Abstract
Background
Although genetic variants may play a key role in development of treatment-resistant depression (TRD), relevant research is scarce.
Methods
To examine whether the polymorphisms of BDNF (rs6265) and NTRK2 (rs1387923, rs2769605 and rs1565445) genes confer risk for TRD in major depressive disorder (MDD), a total of 948 MDD patients were recruited in a 12-week, multi center, prospective longitudinal study.
Results
Our study showed a significant allelic association between rs1565445 and TRD with an excess of the T allele in the TRD group, compared to non-TRD group (OR = 1.43, 95%CI: 1.16–1.76, p = 0.0008); while patients with genotype C/C and T/C in rs1565445 were less likely to develop TRD than those carrying T/T (OR = 0.52, 95%CI: 0.33–0.82; OR = 0.72, 95%CI: 0.54–0.97, respectively; p = 0.005). Haplotype T–T (rs1565445 and rs1387923) had 1.41-fold increased risk of TRD (p = 0.0014). Furthermore, significant four-locus (rs1387923-rs1565445-rs2769605-rs6265) gene-gene interactions were detected by the Multifactor-dimensionality reduction (MDR) method.
Discussion
These results suggest that the interactions of BDNF (rs6265) with NTRK2 (rs1387923, rs2769605 and rs1565445) gene polymorphisms likely play an essential role in the development of TRD in Han Chinese MDD patients.
Keywords: BDNF, NTRK2, Interaction, Treatment-resistant depression
1. Introduction
Major depressive disorder (MDD) is a recurrent, disabling and heterogeneous mental illness with a high cost to society and individuals across theworld (Fang et al., 2010; Rush et al., 2006). Although there are a number of available antidepressants targeting monoamine systems, it is estimated that one to two thirds of patients treated for MDD do not respond satisfactorily to an initial antidepressant treatment (Rush et al., 2006), and up to 15–40% of patients do not respond to multiple pharmacological treatments (or psychotherapy in combination) (Berlim and Turecki, 2007; Kayser et al., 2011; Rush et al., 2003). Patients who do not respond to at least two distinctly different classes of antidepressants with adequate treatment durations and dosages are classified as treatment-resistant depression (TRD) (Fang et al., 2010). It is well known that TRD is more likely to cause suicide or a suicide attempt, frequent relapses, poor prognosis, and accounts for a large proportion of the overall cost of depression (Gibson et al., 2010; Scott and Dickey, 2003).
Currently, the underlying mechanism of TRD is still poorly understood. Multiple factors, including environmental and genetic, are likely involved in the pathophysiology of TRD. However, mounting evidence has demonstrated that genetic variations associated with antidepressant responses appear to cluster in families, supporting a more critical role for genetic variations in mechanisms underlying TRD (Laje and McMahon, 2007; O’Reilly et al., 1994). Recently, the neurotrophin hypothesis of MDD has attracted more attention (Dunham et al., 2009; Fernandes et al., 2011; Sillaber et al., 2008). Previous studies found decreased gray matter volume (particularly in the hippocampus) in patients with TRD, and patients who received more effective antidepressant treatment displayed less gray matter loss and were less liable to develop TRD (Obergriesser et al., 2003; Shah et al., 2002). It has been suggested that hippocampal volumes are correlated with decreased expression of neurotrophic factor (most notably, brain derived neurotrophic factor [BDNF]) (Blugeot et al., 2011; Taylor, 2008). Furthermore, previous studies demonstrated that BDNF levels were decreased in patients with TRD, and elevated after successful treatment with electroconvulsive therapy (ECT), high-frequency repetitive transcranial magnetic stimulation (rTMS), antidepressants, and atypical antipsychotic drugs and lithium (Piccinni et al., 2009; Taylor, 2008; Xu et al., 2006; Yukimasa et al., 2006). In addition, pre-clinical studies suggested that BDNF was required for an antidepressant effect in animal models of depression and could induce antidepressant-like behavioral effects by itself (Duman, 2004; Hashimoto et al., 2004). Toups et al. (2011) recently found that BDNF may improve the efficacy of exercise treatment of MDD. All these evidence suggests that BDNF may be involved in the pathophysiology underlying TRD.
BDNF is a member of the neurotrophin super family, which mediates neuronal differentiation and survival, modifies synaptic connections, and plays an important role in the neurodevelopment of the central nervous system(CNS). Position 196 in exon 5 of the BDNF gene contains a G to A transition (dbSNP: rs6265) that results in an amino acid substitution (valine to methionine) at codon 66 in the precursor BDNF peptide sequence. This polymorphism can effect activity dependent secretion of BDNF as well as hippocampal function and morphology (Egan et al.,2003; Pezawas et al.,2004). Furthermore, this functional Val66Met polymorphism has been associated with antidepressants response (Zou et al., 2010a,b). However, Anttila et al. (2007) reported no association between BDNF Val66Met poly-morphism and risk of TRD. It’s notable that BDNF most likely functions through its high-affinity receptor, neurotrophic tyrosine kinase receptor 2 (NTRK2) (Squinto et al., 1991). It has been suggested that gene-gene interaction is critical to describe a phenotypic effect, especially when a specific individual genetic variant displays a minor marginal effect in a complex psychiatric disease (Burmeister et al., 2008). NTRK2 has also been found to be associated with major depression and antidepressant-like effects (Blugeot et al., 2011; Saarelainen et al., 2003). Furthermore, augmentation or combination with lithium has proven to be an effective alternative for patients with TRD in number of clinical studies (Shelton et al., 2010; Vieta and Colom, 2011). Previous studies have shown that NTRK2 gene polymorphisms rs2769605, rs1387923, and rs1565445 were associated with mood disorders and lithium response (Bremer et al., 2007; Smith et al.,2009;Wang et al., 2012). Polymorphismrs1565445isanintronic SNP located in intron 19, rs1387923 and rs2769605 are ~2 kb and 274 kb respectively downstream of NTRK2 gene on chromosome 9.
