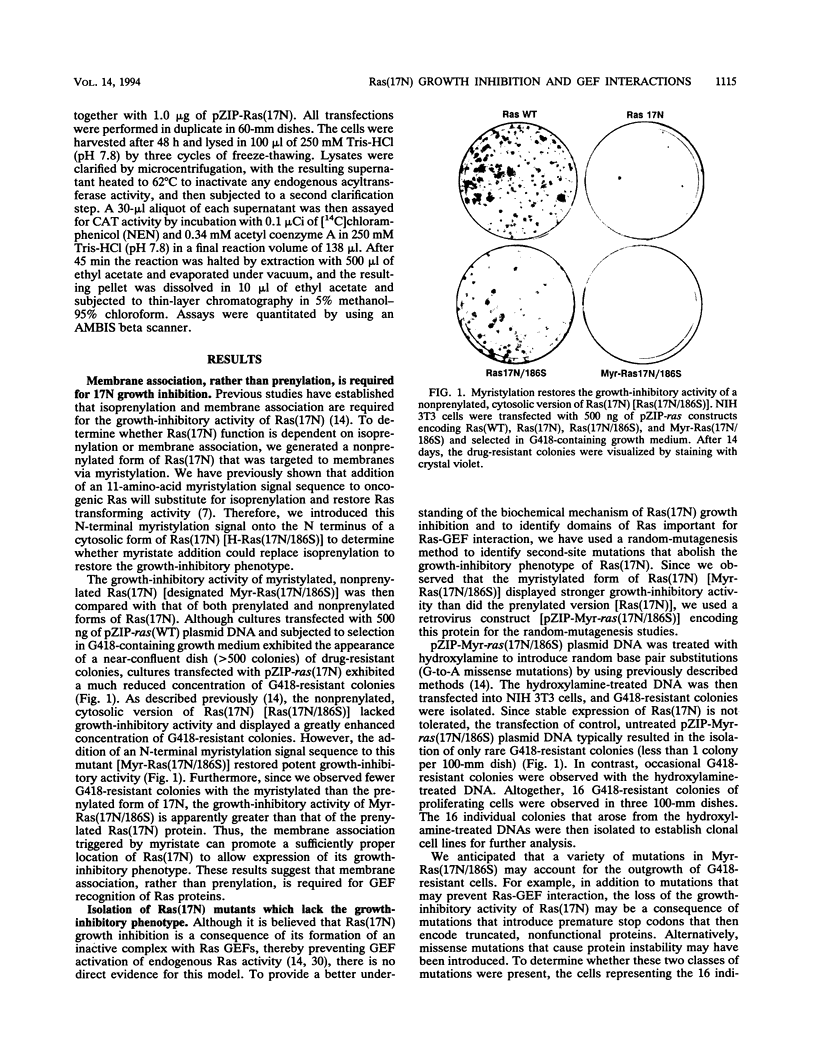
Abstract
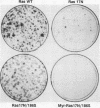
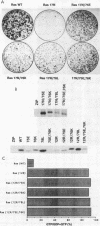
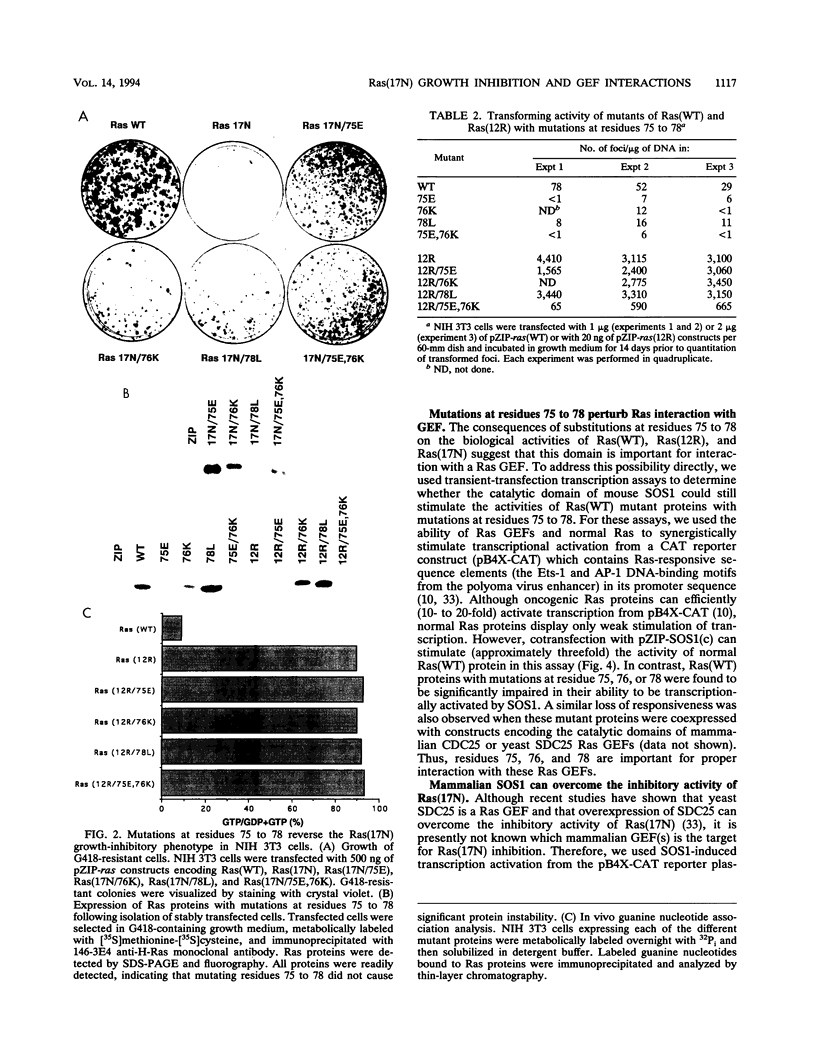
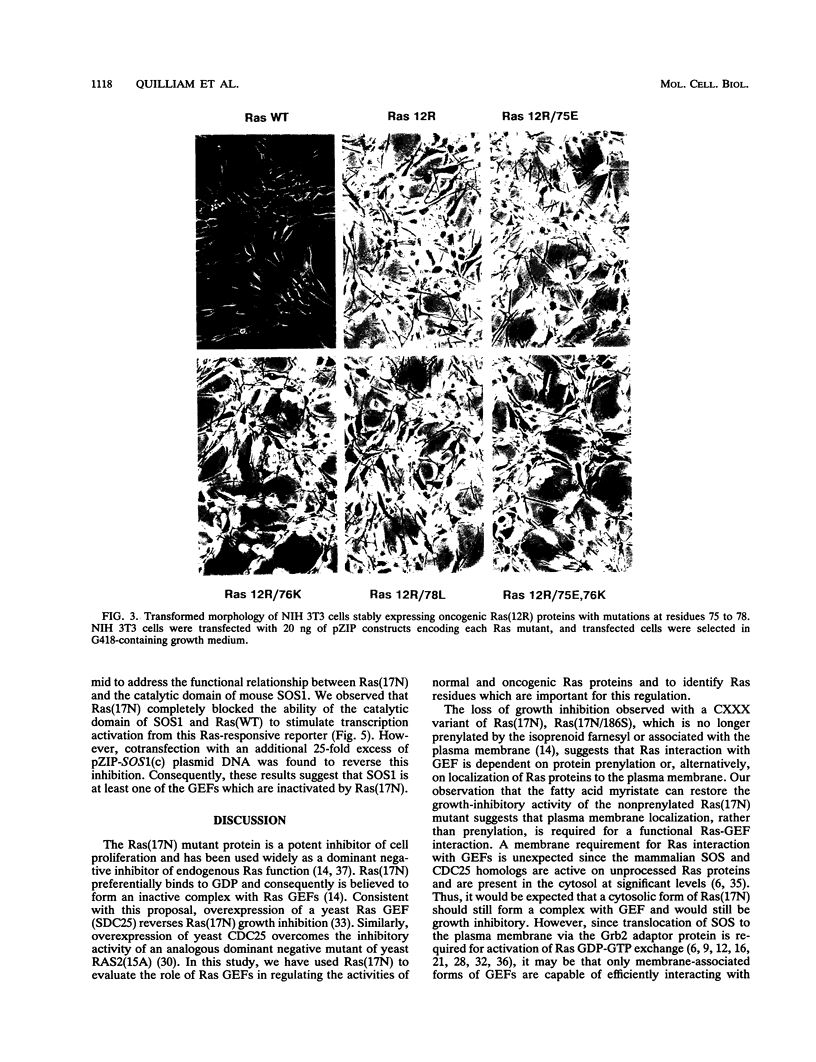
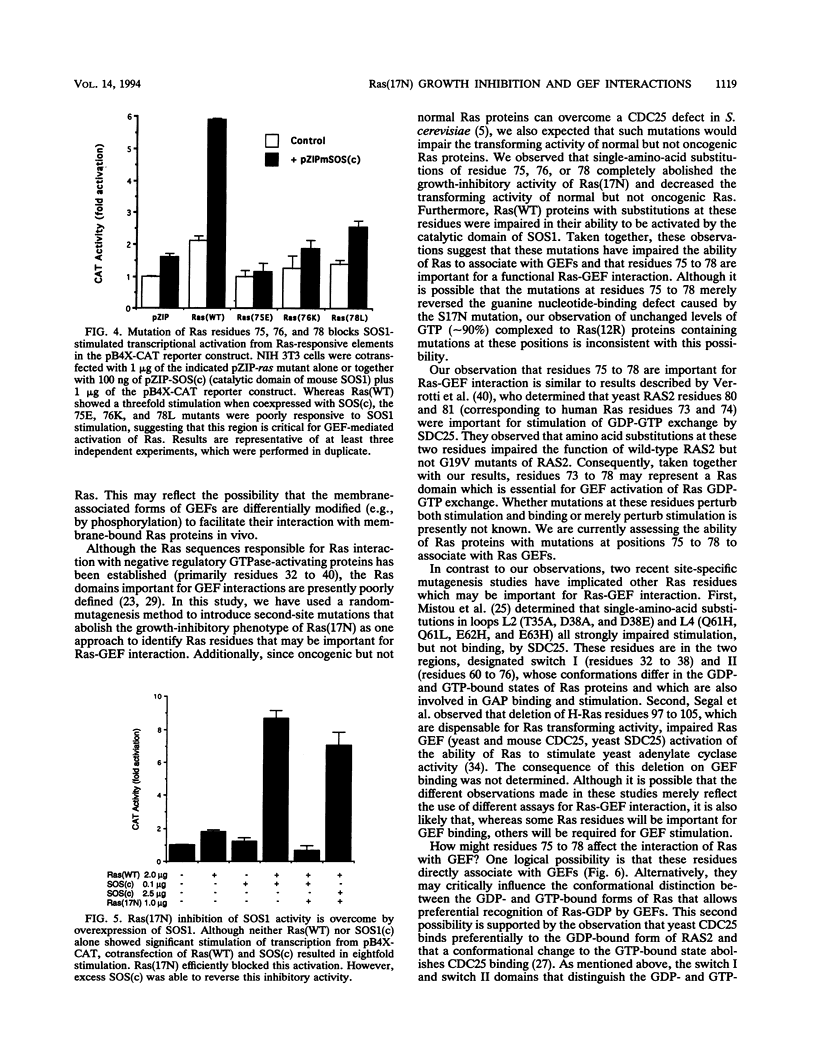
The Ras(17N) dominant negative antagonizes endogenous Ras function by forming stable, inactive complexes with Ras guanine nucleotide exchange factors (GEFs; e.g., SOS1). We have used the growth-inhibitory phenotype of Ras(17N) to characterize two aspects of Ras interaction with GEFs. First, we used a nonprenylated version of Ras(17N), designated Ras(17N/186S), which no longer associates with the plasma membrane and lacks the growth-inhibitory phenotype, to address the importance of Ras subcellular location and posttranslational modification for its interaction with GEFs. We observed that addition of an N-terminal myristylation signal to Ras(17N/186S) restored the growth-inhibitory activity of nonprenylated Ras(17N). Thus, membrane association, rather than prenylation, is critical for Ras interaction with Ras GEFs. Second, we used a biological selection approach to identify Ras residues which are critical for Ras(17N) growth inhibition and hence for interaction with Ras GEFs. We identified mutations at residues 75, 76, and 78 that abolished the growth-inhibitory activity of Ras(17N). Since GEF interaction is dispensable for oncogenic but not normal Ras function, our demonstration that single-amino-acid substitutions at these three positions impaired the transforming activity of normal but not oncogenic Ras provides further support for the role of these residues in Ras-GEF interactions. Finally, Ras(WT) proteins with mutations at these residues were no longer activated by mammalian SOS1. Altogether, these results suggest that the Ras intracellular location and Ras residues 75 to 78 are critical for Ras-GEF interaction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bokoch G. M., Der C. J. Emerging concepts in the Ras superfamily of GTP-binding proteins. FASEB J. 1993 Jun;7(9):750–759. doi: 10.1096/fasebj.7.9.8330683. [DOI] [PubMed] [Google Scholar]
