Abstract
Abscisic acid (ABA) is an essential hormone for plants to survive environmental stresses. At the center of the ABA signaling network is a subfamily of type 2C protein phosphatases (PP2Cs), which form exclusive interactions with ABA receptors and subfamily 2 Snfl-related kinase (SnRK2s). Here, we report a SnRK2-PP2C complex structure, which reveals marked similarity in PP2C recognition by SnRK2 and ABA receptors. In the complex, the kinase activation loop docks into the active site of PP2C, while the conserved ABA-sensing tryptophan of PP2C inserts into the kinase catalytic cleft, thus mimicking receptor-PP2C interactions. These structural results provide a simple mechanism that directly couples ABA binding to SnRK2 kinase activation and highlight a new paradigm of kinase-phosphatase regulation through mutual packing of their catalytic sites.
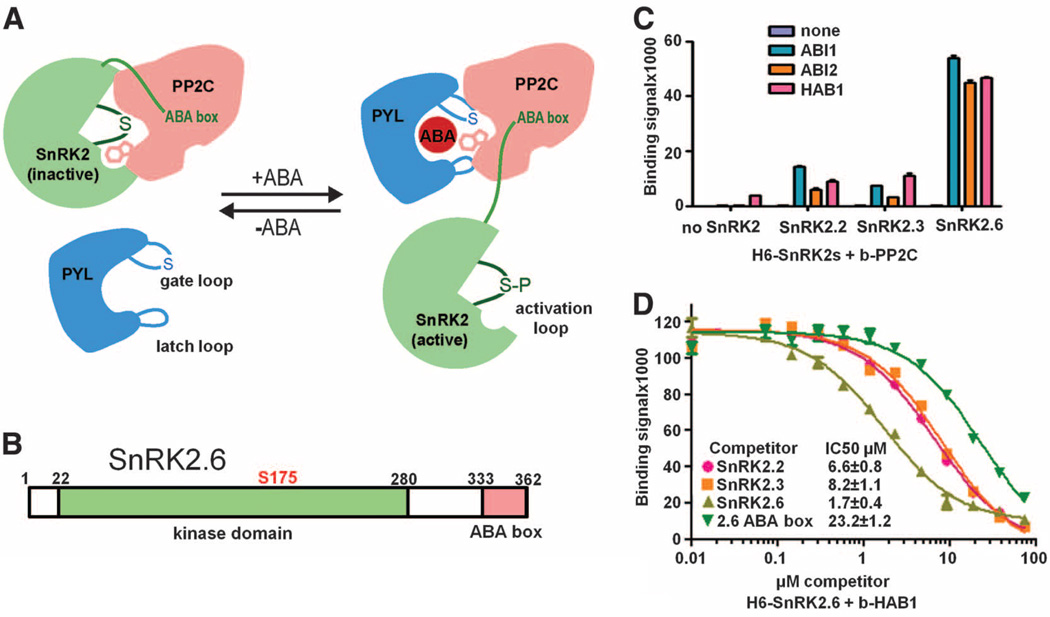
Abscisic acid (ABA) is a vital plant hormone and a central regulator that protects plants against abiotic stresses such as drought and salinity. The core of the ABA signaling network comprises a subfamily of type 2C protein phosphatases (PP2Cs) and three Snf1-related kinases, SnRK2.2, 2.3, and 2.6 (1, 2), whose activities are tightly controlled by ABA. In the absence of ABA, SnRK2 kinases are inactivated by PP2Cs, including ABI1, ABI2, and HAB1 (2–5), which physically interact with SnRK2s and dephosphorylate a serine residue in the kinase activation loop (S175 in SnRK2.6) whose phosphorylation is required for kinase activity (Fig. 1A) (6–8). ABA binding to the PYR/PYL/RCAR family of ABA receptors promotes the receptors to bind to the catalytic site of PP2Cs and inhibit their enzymatic activity (Fig. 1A) (3–5, 9–12). In turn, ABA-induced inhibition of PP2Cs leads to SnRK2 activation by activation loop autophosphorylation (6, 7), which allows the SnRK2s to relay the ABA signal to downstream effectors (13, 14). In plants, SnRK2 activation loop phosphorylation may also involve unidentified upstream kinases (7, 15).
Fig. 1.