To further investigate the role of BDNF and NTRK2 genes in pathophysiology underlying TRD, the BDNF functional Val66Met polymorphism (rs6265) and three polymorphisms in NTRK2 gene (rs1387923, rs2769605 and rs1565445) were examined in a large sample of MDD patients from our multicenter, longitudinal studies. To our knowledge this is the first study to investigate the genetic risk factors for the development of TRD in MDD patients from the Han Chinese population.
2. Materials and methods
2.1. Subjects
The participants in the study were inpatients and outpatients from our multicenter, longitudinal “OPERATION” (OPtimized trEatment stRAtegies for Treatment-resIstant depressiON) study, “OPERATION-ECMA” (OPERATION Extended-Climbing Mountain Action plan) study, and “CARE-SSD/MDD” (Construct An Rough Evaluation index system for subsyndromal symptomatic depression and major depressive disorder) study, during January 2005–October 2010. The aims of the “OPERATION” and “OPERATION-ECMA” studies were to clarify and investigate optimized treatment strategies, health economy and factors associated with TRD. The aim of the “CARE-SSD/MDD study” was to study the outcome of SSD and MDD with antidepressants treatment, as well as the biological relationship between them.
Inclusion criteria were as follow: (1) Between the ages of 18 and 65; (2) Han Chinese in origin; (3) Met the criteria of the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSMIV) for MDD (not secondary to any other Axis I disorder); (4) Blood samples were available; (5) Completed 12-week follow-up study to identify TRD or non-TRD.
Exclusion criteria were as follows: (1) Had a lifetime diagnosis of bipolar disorder, schizoaffective disorder, schizophrenia, or other psychotic disorders; (2) Had an imminent risk for suicide or homicide judged by a research psychiatrist; (3) Had any medical contraindication to antidepressants or other psychotropic medication; (4) Had an unstable general medical condition or a condition that required the combination treatment of an antidepressant and any other psychotropic medication including antipsychotic agents, mood stabilizers, anticonvulsants, and stimulants; (5) Had modified electroconvulsive therapy within 1 month of the study screening; (6) Female patients who were pregnant, planning to become pregnant, or breast-feeding during the study period.
All procedures were reviewed and approved by Institutional Review Boards of Shanghai Mental Health Center and other participating institutions. This study was conducted in accordance with the Helsinki Declaration as revised 1989 and written informed consent was obtained from each participant before any study related procedures were performed.
2.2. Procedures and primary outcome measures
In the present study, the definition of TRD was MDD patients who failed 2 or more antidepressants from different classes with adequate treatment durations and dosages according to the stage II TRD criteria described by Thase and Rush (Rush et al., 2006). The procedure for identifying TRD patients was previously described elsewhere in detail (Fang et al., 2010, 2011). Briefly, MDD patients were treated with adequate antidepressant treatment for 6 weeks according to their previous treatment and family histories and other clinical considerations. From the total number of patients, 603 were treated with SSRI, 267 were treated with SNRI, and 78 were treated with other classical antidepressants. Participants who failed the initial 6-week treatment trial were treated with a second antidepressant from a different class for an additional 6 weeks based on the clinician’s judgment. Failure to respond was determined as the reduction rate of HRSD-17 score less than 50%, while response to antidepressants was determined as the reduction rate in HRSD-17 score higher or equal to 50% (Li et al., 2012). All participants were diagnosed with the Structured Clinical Interview for DSM-IV-TR Axis I Disorders-Patient Edition (SCID-I/P) and demographic data was collected. HRSD-17 assessments were conducted by two experienced psychiatrists independently (inter-rater reliability, Kappa = 0.84) to assess the depressive symptoms of MDD patients. The outcome measure was the occurrence of TRD.
2.3. DNA extraction and SNP genotyping
Peripheral blood samples were collected from the MDD patients in 5ml EDTA vacuum tube. Genomic DNA was extracted from peripheral blood according to standard laboratory procedures (Blood genomic DNA extraction kit, Tiangen, Beijing) and stored in at −80°C until genotyping. After quality assessment, DNA samples (250 ng each)were genotyped with TaqMan genotyping assay on an ABI PRISM 7900 Sequence Detection System instrument (Applied Biosystems, Foster City, CA).Primers and probes were purchased from Applied Biosystems.
For quality control, all genotypes were called blind to the case or control status in the genotyping process. Of the samples collected, 10% were repeated for the genotyping assay, and the results were more than 99% concordant.
2.4. Statistical analyses
Differences in age and gender between patients with TRD and non-TRD were compared by using chi-square (χ2) test or t-test. Hardy-Weinberg Equilibrium, allele and genotype frequencies of individual SNPs, pair-wise linkage disequilibrium of all pairs of SNPs and haplotype analysis were calculated by using the Haploview 4.1 software and the extent of linkage disequilibrium (LD) was measured by the standardized D′. Those haplotypes with a frequency under 3% were ignored. For multiple test correction, the Bonferroni correction was applied and the significance level α was set at 0.0125(0.05/4). The power to detect significant association was estimated using the online software Quanto (Version1.2.3, available at http://hydra.usc.edu/GxE/) (Gauderman and Morrison, 2006). The statistical power of our study was more than 93% under the assumption of a moderate effect size (Odds Ratio [OR] = 1.5), a log additive model and TRD occurrence in MDD patients of 15–30%.
Multifactor-dimensionality reduction (MDR)method was applied to investigate gene-gene interactions (Ritchie et al., 2001). Briefly, MDR used constructive induction of information gain, in which the n dimensions corresponding to n genotypes are collapsed into one dimension (genetic attribute of efficacy) by combining higher and lower predisposing genotypes into two different groups (high-risk or low-risk); and the combination with the lowest misclassification error is selected. Moreover, 10-fold cross-validation was used to evaluate the predictive ability of the model by calculating the prediction error. The best model was selected based on the maximization of cross-validation consistency. P-values of prediction accuracy were determined empirically by permuting the case and control labels 1000 times. Hierarchical interaction graphs and interaction dendrogram of MDR were used to show the SNPs interaction of the best model (Dervieux et al., 2012). In addition, traditional statistical methods were performed to examine the results from MDR analyses. A p-value of less than 0.05 was considered statistically significant.
