Abstract
Neurotrauma, as in the case of traumatic brain injury, promotes protease over-activation characterized by the select fragmentation of brain proteins. The resulting polypeptides are indicators of biochemical processes, which can be used to study post-injury dynamics and may also be developed into biomarkers. To this end, we devised a novel mass spectrometry approach to characterize post-injury calpain proteolytic processing of myelin basic protein (MBP), a biomarker of brain injury that denotes white matter damage and recovery. Our approach exceeds conventional immunological assays in its deconvolution of multiple protein isoforms, its absolute quantification of proteolytic fragments and its polypeptide selectivity. We quantified and characterized post-injury proteolytic processing of all MBP isoforms identified in adult rat cortex. Further, the translation of calpain-cleaved MBP into CSF was verified following brain injury. We ascertained that the exon-6 sequence of MBP resulted in a characteristic shift in gel migration for intact and fragmented protein alike. We also found evidence for a second post-TBI cleavage event within exon-2 and for the dimerization of the post-TBI 4.3 kDa fragment. Ultimately, the novel methodology described here can be used to study MBP dynamics and other similar proteolytic events of relevance to brain injury and other CNS processes.
Keywords: biomarkers, mass spectrometry, myelin basic protein, neurotrauma, proteolysis, quantification
Myelin is a multilamellar membrane that sheaths axons in the white matter of the CNS, which serves to protect and improve signal conductance (Arroyo and Scherer 2000). The membrane is composed of lipid layers interspersed between protein layers comprised near equally by myelin basic protein (MBP), proteolipid protein, and myelin-oligodendrocyte glycoprotein (Norton and Cammer 1984). Each of the three myelin proteins is expressed as multiple isoforms (Voskuhl et al. 1994) that speculatively have unique functions in the formation and maintenance of myelin. In particular, MBP isoform dynamics vary in association with myelin formation, compaction and stabilization from embryonic development on throughout life (Kruger et al. 1999), and is of interest to the study of neurological development (Boggs 2006), neurodegenerative disease (Schaecher et al.2001; Boggs 2006) and neurotrauma (Liu et al. 2006a).
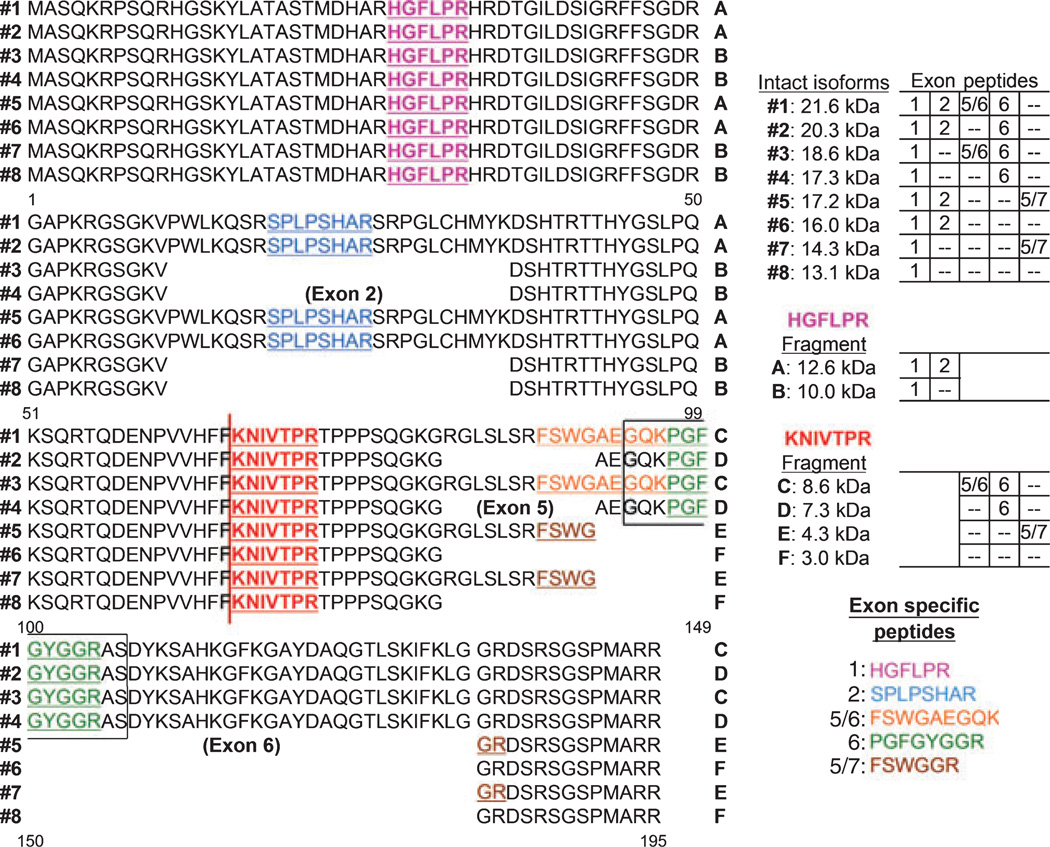
Myelin basic protein isoforms are derived from differential splicing of the MBP gene (Harauz et al. 2004; Boggs 2006) with variations in the translation of exons 2, 5, and 6 (Kimura et al. 1998). Eight possible isoforms (Fig. 1) are derived, of which all have been identified in mice at the mRNA level (Nakajima et al. 1993), and five (21.6, 18.6, 17.3, 17.2, and 14.0 kDa) at the protein level (Givogri et al. 2000). Rat and mouse MBP are 98% homologous, though only four rat isoforms (21.5, 18.5, 17, and 14 kDa) are reported at the protein level (Boggs et al. 2000; Akiyama et al. 2002). It is known that the isoform profile varies temporally with respect to the functional relevance of the spliced regions (Boggs 2006), as classified into three groups. The first group of isoforms (2, 4, 6, and 8 in Fig. 1) lack exon 5 and are linked with embryonic myelin formation (Nakajima et al. 1993). Isoforms in the second group (1, 2, 5, and 6) contain exon 2 and are linked with postnatal myelin development and with remyelination in aged (Kruger et al. 1999; Boggs et al. 2000; Akiyama et al. 2002) and diseased rats (Norton and Cammer 1984; Capello et al. 1997). Isoforms of the third group (3, 4, 7, and 8) lack exon 2 and are associated with maturation of myelin up through adolescence. The exon-2 sequence also contains a phosphorylation site that has been linked with MBP trafficking into cells and is a possible control for temporal and spatial dynamics (Boggs et al. 2000; Boggs 2006). The functional significance of the exon-6 sequence is not well understood. However, isoforms that lack the exon-6 sequence are less basic (Kruger et al. 1999; Akiyama et al. 2002) and thereby interact with proteolipid protein (Wood and Moscarello 1989) for the maintenance of myelin integrity. This may also explain the greater abundance of those isoforms that lack the exon-6 sequence in the developed CNS.
Fig. 1.
Rat MBP isoforms and marker peptides. Indicated are the sequences for each isoform (#1–8), the calpain cleavage site (vertical red line) with associated proteolytic fragments (A–F), and the select marker peptides of myelin basic protein (underlined and colored). The molecular mass and associated variable transcription regions of the eight known isoforms and six predicted calpain fragments of MBP are tabulated. Two peptides used for IDA quantification analysis, HGFLPR (magenta) and KNIVTPR (red), are shown in bold. Five exon specific peptides used to differentiate the MBP isoforms are listed. The boxed sequence indicates the epitope for the MBP antibody used in this and earlier studies (Chemicon MAB381).