SnRK2–PP2C interactions. (A) Summary model for the interactions between SnRK2, PP2C, and the ABA receptor PYR/PYL/RCAR in ABA signaling. In the absence of ABA, PP2C binds to the SnRK2 kinase domain and inhibits the kinase activity by dephosphorylating the activation loop serine and blocking the catalytic cleft. In the presence of ABA, ABA-receptor complex binds to PP2C and inhibits PP2C’s catalytic activity by inserting the gate loop into the PP2C active cleft. PYL-mediated inhibition of PP2C allows activation of the kinase by activation loop autophosphorylation. The activated kinase then transmits the ABA signal by phosphorylating downstream factors. (B) Schematic presentation of the domain structure of SnRK2.6 with amino acid residue numbers shown on top. (C) Interactions of ABI1, ABI2, and HAB1 with SnRK2.2, 2.3, and 2.6. Binding of recombinant H6GST-SnRK2s to biotin-PP2Cs was determined by AlphaScreen assays (see fig. S2 and Materials and Methods). Error bars indicate SD (n = 3). (D) Inhibition of the interaction between H6GST-SnRK2.6 and biotin-HAB1 by untagged SnRK2s and SnRK2.6 ABA box peptide (amino acids 333 to 362). The IC50 values were derived from curve fitting. Error bars indicate SD (n = 3).
Recent structural studies have revealed a gate-latch-lock mechanism for PP2C inhibition by ABA receptors (9–12, 16). The ABA-binding pocket of receptors is flanked by two highly conserved loops that serve as a gate and latch. The gate in apo receptors is in open conformation. ABA binding induces the closure of the gate onto the latch and allows the receptor to dock into the PP2C active site, thus inhibiting PP2C activity by blocking substrate access. A conserved tryptophan of PP2C, which inserts between the gate and latch, functions as a molecular lock to further stabilize the receptor-PP2C complex (Fig. 1A). This tryptophan also functions as an ABA sensor through a water-mediated contact with the bound ABA in the receptor pocket. Thus, the gate of the receptors and the tryptophan lock of PP2Cs are both required for ABA-dependent receptor-PP2C interactions.
A fundamental but currently unresolved question is how SnRK2 activation is orchestrated with ABA binding and PP2C inhibition. To address this, we have solved the crystal structure of a SnRK2.6-HAB1 complex, which reveals a striking similarity between the SnRK2.6-HAB1 interface and the receptor-PP2C interface. The activation loop of SnRK2 is packed against the active site of PP2C in a manner mimicking the gate of ABA receptors. The conserved tryptophan of PP2Cs, which locks the gate of ABA receptors, inserts deeply into the kinase active cleft and completely blocks its activity. These shared structural elements for PP2C binding by ABA receptors and SnRK2 thus provide a direct mechanism that links ABA binding to kinase activation and highlights a new regulatory paradigm for phosphatase-kinase regulation through mutual packing of their catalytic sites.
SnRK2s contain a Snf1-related kinase domain and a highly acidic C-terminal segment termed ABA box, which is important for their interactions with PP2Cs (Fig. 1B and fig. S1) (6, 7). To characterize SnRK2-PP2C interactions, we purified the three SnRK2s (SnRK2.2, 2.3, and 2.6) and three PP2Cs (ABI1, ABI2, and HAB1). As determined by AlphaScreen (PerkinElmer) assays (fig. S2), each of the three SnRK2s was able to bind to the three PP2Cs with half maximal inhibitory concentration (IC50) values of ~2 to 8 µM, indicating a relatively weak binding between SnRK2s and PP2Cs (Fig. 1, C and D). The binding of the SnRK2.6 ABA box alone is 10- to 15-fold weaker compared with that of full-length SnRK2.6 (Fig. 1D), suggesting additional PP2C binding site(s) in SnRK2s.