3. Results
3.1. Participant characteristics
948 MDD patients were recruited in the present genetic study. After 12-week follow-up, 304 patients were identified as TRD with a mean age of 34.5 ± 5.8 years (121 males and 183 females) whose mean reduction rate ofHRSD-17 score (39.4±7.1%) is lower than 50%, while 644 patients were identified as non-TRD with a mean age of 35.0 ±5.3 years (249males and 395 females) whose mean reduction rate of HRSD-17 score (62.3 ± 7.2%) higher or equal to 50%. No significant difference was observed in demographic and clinical features of TRD patients and non-TRD patients (shown in Table 1).
Table 1.
Demographic and clinical variables of TRD and non-TRD patients.
| Variables | TRD | Non-TRD | Statistics | p |
|---|---|---|---|---|
| Age (year) | 34.5 (5.8) | 35.0 (5.3) | 1.35 | 0.18 |
| Gender | ||||
| Male | 121 (39.8%) | 249 (38.7%) | 0.11 | 0.74 |
| Female | 183 (60.2%) | 395 (61.3%) | ||
| Education (N, %) | ||||
| Primary school | 63 (20.7%) | 139 (21.6%) | 2.49 | 0.48 |
| Secondary school | 125 (41.1%) | 232 (36.0%) | ||
| Junior or regular college | 106 (34.9%) | 252 (39.1%) | ||
| Master or above | 10 (3.3%) | 21 (3.3%) | ||
| Smoker (N, %) | 22 (7.2%) | 36 (5.6%) | 0.98 | 0.32 |
| Drinker (N, %)a | 4 (1.3%) | 16 (2.5%) | 1.37 | 0.24 |
| Duration of diseaseb (Month) | 3.7 (1.5) | 3.9 (1.9) | 1.49 | 0.14 |
| Number of episode (Mean, SD) | 2.9 (0.7) | 2.95 (0.7) | 0.60 | 0.55 |
| Pre-treatment HRSD-17c | 23.8 (2.6) | 24.1 (2.6) | 1.63 | 0.10 |
| Post-treatment HRSD-17d | 14.3 (1.3) | 9.0 (1.7) | 47.95 | <0.0001 |
Drinker was defined as habitual drinker, harmful drinker, alcohol abuse, alcohol dependence and alcoholism.
Duration of disease prior to admission.
HRSD-17 on admission.
HRSD-17 was assessed at the end of the 12 weeks.
3.2. Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD)
The distributions of genotypes in TRD and non-TRD participants were consistent with Hardy-Weinberg equilibrium (P > 0.05) respectively. Analysis of pair wise LD was conducted for three SNPs in NTRK2 gene. The D′ and r2 of three SNPs in NTRK2 gene are shown in Supplementary Table 1. Strong LD (D′>0.7) was observed between rs1565445 and rs1387923 of NTRK2 (D = 0.926, r2 = 0.569) (detailed in Supplementary Table 1).
3.3. Association analysis of four polymorphisms with treatment resistant depression
As shown in Table 2, statistically significant differences in allele and genotype frequencies were detected between TRD and non- TRD participants for the NTRK2 gene polymorphism rs1565445. There was a significant allelic association between rs1565445 and TRD with an excess of the T allele in the TRD group, compared to non-TRD group (OR = 1.43, 95%CI: 1.16–1.76, p = 0.0008). MDD patients with genotype C/C and T/C in rs1565445 were less likely to develop TRD compared with patients carrying the genotype T/T (OR = 0.52, 95%CI: 0.33–0.82 and OR = 0.72, 95%CI: 0.54–0.97, respectively; p = 0.005). No statistically significant differences were found in allele or genotype frequencies between TRD and non-TRD participants for other individual SNPs (rs276905 and rs1387923 in NTRK2 gene, rs6265 in BDNF gene). Haplotype-based analysis showed that NTRK2 gene polymorphisms rs1565445- rs1387923 (T-T) haplotype was significantly associated with the risk of TRD (χ2 = 10.24, OR = 1.41, 95%CI: 1.14–1.74, p = 0.0014, global p = 0.0042) (detailed in Supplementary Table 2).
Table 2.
Allele and genotype distributions of SNPs and association analysis of each SNP between TRD and non-TRD samples.
| SNPs | Sample | N | Genotype (%) | χ2 | P a | Allele (%) | χ2 | P a | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2769605 | G/G | G/A | A/A | G | A | |||||||
| TRD | 304 | 189(62.0) | 94(30.9) | 21(6.9) | 3.43 | 0.18 | 472(77.6) | 136(22.4) | 0.15 | 0.70 | 0.96 | |
| Non-TRD | 644 | 394(61.2) | 222(34.5) | 28(4.3) | 1010(78.4) | 278(21.6) | (0.76–1.21) | |||||
| rs1387923 | T/T | T/C | C/C | T | C | |||||||
| TRD | 304 | 183(60.2) | 105(34.5) | 16(5.3) | 3.48 | 0.18 | 471(77.5) | 137(22.5) | 3.50 | 0.06 | 1.25 | |
| Non-TRD | 644 | 349(54.2) | 247(38.2) | 48(7.5) | 945 (73.4) | 343(26.6) | (0.99–1.56) | |||||
| rs1565445 | T/T | T/C | C/C | T | C | |||||||
| TRD | 304 | 156(51.3) | 120(39.5) | 28(9.2) | 10.61 | 0.005 | 432(71.1) | 176(28.9) | 11.25 | 0.0008 | 1.43 | |
| Non-TRD | 644 | 266(41.3) | 282(43.8) | 96(14.9) | 814(63.2) | 474(36.8) | (1.16–1.76) | |||||
| rs6265 | G/G | G/A | A/A | G | A | |||||||
| TRD | 304 | 86(28.3) | 134(44.1) | 84(27.6) | 2.96 | 0.23 | 306(50.3) | 302(49.7) | 1.29 | 0.26 | 0.89 | |
| Non-TRD | 644 | 185(28.7) | 313(48.6) | 146(22.7) | 683(53.0) | 605(47.0) | (0.74–1.09) |
Significant difference was presented in bold.