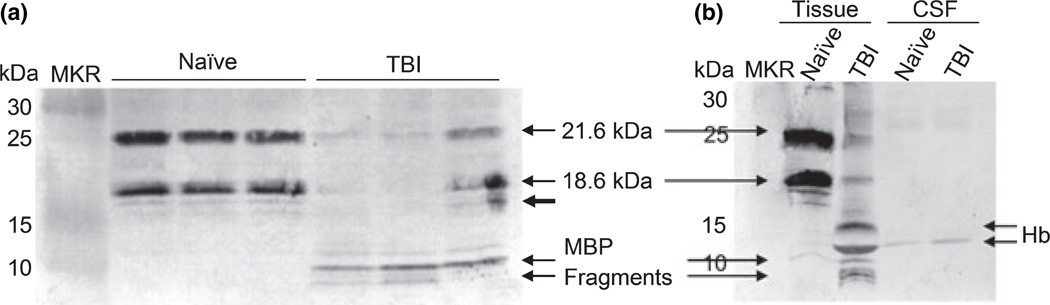
Myelin basic protein has been considered a biomarker of head injury for close to 30 years (Thomas et al. 1978), yet it has not been widely studied in neurotrauma because of a lack in assay sensitivity (Berger et al. 2006). Recently, colleagues at the University of Florida reported fragmentation of MBP following traumatic brain injury (TBI), and linked the biochemical process with the protease calpain (Liu et al. 2006b), already known to proteolyze MBP in association with multiple sclerosis (MS) (Schaecher et al. 2001). In their study, they identified a major calpain cleavage site (between Phe114 and Lyc115), but the characterization of post-TBI MBP fragmentation by immunoblotting analysis was complicated by the multiple isoforms present (Fig. 2a). Further, the assay used was either not sensitive or selective enough to detect the resulting cleavage products in CSF, an important next step for biomarker validation (Fig. 2b and unpublished data). To this end, we recruited mass spectrometry as an alternative technology for the characterization of MBP proteolysis following brain injury. The ensuing methodology would also hold potential for characterization of other proteolytic processes common in CNS injury. Products of proteolysis are a particularly useful class of markers as they often transition effectively into biofluids and denote mechanisms of cell stress and death.
Fig. 2.
Immunoblot analysis of proteolyzed MBP following TBI. (a) Naïve and TBI rat cortex samples resolved by gel electrophoresis were probed with an antibody (MAB381; Chemicon) selective for the exon-6 coded region of MBP. The Western blot illustrates the dramatic reduction of the 21.6 and 18.6 kDa isoforms of MBP following TBI, with a concomitant rise in MBP fragments at and below an Ma of 10. This data correlates with that ofAkiyama et al. (2002) and Liu et al. (2006a), where the same antibody was used to characterize MBP in rat cortex. (b) CSF and TBI samples were resolved along side one another by gel electrophoresis and were probed with the MBP antibody. The MBP fragments observed in the TBI tissue were not detected in TBI CSF. The only band (Ma of 12) in the CSF samples is adjacent to a non-specifically detected band of hemoglobin (Hb) observed in TBI tissue (blood contamination).
In this study, we adapted isotope dilution analysis (IDA) (Barr et al. 1996; Gerber et al. 2003) for the novel purpose of quantifying proteolytic processes, in particular the calpain-induced fragmentation of MBP following TBI in rats. We also devised a second tandem mass spectrometry (MSMS) strategy to selectively characterize the complement of MBP isoforms and associated fragments present in naïve (uninjured) and TBI adult rat cortex. Correlation of the resulting quantitative and MBP isoform data provided a detailed profile of MBP dynamics in rat brain 2 days post-TBI. Further, we demonstrated femtomole (fmol) quantification of a calpain-specific MBP fragment in CSF of TBI injured rats, identified with definitive tandem mass spectra.
Materials and methods
Traumatic brain injury model
All procedures complied with the guidelines of the University of Florida Institutional Animal Care and Use Committee. Ten-week-old male Sprague–Dawley rats (Harlem, Indianapolis, IN, USA) were anesthetized with 4% and maintained at 2% isoflurane while an ipsilateral 0.5 mm diameter craniotomy was performed. Severe TBI was induced in the injured group via a pneumatically controlled cortical impact device, penetrating 1.6 mm into the exposed cortex at a velocity of 3.5 m/s. The animals were killed 2 days following injury, with CSF and ipsilateral cortical tissue collected (snap frozen immediately). Clarified CSF was pooled (n = 7, 12 µL each) and used directly for this study. Frozen cortex tissue was ground to a powder on dry ice and suspended in 300 µL of a 1% Triton X-100 lysis buffer containing 150 mmol/L NaCl, 50 mmol/L EDTA, 50 mmol/L EGTA, 1 mmol/L dithiothreitol, 1 mmol/L NaVO3, and a Complete Mini Protease Inhibitor Cocktail tablet (Roche, Indianapolis, IN, USA). Cells were lysed for 3 h at 4°C. Lysates were centrifuged at 14 000 g and 4°C for 10 min, and then passed through a 0.1 µm Ultrafree filter unit (Millipore, Billerica, MA, USA) to remove DNA, lipids, and particulates. Protein concentration was determined by Bio-Rad DC Protein Assay (Hercules, CA, USA).
MBP Western blotting
Naïve and TBI tissue (20 µg) and CSF (20 µL) were separated on a Novex (Invitrogen, Carlsbad, CA, USA) 10-20% Tris–glycine (1 mm, 10-well) gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad) by the semi-dry method. Following blocking in 5% milk, the membrane was coated with 200 µL of monoclonal anti-MBP (MAB381; Chemicon, Temecula, CA, USA) in 10 mL of 5% milk. Biotin conjugated anti-mouse IgG (Sigma-Aldrich, St Louis, MO, USA) secondary and alkaline phosphatase conjugated streptavadin (GE Healthcare, Piscataway, NJ, USA) tertiary antibodies were also applied in 5% milk. The Western blot was developed by nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (KPL, Gaithersburg, MD, USA).
Peptide synthesis
Native and isotopically labeled versions of the peptides HGFLPR and KNIVTPR were prepared using FMOC chemistry (University of Florida Interdisciplinary Center for Biotechnology Research, Gainesville, FL, USA). The isotopic amino acids 13C6/15N L-leucine (Δm/z = 7) and 13C5 L-valine (Δm/z = 5) were purchased at 98% purity from Cambridge Isotope Laboratories (Andover, MA, USA), and were used to label HGFL*PR and KNIV*TPR, respectively. Peptides were purified by reversed-phase liquid chromatography (RPLC) to 99% purity as confirmed by mass spectrometry.
Gel electrophoresis
Samples mixed with Laemmli buffer (Bio-Rad) and 10 mmol/L dithiothreitol were heated to 80°C for 2 min prior to separation on Invitrogen Tris–glycine Novex polyacrylamide gels (1 mm, 10-well). Rainbow molecular mass marker (GE Healthcare) was used to indicate apparent molecular mass (Ma), as calculated for extracted gel slices using Phoretix 1D gel processing software (Nonlinear USA, Durham, NC, USA). Bovine MBP (Sigma Aldrich) was purified by cation exchange chromatography to isolate the 18.6 kDa MBP isoform as a marker and positive control. Gels were visualized by staining with concentrated Bio-Rad Coomassie Blue R250 for 20 min and destained in 40% ethanol/10% acetic acid in water for 2 h.
In-gel clostripain digestion
Excised gel slices were cut into 1-mm cubes and washed in HPLC grade water (Burdick & Jackson, Muskegon, MI, USA), destained in 50 : 50 100 mmol/L NH4HCO3/spectroscopy grade acetonitrile (Burdick & Jackson) and dried in 100% acetonitrile followed by speed vacuum. Dried gel pieces were re-hydrated with 20 µL of a 25 ng/µL clostripain solution (Worthington Biochemical, Lakewood, NJ, USA) made with an NH4HCO3 buffer (100 mmol/L) with CaCl2 (20 mmol/L) and the two isotopically labeled internal standard peptides (10 nmol/L). Digestion was performed overnight at 37°C. The resulting peptide supernatant and two 50% acetonitrile extractions were pooled and dried by speed vacuum. Peptides were resuspended in 20 µL of HPLC grade 4% acetonitrile/0.1% formic acid (J. T. Baker, Phillipsburg, NJ, USA).
Capillary reversed-phase liquid chromatography
Two microliters of each sample was loaded by autosampler onto a 100 µm diameter × 5 cm long column of 3-µm C-18 reversed-phase particles (Agilent, Palo Alto, CA, USA) packed behind an Upchurch (Oak Harbor, WA, USA) 0.5-µm polyetherketone microfilter assembly operated at a flow rate of 600 nL/min. A rapid 10 min gradient separation from 4% to 30% acetonitrile (0.1% formic acid) was performed for quantitative analysis. A longer 40 min gradient separation from 4% to 60% acetonitrile was performed for MBP isoform characterization. Chromatographic effluent was directly electrosprayed into a Thermo Electron (San Jose, CA, USA) LCQ Deca XP Plus™ ion trap mass spectrometer.
