin Cancer Res. 2003;9:1705–1709. [PubMed] [Google Scholar]
- 12.Iscan M, Klaavuniemi T, Coban T, Kapucuoğlu N, Pelkonen O, Raunio H. The expression of cytochrome P450 enzymes in human breast tumours and normal breast tissue. Breast Cancer Res Treat. 2001;70:47–54. doi: 10.1023/a:1012526406741. [DOI] [PubMed] [Google Scholar]
- 13.Kapucuoglu N, Coban T, Raunio H, Pelkonen O, Edwards RJ, Boobis AR, Iscan M. Immunohistochemical demonstration of the expression of CYP2E1 in human breast tumour and non-tumour tissues. Cancer Letters. 2003;196:153–159. doi: 10.1016/s0304-3835(03)00277-5. [DOI] [PubMed] [Google Scholar]
- 14.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trial. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 15.Thompson FE, Subar AF, Brown CC, et al. Cognitive research enhances accuracy of food frequency questionnaire reports: results of an experimental validation study. J Am Diet Assoc. 2002;102:212–225. doi: 10.1016/s0002-8223(02)90050-7. [DOI] [PubMed] [Google Scholar]
- 16.Subar AF, Ziegler RG, Thompson FE, et al. Is shorter always better? Relative importance of questionnaire length and cognitive ease on response rates and data quality for two dietary questionnaires. Am J Epidemiol. 2001;153:404–409. doi: 10.1093/aje/153.4.404. [DOI] [PubMed] [Google Scholar]
- 17.Subar AF, Thompson FE, Smith AF, et al. Improving food frequency questionnaires: a qualitative approach using cognitive interviewing. J Am Diet Assoc. 1995;95:781–788. doi: 10.1016/s0002-8223(95)00217-0. [DOI] [PubMed] [Google Scholar]
- 18.Stolzenberg-Solomon RZ, Chang S-C, Leitzmann MF, Johnson KA, Johnson C, Buys SS, et al. Folate intake, alcohol use, postmenopausal breast cancer risk in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Am J Clin Nutr. 2006;83:895–904. doi: 10.1093/ajcn/83.4.895. [DOI] [PubMed] [Google Scholar]
- 19.Visvanathan K, Crum RM, Strickland PT, You X, Ruczinski I, Berndt SI, et al. Alcohol dehydrogenase genetic polymorphisms, low-to-moderate alcohol consumption and risk of breast cancer. Alcohol Clin Exp Res. 2007;31:467–476. doi: 10.1111/j.1530-0277.2006.00334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hines LM, Hankinson SE, Smith-Warner SA, Spiegelman D, Kelsey KT, Colditz GA, Willett WC, Hunter DJ. A prospective study of alcohol consumption and ADH3 genotype on plasma steroid hormone levels and breast cancer risk. Cancer Epidemiol Biomarker Prevent. 2000;9:1099–1105. [PubMed] [Google Scholar]
- 21.Terry MB, Gammon MD, Zhang FF, Knight JA, Wang Q, Britton JA, et al. ADH3 genotype, alcohol intake and breast cancer risk. Carcinogenesis. 2006;27:840–847. doi: 10.1093/carcin/bgi285. [DOI] [PubMed] [Google Scholar]
- 22.Fan S, Meng Q, Gao B, Grossman J, Yadegari M, Goldberg ID, Rosen EM. Alcohol stimulates estrogen receptor signaling in human breast cancer cell lines. Cancer Res. 2000;60:5635–5639. [PubMed] [Google Scholar]
- 23.Nasca PC, Liu S, Baptiste MS, Kwon CS, Jacobson H, Metzger BB. Alcohol consumption and breast cancer: estrogen receptor status and histology. Am J Epidemiol. 1994;140:980–987. doi: 10.1093/oxfordjournals.aje.a117205. [DOI] [PubMed] [Google Scholar]
- 24.Enger SM, Paganini-Hill A, Longnecker MP, Bernstein L. Alcohol consumption and breast cancer oestrogen and progesterone receptor status. Br J Cancer. 1999;79:1308–1314. doi: 10.1038/sj.bjc.6690210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li CI, Malone KE, Porter PL, Weiss NS, Tang M-TC, Daling JR. The relationship between alcohol use and risk of breast cancer by histology and hormone receptor status among women 65–79 years of age. Cancer Epidemiol Biomarker Prevent. 2003;12:1061–1066. [PubMed] [Google Scholar]
- 26.Suzuki R, Ye W, Rylander-Rudqvist T, Saji S, Colditz GA, Wolk A. Alcohol and postmenopausal breast cancer risk defined by estrogen and progesterone receptor status: a prospective study. J Natl Cancer Inst. 2005;97:1601–1608. doi: 10.1093/jnci/dji341. [DOI] [PubMed] [Google Scholar]
- 27.Gapstur SM, Potter JD, Drinkard C, Folsom AR. Synergistic effect between alcohol and estrogen replacement therapy on risk of breast cancer differs by estrogen/progesterone receptor status in the Iowa Womens Health Study. Cancer Epidemiol Biomarker Prevent. 1995;4:313–318. [PubMed] [Google Scholar]
- 28.Jasmine QL, Freedman ND, Leitzmann MF, Brinton LA, Hoover RN, Hollenbeck AR. Alcohol and risk of breast cancer by histologic type and hormone receptor status in postmenopausal women. The NIH-AARP Diet and Health Study. Am J Epidemiol. 2009;170:308–317. doi: 10.1093/aje/kwp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crabb DW, Dipple KM, Thomasson HR. Alcohol sensitivity, alcohol metabolism, risk of alcohol metabolism, and the role of alcohol and aldehyde dehydrogenase genotypes. J Lab Clin Med. 1993:234–240. [PubMed] [Google Scholar]