P-values are adjusted by Bonferroni method for the number of tests performed, the level of signi.cance was set at 0.0125.
3.4. MDR analysis of gene–gene interaction
Table 3 summarizes the cross-validation consistency and the prediction error obtained from MDR (Ritchie et al., 2001) analysis for each number of loci evaluated. One four-locus model had a maximum testing accuracy of 57.37% and a maximum cross-validation consistency (10/10) that was significant at P < 0.0001 level, as determined empirically by permutation testing. Thus, under the null hypothesis of no association, it is highly unlikely that a testing accuracy of 57.37% will be observed by chance in randomized data.
Table 3.
The best model for predicting the occurrence of the TRD in MDD patients.
| Best model | Training accuracy (%) |
Testing accuracy (%) |
CVC | χ2 | p value |
|---|---|---|---|---|---|
| NTRK2 (rs1565445) | 55.01 | 55.01 | 10/10 | 8.38 | 0.004 |
| NTRK2 (rs1565445, rs2769605) | 58.57 | 55.61 | 5/10 | 23.49 | p < 0.0001 |
| NTRK2 (rs1565445, rs2769605), BDNF (rs6265) | 61.11 | 55.90 | 9/10 | 42.67 | p < 0.0001 |
| NTRK2 (rs1387923, rs1565445, rs2769605), BDNF (rs6265) | 63.97 | 57.37 | 10/10 | 64.14 | p < 0.0001 |
VC = Cross-validation consistency.
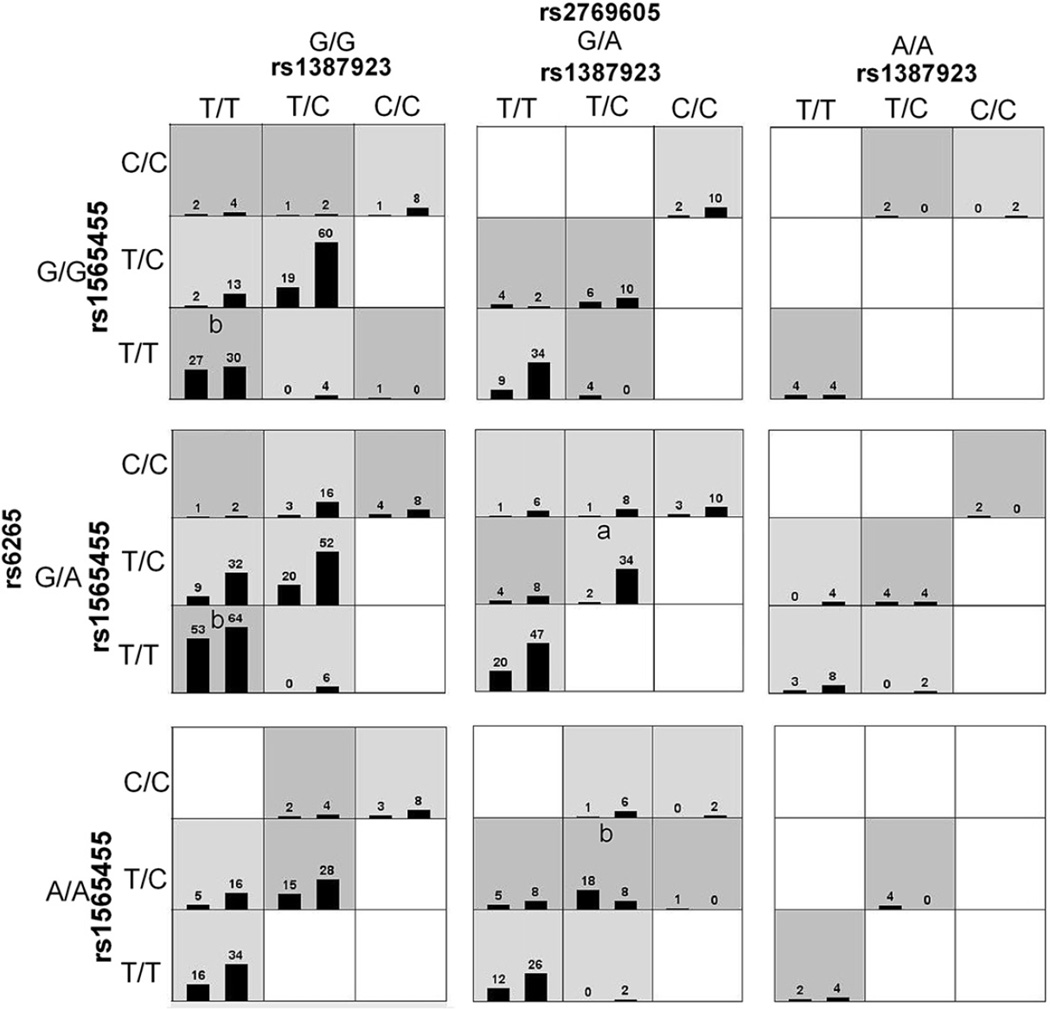
Fig. 1 summarizes the four-locus genotype combinations associated with high risk and with low risk for each multilocus-genotype combination. The varied patterns of high-risk and low risk cells across each of the different multilocus dimensions provide evidence of epistasis, or gene-gene interaction; that is, the influence that each genotype at a particular locus has on disease risk is dependent on the genotypes at each of the other three loci (Ritchie et al., 2001). As many genotype combinations are very rare (as shown in Fig. 1), it is difficult to classify these rare genotype combinations into high- or low-risk groups.
Fig. 1.
Distribution of high-risk and low-risk genotypes in the best four-locus model. Dark gray and light gray boxes presented the high- and low-risk factor combinations, respectively. Left bars within each box represented TRD while the right bars represented non-TRD. The heights of the bars are proportional to the sum of samples in each group. Note that the patterns of high-risk and low-risk cells differ across each of the different multilocus dimensions. This is evidence of epistasis, or gene-gene interaction. “a” and “b” represented low-risk and high-risk genotype combinations respectively which were also validated by traditional statistical analysis.