Quantitative isotope dilution analysis
Control and TBI samples (50 µg for cortical tissue and 20 µL for CSF) were resolved by gel electrophoresis. Each gel lane was sliced into 1-mm bands below a relative molecular mass of 30. For each gel slice, we performed an in-gel digestion using clostripain to which a fixed amount of isotopically labeled HGFLPR and KNIVTPR peptides was added as internal standards. A custom mass spectrometry method was developed to sequentially perform MSMS analysis for the endogenous and isotopically labeled HGFLPR and KNIVTPR peptides. Chromatographic peaks were identified after filtering for the doubly charged precursor ion and the respective dominant singly charged fragment ion (the y5 ion of HGFLPR and y6 ion of KNIVTPR). Peptide selectivity was confirmed by the observed tandem mass spectrum. We then multiplied the peak area ratio of endogenous-to-isotopically labeled peptide by the amount of internal standard (14 fmol for tissue and 18.6 fmol for CSF) to determine the quantity of endogenous peptide sampled (one-tenth of total digest). Data was reported as the mean with the range indicated by error bars. Statistical evaluation was performed with a non-parametric one-tailed Mann–Whitney U-test (n of 3), anticipating a decrease in intact MBP following injury.
MBP isoform characterization
As above, 50 µg of control and TBI samples were resolved by gel electrophoresis, with gel slices extracted below an Ma of 30. In-gel trypsin (Promega, Madison, WI, USA) digestion was performed as described for clostripain, but without CaCl2 and internal standard peptides. Five of 30 µL for each digest was analyzed by RPLC-MSMS. A custom MSMS method was devised to sequentially analyze five target peptides selective for the variable transcription regions of MBP: exon encoding sequences 1, HGFLPR; 2, SPLPSHAR; tandem 5/6, FSWGAEGQK; 6, PGFGYGGR; tandem 5/7, FSWGGR (the short exon-5 coded region was best characterized by peptides overlapping with adjacent sequences). Chromatographic peaks were identified after filtering for the precursor ion and dominant fragment ion of each peptide (confirmed by tandem mass spectra). Peak areas for each target peptide were averaged from replicate digests (same Ma, n of 3) and normalized across all samples to adjust for differences in extraction, separation, and ionization efficiencies between peptides.
Determination of peptide specificity and tandem mass spectrum selectivity
Peptide sequence specificity was evaluated by BLAST analysis (NCBI, Bethesda, MD, USA) with a word size of 2 and an expect value of 20 000 searched against a non-redundant RefSeq (NCBI) database (Release 22) filtered for rattus norvegicus proteins. Separately, tandem mass spectrum selectivity was evaluated by Mascot search (Matrix Science, Boston, MA, USA). Raw MSMS data were converted to DTA file format (extract_msn.exe; Thermo Electron) for each peptide. Mascot parameters included Arg-C (clostripain) enzyme selectivity and a mass tolerance of 0.75 Da for peptide and MSMS values. Searches were performed against an Integr8 non-redundant rattus norvegicus (species 122) protein database (Release 59).
Results
Isotope dilution mass spectrometry quantification
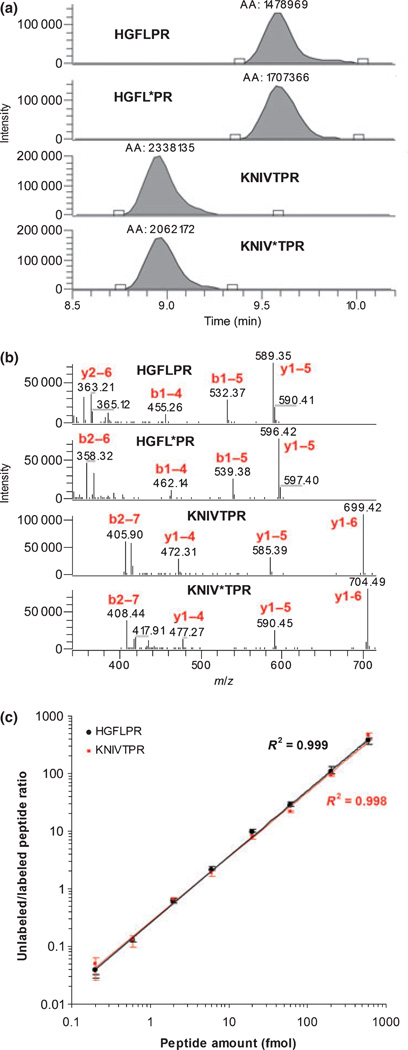
We adapted IDA to selectively quantify proteolytic cleavage events, in our case calpain-induced MBP fragmentation, by targeting the resulting N-terminal peptide (KNIVTPR). It is first important to demonstrate that the target peptide is readily detected. We analyzed tryptic digests of purified MBP by data-dependent mass spectrometry, which biases detection of peptides that ionize efficiently (Dongré et al. 1997; Aebersold and Mann 2003). We observed a strong signal and detailed tandem mass spectrum for the related NIVTPR peptide. HGFLPR was another peptide that was readily detected, which was common to all isoforms of MBP(exon-1). Pre-evaluation, however, may prove more difficult for other cleavage specific peptides, as discussed later. Native and isotopically labeled forms of the KNIVTPR peptide were synthesized with a mass difference of 5 Da (easily resolved by the mass spectrometer despite simultaneous chromatographic elution) (Gerber et al. 2003). We also synthesized native and isotopically labeled forms (7 Da mass difference) of the peptide HGFLPR to quantify intact MBP and MBP fragments complementary to those detected with KNIVTPR. We used the native peptides for assay development, while the isotopically labeled peptides served as internal standards for quantification. RPLC and MSMS parameters were optimized with mixtures of the four synthesized peptides to produce a rapid 10 min RPLC gradient that resolved HGFLPR and KNIVTPR (Fig. 3a) with subsequent selective detection (Fig. 3b). Data were filtered by the mass-to-charge (m/z) values for the intact peptide ion (precursor ion) and most abundant fragment ion to generate a chromatographic peak for each peptide. The moles of native peptide were quantified by multiplying the peak area ratio (native to internal standard) by the known concentration of internal standard. The method also assumed a linear dynamic range, which was validated across four orders in magnitude and with a detection limit of less than 1 fmol (Fig. 3c).
Fig. 3.
Data for peptide quantification by tandem mass spectrometry. (a) Chromatographic peak data for the peptides HGFLPR and KNIVTPR and their corresponding isotopically labeled forms (denoted by *) were generated from corresponding tandem mass spectra. Chromatographic peaks were generated by plotting the signal of the most intense fragment ion (y1–5 for HGFLPR and y1–6 for KNIVTPR) from tandem mass spectra filtered by the m/z of the precursor ion, with a seven-point Gaussian smoothing algorithm applied. (b) Collected tandem mass spectra were used to confirm peptide identity. Data were acquired with a mixture of the four peptides, each at a concentration of 1 nmol/L. (c) Dynamic range for MBP peptide quantification by tandem mass spectrometry. From 0.2 to 600 fmol of HGFLPR and KNIVTPR was quantified relative to 20 fmol of their respective isotopically labeled internal standards. The chromatographic peak ratio remained linear across four orders of magnitude, with a detection limit in the attomole range. Mean values are presented with the range denoted by error bars (n = 3).