To obtain the structure of a SnRK2-PP2C complex, we coexpressed and purified SnRK2s and PP2Cs in all nine combinations with an apparent 1:1 molar ratio, yet none of these complexes crystallized under any of the conditions tested. We hypothesized that the recalcitrance of these complexes to crystallization is due to their dynamic flexibility arising from the high dissociation rates associated with the low SnRK2-PP2C binding affinities. We therefore generated various SnRK2.6-HAB1 fusion proteins [SnRK2.6 is the strongest PP2C binder among the SnRK2s (Fig. 1C)] in which SnRK2.6 and HAB1 were separated by flexible linkers of various lengths (see Materials and Methods). A SnRK2.6–11 amino acid linker–HAB1 fusion protein produced poorly diffracting crystals under a number of different conditions. Introduction of two adjacent surface-entropy-reduction mutations (Asp296→Ala296 and Glu297→Ala297, fig. S1) into the SnRK2.6 moiety greatly improved the quality of crystals and allowed us to solve the structure of the SnRK2.6–HAB1 complex at a resolution of 2.6 Å (structure statistics in table S1). Kinase activity was strongly inhibited in the fusion protein (fig. S3), and this inhibition was fully reversed in the presence of ABA-bound PYL2 receptor (fig. S3), indicating that both SnRK2.6 and HAB1 were active and functionally interacted with each other in the context of the fusion protein. Moreover, hydrogen/deuterium exchange (HDX) protection of this fusion protein was indistinguishable from that of a SnRK2.6-HAB1 complex formed with nonfused proteins (fig. S4), supporting that the fusion complex represents the functional complex formed by isolated proteins.
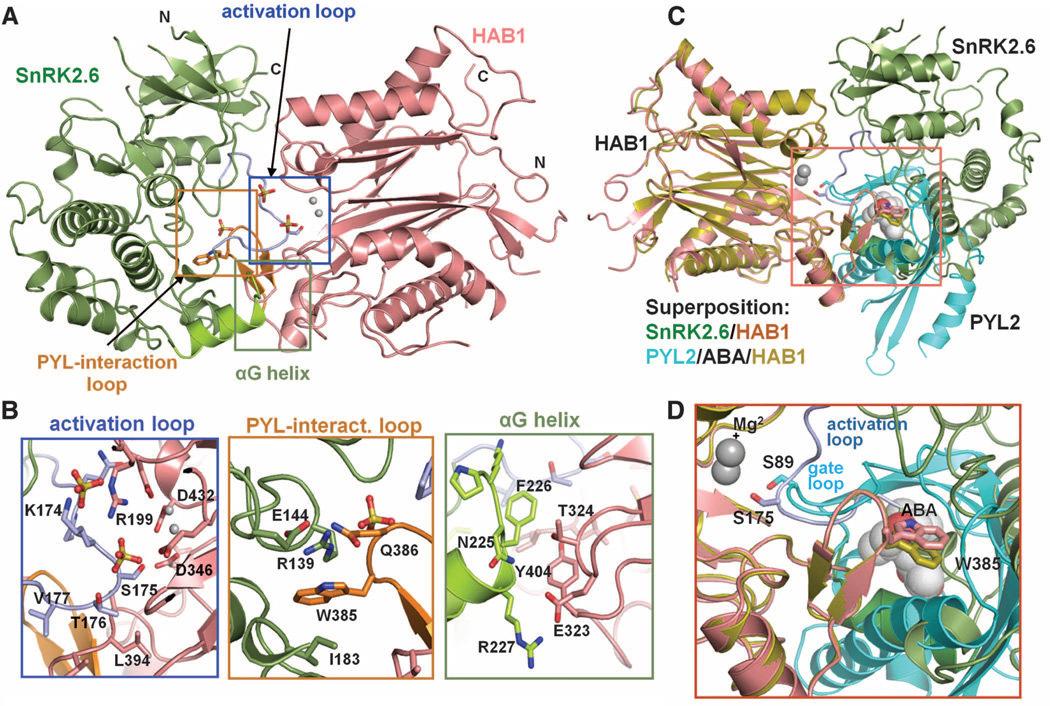
The overall structure of the SnRK2.6-HAB1 complex reveals a monomeric kinase-phosphatase complex with 1:1 stoichiometry (Fig. 2A). The salient feature of the SnRK2.6-HAB1 complex is the mutual packing of the kinase and phosphatase active sites, which form the major binding interface (Fig. 2A). SnRK2.6 contributes three separate regions within the kinase domain for binding to HAB1 (Fig. 2B). The first region is the SnRK2.6 activation loop, which is fully visible in the electron density map (fig. S5). This activation loop inserts deeply into the catalytic cleft of HAB1 and thus mimics the gate loop of ABA receptors in receptor-PP2C complexes (Fig. 2 and fig. S6A) (3, 9, 10). In this orientation, S175 from the activation loop is directed toward the catalytic site of HAB1, which is consistent with the fact that pS175 of SnRK2.6 is preferentially dephosphorylated by HAB1 when compared with pS29 (fig. S7). The second region is the kinase catalytic cleft near residues Arg139, Ile183, and Glu144 of SnRK2.6. This region mimics the cleft formed by the gate and latch regions of ABA receptors (3, 9, 10) and is occupied by the conserved ABA-sensing tryptophan [Trp385 (W385) in HAB1] (fig. S6B). The third region is the SnRK2.6 αG helix, which binds to the HAB1 loop region adjacent to the W385-containing HAB1 PYL-interaction site (Fig. 2B). The extensive interactions between the catalytic sites of kinase and phosphatase suggest that their activities have coevolved and are co-regulated.