Traditional statistical methods were applied to this four-locus model to aid in interpretation, which identified three high-risk genotype combinations and one low-risk genotype combination from all possible genotype combinations. In this four-locus (rs1387923-rs1565445-rs2769605-rs6265) model, the OR for the low-risk genotype combination (TC)-(TC)-(GA)-(GA) was 0.12 (95%CI: 0.029-0.50, P = 0.001), the ORs for the three high-risk genotype combinations (TT)-(TT)-(GG)-(GG), (TT)-(TT)-(GG)-(GA), and (TC)-(TC)-(GA)-(AA) were 2.0 (95%CI: 1.1–3.6, P = 0.014), 1.95 (95%CI: 1.3–2.9, P < 0.001), and 5.1 (95%CI: 2.1–12.9, P < 0.0001) respectively.
3.5. Hierarchical interaction graphs
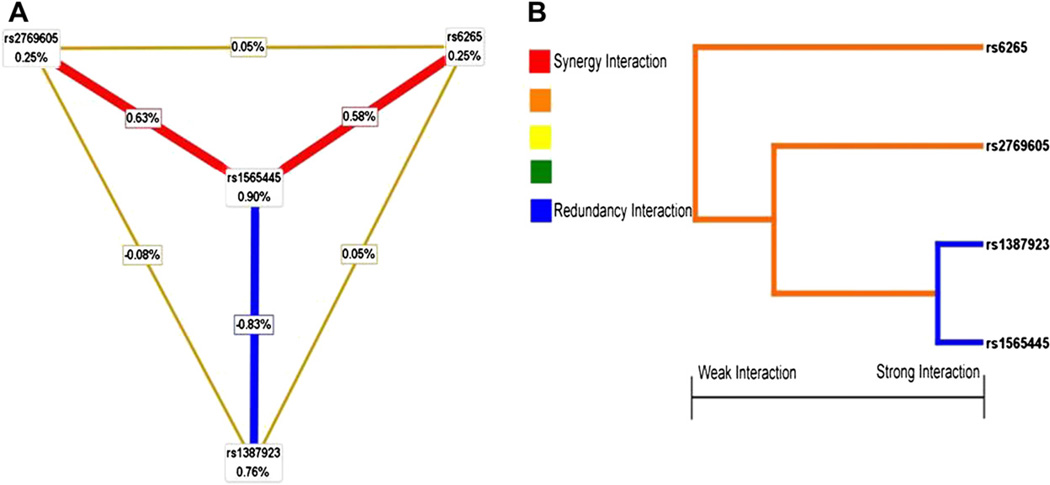
After identifying a high-risk combination of SNPs using MDR, we used the theory of information gain to interpret the relationship between these four SNPs and drew the hierarchical interaction graphs. As shown in Fig. 2A, we found a clear negative interaction effect of rs1565445 and rs1387923 in NTRK2 gene with interaction entropy of −0.83%. Although polymorphisms rs276905 in NTRK2 gene and rs6265 in the BDNF gene had the lowest independent effect (0.25%), both of them had the positive interaction entropy between NTRK2-rs1565445 (0.63% and 0.58%, respectively).
Fig. 2.
Hierarchical interaction graphs and interaction dendrogram. (A) Hierarchical interaction graphs showed that the percentage at the bottom of the each polymorphism represented entropy of it, and the percentage on each line represented the interaction percentage of entropy between two polymorphisms. The red line represented synergy redundancy interaction and the blue line represented redundancy interaction. (B) Interaction dendrogram showed that the red line represented synergy redundancy interaction and the blue line represented redundancy interaction. From left to right the interaction was more intensive. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Interaction dendrogram
Interaction dendrogram demonstrated that rs1565445 in NTRK2 and rs1387923 in NTRK2 located on the same branch (Fig. 2B). These two SNPs were estimated to have the strongest redundancy interaction, as indicated visually by the blue line. rs6265 in BDNF gene was on a different branch, demonstrating a synergy interaction with its receptor’s three polymorphisms (rs1565445, rs1387923, and rs276905), as indicated visually by the red line.
4. Discussion
Recently, BDNF and its high-affinity receptor, NTRK2, widely expressed in the adult brain, were both reported to be involved in antidepressant response (Blugeot et al., 2011; Saarelainen et al., 2003; Squinto et al., 1991); and mounting data indicated that decreased BDNF levels were likely to develop to TRD (Obergriesser et al., 2003; Piccinni et al., 2009; Shah et al., 2002). Although previous studies have suggested that genetic variants may play a key role in mechanism of TRD (Laje and McMahon, 2007; O’Reilly et al., 1994), attempts to identify risk genes for TRD have had limited success thus far.
In the present study, we recruited 948 MDD patients from several multicenter, longitudinal projects to examine the putative association between TRD and polymorphisms in BDNF (rs6265) and NTRK2 genes (rs1387923, rs2769605, and rs1565445). Statistically significant differences in allele and genotype frequencies were observed between TRD and non-TRD participants for rs1565445 polymorphism in NTRK2 gene. Our analyses revealed a significant allelic association between rs1565445 and TRD with an excess of the T allele in the TRD group, and genotype T/C and C/C of rs1565445 were protective factors for TRD with 0.72-fold and 0.52- fold risk respectively comparing with patients with T/T genotype. Haplotype T-T of rs1565445 and rs1387923 in NTRK2 gene was also found to be associated with 1.41-fold increased risk of the occurrence of TRD in MDD patients. To the best of our knowledge, this is the first study reporting an association between genetic polymorphisms within NTRK2 and TRD occurrence in MDD patients. Previous clinical studies had shown that augmentation or combination with lithium effect TRD patients (Shelton et al., 2010; Vieta and Colom, 2011), while polymorphisms (rs1565445 and rs1387923) in the NTRK2 gene had been previously related to lithium response in Caucasian patients with Post-traumatic Stress Disorder (PTSD) (Bremer et al., 2007).