MBP quantification in brain
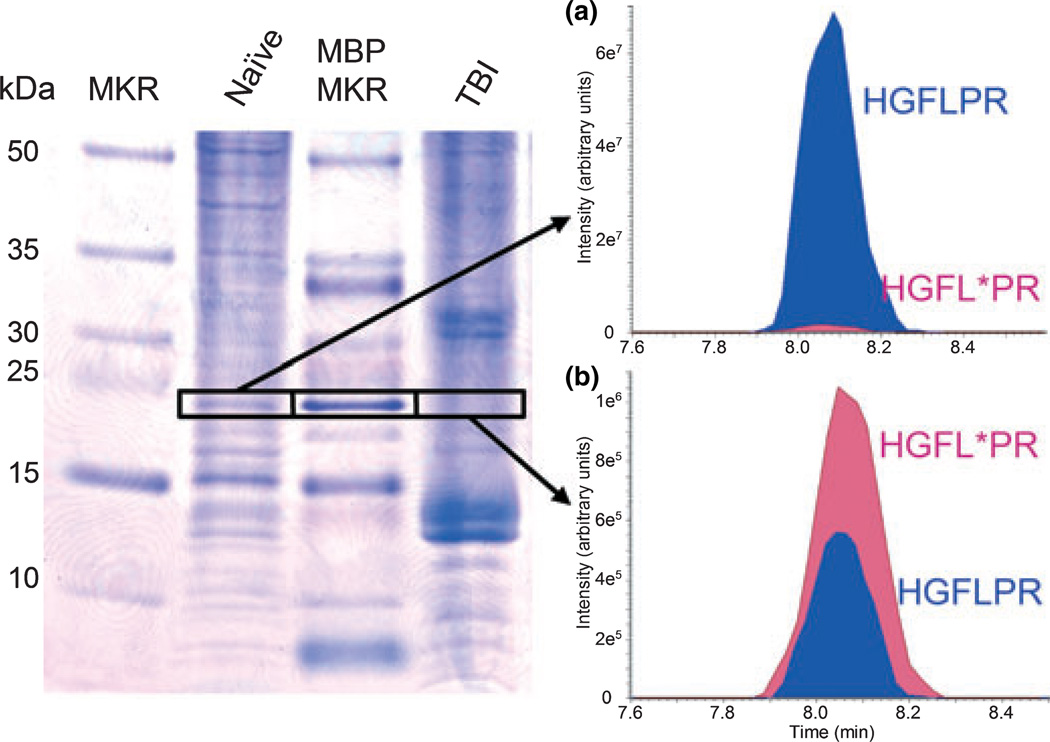
Our next step was to validate our IDA method with peptide endogenous to brain samples. Protein lysates of naïve and TBI rat cortex were resolved by polyacrylamide gel electrophoresis. Purified MBP in a separate gel lane served to mark the location of the endogenous 18.6 kDa MBP isoform and as a quality control sample (Fig. 4). For our application, we used the enzyme clostripain for in-gel digestion, which unlike conventional trypsin would preserve the N-terminal lysine of the KNIVTPR peptide. We quantified a reduction in endogenous HGFLPR from 510 (naïve) to 6.8 fmol (TBI) in gel slices adjacent to the purified MBP marker (Fig. 4). With one peptide copy per protein, the data denoted a 75-fold reduction in the amount of the 18.6 kDa MBP isoform at 2 days following TBI.
Fig. 4.
Quantifying the loss of the 18.6 kDa MBP isoform after TBI by isotope dilution mass spectrometry. In-gel clostripain digestion of the boxed gel slices was performed in a 7 nmol/L solution of the isotopically labeled HGFL*PR peptide (internal standard). Based on the ratio of endogenous HGFLPR to isotopically labeled standard, we calculated that the endogenous peptide (and associated intact MBP 18.6 kDa protein) was reduced 75-fold from (a) 510 fmol in naïve rat cortex (peak ratio of 37) to (b) 6.8 fmol in TBI rat cortex (peak ratio of 0.48).
Profiling neurotrauma induced MBP proteolysis
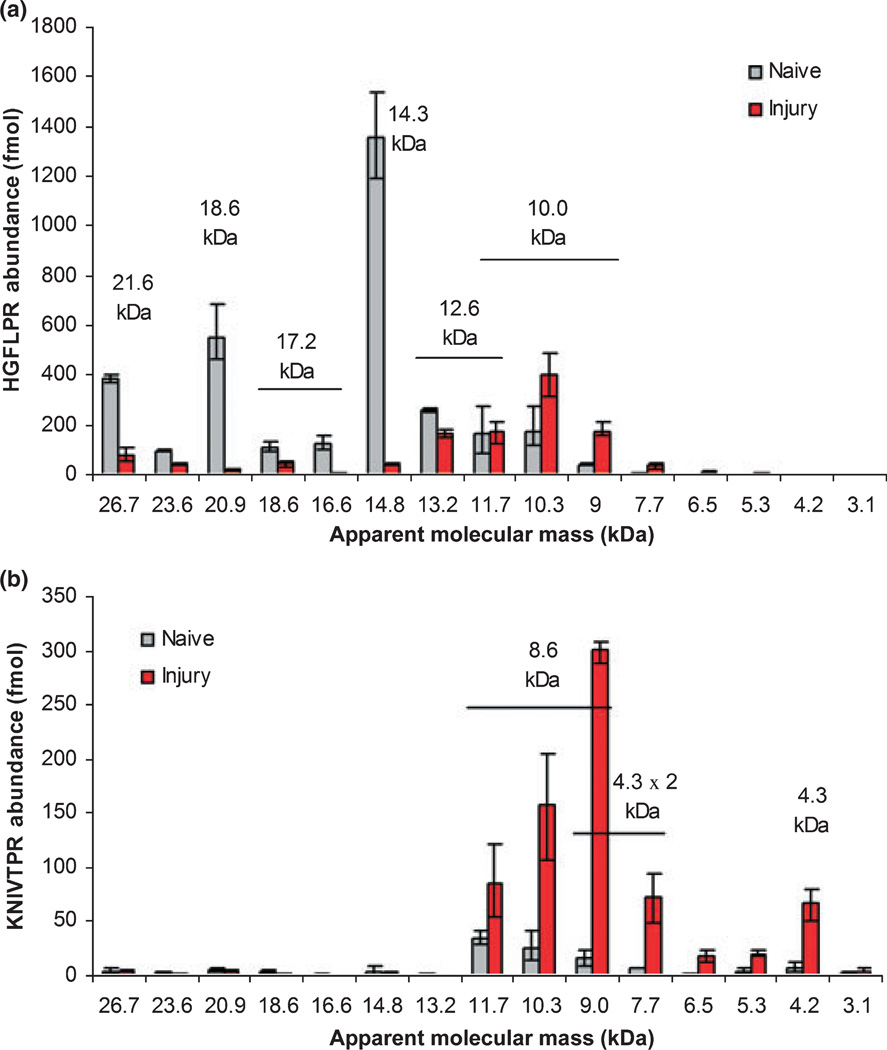
A set of naïve and TBI samples were resolved by polyacrylamide gel electrophoresis (n of 3), with uniform gel slices excised below the 30 kDa molecular mass marker to the bottom of the gel (Fig. 5). Endogenous HGFLPR and KNIVTPR were quantified by IDA analysis as graphed in Fig. 6 for the Ma of each gel slice. Following TBI, the amount of HGFLPR at Ma values corresponding to intact MBP isoforms was several-fold less than for naïve samples, while HGFLPR levels increased at lower Ma values as indicative of MBP fragmentation. In comparison, KNIVTPR was not present at Ma values for intact MBP, speaking to its selectivity of calpain-induced cleavage products. Indeed, calpain-cleavage was necessary to bisect the native NPVVHFFKNIVTPR clostripain peptide to form KNIVTPR. KNIVTPR was increased several-fold at lower Ma values where MBP fragments were expected.