- Bollag G., McCormick F. Regulators and effectors of ras proteins. Annu Rev Cell Biol. 1991;7:601–632. doi: 10.1146/annurev.cb.07.110191.003125. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bowtell D., Fu P., Simon M., Senior P. Identification of murine homologues of the Drosophila son of sevenless gene: potential activators of ras. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6511–6515. doi: 10.1073/pnas.89.14.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Buday L., Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993 May 7;73(3):611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- Buss J. E., Solski P. A., Schaeffer J. P., MacDonald M. J., Der C. J. Activation of the cellular proto-oncogene product p21Ras by addition of a myristylation signal. Science. 1989 Mar 24;243(4898):1600–1603. doi: 10.1126/science.2648572. [DOI] [PubMed] [Google Scholar]
- Casey P. J., Solski P. A., Der C. J., Buss J. E. p21ras is modified by a farnesyl isoprenoid. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8323–8327. doi: 10.1073/pnas.86.21.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P., Camonis J. H., Gale N. W., van Aelst L., Schlessinger J., Wigler M. H., Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993 May 28;260(5112):1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- Clark G. J., Quilliam L. A., Hisaka M. M., Der C. J. Differential antagonism of Ras biological activity by catalytic and Src homology domains of Ras GTPase activation protein. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):4887–4891. doi: 10.1073/pnas.90.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. D., Der C. J. Protein prenylation: more than just glue? Curr Opin Cell Biol. 1992 Dec;4(6):1008–1016. doi: 10.1016/0955-0674(92)90133-w. [DOI] [PubMed] [Google Scholar]