Previous clinical and psychopharmacological studies have implicated that BDNF functional Val66Met polymorphism (rs6265) may play a critical role in the pathophysiology of antidepressant responses. However, results in Asian populations were highly inconsistent. Choi et al. (2006) and Zou et al. (2010a, b) found significant associations between the rs6265 variant and response to citalopram and fluoxetine respectively in MDD patients from Asian populations. However, Tsai et al. (2003) and Kang et al. (2010) reported no association between the rs6265 variant and response to fluoxetine and mirtazapine respectively. Many factors can contribute to the variability in the study findings above, such as small sample size, varying follow-up duration (4–8 weeks), and different assessment methods. Recently, Anttila et al. (2007) reported no significant association between BDNF gene Val66Met polymorphism and risk of TRD in a sample of Caucasian origin, although their study was carried out in samples of TRD patients and healthy controls. In our present multicenter, 12-week, longitudinal prospectively study, there is no significant association with the BDNF gene functional polymorphism rs6265 (Val66Met) and occurrence of TRD in a cohort of MDD patients, which indicated that BDNF functional polymorphism Val66Met is unlikely to play a major role in the risk of TRD occurrence in MDD patients in the Han Chinese population.
In addition, we further analyzed the role of interaction of BDNF (rs6265) and NTRK2 (rs1387923, rs2769605 and rs1565445) gene polymorphisms in development of TRD. Using the MDR method followed by conventional statistical analysis, the best gene-gene interaction model identified was a four-locus model. In this model, one low-risk and three high-risk four-locus genotype combinations were identified. By contrast, the effect sizes (ORs) of our-locus genotype combinations were larger than the effects observed in single marker association analysis, which implies a genetic interaction between these four loci in the susceptibility to TRD. However, the nature of the interaction between these four loci is not clear, as suggested previously when the MDR method was applied to breast cancer (Ritchie et al., 2001). A consistent trend of high-risk or low-risk genotype combinations would indicate that a particular locus had a main effect. As TRD is most likely caused by interactions among multiple genes and environment factors, the lack of such a trend in our best four-locus model could be regarded as quite natural.
In the present study, a total trial of 12-week (including two 6-week trials using two different classes of antidepressants) at an adequate dose was enough to identify the TRD patients, who were distinguished from “difficult-to-treat” depression (Difficult-to-treat depression included TRD and depression that did not respond satisfactorily to one or more classes of antidepressants for insufficient doses or duration) (Rush et al., 2003). However, several concerns or limitations still need to be addressed. Firstly, it is noteworthy that our follow-up study used a naturalistic design and patients were treated with different classical antidepressants. Since it was unclear whether such heterogeneity could affect our findings, the potential effects of specific antidepressants should undergo further investigation. Secondly, other critical genes in BDNF/NTRK2 signaling pathway were not examined in our current study. A more recent study suggests that genetic polymorphisms within cyclic adenosine monophosphate response element binding gene (CREB), a downstream signal in BDNF/NTRK2 signaling pathway, is associated with TRD (Serretti et al., 2011). Thus, the possible role of other genes in the BDNF/NTRK2 signaling pathway and their interactions on development of TRD should be further examined. Thirdly, the allele frequencies of the four polymorphisms examined in the present study showed significant global variation raising concerns of possible population stratification among case-control studies (The International HapMap Consortium, 2010). However, the current study utilized participants from the same geographic region and ethnic origin, which should help to reduce the potential effects of stratification, but formal assessments such as genomic control were not performed. Finally, it should be noted that due to the moderate sample size and lack of independent replication, the current results should be interpreted with caution and further studies of independent, largescale, well-characterized clinical samples should be conducted in order to understand the role of these two genes in TRD susceptibility.
In conclusion, we found significant evidence for an association between the NTRK2 gene polymorphism rs1565445 and TRD in MDD patients. Our data suggests a modest but statistically significant role of interaction of BDNF (rs6265) and NTRK2 (rs1387923, rs2769605 and rs1565445) genes polymorphisms in the development of TRD in Chinese Han MDD patients.
Supplementary Material
Acknowledgments
The authors are very grateful to all participants. We especially thank Naihua Duan, Director of the Division of Biostatistics and Data Coordination, Department of Psychiatry, Columbia University for his constructive comments.
Role of funding source
Funding for this study was provided by the National Natural Science Foundation of China (30971047, 81000581, 81171272), National High-tech R&D Program (863 Program, 2006AA02Z430, Ministry of Science and Technology of China), the “Climbing Mountain Action Plan” Program (064119533, Science and Technology Commission of Shanghai Municipality), the “12th Five-year Plan” of National Key Technologies R&D Program (2012BAI01B04, Ministry of Science and Technology of China), and National Key Clinical Disciplines at Shanghai Mental Health Center (OMA-MH, 2011-873).
Janssen Pharmaceutical Ltd; China, Wyeth, China; Organon, China; GlaxoSmithKline, China; Huahai Pharmaceutical, China; WATSON, China; and Southwest Synthetic Pharmaceutical Corp Ltd, China partly sponsored the study.
Footnotes
Contributors
Zezhi Li, Shunying Yu and Yiru Fang all contributed to the overall design of the study. Zuowei Wang, Yangtai Guan and Shunying Yu selected the SNPs and wrote the protocol for the genotyping. Zezhi Li, Yanxia Zhang, and Zuowei Wang carried out the majority of the studies under the supervision of Shunying Yu and Yiru Fang. Jun Chen, Chen Zhang, Chengmei Yuan, Wu Hong, Yong Wang, Zhiguo Wu, Jia Huang, Yingyan Hu, Lan Cao, and Zhenghui Yi got involved sample collection. Zezhi Li, Jinbo Fan and Donghong Cui undertook the statistical analysis and interpretation of data. Zezhi Li wrote the first draft of the manuscript. All authors contributed to have approved the final manuscript.
Conflict of interest
The authors of this paper do not have any financial or non-financial associations that might pose a conflict of interest in connection with this manuscript.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jpsychires.2012.10.003.