Fig. 2.
Structures of the SnRK2.6-HAB1 complex. (A) A structure overview. SnRK2.6 is shown in green, the activation loop in light blue, and the αG helix in light green. HAB1 is colored pink, and its PYL2-interaction loop orange. The two catalytic Mg2+ ions are presented as gray spheres, and three sulfate ions as ball-stick models. (B) Details of the SnRK2.6-HAB1 interface with key residues shown in stick presentation. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; D, Asp; E, Glu; F, Phe; I, Ile; K, Lys; L, Leu; N, Asn; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (C and D) Overlay of the interaction surfaces in the SnRK2.6-HAB1 and PYL2-ABA-HAB1 complexes. SnRK2.6 S175 and HAB1 W385 are shown in stick presentation, ABA in ball presentation, and the HAB1 Mg2+ ions as spheres.
The kinase domain-PP2C interface largely overlaps with the ABA receptor binding interface of HAB1 (Fig. 2, C and D, and fig. S6). The HAB1 structure in the SnRK2.6-PP2C complex is nearly identical to the HAB1 structure in the ABA receptor complex (root mean square deviation = 0.6 Å between the entire sets of Cα atoms of both HAB1 structures). We also solved the structures of the PP2C ABI2 in its apo state and as an ABI2-ABA-PYL2 complex (fig. S8). The PP2C components in these structures (apo PP2C, PP2C-ABA-receptor, and PP2C-SnRK2) are all super-imposable, indicating that PP2Cs have a fairly rigid fold (fig. S9). ABA-bound receptors and SnRK2s have thus adopted remarkably similar surface features that enable them to dock into the rigid PP2C scaffold (Fig. 2C). Consistently, the PYL2 ABA receptor–HAB1 interaction can be inhibited by increasing concentrations of the SnRK2.6 kinase domain (fig. S10). The shared use of HAB1 W385 and its catalytic site by ABA receptors and SnRK2.6 thus provides a direct mechanism that links ABA binding to kinase activation.
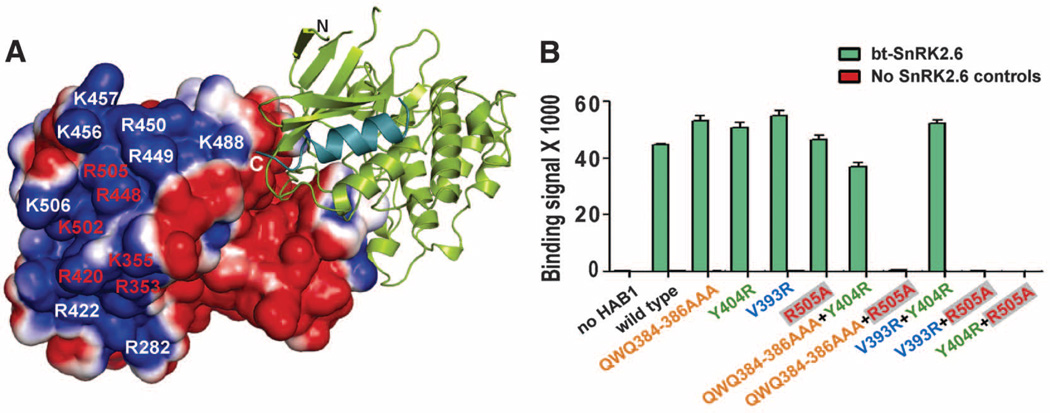
In addition to the kinase domain, SnRK2.6 interacts with PP2Cs via its highly acidic C-terminal ABA box (6, 7). However, we could not unambiguously resolve the ABA box. Consistent with its lack of clear electron density, the ABA box was not protected against HDX upon HAB1 binding (fig. S11). We hypothesized that this highly acidic ABA box (net charge: −14) should associate with a highly positive region of HAB1, which is at the proximity of the C terminus of SnRK2.6 (Fig. 3A and fig. S1). We therefore mutated all lysine and arginine residues within the positively charged HAB1 surface (Fig. 3A). Mutation of single residues within a tight cluster of six positively