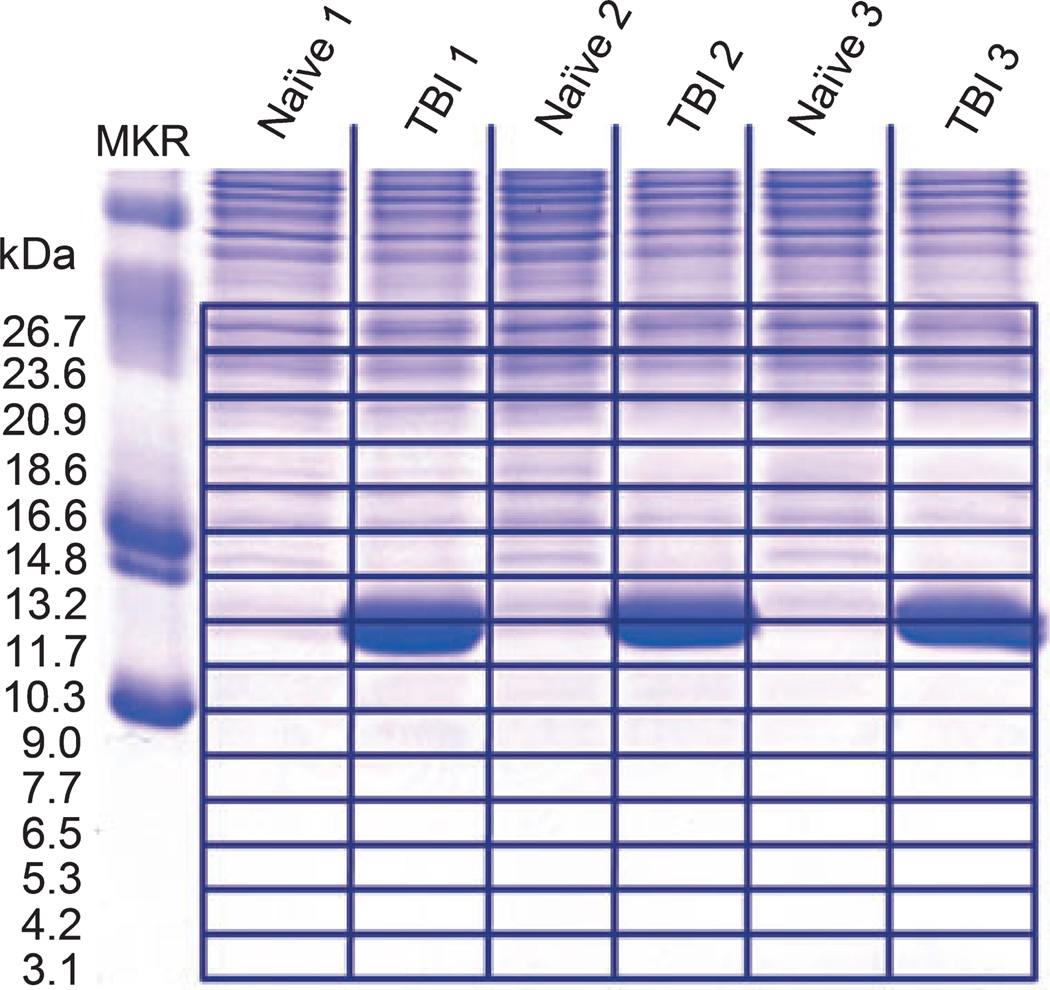
Fig. 5.
Gel electrophoresis separation of naïve and TBI samples. Prior to mass spectrometry analysis, we separated our brain samples by polyacrylamide gel electrophoresis to resolve MBP polypeptides measured against molecular mass markers (MKR). Slices were excised along each gel lane as indicated, from below an Ma of 30 to the bottom of the gel. The large band found in TBI lanes (Ma 12.5) is from hemoglobin that infiltrated the injured tissue by blood–brain barrier leakage.
Fig. 6.
Quantitative profile of MBP in naïve and TBI samples. Profiles of HGFLPR (a) and KNIVTPR (b) abundance by Ma value for naïve and TBI rat cortex, overlaid with molecular mass labels demarking the correlating MBP isoforms and proteolytic fragments derived from data in Fig. 4. Mean values are presented with the measured range denoted by error bars (n = 3).
Profiling MBP isoforms in naïve and TBI adult rat cortex
We subsequently employed the selectivity of MSMS to discriminate MBP isoforms. We devised a MSMS method to target five peptides each with specificity to a different variable transcription region of MBP (Fig. 1). The combined profile of the five peptides was thereby used to decipher the MBP isoforms or fragments present at each Ma value (Fig. 7). First, we verified our observation that the 21.6 and 18.6 kDa isoforms were shifted to greater Ma values while the 17.2 and 14.3 kDa isoforms were not. The phenomenon was thereby associated with the exon-6 coded sequence common to those isoforms that shifted and absent from those that did not. The exon-6 sequence contains two protein kinase C phosphorylation sites and a greater lysine density (more basic) that would affect electrophoretic migration (Kishimoto et al. 1985).
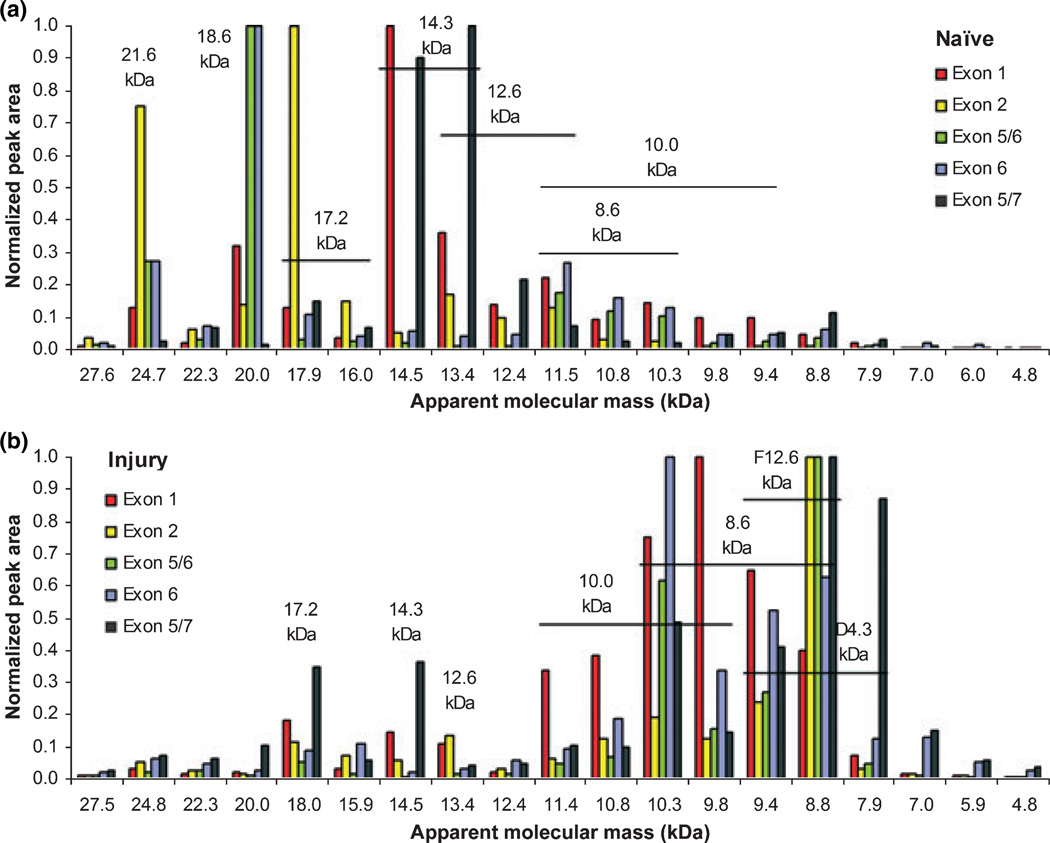
Fig. 7.
Profile of MBP isoforms and proteolytic fragments in naïve and TBI cortex samples. Shown are profiles of the variably spliced exon sequence-specific peptides for naïve (a) and TBI (b) samples. Chromatographic peaks were generated for the most intense fragment ion of each peptide. Peak area means for each peptide (n = 3) were normalized across all Ma values. The presence of a peptide (and correlating exon-coded sequence) was determined when the normalized area at a given Ma was greater than three times the minimum for that peptide. Molecular mass labels demarking the identified MBP isoforms and proteolytic fragments are shown for each gel slice per the pattern of exon-specific peptides and given Ma value. ‘F12.6 kDa’ indicates a truncated 12.6 kDa fragment and ‘D4.3 kDa’ indicates a dimer of the 4.3 kDa fragment.