References
- Anttila S, Huuhka K, Huuhka M, Rontu R, Hurme M, Leinonen E, et al. Interaction between 5-HT1A and BDNF genotypes increases the risk of treatment-resistant depression. Journal of Neural Transmission. 2007;114:1065–1068. doi: 10.1007/s00702-007-0705-9. [DOI] [PubMed] [Google Scholar]
- Berlim MT, Turecki G. Definition, assessment, and staging of treatment resistant refractory major depression: a review of current concepts and methods. Canadian Journal of Psychiatry. 2007;52:46–54. doi: 10.1177/070674370705200108. [DOI] [PubMed] [Google Scholar]
- Blugeot A, Rivat C, Bouvier E, Molet J, Mouchard A, Zeau B, et al. Vulnerability to depression: from brain neuroplasticity to identification of biomarkers. Journal of Neuroscience. 2011;31:12889–12899. doi: 10.1523/JNEUROSCI.1309-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer T, Diamond C, McKinney R, Shehktman T, Barrett TB, Herold C, et al. The pharmacogenetics of lithium response depends upon clinical co-morbidity. Molecular Diagnosis & Therapy. 2007;11:161–170. doi: 10.1007/BF03256238. [DOI] [PubMed] [Google Scholar]
- Burmeister M, McInnis MG, Zollner S. Psychiatric genetics: progress amid controversy. Nature Reviews Genetics. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- Choi MJ, Kang RH, Lim SW, Oh KS, Lee MS. Brain-derived neurotrophic factor gene polymorphism (val66met) and citalopram response in major depressive disorder. Brain Research. 2006;1118:176–182. doi: 10.1016/j.brainres.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Dervieux T, Wessels JA, Kremer JM, Padyukov L, Seddighzadeh M, Saevarsdottir S, et al. Patterns of interaction between genetic and nongenetic attributes and methotrexate efficacy in rheumatoid arthritis. Pharmacogenetics and Genomics. 2012;22:1–9. doi: 10.1097/FPC.0b013e32834d3e0b. [DOI] [PubMed] [Google Scholar]
- Duman RS. Role of neurotrophic factors in the etiology and treatment of mood disorders. Neuromolecular Medicine. 2004;5:11–25. doi: 10.1385/NMM:5:1:011. [DOI] [PubMed] [Google Scholar]
- Dunham JS, Deakin JF, Miyajima F, Payton A, Toro CT. Expression of hippocampal brain-derived neurotrophic factor and its receptors in Stanley consortium brains. Journal of Psychiatric Research. 2009;43:1175–1184. doi: 10.1016/j.jpsychires.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Fang YR, Yuan CM, Xu YF, Chen J, Wu ZG, Cao L, et al. Comparisons of the efficacy and tolerability of extended-release venlafaxine, mirtazapine, and paroxetine in treatment-resistant depression a double-blind, randomized pilot study in a Chinese population. Journal of Clinical Psychopharmacology. 2010;30:357–364. doi: 10.1097/JCP.0b013e3181e7784f. [DOI] [PubMed] [Google Scholar]
- Fang YR, Yuan CM, Xu YF, Chen J, Wu ZG, Cao L, et al. A pilot study of the efficacy and safety of paroxetine augmented with risperidone, valproate, buspirone, trazodone, or thyroid hormone in adult Chinese patients with treatment-resistant major depression. Journal of Clinical Psychopharmacology. 2011;31:638–642. doi: 10.1097/JCP.0b013e31822bb1d9. [DOI] [PubMed] [Google Scholar]
- Fernandes BS, Gama CS, Ceresér KM, Yatham LN, Fries GR, Colpo G, et al. Brain-derived neurotrophic factor as a state-marker of mood episodes in bipolar disorders: a systematic review and meta-regression analysis. Journal of Psychiatric Research. 2011;45:995–1004. doi: 10.1016/j.jpsychires.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ, Morrison JM. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. 2006 http://hydra.usc.edu/gxe.
- Gibson TB, Jing Y, Smith Carls G, Kim E, Bagalman JE, Burton WN, et al. Cost burden of treatment resistance in patients with depression. American Journal of Managed Care. 2010;16:370–377. [PubMed] [Google Scholar]
- Hashimoto K, Shimizu E, Iyo M. Critical role of brain-derived neurotrophic factor in mood disorders. Brain Research Reviews. 2004;45:104–114. doi: 10.1016/j.brainresrev.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Kang R, Chang H, Wong M, Choi M, Park J, Lee H, et al. Brain-derived neurotrophic factor gene polymorphisms and mirtazapine responses in Koreans with major depression. Journal of Psychopharmacology. 2010;24:1755–1763. doi: 10.1177/0269881109105457. [DOI] [PubMed] [Google Scholar]
- Kayser S, Bewernick BH, Grubert C, Hadrysiewicz BL, Axmacher N, Schlaepfer TE. Antidepressant effects, of magnetic seizure therapy and electroconvulsive therapy, in treatment-resistant depression. Journal of Psychiatric Research. 2011;45:569–576. doi: 10.1016/j.jpsychires.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Laje G, McMahon FJ. The pharmacogenetics of major depression: past, present, and future. Biological Psychiatry. 2007;62:1205–1207. doi: 10.1016/j.biopsych.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Li Z, Qi D, Chen J, Zhang C, Yi Z, Yuan C, et al. Venlafaxine inhibits the upregulation of plasma tumor necrosis factor-alpha (TNF-α) in the Chinese patients with major depressive disorder: a prospective longitudinal study. Psychoneuroendocrinology. 2012 doi: 10.1016/j.psyneuen.2012.05.005. http://dx.doi.org/10.1016/j.psyneuen.2012.05.005; Epub ahead of print. [DOI] [PubMed]
- O’Reilly RL, Bogue L, Singh SM. Pharmacogenetic response to antidepressants in a multicase family with affective disorder. Biological Psychiatry. 1994;36:467–471. doi: 10.1016/0006-3223(94)90642-4. [DOI] [PubMed] [Google Scholar]