- Egan S. E., Giddings B. W., Brooks M. W., Buday L., Sizeland A. M., Weinberg R. A. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature. 1993 May 6;363(6424):45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- Feig L. A., Cooper G. M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988 Aug;8(8):3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L. A. The many roads that lead to Ras. Science. 1993 May 7;260(5109):767–768. doi: 10.1126/science.8484117. [DOI] [PubMed] [Google Scholar]
- Fujioka H., Kaibuchi K., Kishi K., Yamamoto T., Kawamura M., Sakoda T., Mizuno T., Takai Y. Transforming and c-fos promoter/enhancer-stimulating activities of a stimulatory GDP/GTP exchange protein for small GTP-binding proteins. J Biol Chem. 1992 Jan 15;267(2):926–930. [PubMed] [Google Scholar]
- Gale N. W., Kaplan S., Lowenstein E. J., Schlessinger J., Bar-Sagi D. Grb2 mediates the EGF-dependent activation of guanine nucleotide exchange on Ras. Nature. 1993 May 6;363(6424):88–92. doi: 10.1038/363088a0. [DOI] [PubMed] [Google Scholar]
- Gulbins E., Coggeshall K. M., Baier G., Katzav S., Burn P., Altman A. Tyrosine kinase-stimulated guanine nucleotide exchange activity of Vav in T cell activation. Science. 1993 May 7;260(5109):822–825. doi: 10.1126/science.8484124. [DOI] [PubMed] [Google Scholar]
- Howe L. R., Marshall C. J. Identification of amino acids in p21ras involved in exchange factor interaction. Oncogene. 1993 Sep;8(9):2583–2590. [PubMed] [Google Scholar]
- Huang D. C., Marshall C. J., Hancock J. F. Plasma membrane-targeted ras GTPase-activating protein is a potent suppressor of p21ras function. Mol Cell Biol. 1993 Apr;13(4):2420–2431. doi: 10.1128/mcb.13.4.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Kaibuchi K., Masuda T., Yamamoto T., Matsuura Y., Maeda A., Shimizu K., Takai Y. The post-translational processing of ras p21 is critical for its stimulation of mitogen-activated protein kinase. J Biol Chem. 1993 Feb 15;268(5):3025–3028. [PubMed] [Google Scholar]