The most abundant signal for the pan MBP peptide HGFLPR (exon-1) coincided with the greatest abundance of the exon-5/7 sequences, which along with data in Fig. 6 confirmed that the 14.3 kDa was the most prominent MBP isoform in naïve rat cortex. The disparity with the report by Liu et al. (2006a) is explained by their use of two antibodies, differing in sensitivity, to detect the 18.6 and 14.3 kDa isoforms. We also confirmed that the exon-2 containing 17.2 kDa isoform was present over the 17.3 kDa isoform in adult rat cortex as first suggested by Akiyama et al. (2002). In earlier rat MBP studies, isobaric 17 kDa isoforms were indistinguishable (Määttä et al. 1997; Kruger et al. 1999).
The selective MSMS data allowed us to distinguish among the post-injury polypeptide fragments despite overlap below an Ma of 16 (resolution shift from 2 to 1 Ma per gel slice). Only trace amounts of the largest predicted MBP fragment (12.6 kDa) were present at the expected Ma value in TBI samples. It was, however, present as a further fragmented polypeptide, denoted by the dominant exon-2 peptide signal at an Ma of 9. The 12.6 kDa fragment was likely proteolyzed further by calpain or another protease, given the over-activation of proteases following TBI. Conversely, the 10.0 kDa fragment appeared unmodified at an Ma of 10. The results suggest that the secondary proteolytic event reducing the 12.6 kDa fragment occurs within the exon-2 sequence. More specifically, the resulting Ma of 9 suggests that secondary cleavage occurs toward the c-terminal end of the exon-2 sequence. We also noticed basal levels of MBP proteolytic fragments in naïve tissue (Figs 6 and 7); however, this may be a result of postmortem processing. Next, the only exon-6 containing fragment, the 8.6 kDa, was found shifted higher than its molecular mass (up to an Ma of 11). The same phenomenon was observed with intact exon-6 containing isoforms, further linking this variable sequence with the shift behavior. Lastly, the exon-5/7 peptide that denotes the 4.3 kDa fragment was predominantly found between Ma values of 9.4 and 7.0, twice its mass, suggesting the presence of a dimer. Partial dimerization also explained why the greatest KNIVTPR signal was at an Ma of 9 (Fig. 6), which was contrary to that expected with the most abundant 14.3 kDa isoform producing the 4.3 kDa fragment. Intact MBP dimerization was noted elsewhere (Boulias et al. 1995; Määttä et al. 1997), though an exclusive link with the exon-5/7 tandem sequence was not established.
Absolute quantification of calpain proteolyzed MBP in CSF
Polypeptides must be assayable in biofluids to be useful as biomarkers. CSF is the most CNS relevant biofluid accessible from severe TBI patients. Thus, we aimed to demonstrate that our mass spectrometry method could be used to measure a calpain-induced MBP fragment in CSF of TBI rats. We calculated 64 fmol of the calpain-cleavage specific peptide in a TBI CSF (20 µL) gel band at a Ma of 11 (Fig. 8a and b) that was not present in naïve CSF. The data left little doubt that a proteolytic product of MBP was present in the CSF of TBI injured rats, as the measurements were made from tandem mass spectra that distinctly identified the KNIVTPR peptide (Fig. 8c). In contrast, immunoblot analysis lacked either the sensitivity or selectivity necessary to detect a MBP fragment band in CSF (Fig. 2).
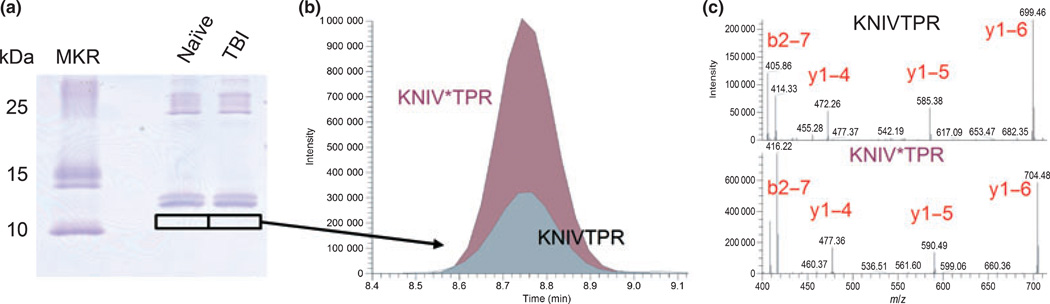
Fig. 8.
Quantification of a calpain-induced MBP fragment in CSF of TBI rats. (a) 20 µL of CSF from naïve and TBI rats (pooled, n = 7 each) were resolved by gel electrophoresis. The KNIVTPR calpain-cleavage selective MBP fragment was detected in TBI and not in naïve for the indicated gel slices at an Ma value of 11. (b) The chromatographic peak ratio of endogenous KNIVTPR to isotopically labeled internal standard (KNIV*TPR) was 0.35, and when multiplied by 18.6 fmol of standard would denote a nominal 6.4 fmol of endogenous peptide with one-tenth of the sample analyzed. (c) Detection of the endogenous peptide was confirmed by the tandem mass spectrum, shown to match that of the isotopically labeled peptide standard (shifted by 5 m/z).
Discussion
In this study, we employed a new MSMS approach to profile MBP dynamics, an integral protein component of the myelin sheath protecting vulnerable white matter in the brain (Smith et al. 2003). We assessed what MBP isoforms were present in normal adult rat cortex, and their degradation by calpain proteolysis following neurotrauma. MSMS facilitated femtomole quantification and the identification of isoforms and related proteolytic fragments. The resulting data depict near complete degeneration at 2 days post-injury of the four MBP isoforms normally present, with correlating increases in calpain proteolytic fragments detectable at the tissue and CSF levels. Indeed, a statistically significant decrease in the 21.6, 18.6, and 14.3 kDa isoforms was determined despite low statistical power (p of 0.04, n of 3, non-parametric Mann–Whitney U-test). These results correlate with previous reports using the same model of large-scale axonal and neuronal degeneration at 2 days post-TBI (Ringger et al. 2004; Kobeissy et al. 2006; Liu et al. 2006a).
Tandem mass spectrometry data provided greater detail over previous reports to establish the following findings. First, we determined that the isoform of greatest abundance in adult rat cortex was the 14.3 kDa, as denoted by the corresponding tandem exon-5/7 sequence signal. The report by Liu et al. (2006b) was confounded by use of two different antibodies, which, as a result of a difference in sensitivity, made the 18.6 kDa isoform appear more abundant than the 14.3 kDa. The same issue limited their evaluation of the detected fragment bands. We were able to ascertain that the 14.3 kDa isoform was proteolyzed to a 4.3 kDa fragment as predicted from the sequence (Fig. 3b, Ma of 4.2). Further, the fragment predominantly appeared as a dimer (Fig. 6), which would explain the two bands observed by Liu et al. (2006a) (reported at 8 and 6 kDa) found with an antibody to the exon-5/7 sequence.
Second, isoforms of MBP appeared at an elevated Ma (as high as 27) and considerably greater than the largest isoform (21.6 kDa), a phenomenon mentioned though not described elsewhere (Boulias et al. 1995; Akiyama et al. 2002). Our data show that the phenomenon is limited to the two largest isoforms, which are mutually distinct from the other isoforms by the exon-6 sequence. The shift to greater Ma was also observed for the exon-6 containing 8.6 kDa MBP fragment, present between 10.3 and 8.8 in our data. We were unable to corroborate a shift in intact MBP against the report by Liu et al. (2006b) (their data do not include molecular mass markers); however, the two fragment bands they reported at 10 and 8 kDa using an antibody specific to exon-6 overlap with our data. The presence of two distinct bands related to the 8.6 kDa fragment suggests a differential modification, such as phosphorylation following TBI. Indeed, the intact 18.6 kDa isoform would appear to exist in two modification states as well following TBI (see bold arrow in Fig. 2a). Further study will be conducted to confirm the presence of differential phosphorylation following injury.