charged HAB1 surface amino acids disrupted interaction with the ABA box but had little effect on HAB1 catalytic activity (Fig. 3A and fig. S12), demonstrating that this region is critical for ABA box binding. On the other hand, alanine scanning mutagenesis of the SnRK2.6 ABA box identified many residues important for HAB1 interaction, particularly the negatively charged residues (fig. S13). Together, these data suggest that ABA box–PP2C interactions are primarily mediated by charge interactions between the negatively charged ABA box and the positively charged PP2C surface. The ABA-box binding site in HAB1 is opposite of the kinase domain/ABA receptor binding site (Figs. 2A and 3A), consistent with the fact that ABA-box binding to PP2Cs is not affected by ABA or its receptors (fig. S14), further supporting the previous model that the ABA box provides a constant interaction for SnRK2.6 with PP2Cs (2).
Fig. 3.
SnRK2.6 and HAB1 associate with each other via two modular interactions. (A) Charge-distribution surface of HAB1 in the HAB1-SnRK2.6 complex with an electrostatic scale from –1 to +1 ev, corresponding to red and blue colors. The positively charged residues required for ABA box interaction are labeled in red. (B) Mutational analysis of the HAB1-SnRK2.6 interaction by AlphaScreen luminescence proximity assay. Error bars indicate SD (n = 3). HAB1 mutant proteins are color-coded on the basis of the mutated regions that bind to SnRK2.6. Q384A/W385A/Q386A, which is the “lock” that inserts into both SnRK2.6 and PYL2 ABA receptor, is colored in orange; V393R, which interacts with SnRK2.6 activation loop interaction, is in blue; Y404R, which binds to αG, is in green; R505A, which is required for ABA box interaction, is in red within gray boxes.
The separation of the ABA box binding site from the kinase domain/ABA receptor binding site in HAB1 suggests that the two sites function modularly. As shown in Fig. 3B, the Arg505→Ala505 (R505A) mutation in HAB1, which abolished the interaction between HAB1 and the ABA box (fig. S12), did not significantly weaken the interaction of HAB1 with full-length SnRK2.6. Similarly, mutations designed to disrupt each of the three kinase domain-interaction regions of HAB1 have little effect on the HAB1-SnRK2.6 interaction (Fig. 3B). In contrast, combination of the R505A mutation, which disrupted the ABA box interaction (fig. S12), with any mutation in the three kinase-interaction sites completely abolished SnRK2.6 binding (Fig. 3B). Together, these data further support that SnRK2.6 has two modular HAB1-binding sites.