- Kavounis C., Verrotti A. C., De Vendittis E., Bozopoulos A., Di Blasi F., Zahn R., Crechet J. B., Parmeggiani A., Tsernoglou D., Fasano O. Role of glycine-82 as a pivot point during the transition from the inactive to the active form of the yeast Ras2 protein. FEBS Lett. 1991 Apr 9;281(1-2):235–239. doi: 10.1016/0014-5793(91)80401-n. [DOI] [PubMed] [Google Scholar]
- Li N., Batzer A., Daly R., Yajnik V., Skolnik E., Chardin P., Bar-Sagi D., Margolis B., Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993 May 6;363(6424):85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- Magee T., Newman C. The role of lipid anchors for small G proteins in membrane trafficking. Trends Cell Biol. 1992 Nov;2(11):318–323. doi: 10.1016/0962-8924(92)90172-j. [DOI] [PubMed] [Google Scholar]
- Marshall M. S. The effector interactions of p21ras. Trends Biochem Sci. 1993 Jul;18(7):250–254. doi: 10.1016/0968-0004(93)90175-m. [DOI] [PubMed] [Google Scholar]
- Martegani E., Vanoni M., Zippel R., Coccetti P., Brambilla R., Ferrari C., Sturani E., Alberghina L. Cloning by functional complementation of a mouse cDNA encoding a homologue of CDC25, a Saccharomyces cerevisiae RAS activator. EMBO J. 1992 Jun;11(6):2151–2157. doi: 10.1002/j.1460-2075.1992.tb05274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistou M. Y., Jacquet E., Poullet P., Rensland H., Gideon P., Schlichting I., Wittinghofer A., Parmeggiani A. Mutations of Ha-ras p21 that define important regions for the molecular mechanism of the SDC25 C-domain, a guanine nucleotide dissociation stimulator. EMBO J. 1992 Jul;11(7):2391–2397. doi: 10.1002/j.1460-2075.1992.tb05303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Kaibuchi K., Yamamoto T., Kawamura M., Sakoda T., Fujioka H., Matsuura Y., Takai Y. A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-Ki-ras p21 and rhoA p21. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6442–6446. doi: 10.1073/pnas.88.15.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder T., Fürst P. The Saccharomyces cerevisiae CDC25 gene product binds specifically to catalytically inactive ras proteins in vivo. Mol Cell Biol. 1992 May;12(5):2091–2099. doi: 10.1128/mcb.12.5.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier J. P., Raabe T., Henkemeyer M., Dickson B., Mbamalu G., Margolis B., Schlessinger J., Hafen E., Pawson T. A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell. 1993 Apr 9;73(1):179–191. doi: 10.1016/0092-8674(93)90170-u. [DOI] [PubMed] [Google Scholar]
- Polakis P., McCormick F. Structural requirements for the interaction of p21ras with GAP, exchange factors, and its biological effector target. J Biol Chem. 1993 May 5;268(13):9157–9160. [PubMed] [Google Scholar]
- Powers S., O'Neill K., Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey I., Schweighoffer F., Barlat I., Camonis J., Boy-Marcotte E., Guilbaud R., Jacquet M., Tocque B. The COOH-domain of the product of the Saccharomyces cerevisiae SCD25 gene elicits activation of p21-ras proteins in mammalian cells. Oncogene. 1991 Feb;6(2):347–349. [PubMed] [Google Scholar]
- Rozakis-Adcock M., Fernley R., Wade J., Pawson T., Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993 May 6;363(6424):83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- Schweighoffer F., Cai H., Chevallier-Multon M. C., Fath I., Cooper G., Tocque B. The Saccharomyces cerevisiae SDC25 C-domain gene product overcomes the dominant inhibitory activity of Ha-Ras Asn-17. Mol Cell Biol. 1993 Jan;13(1):39–43. doi: 10.1128/mcb.13.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Willumsen B. M., Levitzki A. Residues crucial for Ras interaction with GDP-GTP exchangers. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5564–5568. doi: 10.1073/pnas.90.12.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou C., Farnsworth C. L., Neel B. G., Feig L. A. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature. 1992 Jul 23;358(6384):351–354. doi: 10.1038/358351a0. [DOI] [PubMed] [Google Scholar]