Third, the majority of the 12.6 kDa fragment (containing exon-2) was found further degraded at an Ma of 9, likely by calpain or another protease following injury. The observed mass would place the cleavage site between R75SRPGLCHMYKD86 and after the exon-2 specific marker peptide. We tabulated a score for the 11 possibilities by adding occurrence values for the amino acid in positions P4 to P7′. The occurrence values were taken from a study by Tompa et al. (2004) of amino acid position among 106 known calpain proteolytic events. One site stood out, SHAR75–S76RPGLCH, with the preferred positioning of arginine at P1, serine at P1′ and proline at P3′ and a score 50% greater than the next highest. Further analysis is required to confirm this site, which is beyond the scope of this study. We also verified the presence of two exon-2 containing intact isoforms (21.6 and 17.2 kDa) in uninjured adult rats. As reviewed earlier, exon-2 is associated with myelin formation and remyelination. The presence of exon-2 containing isoforms raises the possibility that remyelination occurs naturally throughout adulthood. While all isoforms were degraded after TBI, we observed that a greater percentage of the exon-2 containing isoforms remained intact relative to control levels (Fig. 6). Perhaps an artifact, the observation does raise the possibility that degradation or regeneration rates may differ among isoforms. To address these issues, temporal profile studies are planned to further characterize MBP dynamics following TBI, to include in vivo calpain-inhibitor treatment as a cohort. Calpain inhibitors have been demonstrated to modulate MBP degradation (Banik et al. 1997, 2000) and transcription (Ray et al. 2003), though the effect on discrete isoforms, to our knowledge, has not been characterized. The described mass spectrometry methods are also useful to study the dynamics of MBP isoforms following other disease paradigms. In particular, MBP degradation by calpain has long been implicated in the pathobiology of MS (Smith 1977); Sato et al. 1984; Shields et al. 1999). MBP and its antibodies are included among CSF biomarker of MS and other similar neurodegenerative diseases (reviewed elsewhere, Massaro and Tonali 1998; Michalowska-Wender et al. 2001), though the particular proteolytic fragment quantified in our study has not been described. Future efforts might ascertain whether this fragment is present in the CSF of MS patients, for potential utility as a alternative biomarker of degradative myelin processing.
The presented methods hinge on the use of MSMS spectra to selectively characterize peptides specific to the calpain cleavage site and alternative spliced coding regions of MBP. All target peptides are unique to MBP among all other proteins in a rat proteome database as determined by BLAST analysis. We evaluated the selectivity of corresponding MSMS spectra by using Mascot software to calculate the probability (p) that the match of the spectra with the correct peptide sequences would occur by chance (Perkins et al. 1999). For example, the probability for a chance match of the HGFLPR and KNIVTPR MSMS data was p = 0.00050 (1 in 2000)) and p = 0.00016 (1 in 6250)), respectively, when evaluated against a rat protein database. In contrast, the next closest peptide sequences had a greater, 1 in 8, probability of being a chance match of the MSMS spectra (p = 0.13). With significance often defined as a less than 1 in 20 chance occurrence (i.e. p < 0.05), the collected MSMS spectra are highly significant in their selectivity of the target peptide.
Presently, our application of the reported approach is restricted by the speed of our mass spectrometer in that chromatographic peaks are defined only by four or five data points, and IDA and isoform characterization were performed separately. More current equipment, such as the faster, more sensitive linear ion trap (Blackler et al. 2006), would improve peak characterization and still allow for rapid collection of tandem mass spectra. Parameters were optimized in our study to quantify MBP peptides; however, the approach is generally applicable to other proteolyzed proteins. Likewise, isoforms of other proteins can be delineated using the presented method, in combination or separate from IDA quantification, which is significant in that we now anticipate most proteins to have multiple isoforms with unique functions or spatial/temporal dynamics important for the regulation of cellular processes (Godovac-Zimmermann et al. 2005; Yura et al. 2006). The general applicability of the approach is nonetheless expressed in light of limitations to peptide analysis by mass spectrometry. Peptide coelution and poor ionization efficiency are confounds that limit sensitivity and tandem mass spectrum quality. Application of our mass spectrometry approach to other peptides would thereby require individual consideration and optimization of gel electrophoresis or RPLC separations (possibly adding analysis time). To this end, separation and detection efficiencies could be checked quickly through test analysis of a synthesized target peptide(s) within the complex mixture of interest. Overall, the upfront time and cost for this test is less than required for novel antibody generation as the alternative.
In summary, we present mass spectrometry methodology to characterize and quantify proteolytic processing of multiisoform proteins, exemplified in this study by calpain proteolysis of MBP after neurotrauma. Intact and fragmented MBP isoforms were deconvoluted using sequence specific peptide MSMS data, and quantified at femtomole amounts in tissue and CSF using IDA. We demonstrated by the selectivity of MSMS that the 14.3 kDa MBP isoform was predominant in our adult rats and quantified the degradation of four predominant isoforms (21.6, 18.6, 17.2, and 14.3 kDa) following TBI. We ascertained that the presence of the MBP exon-6 sequence resulted in a characteristic shift in gel migration for intact and fragmented MBP alike. We also found evidence for a second post-TBI cleavage event within the exon-2 MBP sequence, and that the post-TBI 4.3 kDa fragment forms a dimer. Overall, we expect that the presented methodology will be broadly applicable for monitoring dynamics of other multi-isoform proteins and proteolytic events of significance in the study of the CNS, with particular relevance to biomarker research.
Acknowledgements
This work was performed with support of the Department of Defense grant DAMD17-03-1-0066 and the National Institute for Neurological Disorders and Stroke grant K25NS055012. We thank W. Harrison for critical reading of the manuscript, A. Chung for peptide synthesis, M. C. Liu for insightful discussions on MBP immunoblot analysis, and R. Tivis, SJ. Nixon, and M. Lewis for assistance with statistical evaluation. RLH, NDD, and KKWW hold equity in Banyan Biomarkers, Inc, a company commercializing biomarker technology in brain injury.
Abbreviations
- IDA
isotope dilution analysis
- Ma
apparent molecular mass
- MBP
myelin basic protein
- MS
multiple sclerosis
- MSMS
tandem mass spectrometry
- RPLC
reversed-phase liquid chromatography
- TBI
traumatic brain injury.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Akiyama K, Ichinose S, Omori A, Sakurai Y, Asou H. Study of expression of myelin basic proteins (MBPs) in developing rat brain using a novel antibody reacting with four major isoforms of MBP. J Neruosci. Res. 2002;68:19–28. doi: 10.1002/jnr.10188. [DOI] [PubMed] [Google Scholar]
- Arroyo EJ, Scherer SS. On the molecular architecture of myelinated fibers. Histochem. Cell Biol. 2000;113:1–18. doi: 10.1007/s004180050001. [DOI] [PubMed] [Google Scholar]
- Banik NL, Matzelle D, Terry E, Hogan EL. A new mechanism of methylprednisolone and other corticosteroids action demonstrated in vitro: inhibition of a proteinase (calpain) prevents myelin and cytoskeletal protein degradation. Brain Res. 1997;748:205–210. doi: 10.1016/s0006-8993(96)01302-9. [DOI] [PubMed] [Google Scholar]