- Obergriesser T, Ende G, Braus DF, Henn FA. Long-term follow-up of magnetic resonance-detectable choline signal changes in the hippocampus of patients treated with electroconvulsive therapy. Journal of Clinical Psychiatry. 2003;64:775–780. doi: 10.4088/jcp.v64n0706. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. Journal of Neuroscience. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinni A, Del Debbio A, Medda P, Bianchi C, Roncaglia I, Veltri A, et al. Plasma brain-derived neurotrophic factor in treatment-resistant depressed patients receiving electroconvulsive therapy. European Neuropsychopharmacology. 2009;19:349–355. doi: 10.1016/j.euroneuro.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. American Journal of Human Genetics. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush AJ, Thase ME, Dube S. Research issues in the study of difficult-to-treat depression. Biological Psychiatry. 2003;53:743–753. doi: 10.1016/s0006-3223(03)00088-x. [DOI] [PubMed] [Google Scholar]
- Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. American Journal of Psychiatry. 2006;163:1905–1917. doi: 10.1176/ajp.2006.163.11.1905. [DOI] [PubMed] [Google Scholar]
- Saarelainen T, Hendolin P, Lucas G, Koponen E, Sairanen M, MacDonald E, et al. Activation of the TrkB neurotrophin receptor is induced by antidepressant drugs and is required for antidepressant-induced behavioral effects. Journal of Neuroscience. 2003;23:349–357. doi: 10.1523/JNEUROSCI.23-01-00349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J, Dickey B. Global burden of depression: the intersection of culture and medicine. British Journal of Psychiatry. 2003;183:92–94. doi: 10.1192/bjp.183.2.92. [DOI] [PubMed] [Google Scholar]
- Serretti A, Chiesa A, Calati R, Massat I, Linotte S, Kasper S, et al. A preliminary investigation of the influence of CREB1 gene on treatment resistance in major depression. Journal of Affective Disorders. 2011;128:56–63. doi: 10.1016/j.jad.2010.06.025. [DOI] [PubMed] [Google Scholar]
- Shah PJ, Glabus MF, Goodwin GM, Ebmeier KP. Chronic, treatment-resistant depression and right fronto-striatal atrophy. British Journal of Psychiatry. 2002;180:434–440. doi: 10.1192/bjp.180.5.434. [DOI] [PubMed] [Google Scholar]
- Shelton RC, Osuntokun O, Heinloth AN, Corya SA. Therapeutic options for treatment-resistant depression. CNS Drugs. 2010;24:131–161. doi: 10.2165/11530280-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Sillaber I, Panhuysen M, Henniger MS, Ohl F, Kühne C, Pütz B, et al. Profiling of behavioral changes and hippocampal gene expression in mice chronically treated with the SSRI paroxetine. Psychopharmacology (Berlin) 2008;200:557–572. doi: 10.1007/s00213-008-1232-6. [DOI] [PubMed] [Google Scholar]
- Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Molecular Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squinto SP, Stitt TN, Aldrich TH, Davis S, Bianco SM, Radziejewski C, et al. trkB encodes a functional receptor for brain-derived neurotrophic factor and neurotrophin-3 but not nerve growth factor. Cell. 1991;65:885–893. doi: 10.1016/0092-8674(91)90395-F. [DOI] [PubMed] [Google Scholar]
- Taylor SM. Electroconvulsive therapy, brain-derived neurotrophic factor, and possible neurorestorative benefit of the clinical application of electroconvulsive therapy. Journal of ECT. 2008;24:160–165. doi: 10.1097/YCT.0b013e3181571ad0. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups MS, Greer TL, Kurian BT, Grannemann BD, Carmody TJ, Huebinger R, et al. Effects of serum brain derived neurotrophic factor on exercise augmentation treatment of depression. Journal of Psychiatric Research. 2011;45:1301–1306. doi: 10.1016/j.jpsychires.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SJ, Cheng CY, Yu YW, Chen TJ, Hong CJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and major depressive disorders, symptomatology, and antidepressant response. American Journal of Medical Genetics. 2003;123B:19–22. doi: 10.1002/ajmg.b.20026. [DOI] [PubMed] [Google Scholar]
- Vieta E, Colom F. Therapeutic options in treatment-resistant depression. Annals of Medicine. 2011;43:512–530. doi: 10.3109/07853890.2011.583675. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li Z, Gao K, Fan J, Wang L, Huang J, et al. Association of BDNF gene polymorphism with bipolar disorder in Han Chinese population. Genes, Brain and Behavior. 2012 Apr 30; doi: 10.1111/j.1601-183X.2012.00797.x. http://dx.doi.org/10.1111/j.1601-183X.2012.00797.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Xu H, Chen Z, He J, Haimanot S, Li X, Dyck L, et al. Synergetic effects of quetiapine and venlafaxine in preventing the chronic restraint stress-induced decrease in cell proliferation and BDNF expression in rat hippocampus. Hippocampus. 2006;16:551–559. doi: 10.1002/hipo.20184. [DOI] [PubMed] [Google Scholar]
- Yukimasa T, Yoshimura R, Tamagawa A, Uozumi T, Shinkai K, Ueda N, et al. Highfrequency repetitive transcranial magnetic stimulation improves refractory depression by influencing catecholamine and brain-derived neurotrophic factors. Pharmacopsychiatry. 2006;39:52–59. doi: 10.1055/s-2006-931542. [DOI] [PubMed] [Google Scholar]
- Zou YF, Wang Y, Liu P, Feng XL, Wang BY, Zang TH, et al. Association of brain-derived neurotrophic factor genetic val66met polymorphism with severity of depression, efficacy of fluoxetine and its side effects in Chinese major depressive patients. Neuropsychobiology. 2010a;61:71–78. doi: 10.1159/000265132. [DOI] [PubMed] [Google Scholar]
- Zou YF, Ye DQ, Feng XL, Su H, Pan FM, Liao FF. Meta-analysis of BDNF val66met polymorphism association with treatment response in patients with major depressive disorder. European Neuropsychopharmacology. 2010b;20:535–544. doi: 10.1016/j.euroneuro.2009.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.