- Simon M. A., Dodson G. S., Rubin G. M. An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to sevenless and Sos proteins in vitro. Cell. 1993 Apr 9;73(1):169–177. doi: 10.1016/0092-8674(93)90169-q. [DOI] [PubMed] [Google Scholar]
- Stacey D. W., Feig L. A., Gibbs J. B. Dominant inhibitory Ras mutants selectively inhibit the activity of either cellular or oncogenic Ras. Mol Cell Biol. 1991 Aug;11(8):4053–4064. doi: 10.1128/mcb.11.8.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouten P. F., Sander C., Wittinghofer A., Valencia A. How does the switch II region of G-domains work? FEBS Lett. 1993 Mar 29;320(1):1–6. doi: 10.1016/0014-5793(93)81644-f. [DOI] [PubMed] [Google Scholar]
- Valencia A., Chardin P., Wittinghofer A., Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991 May 14;30(19):4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- Verrotti A. C., Créchet J. B., Di Blasi F., Seidita G., Mirisola M. G., Kavounis C., Nastopoulos V., Burderi E., De Vendittis E., Parmeggiani A. RAS residues that are distant from the GDP binding site play a critical role in dissociation factor-stimulated release of GDP. EMBO J. 1992 Aug;11(8):2855–2862. doi: 10.1002/j.1460-2075.1992.tb05353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Mosteller R. D., Sanyal P., Gonzales E., McKinney D., Dasgupta C., Li P., Liu B. X., Broek D. Identification of a mammalian gene structurally and functionally related to the CDC25 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7100–7104. doi: 10.1073/pnas.89.15.7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer F. Three-dimensional structure of p21H-ras and its implications. Semin Cancer Biol. 1992 Aug;3(4):189–198. [PubMed] [Google Scholar]