- Banik NL, Matzelle D, Terry E, Gantt-Wilford G, Hogan EL. Inhibition of proteolysis by a cyclooxygenase inhibitor, indomethacin. Neurochem. Res. 2000;25:1509–1515. doi: 10.1023/a:1007684311023. [DOI] [PubMed] [Google Scholar]
- Barr JR, Maggio VL, Patterson DG, Jr, et al. Isotope dilution-mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin. Chem. 1996;42:1676–1682. [PubMed] [Google Scholar]
- Berger RP, Adelson PD, Richichi R, Kochanek PM. Serum biomarkers after traumatic and hypoxemic brain injuries: insight into the biochemical response of the pediatric brain to inflict brain injury. Dev. Neurosci. 2006;28:327–335. doi: 10.1159/000094158. [DOI] [PubMed] [Google Scholar]
- Blackler AR, Klammer AA, MacCoss MJ, Wu CC. Qualitative comparison of proteomic data quality between a 2D and 3D quadrupole ion trap. Anal. Chem. 2006;78:1337–1344. doi: 10.1021/ac051486a. [DOI] [PubMed] [Google Scholar]
- Boggs JM. Myelin basic protein: a multifunctional protein. Cell. Mol. Life Sci. 2006;63:1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs JM, Rangaraj G, Koshy KM, Mueller JP. Adhesion of acidic lipid vesicles by 21.5 kDa (recombinant) and 18.5 kDa isoforms of myelin basic protein. Biochem. Biophys. Acta. 2000;1463:81–87. doi: 10.1016/s0005-2736(99)00181-9. [DOI] [PubMed] [Google Scholar]
- Boulias C, Pang H, Mastronardi F, Moscarello MA. The isolation and characterization of four myelin basic proteins from the unbound fraction during CM52 chromatography. Arch. Biochem. Biophys. 1995;322:174–182. doi: 10.1006/abbi.1995.1449. [DOI] [PubMed] [Google Scholar]
- Capello E, Voskuhl RR, Mctarland HF, Rane CS. Multiple sclerosis: re-expression of developmetnal gene in chromic lesions correlates with remyelination. Ann. Neurol. 1997;41:797–805. doi: 10.1002/ana.410410616. [DOI] [PubMed] [Google Scholar]
- Dongré AR, Eng JK, Yates III JR. Emerging tandem-mass-spectrometry techniques for the rapid identification of proteins. Trends Biotechnol. 1997;15:418–425. doi: 10.1016/S0167-7799(97)01110-4. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givogri MI, Bongarzone ER, Campagnoni AT. New insights on the biology of myelin basic protein gene: the neuralimmune connection. J. Neurosci. Res. 2000;59:153–159. [PubMed] [Google Scholar]
- Godovac-Zimmermann J, Kleiner O, Brown LR, Drukier AK. Perspectives in spicing up proteomics with splicing. Proteomics. 2005;5:699–709. doi: 10.1002/pmic.200401051. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C. Myelin basic protein – diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Kimura M, Sato M, Akatsuka A, Saito S, Ando K, Yokoyama M, Katsuki M. Overexpression of a minor component of myelin basic protein isoform (17.2 kDa) can restore myelinogenesis in transgenic shiverer mice. Brain Res. 1998;785:245–252. doi: 10.1016/s0006-8993(97)01383-8. [DOI] [PubMed] [Google Scholar]
- Kishimoto A, Nishiyama K, Nakanishi H, Uratsuji Y, Nomura H, Takeyama Y, Nishizuka Y. Studies on the phosphorylation of myelin basic protein by protein kinase C and adenosine 3′ 5′-monophosphate-dependent protein kinase. J. Biol. Chem. 1985;260:12492–12499. [PubMed] [Google Scholar]
- Kobeissy FH, Ottens AK, Zhang Z, Liu MC, Denslow ND, Dave JR, Tortella FC, Hayes RL, Wang KK. Novel differential neuroproteomics analysis of traumatic brain injury in rats. Mol. Cell. Protoemics. 2006;5:1887–1898. doi: 10.1074/mcp.M600157-MCP200. [DOI] [PubMed] [Google Scholar]
- Kruger GM, Diemel LT, Copelman CA, Cuzner ML. Myelin basic protein isoforms in myelinating and remyelinating rat brain aggregate cultures. J. Neurosci. Res. 1999;56:241–247. doi: 10.1002/(SICI)1097-4547(19990501)56:3<241::AID-JNR3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Liu MC, Akle V, Zheng W, et al. Extensive degradation of myelin basic protein isoforms by calpain following traumatic brain injury. J. Neurochem. 2006a;98:700–712. doi: 10.1111/j.1471-4159.2006.03882.x. [DOI] [PubMed] [Google Scholar]
- Liu MC, Akle V, Zheng W, Dave JR, Tortella FC, Hayes RL, Wang KK. Comparing calpain- and caspase-3-mediated degradation patterns in traumatic brain injury by differential proteome analysis. Biochem J. 2006b;394:715–725. doi: 10.1042/BJ20050905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttä JA, Coffey ET, Hermonen JA, Salmi AA, Hinkkanen AE. Detection of myelin basic protein isoforms by organic concentration. Biochem. Biophys. Res. Commun. 1997;238:498–502. doi: 10.1006/bbrc.1997.7318. [DOI] [PubMed] [Google Scholar]
- Massaro AR, Tonali P. Cerebrospinal fluid markers in multiple sclerosis: an overview. Mult. Scler. 1998;4:1–4. doi: 10.1177/135245859800400101. [DOI] [PubMed] [Google Scholar]
- Michalowska-Wender G, Losy J, Wender M. Biological markers to confirm diagnosis and monitor the therapy in multiple sclerosis patients. Folia Neuropathol. 2001;39:1–5. [PubMed] [Google Scholar]
- Nakajima K, Ikenaka K, Kagawa T, Aruga J, Nakao J, Nakahira K, Shiota C, Kim SU, Mikoshiba K. Novel isoforms of mouse myelin basic protein predominantly expressed in embryonic stage. J. Neurochem. 1993;60:1554–1563. doi: 10.1111/j.1471-4159.1993.tb03321.x. [DOI] [PubMed] [Google Scholar]
- Norton WT, Cammer W. In: Isolation and characterization of myelin, in Myelin. 2nd ed. Morell P, editor. New York: Plenum Press; 1984. pp. 147–180. [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Ray SK, Matzelle DD, Sribnick EA, Guyton K, Wingrave JM, Banik NL. Calpain inhibitor prevented apoptosis and maintained transcription of proteolipid protein and myelin basic protein genes in rat spinal cord injury. J. Chem. Neuroanat. 2003;26:119–124. doi: 10.1016/s0891-0618(03)00044-9. [DOI] [PubMed] [Google Scholar]
- Ringger NC, O’Steen BE, Brabham JG, Silver X, Pineda J, Wang KK, Hayes RL, Papa L. A novel marker of traumatic brain injury: CSF alphaII-spectrin breakdown product levels. J Neurotruama. 2004;21:1443–1456. doi: 10.1089/neu.2004.21.1443. [DOI] [PubMed] [Google Scholar]
- Sato S, Quarles RH, Brady RO, Tourtellotte WW. Elevated neutral protease activity in myelin from brains of patients with multiple sclerosis. Ann. Neurol. 1984;15:264–267. doi: 10.1002/ana.410150310. [DOI] [PubMed] [Google Scholar]
- Schaecher KE, Shields DC, Banik NL. Mechanism of myelin breakdown in experimental demyelination: a putative role for calpain. Neurochem. Res. 2001;26:731–737. doi: 10.1023/a:1010903823668. [DOI] [PubMed] [Google Scholar]
- Shields DC, Schaecher KE, Saido TC, Banik NL. A putative mechanism of demyelination in multiple sclerosis by a proteolytic enzume, calpain. Proc. Natl Acad. Sci. 1999;96:11486–11491. doi: 10.1073/pnas.96.20.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ME. The role of proteolytic enzymes in experimental allergic encephalomyelitis. Neurochemistry. 1977;2:233–246. doi: 10.1007/BF00969354. [DOI] [PubMed] [Google Scholar]
- Smith DH, Meaney DF, Shull WH. Diffuse axonal injury in head trauma. J Head Trauma Rehabil. 2003;18:307–316. doi: 10.1097/00001199-200307000-00003. [DOI] [PubMed] [Google Scholar]
- Thomas DG, Palfreyman JW, Ratcliffe JG. Serummyelin-basic-protein assay in diagnosis and prognosis of patients with head injury. Lancet. 1978;1:113–115. doi: 10.1016/s0140-6736(78)90415-4. [DOI] [PubMed] [Google Scholar]
- Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilágyi A, Bánóczi Z, Hudecz F, Friedrich P. On the sequential determinants of calpain cleavage. J. Biol. Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- Voskuhl RR, Robinson ED, Segal BM, Tranquill L, Camphausen K, Albert PS, Richert JR, McFarland HF. HLA restriction and TCR usage of T lymphocytes specific for a novel candidate autoantigen, X2 MBP, in multiple sclerosis. J Immunol. 1994;153:4834–4844. [PubMed] [Google Scholar]
- Wood DD, Moscarello MA. The isolation, characterization, and lipid-aggregating properties of citrulline containing myelin basic protein. J. Biol. Chem. 1989;25:5121–5127. [PubMed] [Google Scholar]
- Yura K, Shionyu M, Hagino K, et al. Alternative splicing in human transcriptome: functional and structural influence on proteins. Gene. 2006;380:63–71. doi: 10.1016/j.gene.2006.05.015. [DOI] [PubMed] [Google Scholar]