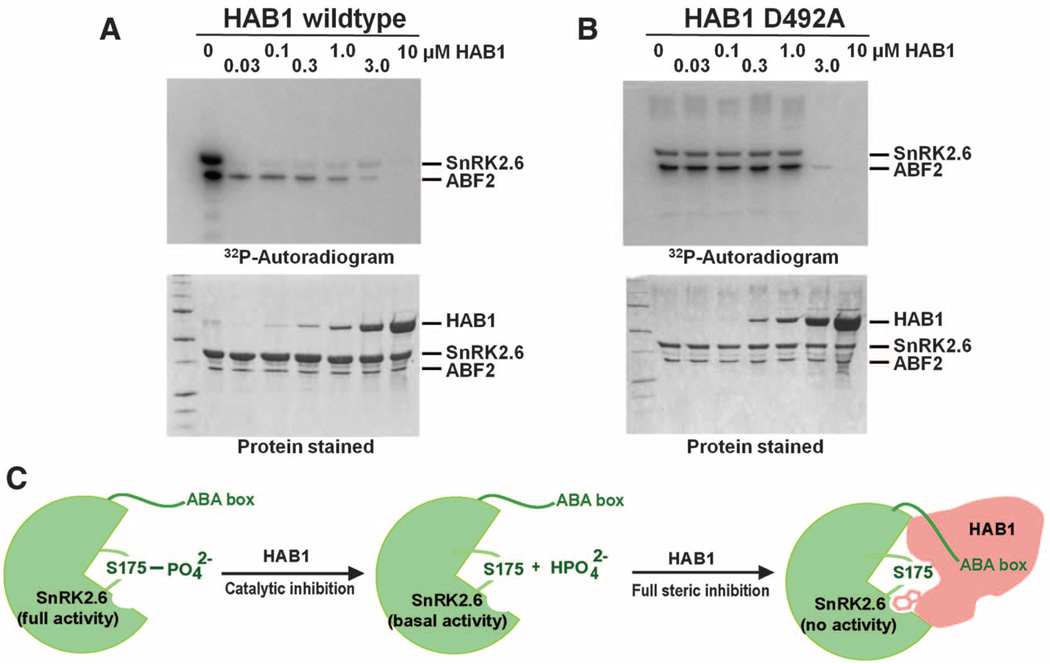
The purified SnRK2.6 is intrinsically active as a kinase toward itself or its physiological substrate ABF2 and can be inactivated by PP2C (Fig. 4A). The SnRK2.6-HAB1 structure suggests a two-step mechanism by which HAB1 completely inactivates SnRK2.6 (Fig. 4, A and B). The first step is mediated by the catalytic activity of HAB1, which dephosphorylates S175 from the activation loop, thus reducing SnRK2.6 activity to the basal level (Fig. 4C). The second step is the physical inhibition of the SnRK2.6 kinase domain by HAB1 (Fig. 4C). As shown in Fig. 2, insertion of HAB1 W385 into the SnRK2.6 catalytic cleft could physically block the access of substrate to the kinase active site, thus completely blocking the kinase activity. We have validated this two-step mechanism by a series of biochemical and mutational experiments (Fig. 4, A and B, and fig. S15). In these experiments, we show that catalytically inactive HAB1 is still capable of inhibiting SnRK2.6 (Fig. 4B and additional controls in fig. S15), in agreement with the observation that catalytic activities of PP2Cs are not required to inhibit SnRK2.6 activity (17, 18). Taken together, these data demonstrate that HAB1 inactivates SnRK2.6 by dephosphorylating S175 in the activation loop and by physically blocking the SnRK2.6 active site.
Fig. 4.
Enzymatic and physical inhibition of SnRK2 activity by HAB1. (A) Inhibition of SnRK2.6 activity by HAB1. Increasing amounts of HAB1 were added to a kinase reaction with 5 µM SnRK2.6 and 1 µM of the SnRK2.6 substrate ABF2. At low concentrations, HAB1 inactivates SnRK2.6 catalytically (>50% kinase inhibition by HAB1 at a 100-fold molar excess of SnRK2.6). SnRK2.6 activity is only completely inhibited when HAB1 is at or above the molar amount of SnRK2.6. (B) Phosphatase activity of HAB1 is not required for its ability to inhibit SnRK2 kinase activity. Increasing amounts of the catalytically inactive HAB1 mutant D492A were added to a kinase reaction with constant concentrations of SnRK2.6 and ABF2. (C) Cartoon model for the two-step mechanism of inhibition of SnRK2.6 activity by HAB1. The first step is that HAB1 catalytically dephosphorylates S175 in the activation loop, thus reducing SnRK2.6 activity to the basal level. The second step is that at stoichiometric levels HAB1 physically inhibits SnRK2.6 activity by blocking its catalytic cleft.
Our structural studies have revealed a molecular mimicry between ABA receptors and SnRK2 kinases with respect to how they bind to PP2Cs. Both SnRK2.6 and ABA receptors use a similar gate-and-lock mechanism to recognize PP2Cs. Given the large number of PP2Cs and SnRK2s encoded by plant genomes, of which only restricted sets are involved in ABA signaling, the mutual recognition system of gate and lock likely ensures signal integrity by preventing the wrong players from being inappropriately regulated. The repeated use of gate-and-lock recognition as reported here thus provides a basis for the exquisite control of specificity in the ABA signaling network.
The packing interactions between the catalytic active sites of SnRK2.6 and HAB1 may also have important implications in other kinase– phosphatase signaling systems. PP2Cs are known to interact and regulate mammalian adenosine monophosphate–activated kinases (AMPK) (19). Furthermore, the recent discovery that the catalytically inactive pseudophosphatases EGG-4 and EGG-5 recognize the activation loop of the auto-phosphorylating MBK-2 kinase and inhibit MBK-2 activity in the absence of any other protein (20, 21) suggests that kinase inactivation by phosphatase as observed in the SnRK2.6-HAB1 complex may represent a widespread mechanism for phosphatase-kinase regulation.
Supplementary Material
Acknowledgments
The Protein Data Bank (PDB) code for SnRK2.6–HAB1 is 3UJG, for ABI2 is 3UJK, and for PYL2–ABA–ABI2 is 3UJL. We thank the staff of LS-CAT for assistance in data collection at the beam lines of sector 21, which is in part funded by Michigan Economic Development Corporation. Use of the Advanced Photon Source was supported by the Office of Science of the U.S. Department of Energy. This work was supported by the Jay and Betty Van Andel Foundation and Amway (China) Limited (H.E.X.), NIH (H.E.X., P.R.G., and J.-K.Z.), NSF (S.C. and J.-K.Z.), and the Singapore Biomedical Research Council (E.-L.Y.). L.-M.N., F.-F.S., and M.H.E.T. were supported by an overseas Ph.D. scholarship from the NUS Graduate School for Integrative Sciences and Engineering (NGS). S.C. received funding from the NSF (grant IOS-0820508), and P.R.G. received funding from National Institute for General Medical Studies (grant GM084041).
Footnotes
Author contributions: J.L., E.-L.Y., J.-K.Z., P.R.G., K.M., and H.E.X. conceived the project and designed research; F.-F.S., L.-M.N., X.E.Z., G.W., A.K., M.H.E.T., K.M.S.-P., Y.H., Y.X., M.C., J.S.B., H.Z., H.Y., H.J., K.M., and H.E.X. performed research; F.-F.S., L.-M.N., X.E.Z., G.W., A.K., M.H.E.T., K.M.S.-P., M.C., J.S.B., P.R.G., K.M., and H.E.X. analyzed data; and K.M. and H.E.X. wrote the paper with contributions from all authors.
Supporting Online Material
www.sciencemag.org/cgi/content/full/science.1215106/DC1
Materials and Methods
Figs. S1 to S15
Table S1
References (22–34)
References and Notes
- 1.Fujii H, et al. Nature. 2009;462:660. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umezawa T, et al. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17588. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin P, et al. Nat. Struct. Mol. Biol. 2009;16:1230. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, et al. Science. 2009;324:1064. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 5.Park SY, et al. Science. 2009;324:1068. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belin C, et al. Plant Physiol. 2006;141:1316. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C. Plant Mol. Biol. 2007;63:491. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- 8.Vlad F, et al. Plant Cell. 2009;21:3170. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melcher K, et al. Nature. 2009;462:602. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazono K, et al. Nature. 2009;462:609. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura N, et al. Science. 2009;326:1373. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santiago J, et al. Nature. 2009;462:665. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 13.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Annu. Rev. Plant Biol. 2010;61:651. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Genes Dev. 2010;24:1695. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burza AM, et al. J. Biol. Chem. 2006;281:34299. doi: 10.1074/jbc.M601977200. [DOI] [PubMed] [Google Scholar]
- 16.Melcher K, Zhou XE, Xu HE. Curr. Opin. Struct. Biol. 2010;20:722. doi: 10.1016/j.sbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SC, Lan W, Buchanan BB, Luan S. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21419. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiger D, et al. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21425. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Biochem. J. 2007;403:139. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng KC, Klancer R, Singson A, Seydoux G. Cell. 2009;139:560. doi: 10.1016/j.cell.2009.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parry JM, et al. Curr. Biol. 2009;19:1752. doi: 10.1016/j.cub.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.